Abstract
Imaging mass spectrometry (IMS) has emerged as an important imaging modality because of its broad non-specific nature for molecular detection from highly complex samples. Within this broad field, new sub-categories of technologies have been developed incorporating many different molecular ionization processes. This article will focus on one of the major ionization processes, matrix-assisted laser desorption ionization (MALDI). IMS provides a critically important technology that brings new insight into complex biological samples and opens the door to new discoveries. Applications range widely, from fundamental studies in biology to specific clinical issues, all addressing the need to understand molecular spatial distributions at the tissue and cellular levels.
Keywords: molecular images, image fusion
INTRODUCTION
Imaging technologies pervade our lives ever more so in the 21st century, both in our personal and professional activities. In the field of science, many imaging modalities exist that provide enormous amounts of information for the human intellect to digest, from simple visual observations dating back several hundreds of years to more detailed observations through instruments, such as the first compound microscope introduced by Galileo in 1609, to modern day technologies such as MRI and computed tomography instruments, among others. In recent decades, microscopies based on molecular detection have been developed to aid us in our understanding of the natural world, especially our grand attempt to understand the detailed molecular basis of life. Many of these modalities are targeted, for example, using specific isotopes or chemical and biologic reagents aimed at imaging specific molecules in a complex biologic medium.
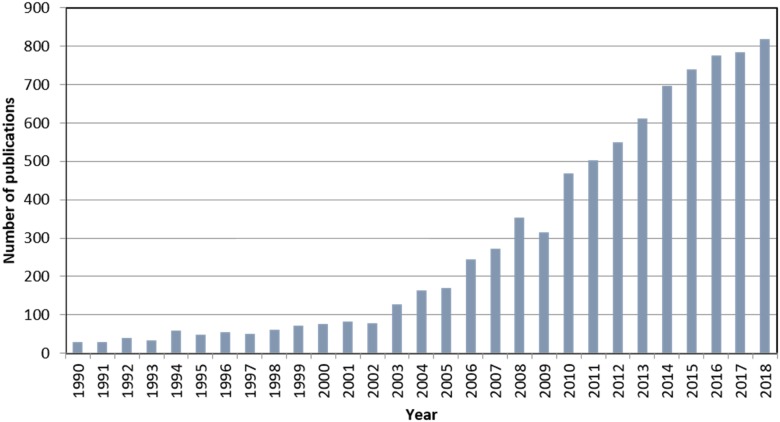
Imaging mass spectrometry (IMS) has emerged strongly in the last decade or so because of its broad nonspecific nature for molecular detection even in highly complex samples. It can image most biomolecules, including peptides, proteins, lipids, sugars, metabolites, oligonucleotides, and xenobiotics such as drugs and their metabolites. Within the field of IMS, dozens of new technologies have emerged that encompass a wide variety of ionization methods to produce ion maps of a particular sample. Figure 1 shows the growth of this field in terms of papers published over the past 28 yr or so, with data obtained through a search for the terms “imaging” and “mass spectrometry.” Of course, many, many more applications and reports that utilize this technology in research labs and industry labs exist but are not public, and so they are practically impossible to capture. Because the breadth of this field today is truly wide, the examples chosen for illustration in this article will be work from the author’s lab, where matrix-assisted laser desorption/ionization (MALDI) is the primary ionization process used for IMS.
FIGURE 1.
The number of publications on the topic of IMS obtained from an Internet search using the key words “imaging” and “mass spectrometry.”
Commonly, MALDI IMS is performed in the microprobe mode; that is, each desorption spot or pixel on the target is recorded individually.1 Although early on this led to very long acquisition times for a typical sample, modern laser technologies used on imaging mass spectrometers can acquire from 30 to 50 pixels per second. Thus, for a 5 mm square sample in which the laser spot size diameter is 10 μm with adjacent ablation spots, assuming 10 shots per pixel, an acquisition time of this area would be about 1.5 hours. From this data set, after removing multiple bins across a single peak and removing isotope peaks, several hundreds of images can be produced each at a different m/z value. This molecular specificity is enormous and is unique to image analysis using mass spectrometry (MS) technology.
The highest spatial resolution routinely achievable with today’s off-the-shelf technology is about 10 microns. In labs where specialized research instrumentation is used, resolutions of 1 micron or less have been achieved.2 However, spatial resolution is but only one of the parameters that are important in setting up an imaging experiment because there are several trade-offs that should be carefully considered. In going to higher and higher spatial resolutions, the sensitivity per pixel decreases as a result of the smaller area ablated, and the acquisition time and data file size increases, all as a square function. Thus, the spatial resolution chosen should be set no higher than what is needed to answer the question being asked.
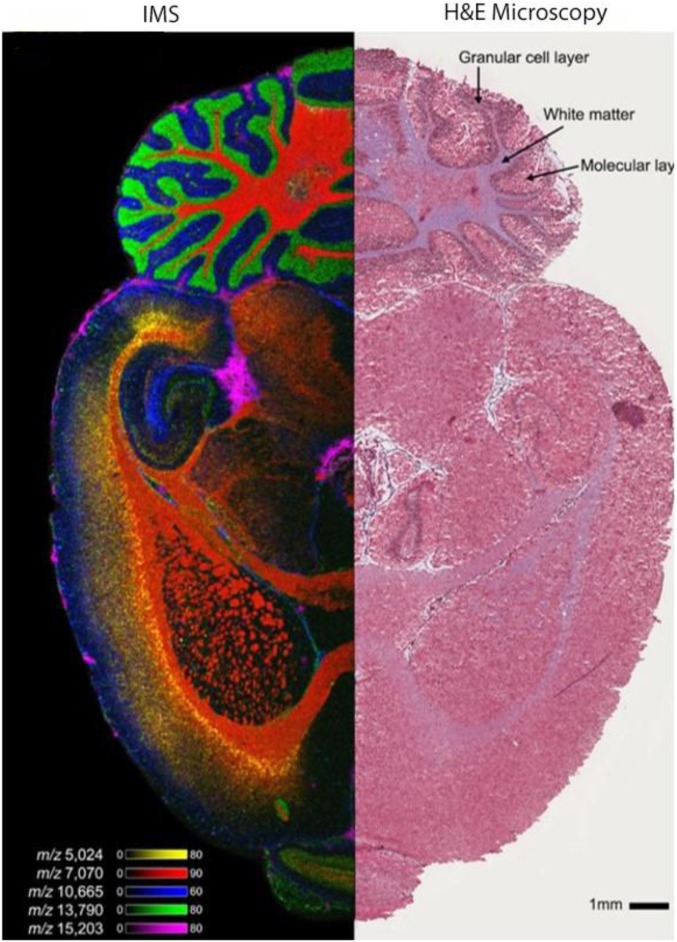
Perhaps the greatest impact of the technology comes in the area of imaging biologically relevant specimens, particularly tissues. Applications in this area have included the mapping of molecular distributions in basic biologic research to better understand the spatiotemporal aspects of biology and in clinically relevant areas to provide better diagnoses and prognoses and to assess treatment in disease. However, tissue samples represent some special considerations for the imaging experiment, with some being more or less critical depending on the spatial resolution required. Such issues as flatness of the tissue sample, flatness of the sample holder in the MS ion source, the matrix used, the homogeneity of the matrix application, and size of the solid matrix crystals all play a part in achieving images of optimal quality. Nevertheless, standard protocols have been developed that make obtaining high-quality images relatively straightforward. Figure 2 shows an ion image of a mouse brain with 1 side imaged by MALDI IMS and the other by microscopy of a hematoxylin and eosin (H&E)–stained half section. The IMS data were acquired on a Bruker Rapiflex MALDI Time-of-Flight (TOF) (Bruker, Billerica, MA, USA) instrument at a spatial resolution of 15 μm. Several color ion images are shown that were taken from the several hundreds of individual ion images produced from this data set. The raw image data set is just over 100 GB in size.
FIGURE 2.
Left panel: IMS image of a section of mouse brain showing the location of 5 different low MW proteins. The data were obtained on a Bruker Rapiflex MALDI TOF MS at a spatial resolution of 15 microns and the UV laser set at 10-kHz repetition rate. The data file size is ∼100 GB. Right panel: Microscopy image of an H&E stained section of mouse brain.
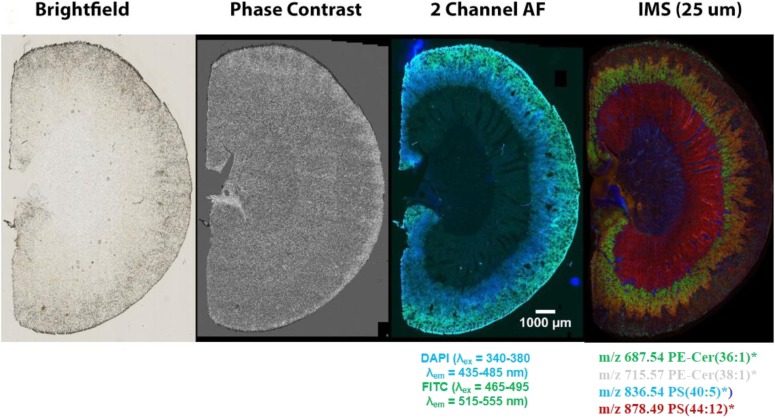
Often, investigators wish to match an MS image to one that was obtained from another imaging modality, for example, commonly from microscopy. In the early development of IMS, this was performed by using 2 serial sections of a tissue sample cut on a cryostat, 1 imaged by each modality, and subsequent overlaying of the 2 images with appropriate software such as Photoshop or a similar program. Although this process is acceptable for a rough comparison, it has some drawbacks. First, 2 adjacent sections by definition are not identical and, in some tissues, may show significant differences in shape and structural and molecular content. Of course, the best solution is to perform both image acquisitions on the same piece of tissue. However, because stain applications often compromise the integrity of the tissue, we have implemented another approach that involves fluorescence imaging of a single section followed by MS image analysis of that same section.3 Figure 3 shows the comparison of images of mouse kidney sections from several different modalities. Clearly, the fluorescence image provides great structural detail so that it can accurately be overlaid with MS ion images.
FIGURE 3.
Images of a mouse kidney section taken with different imaging modalities. The combination of the fluorescence image, which clearly shows tissue morphology, with IMS images, which gives unparalleled molecular specificity, is an excellent combination for image overlay to extract maximum spatial molecular information from discrete areas within the tissue.
Another application that has special considerations and requires a different set of protocols are tissue biopsies that are obtained from clinical cases. First, most samples of this type come through pathology as formalin fixed paraffin embedded (FFPE) samples. In addition, it is common that the histologist or pathologist does not necessarily need an image of the entire biopsy but rather discrete areas of the tissue that are of concern following normal microscopic examination. We term this type of analysis as “histology guided imaging” because it targets specific areas of the tissue for MS image analysis discerned from a microscopy image.4 For FFPE preserved tissue, protein analysis is achieved by deposition of microdroplets of trypsin in the designated areas to perform in situ trypsinolysis. Free peptides are produced from domains within the proteins between crosslinks. Peptides in these digested areas are desorbed by the laser and are then directly sequenced to identify the protein parent proteoform following a search of a protein database, as performed in a typical liquid chromatography–tandem MS analysis. With respect to the analysis of lipids and metabolites in FFPE tissue, although some of these compounds remain after the fixation protocols, the amount remaining is unknown, so any quantitative conclusions should be made with caution. The analyses of fresh frozen biopsies do not suffer this disadvantage but generally are not as easily obtained in the normal clinical biopsy processing protocol.
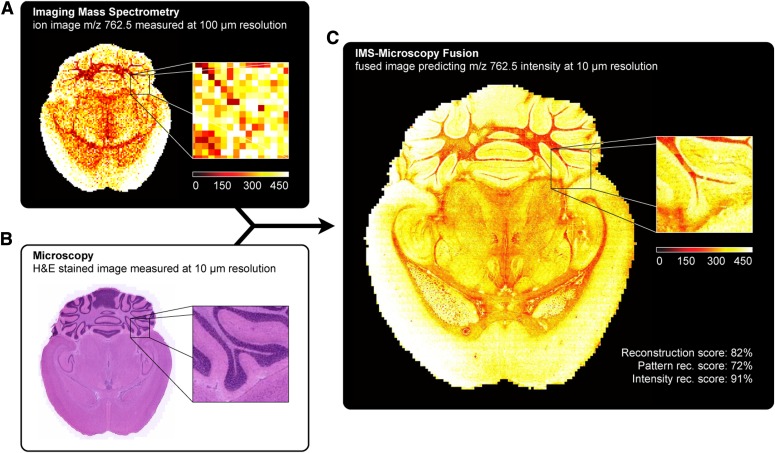
We have developed image fusion as a means of mining relationships between different imaging modalities and have demonstrated its use for predictive applications.5 In this process, IMS-generated molecular maps, rich in chemical information but having lower spatial resolution, are combined with optical microscopy maps, which have relatively low chemical specificity but higher spatial resolution. The resulting mathematically generated “predicted” images combine the advantages of both technologies, enabling an image of a molecular distribution both at high spatial resolution and at high chemical specificity. Multivariate regression is used to model variables in 1 technology, using variables from the other technology. In order to validate the quality of the predictions, each ion is scored based on the strength of the correlation found between the 2 modalities. Several applications have demonstrated the remarkable potential of image fusion in several different areas. First, “sharpening” of IMS images (i.e., the process of transforming a lower-spatial-resolution MS image into a higher-resolution predicted ion image) uses microscopy measurements to predict ion distributions at a spatial resolution that exceeds that of measured ion images by 10 times or more. Second, out-of-sample prediction predicts the ion image of like tissue that has not actually been imaged. Third, implementing enrichment of biologic signals and attenuation of instrumental artifacts reveals insights that are not easily extracted from either microscopy or IMS separately. Figure 4 shows an example of fusion-driven sharpening of an ion image. This example demonstrates the remarkable fidelity that can be achieved by image fusion for selected ions. In short, image fusion enables a new multimodality paradigm for tissue exploration whereby mining relationships between different image sources yields novel insights that cannot be gleaned from the use of the individual technologies alone.
FIGURE 4.
Prediction of the ion distribution of m/z 762.5 in mouse brain at 10-μm resolution from 100-μm IMS and 10-μm microscopy measurements (sharpening). This figure fuses a measured ion image for m/z 762.5 [identified as lipid phosphatidyl ethanolamine (PE) (16:0/22:6)] at 100-μm spatial resolution (A) with a measured H&E-stained microscopy image at 10-μm resolution (B), predicting the ion distribution of m/z 762.5 at 10-μm resolution (C).
CONCLUSIONS
Science has witnessed remarkable gains in our understanding of our natural environment in recent decades through the eyes of many different types of image generation and capture technologies. MS has now taken its place among these technologies, bringing unparalleled molecular specificity to the scientific imaging world. Over many decades, we have discovered the complex nature of living systems through their inherent biologic processes. However, these processes in themselves are complex in another sense in that they do not necessarily take place at the same time or the same place in multicellular organisms but nevertheless exist in concert and harmony with each other. IMS provides a critical tool that brings new insight into these processes and opens doors to new discoveries. Moreover, this technology is innovative in clinical areas, helping define molecular mechanisms of compromised or diseased areas of tissue. Specifically, it provides a new molecularly specific view for pathologists to better diagnose disease and helps bring us closer to individual patient care and treatment. Of course, there are remaining challenges, but what has occurred thus far reinforces our vision of where this technology will take us through continued innovation and application of IMS to problems critical to our survival.
ACKNOWLEDGMENTS
Financial support from the National Institute of General Medical Sciences, National Institutes of Health, Grant 5P41GM103391‐08 is gratefully acknowledged.
REFERENCES
- 1.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. [DOI] [PubMed] [Google Scholar]
- 2.Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM. Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal Bioanal Chem. 2015;407:2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patterson NH, Tuck M, Lewis A, et al. . Next generation histology-directed imaging mass spectrometry driven by autofluorescence microscopy. Anal Chem. 2018;90:12404–12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornett DS, Mobley JA, Dias EC, et al. . A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5:1975–1983. [DOI] [PubMed] [Google Scholar]
- 5.Van de Plas R, Yang J, Spraggins J, Caprioli RM. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat Methods. 2015;12:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]