Abstract
Molecular analysis of circulating and disseminated tumor cells (CTCs/DTCs) has great potential as a means for continuous evaluation of prognosis and treatment efficacy in near-real time through minimally invasive liquid biopsies. To realize this potential, however, methods for molecular analysis of these rare cells must be developed and validated. Here, we describe the integration of imaging mass cytometry (IMC) using metal-labeled antibodies as implemented on the Fluidigm Hyperion Imaging System into the workflow of the previously established High Definition Single Cell Analysis (HD-SCA) assay for liquid biopsies, along with methods for image analysis and signal normalization. Using liquid biopsies from a metastatic prostate cancer case, we demonstrate that IMC can extend the reach of CTC characterization to include dozens of protein biomarkers, with the potential to understand a range of biological properties that could affect therapeutic response, metastasis and immune surveillance when coupled with simultaneous phenotyping of thousands of leukocytes.
Introduction
Enabling precision medicine in cancer care requires the ability to both deconvolute heterogeneity in the primary and metastatic tissues, and to characterize the liquid phase of the disease. In settings of chemotherapy, molecularly targeted and immune-system engaging treatment approaches, the high-resolution characterization of the disease along its evolutionary path at the time of the decision making is a primary challenge that can be addressed by the integration of quantitative single cell technologies within the solid and liquid biopsy workflows. The recognition of the complex interactions between the cancer and the immune system are particularly important drivers for the detailed analysis at the cellular, proteome and genome levels to both establish a treatment strategy and monitor its efficacy.
Since its commercial introduction in 2011, cytometry by time-of-flight (CyTOF) mass spectrometry using metal-labeled antibodies, has rapidly entered the basic and clinical research laboratory settings [1-3]. Its main distinction with respect to standard fluorescence-based cytometry is the capacity to assay the binding of 35 or more specific antibodies on each cell simultaneously. Bodenmiller and Günther et al have developed methods for coupling laser ablation with CyTOF technology, enabling multiplexed image-based proteomic analysis of formalin-fixed paraffin-embedded (FFPE) tissue sections or cultured cells mounted on glass slides [4, 5]. The technology is now commercially available from Fluidigm Corporation (South San Francisco) as the Hyperion Imaging System for imaging mass cytometry, or IMC. In the IMC process, the target tissue or cell preparation is treated with a cocktail of antibodies, each labeled with a specific rare earth isotope. Regions of interest (ROI) on the slides are scanned in situ with a highly focused, pulsed laser, such that each pulse vaporizes a 1μm2 bloc of the sample and the resulting ions are introduced into the inductively coupled plasma time-of-flight mass spectrometer (ICP-TOF-MS) with helium as a carrier gas. The ion counts for each pulse can then be assembled into a protein expression image with a resolution of 1 μm2 across the ROI allowing for limited characterization of sub-cellular localization.
In this report, we describe the integration of imaging mass cytometry using the Hyperion instrument into the previously validated High Definition Single Cell Analysis (HD-SCA) workflow [6], with the goal to incorporate morphology, proteomics and genomics of rare single cells in a single streamlined process. The HD-SCA method was designed to identify and characterize ultra-rare (<0.0001%) cancer cells in in liquid biopsies from blood and bone marrow aspirates, or high-complexity samples such as tissue ‘touch preparations’ on glass slides. In contrast to other CTC technologies, the HD-SCA workflow uses a direct analysis, 'no cell left behind' strategy in which the entire population of nucleated cells from a blood draw is spread on glass slides at 3 million cells per slide, stained, and imaged in situ. Fresh samples must be fractionated within 48 hours following blood collection, which enables simple shipping from clinical sites. Candidate tumor cells are identified among the millions of leukocytes using immunofluorescent markers. In addition to the morphometric information obtained by imaging, the candidate circulating tumor cells (CTC) from blood or disseminated tumor cells (DTC) from bone marrow, or other cells of interest can subsequently be relocated and physically isolated for single cell genomic analysis [7, 8]. The data derived from the HD-SCA workflow has been demonstrated to generate clinically actionable information [9]. Further, due to their minimally invasive nature, liquid biopsies can be repeated frequently during treatment to assess response and provide early indications of changes that could lead to disease progression. Despite its high sensitivity, up to now the phenotyping of cells detected by HD-SCA has been largely limited to morphometry and four fluorescent markers. Targeted single cell proteomic analysis can complement genomic and whole plasma characterization to assign functional relevance to the molecular observations as part of an integrated signature. The longitudinal characterization of the disease using cancer cells from blood and bone marrow liquid biopsies provides insight into the evolution of the tumor, and the significance of sub-populations of circulating tumor cells present in specific environments.
Here, we present experimental procedures along with data analysis methods, specifically adapted for the HD-SCA workflow to incorporate multiplex targeted proteomic analysis on intact single cells using the Hyperion instrument. The functionality and performance of the workflow is demonstrated, including methods for validating metal conjugated antibodies for use specifically in the combined HD-SCA/IMC workflow. In addition, the robustness of the method allows for immunophenotyping of the leukocyte populations that are simultaneously imaged along with the CTCs, which is demonstrated here by gating the multi-parametric data from the HD-SCA/IMC workflow in a manner similar to cytometric sorting methods, yielding well separated sub-populations. Finally, we present combined proteo-genomic results relating the HD-SCA/IMC to the HD-SCA/genomics from the analysis of liquid biopsies from a metastatic prostate cancer patient, showing the distribution and subcellular location of characteristic prostate cancer protein markers in a clonally related population of CTCs and DTCs.
Methods
HD-SCA Specimen Preparation
Whole blood samples from primary and metastatic cancer patients were collected in cell-free DNA blood collection tubes (Streck, Omaha, USA) and shipped to the Kuhn laboratory at USC in temperature stabilized shippers (Standard 71, LLC, Los Angeles). Samples were processed within 24 hours of collection using the HD-SCA protocol [6] and archived at −80°C. The sample MDA-42109 was collected from a patient with metastatic prostate cancer in 2013 with informed consent and under IRB approval as previously described [8]. Normal control donor blood samples were obtained from The Scripps Research Institute, La Jolla, California, USA. All specimen collections were performed under IRB approval.
Cell lines spiked in blood
Cell lines were obtained from the American Type of Culture Collection, (ATCC, Manassas, VA, USA). Prostate cancer line LNCaP was cultured in RPMI-1640 medium and breast cancer line MDA-MB-231 was cultured in DMEM/F12 medium (Corning, Manassas, VA, USA). Cells were harvested using Versene solution (Gibco) and washed in PBS. For validation of the metal-labeled antibodies in the combined workflow, cultured cells were added at a ratio 1:10,000 to whole blood from normal blood donors and processed according to the standard HD-SCA protocol [6], including storage at −80°C for a minimum of 24 hours.
HD-SCA fluorescent labelling, imaging and cell identification
To complete the HD-SCA procedure, slides were thawed, stained with DAPI (nuclear stain) and fluorescent antibodies against CD45 (leukocyte marker) and pan-cytokeratin (CK, epithelial marker), and imaged in three colors using a high-speed automated microscope scanner. As part of the normal HD-SCA image analysis, the candidate CK positive cells were identified and coordinates registered. High resolution 40x images of candidate cells were captured using a Nikon 80i microscope. Prior to staining with the metal-labeled antibody cocktail for IMC analysis, the slides were stored at 4°C for several days to parallel normal operation of the HD-SCA protocol. Finally, cover slips were removed in PBS with minimal disruption of the cell layer. Slides were then washed with PBS to dissolve the imaging media in preparation for the metal-labeled antibody incubation. For the HD-SCA/Genomics workflow results presented in Figure 3, samples were stained with 4-colors, adding AR to the DAPI, CD45 and CK panel described above.
Figure 3.
Genomic profiles of cells found in the blood and bone marrow from the prostate cancer patient. (a) Genomic heat map displays the CNV profiles (chromosomal deletions in blue and amplification in red) for the CTC/DTC analyzed from the prostate cancer patient. (b) Representative CNV profiles of a CTC (left) and DTC (center, right) including 40x immunofluorescent composite images (DAPI, blue; CK, red; CD45, green; and AR white). The AR loci on X chromosome is highlighted (b, panel 1).
MaxPar metal-labeled antibody staining
Metal-labeled antibodies were provided by Fluidigm, either from the standard CyTOF catalog (http://maxpar.fluidigm.com/product-catalog-metal.php) or as custom conjugates (Supplementary Table 1). Metal-labeled antibody cocktails were prepared in 0.1% Tween-20, 1% BSA in PBS as per the dilution scheme presented in Supplementary Table 1. All samples, prepared as described above, were first blocked with 1%BSA and 0.2mg/mL mouse IgG Fc fragment (Thermo Scientific) in PBS for 30 min and then incubated with antibody cocktail for 1.5 hours at RT, followed by washing with PBS and staining with DNA intercalator Ir-193 (Fluidigm) and cell membrane counter-stain In-115 (Fluidigm) for 30 minutes. Slides were again washed with PBS and rinsed with ddH20 for 5 seconds and dried overnight at room temperature prior to IMC analysis.
Statistical considerations and data analysis
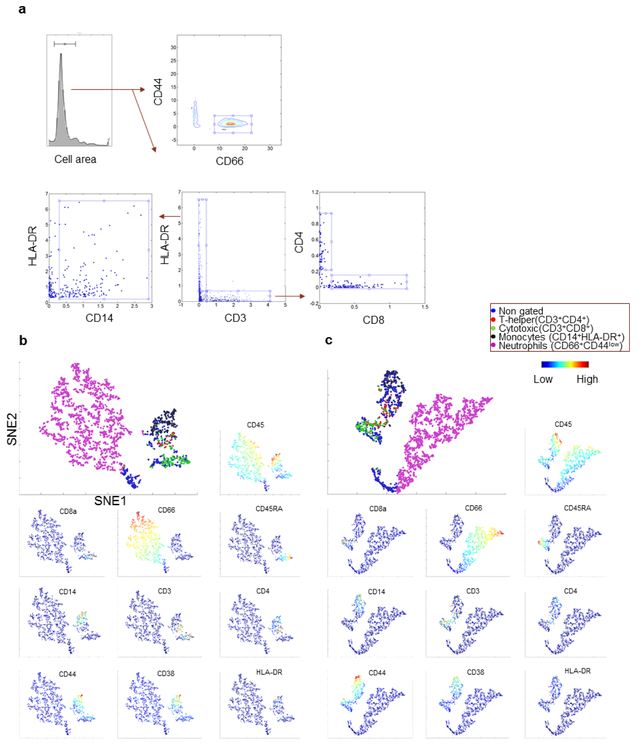
Cell boundaries were determined by the segmentation method described in the results section, using the Bioconductor package EBImage in R [10]. The standard deviation over the mean (SDOM) per cell for each channel was defined as the difference between the average ion count for the cell and the mean signal for all the cells divided by the standard deviation of all cells within each ROI (400 × 400 um). The noise level, used as a denominator for the S/N ratio was approximated as 1.5 × IQR (inter-quartile range) outside the upper quartile of all cells. When applicable, the limit of detection (LOD) for each marker was set as to mean + 3.3 × standard deviation (one sided 95% * 2) or S/N = 3 [11]. For Figure 5, the dataset was exported in a FCS format using the FlowCore package in R and the gating viSNE plots were generated with the CYT graphical software package [12] in MATLAB.
Figure 5.
Immune cell phenotyping. Identification of subpopulations of leukocytes within the ROIs analyzed for sample MDA-42109 showing (a) conventional biaxial gating of cells (after initial size exclusion of non-single cells) to identify neutrophils (CD66+), cytotoxic T-cells (CD3+, CD8+), t-helper cells (CD3+, CD4+) and monocyte populations (HLA-Dr+, CD14+). (b) viSNE plots (SNE1 vs SNE2) of around 2000 cells captured with IMC in blood and (C) bone marrow, respectively. The larger image shows the same populations as gated above, overlaid by color. The smaller images show the individual markers with the expression represented as a gradient from low (blue) to high (red).
Results
The HD-SCA/IMC integrated workflow
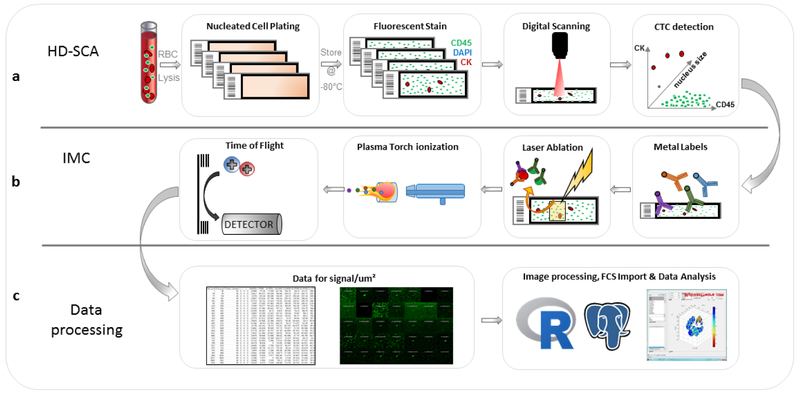
Both the HD-SCA and the IMC were designed to operate on standard glass microscope slides in keeping with general procedures in the clinical pathology laboratory setting. The common slide format facilitated the integration of the two technologies into a single workflow viable for both basic and clinical research applications. The workflow, shown in Figure 1a, begins with processing the blood sample and spreading all nucleated cells on slides, followed by 3- or 4-color immunofluorescent staining and high-speed digital scanning to identify and image cells of interest. The coordinates of each candidate cell are recorded and reimaged at 40x for morphometric measurements, as previously reported[6, 8]. In preparation for the IMC (Fig. 1b), slides are re-incubated with MaxPar™ metal-labeled antibodies. The slide is placed in the IMC instrument and candidate cells are relocated using a previously determined offset between coordinates from the 40× imaging microscope and the IMC stage. A 400×400 μm ROI around the cell of interest undergoes laser ablation aerosolizing a 1 μm2 area/pulse (200 Hz), followed by ionization and quantification in the CyTOF Helios instrument. Ion mass data is collected for each pulse and processed to render images for each individual channel at 1 μm resolution, where the intensity of each pixel corresponds to the ion count value. The images are analyzed using an in-house developed software application in R. Raw data and processed results are stored in a PostgreSQL database enabling export of datasets and integration with HD-SCA assay parameters (Fig. 1c).
Figure 1.
Schematic workflow for protein profiling of rare cells identified with the HD-SCA workflow. (a) Sample processing and fluorescent imaging; (b) IMC staining and data acquisition; (c) Data processing.
Although the basic formats of the two assays are similar, adapting the IMC technology to the HD-SCA workflow required the development and validation of staining and analysis methods consistent with the previously validated HD-SCA protocols. The antibody preparation and staining steps were initially developed and optimized using cell lines spiked into normal donor blood samples (Methods).
For this study, a panel of metal-labeled antibodies was designed to include targets relevant to prostate cancer. The panel included PSA, PSMA and two epitopes for AR, along with epithelial markers EpCAM, E-cadherin, cancer stem cell marker CD44, EMT marker B-catenin, as well as a subset of markers for leukocyte characterization [13-15]. HLA-DR and CD14 were selected for monocytes identification along with CD3, CD8, CD4, CD45RA and CD38 to further characterize the T cell population [16]. CD66 was selected to identify mainly the neutrophil population, however, CD66 or CEA (carcinoembryonic antigen) is also expressed in in tumor cells and is known to have a tumor suppressive role in prostate cancer [17]. All catalog antibodies had been previously validated by Fluidigm for use in the CyTOF/Helios systems, but had not been validated in the IMC modality nor tested in the context of the HD-SCA workflow.
Each metal-labeled antibody was first tested for specificity and sensitivity against the prostate cancer line LNCaP, and metastatic breast cancer line MDA-MB231 (Materials and Methods and Supplemental Fig. S1), using the leukocyte populations surrounding the candidate cells as an additional control. The unique challenge of determining the presence of the various antigens on the single candidate cell within each ROI in comparison to flow-based methods required the development of routines for quantification and scoring as described below.
Cell segmentation and statistical analysis
As the pulsed laser sequentially ablates subcellular areas across the candidate cell along with all of the ~300 leukocytes present in the defined ROI, an ion count value is reported for each isotope at each of the 160,000 acquired pulses. The resulting data matrix is stored in a text file from which false color 1 um resolution images are generated from each array of ion count values. The range of ion count values is internally normalized within each image, usually so that the brightest pixel represents the 98th percentile of the cumulative ion count signal.
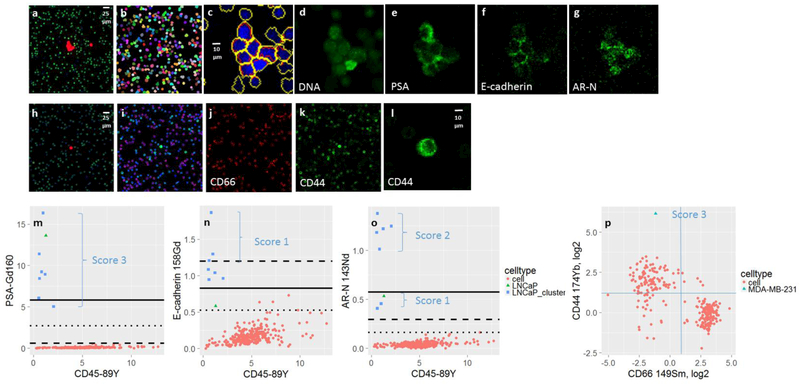
Figure 2 shows an example of the images obtained from the IMC analysis of the two spiked cell lines. In the top row, an aggregate of LNCaP cells is seen in the center of each frame. A single LNCaP cell is also captured within the ROI. The different localized signal for PSA (membrane, Fig. 2e), E-cadherin (cell junctions, Fig. 2f), and androgen receptor (translocated to nuclei upon activation, Fig. 2g), can clearly be seen. For segmentation, the DNA intercalator data (Fig. 2d) is used to identify the objects in each frame. Adaptive thresholding[10] is utilized to generate a binary image, and then applying a watershed algorithm [18] to separate objects that are touching. Finally, the membrane counterstain is used for Voronoi propagation [19]to define the cell boundaries of each object with the primary mask acting as seeds (Fig. 2c). The mask is used to quantify the signals associated with each cell for each channel, by applying the mask on the raw data array of ion count values, rather than the normalized image. The ion count signals within the boundaries of each segmented cell in the ablation field are used to generate biaxial plots showing the average signal per cell for each marker relative to a chosen leukocyte marker, typically CD45 (Fig. 2m-o). This quantitation leads to a heuristic scoring system for each marker as described below.
Figure 2.
Illustrations of segmentation and scoring of individual cells and aggregates. (a) HD-SCA composite image of LNCaP cells spiked in normal blood containing a cluster of cells in the center of the frame along with a single cell to the right. (b) Colored segmented cells in the IMC ROI. (c) Zoomed in LNCaP cluster of cells segmented using the DNA-Intercalator signal (blue) to outline the nuclei (yellow) as primary masks acting as seeds for propagation onto the membrane image to identify the outer cell boundaries (red). (d-g) Images showing the signal for individual IMC channels as labeled. (h) HD-SCA composite image of a single MDA-MB-231 cell spiked in normal blood. (i) Composite image, showing CD66 (red), CD44 (green) and DNA (blue), clearly showing the mutually exclusive populations of leukocytes (j-k). (l) Enlarged image of the MDA-MB-231 cell stained with CD44. (m-o) Biaxial plots of the segmented cells from ROI in Figure 2b with channels specified on the y-axis and CD45 on x-axis. The LNCaP cells are highlighted as green triangles (part of the cluster) or blue squares (single cell), with the associated score. The lines represents the estimated LOD based on either S/N (dashed), SDOM for all cells (solid) or SDOM based on the leukocytes only (dotted). (p) Biaxial plots of CD66 vs. CD44 on a log2 scale of the cells present in the ROI in I along with gating lines in blue. The MDA-MB-231 cell is shown as a blue triangle, scored as a 3.
Design of a scoring system for marker classification
In practice, the quantitation of different markers is hampered by i) subjectivity in adjusting the gain for normalization of images, ii) a wide dynamic range of protein expression, iii) inherent differences in detector sensitivity for the metal ions [20], iv) experimental variation due to instrument fluctuation [21] and differences between staining batches etc., and v) the difficulty of separating low abundant protein signals from background noise. As alluded to above, these challenges exacerbated in the setting of evaluating rare single cells, as compared to the common application of cytometry where larger populations are characterized.
Classification based on ion count alone was deemed unfeasible due to vast differences in dynamic range for each antibody-analyte pair, impeding normalization of the signals and their interpretation in heat maps. Thus, to establish an objective, semi-automated means for evaluating signals, a four level scoring system was created, with 0 being below the limit of detection (LOD), 1 exceeding LOD, and levels 2 to 3 set to discriminate the highest signal levels. The scoring system is based both on standard deviation over the mean (SDOM), and signal to noise (S/N) of the ion count value for the candidate cell to the surrounding leukocyte population, providing a relative measurement that is less sensitive to experimental variation than ion count alone. The limit of detection (LOD) for each marker was set as equal to S/N = 3 or SDOM = 3.3[16]. Cells exceeding the LOD were scored as 1+, signals with S/N of 7-20 or SDOM>6 were scored 2+, and finally score 3+ was assigned to signals of S/N>20 or SDOM>12. In Figure 2m-p, the scores are marked in the plots along with the estimated LOD based on either S/N = 3 (dashed lines) or SDOM = 3.3.
Some of the markers tested, including CD44, CD66, and vimentin, are also present on subpopulations of leukocytes. As seen in Figure 2i-k, most of the cells in the ROI express either CD44 or CD66, comprising two mutually exclusive subpopulations. In such cases, manual or automated gating can be readily applied to score the cell of interest, as demonstrated for a spiked MDA-MB-231 cell shown in Figure 2p. In this case, the CD44 signal was above the gated CD44+ leukocyte population, and was given a score of 3. Additional scoring of the different markers for LNCaP and MDA-MB-231 are presented in Supplemental Figure 1.
Analysis of a liquid biopsy sample from a prostate cancer patient
To test the integration of IMC with the HD-SCA workflow and to demonstrate the added information provided by IMC over 4-color immunofluorescence, the combined HD-SCA/IMC workflow was performed on matched blood and bone marrow aspirate samples from a patient with metastatic castrate resistant prostate cancer (MDA-42109) collected as part of a larger study [8]. In the initial HD-SCA assay of MDA-42109 conducted in 2013, 9 CTCs/ml from blood and 207 DTCs /ml in bone marrow were identified, with 48% of the CTCs and 78% of the DTCs also scoring positive for androgen receptor (AR) expression[22] High resolution (40x) images had been collected for all the CTCs and a subset of DTCs to verify the AR positivity and subcellular localization prior to isolating the cells for whole genome amplification using the HD-SCA genomics workflow [7] The results showed that 41/42 of the successfully amplified cells yielded highly rearranged and closely related copy number profiles with common elements typical of metastatic prostate cancer, indicating a single cancer lineage with 4 distinct sub clones as shown in the heat map and representative profiles (Fig. 3a). The high degree of clonality in the CNV profiled cells provided assurance that the candidate CTCs and DTCs selected for subsequent imaging mass cytometry analysis were highly likely to be bona fide cancer cells from the same lineage. However, we note that despite sharing the same genotype, individual cells exhibit significant phenotypic variability in AR expression and subcellular location, consistent with previous results[7]. In most cases, amplification of the AR gene on chromosome X corresponds to a strong nuclear AR signal in the corresponding fluorescent image (Fig. 3b, panel 3, 4, 5 and 7). In other cases, the gene encoding AR was amplified even though the protein could not be detected by fluorescence (Fig. 3b, panel 1, 2 and 6). Notably, there is also a genomic subclone comprising cases where AR amplification is not observed in the CNV profile (Fig. 3b, panel 9-12) but AR is still detected in some of the fluorescent images (Fig. 3b, panel 11 and 12). Morphological traits, such as cell and nuclear size and CK staining also varied among individual cells as is evident from the fluorescent images.
To evaluate the performance of the combined HD-SCA/IMC analysis in relation to the results from the HD-SCA fluorescent assay alone, we retrieved one unstained slide each from blood and bone marrow of MD42109 from the −80°C archive where they had been stored for 3 years. The slides were stained and scanned using the standard HD-SCA 3-color protocol (DAPI, CD45 and pan-CK) to identify candidate cancer cells (Fig. 1). A total of 21 candidate DTCs and 3 CTCs were identified and their positions located on the respective slides. The stained slides were then stained with metal-labeled antibodies in preparation for imaging on the Hyperion instrument. The combined results of the HD-SCA immunofluorescence and IMC analysis of these cells are presented in Figures 4 and 5.
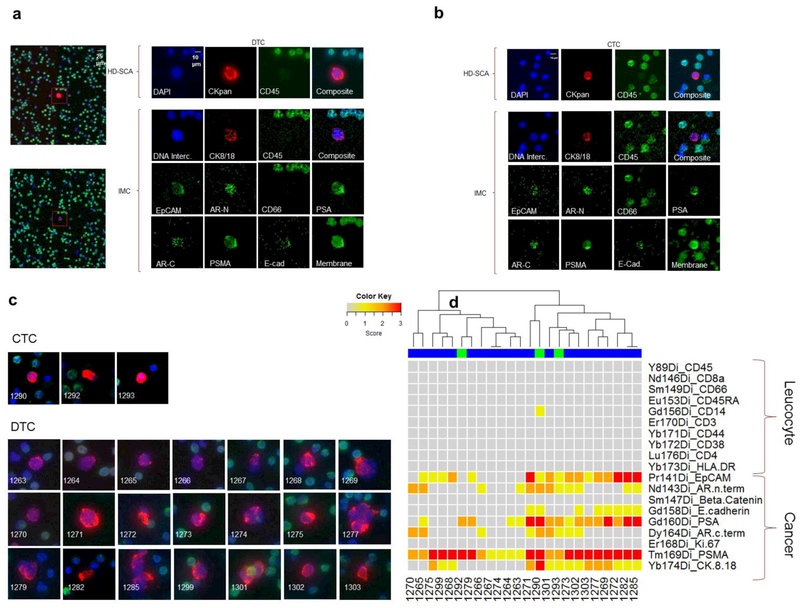
Figure 4.
Combined IMC and HD-SCA characterization of circulating tumor cells from prostate cancer blood and bone marrow. (a) DTC (#1271) from a prostate cancer patient centered in the full frame ROI (400×400) together with cropped/zoomed in fluorescent images (top panel) and the IMC rendered images (bottom panels) as comparison for channel DAPI/Intercalator (blue), CK (red), CD45 (green) and as a 3 color composite image. The IMC generated results for EpCAM, AR-N, CD66, PSA, PSMA, E-cadherin and a lipid membrane counterstain are presented in green below. (b) Images of the CTC (#1290) with a similar expression profile from the same patient as in A. (c) 40x fluorescent images DAPI (blue), CD45 (green), and CK (red) of all the cells found and analyzed in the blood and bone marrow sample taken with optimal exposure for each cell. (d) Heat map of CTCs (green banner) and DTCs (blue banner) analyzed from the same patient colored according to the four step scoring scale; below detection (gray) detectable (yellow), strong (orange) and very strong (red) depending on the detected protein.
Figures 4a and 4b show a selection of images generated in the IMC analysis of two representative cells, DTC #1271 from the bone marrow aspirate and CTC #1290 from blood, along with separate images of the markers being tested in the two modalities. The images from the fluorescence analysis of the representative cells (top panel) show that both cells were strongly CK positive and CD45 negative. The CD45 expression in leukocytes seen in the full ROI composite image generated by the IMC, show a high degree of correlation of CD45 expression ranging from low to high with the same cells in the corresponding ROI captured in the HD-SCA fluorescence assay image (Fig. 4a, Supplementary data, Fig. 1). The composite fluorescence images (Fig. 4c), demonstrate a phenotypic heterogeneity of both the morphology (size) and the variation in the CK signal among the cancer cells.
The heat map (Fig. 4d) provides a summary of the IMC signals in each channel for all 24 cells analyzed. The signal intensity for CK 8/18 varies even among the positive cells, and some of the cells that were positive for CK in the HD-SCA analysis were below the limit of detection in the IMC for CK 8/18, likely due to the fact that more CK epitopes are measured with the pan-CK antibody cocktail used in HD-SCA staining. Fifty percent (12/24) of the cells showed AR positivity in the IMC, in agreement with the parallel HD-SCA assay. Of the AR N-terminal positive cells, the majority were also positive for the C-terminal specific antibody, indicating that the full-length receptor is present in these cells. The rendered images from the IMC show a clear nuclear localization of AR, which follows upon activation of the receptor [23] (Fig. 4a and b). A majority of the cells (18/24) were positive for the epithelial marker EpCAM and 6 cells were also positive for E-cadherin, including the cell clusters 1277 and 1285 (Fig. 4c and d). Importantly, all of the cells were PSMA positive and a majority were also PSA positive, providing independent evidence for the prostate origin of the cells (Fig. 4a, b and d). Thus, the IMC analysis confirmed and complimented the fluorescent HD-SCA and genomic signatures by providing additional markers for phenotypic profiling. Further, these results demonstrate the feasibility of using archived samples for IMC analysis, allowing us to revisit and further characterize long stored and previously analyzed samples in the HD-SCA biobank.
Characterization of leukocyte populations
The HD-SCA assay is unique among the current CTC isolation methods in that it also assays and displays the full complement of leukocytes along with candidate CTC, enabling the analysis of a significant number of immune cells for multiple markers while analyzing CTCs. To demonstrate the feasibility of such an application, the surrounding leukocytes from the prostate cancer samples presented above were characterized by combining data aggregated from multiple ROIs on the slide into one dataset for each of the blood and bone marrow samples. To establish a reproducible analytical workflow, the first step was to remove all poorly segmented cells by size selection. Next, by gating the data (2089 cells) based on the average ion counts for each leukocyte on biaxial plots, the ratio compared to all nucleated cells of neutrophils (CD66+/CD44low, 67%) monocytes (HLA-DR+/CD14+, 7%) and T-cells (CD3+/HLA-DR−, 9%) could be determined (Fig. 5a). The ratio correlated well with the manual differential count performed by a trained clinical lab technician (69% Neutrophils, 14% Lymphocytes, 8% Eosinophils, 8% Monocytes, 1% basophils). Furthermore, the T-helper CD4+ cells (2%) and cytotoxic CD8+ (5%) were also identified within the T-cell population. Amongst each of the T-helper and cytotoxic T-cell populations, we also detected small populations of activated (CD38+, <1%) and naïve/effector cells (CD45RA+/CD4+, 1% and CD45RA+/CD8+, 3.8%). In figure 5b, the signal intensity for different populations are shown overlaid in a 2D-viSNE map [12]. Similar data analysis for the bone marrow sample (2267 cells, Fig. 5c) demonstrated slightly lower ratio of neutrophils (55%), monocytes (4%) and lymphocytes (10%) compared to the blood. Thus, despite lacking markers for precursor cells in this particular analysis, the current results still indicate higher levels of immature cells present in the bone marrow compared to the blood, as expected [24].
In summary, we have developed an integrated approach of CTC and DTC characterization based on the HD-SCA workflow with the subsequent downstream multiplex proteomic analysis using imaging mass cytometry. The method was demonstrated using CTCs, DTCs as well as surrounding leukocytes in samples from a bone metastatic prostate cancer patient, showing added value to the data generated by the HD-SCA assay alone. Results were compared to genomic profiling of matching cells from the same samples, showing a higher degree of heterogeneity on a protein level than on the genomic level.
Discussion
The results presented here provide proof of concept for the application of the combined HD-SCA/IMC workflow for targeted proteomics analysis on rare cancer cells and cell aggregates, as well as associated leukocyte populations, using blood and bone marrow aspirate from a metastatic prostate cancer patient. To achieve these results, we have coupled IMC, using the Fluidigm Hyperion instrument, with the existing HD-SCA method for CTC/DTC identification, morphometrics and single cell genomics. Significantly, this additional downstream high-content analysis was integrated without the need to modify any of the previously validated upstream processes of the HD-SCA. This continuous workflow enables a strategy of deep proteo-genomic analysis applicable for translational science studies within clinical trials. While the technology is currently applied as a discovery tool for markers or new combinations of markers, it has the potential for clinical validity either as a broader pathology platform, or during the development phase of a specific test. Typically, after discovery of a potential biomarker using such a high dimensional technology, a less complex, more focused assay based on a limited number of reagents and suitable for routine clinical implementation would likely be created, as demonstrated by Scher and colleagues [9].
The opportunity to utilize targeted proteomics for research on ultra-rare cells such as CTCs from liquid biopsies lies in the high level of multiplexing available to create ‘smart panels’ with a wide, purposeful spectrum of targets such as tissue specific markers and known markers of therapeutic importance, but also markers of discovery, such as mechanism of immune surveillance and escape [25] and markers for important cell states, including, dormancy, EMT, vascular mimicry and stemness. In practice, unique panels would be constructed for specific clinical studies. In this demonstration study, for example, we included markers for characterization of the surrounding leukocyte population. Patient specific immune signatures in addition to the cancer cells could potentially provide insight for classification and monitoring of cancer for precision medicine purposes [26, 27], and the possibility to do so simultaneously is a unique feature of the HD-SCA assay compared to other CTC technologies. The current panel contained 4 prostate specific markers plus 6 epithelial and cell signaling or cancer-associated markers, and 9 common leukocyte markers, plus a direct conjugated DNA intercalator and membrane counterstain. This combination of markers allowed us to both confirm the tissue of origin for the candidate prostate cancer cells and identify specific populations in the surrounding leukocytes, and further, to test the expression of additional proteins in the candidate cells.
In classifying rare cells it is important to establish normalization and quantification methods to enable cell to cell comparisons as well as comparisons between experiments. This is especially true for those proteins where expression approaches the limit of detection. Each individual antibody assay has to be evaluated in order to estimate the detection limit and linear range. Quantitative scoring of protein expression in individual candidate cells is an ongoing process. The scoring method presented in this study is rooted in that developed for the fluorescent measurements in the HD-SCA, wherein the signals from the surrounding white blood cells are used to establish a negative control baseline for most epithelial tissue markers and a means for normalizing signals from candidate cells. The SDOM measure as applied in the conventional HD-SCA assay is a straightforward means for establishing the background level based on the surrounding leukocytes. SDOM is however sensitive to outliers and is poorly suited for skewed populations, in particular for signals close to zero. SDOM is also limited in the case of CTC clusters, where multiple cells of interest contribute to the overall mean and standard deviation, as illustrated in Figure 2 (m and o) where the LOD (mean + 3.3 σ) is estimated by excluding the 7 cluster cells from the overall calculation (dotted lines) in comparison to estimate based on all cells (solid lines). In contrast, the S/N measure based on the inter-quartile range was introduced because it is less sensitive to outliers, but on the other hand it gives only a rough estimate based on the background signal. Hence, both measures are currently taken into account to provide an objective score.
It should not be overlooked that each type of preparation, whether fresh frozen tissue, FFPE sections or the HD-SCA, requires separate validation of the antibody repertoire [28, 29]In the process of integrating the two platforms, we noted that up to half of the antibody conjugates previously validated for suspension CyTOF did not work well or at all on the glass substrate HD-SCA preparations. We have therefore developed a standardized assay development process for the HD-SCA/IMC workflow using cell lines spiked into normal blood samples. Optimization of sensitivity and specificity for some problematic markers often required iterative testing of multiple antibody clones and metal-conjugate combinations. Expanding the repertoire of validated conjugates in the HD-SCA/IMC setting is a work in progress.
Rare cells from liquid biopsies show great potential for assessing the effectiveness of cancer therapy and detecting resistance or progression in near-real time through a minimally invasive procedure that can be repeated on a time scale of days or weeks, rather than months or years. New technologies such as cell free DNA and single cell sequencing add volumes of new information to what was once limited to enumerating cancer cells in the blood. The HD-SCA workflow is a direct analysis platform that provides the opportunity for a complete characterization of the blood sample without enrichment and that accommodates multiple assay modes including both morphometric and molecular level analysis. The integration of the HD-SCA workflow with single cell targeted protein analysis creates a true multiplex technology for liquid biopsies that is accessible for both basic and translational clinical studies.
Conclusion
This study provides proof of concept that IMC analysis adds another dimension to the established HD-SCA workflow, enabling multiplex proteomic profiling in addition to morphometric and genomic characterization of ultra-rare circulating tumor cells. Importantly, this study provides evidence that samples stored for several years can be revisited and analyzed de novo as new protein targets are identified. With a multiplexing level of (currently) 40 proteins, highly informative biomarker panels can be readily tailored for the scientific question at hand, which gives the HD-SCA/IMC technology immense potential for deriving predictive and prognostic biomarkers from the liquid phase of cancer.
Supplementary Material
Acknowledgements
This work was based wholly or partially on research supported by Breast Cancer Research Foundation 004698-00003; National Cancer Institute and Leidos Biomedical Research, Inc HHSN261200800001E; Prostate Cancer Foundation 16CHAL04; Vassiliadis Research Fellowship, Polak Research Fellowship, the Vicky Joseph Research Fellowship, and the Charles University Research Fund (Progres Q39). USC is an early access partner of Fluidigm for the evaluation of the Hyperion System and Fluidigm provided both expertise and reagents to support this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of these funding agencies and foundations. Competing Financial Interests are PK: Advisor to Epic Sciences, royalty recipient from Epic Sciences, shareholder at Epic Sciences; JBH: On the Clinical Advisory Boards of Epic Sciences, Inc., La Jolla, CA and CelMatix, Inc. of NY, NY.
REFERENCES
- 1.Baca Q, Cosma A, Nolan G, Gaudilliere B. The road ahead: Implementing mass cytometry in clinical studies, one cell at a time. Cytometry Part B, Clinical cytometry 2017;92(1): 10–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell 2016;165(4):780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Analytical chemistry 2009;81(16):6813–22. [DOI] [PubMed] [Google Scholar]
- 4.Wang HA, Grolimund D, Giesen C, Borca CN, Shaw-Stewart JR, Bodenmiller B, Gunther D. Fast chemical imaging at high spatial resolution by laser ablation inductively coupled plasma mass spectrometry. Analytical chemistry 2013;85(21): 10107–16. [DOI] [PubMed] [Google Scholar]
- 5.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature Methods 2014;11(4):417–22. [DOI] [PubMed] [Google Scholar]
- 6.Marrinucci D, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Physical biology 2012;9(1):016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dago AE, et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PloS One 2014;9(8):e101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson A, Kuhn P, Luttgen MS, Dizon KK, Troncoso P, Corn PG, Kolatkar A, Hicks JB, Logothetis CJ, Zurita AJ. Paired High-Content Analysis of Prostate Cancer Cells in Bone Marrow and Blood Characterizes Increased Androgen Receptor Expression in Tumor Cell Clusters. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23(7): 1722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher HI, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncology 2016;2:1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26(7):979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ICH. Q2, Validation of Anlytical Procedures: Test and Methodology. Federal Register 2005. [Google Scholar]
- 12.Amir el-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnology 2013;31(6):545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh A, Heston WD. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. Journal of Cellular Biochemistry 2004;91(3):528–39. [DOI] [PubMed] [Google Scholar]
- 14.J Jiang YG, Luo Y, He DL, Li X, Zhang LL, Peng T, Li MC, Lin YH. Role of Wnt/beta-catenin signaling pathway in epithelial-mesenchymal transition of human prostate cancer induced by hypoxia-inducible factor-1alpha. International Journal of Urology: official journal of the Japanese Urological Association 2007; 14(11): 1034–9. [DOI] [PubMed] [Google Scholar]
- 15.Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. British journal of cancer 2008;98(4):756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zola H, et al. CD molecules 2005: human cell differentiation molecules. Blood 2005;106(9):3123–6. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Tapolsky M, Earley K, Wood CG, Wilson DR, Logothetis CJ, Lin SH. Tumor-suppressive activity of CD66a in prostate cancer. Cancer Gene Therapy 1999;6(4):313–21. [DOI] [PubMed] [Google Scholar]
- 18.Beucher S (1994) Watershed, Hierarchical Segmentation and Waterfall Algorithm In: Serra J, Soille P (eds) Mathematical Morphology and Its Applications to Image Processing. Computational Imaging and Vision, vol 2 Springer, Dordrecht. [Google Scholar]
- 19.Jones TR, Carpenter A, Golland P. Voronoi-based segmentation of cells on image manifolds. Lect Notes Comput Sc 2005;3765:535–43. [Google Scholar]
- 20.Tricot S, et al. Evaluating the efficiency of isotope transmission for improved panel design and a comparison of the detection sensitivities of mass cytometer instruments. Cytometry Part A: The Journal of the International Society for Analytical Cytology 2015;87(4):357–68. [DOI] [PubMed] [Google Scholar]
- 21.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe'er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry Part A: The Journal of the International Society for Analytical Cytology 2013;83(5):483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazar DC, Cho EH, Luttgen MS, Metzner TJ, Uson ML, Torrey M, Gross ME, Kuhn P. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line. Physical Biology 2012;9(1):016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews 2004;25(2):276–308 [DOI] [PubMed] [Google Scholar]
- 24.Brooimans RA, Kraan J, van Putten W, Cornelissen JJ, Lowenberg B, Gratama JW. Flow cytometric differential of leukocyte populations in normal bone marrow: influence of peripheral blood contamination. Cytometry Part B, Clinical cytometry 2009;76(1): 18–26. [DOI] [PubMed] [Google Scholar]
- 25.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nature reviews Clinical oncology 2017;14(3): 155–67. [DOI] [PubMed] [Google Scholar]
- 26.Gruber I, Landenberger N, Staebler A, Hahn M, Wallwiener D, Fehm T. Relationship between circulating tumor cells and peripheral T-cells in patients with primary breast cancer. Anticancer Research. 2013;33(5):2233–8. [PubMed] [Google Scholar]
- 27.Wistuba-Hamprecht K, et al. Establishing High Dimensional Immune Signatures from Peripheral Blood via Mass Cytometry in a Discovery Cohort of Stage IV Melanoma Patients. Journal of Immunology 2017;198(2):927–36. [DOI] [PubMed] [Google Scholar]
- 28.Howat WJ, Lewis A, Jones P, Kampf C, Ponten F, van der Loos CM, Gray N, Womack C, Warford A. Antibody validation of immunohistochemistry for biomarker discovery: recommendations of a consortium of academic and pharmaceutical based histopathology researchers. Methods 2014;70(1):34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, et al. A proposal for validation of antibodies. Nature Methods 2016;13(10):823–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.