Abstract
Non-small-cell lung cancer (NSCLC) can be identified by precise molecular subsets based on genomic alterations that drive tumorigenesis and include mutations in EGFR, KRAS, and various ALK fusions. However, despite effective treatments for EGFR and ALK, promising therapeutics have not been developed for patients with KRAS mutations. It has been reported that one way the RAS-ERK pathway contributes to tumorigenesis is by affecting stability and localization of FOXO3a protein, an important regulator of cell death and the cell cycle. This is through regulation of apoptotic proteins BIM and FASL and cell cycle regulators p21Cip1 and p27 Kip1. We now show that a HDAC inhibitor affects the expression and localization of FOXO proteins and wanted to determine if the combination of a MEK inhibitor with a HDAC inhibitor would increase the sensitivity of NSCLC with KRAS mutation. Combined treatment with a MEK inhibitor and a HDAC inhibitor showed synergistic effects on cell metabolic activity of RAS mutated lung cancer cells through activation of FOXOs, with a subsequent increase in BIM and cell cycle inhibitors. Moreover, in a mouse xenograft model, the combination of belinostat and trametinib significantly decreases tumor formation through FOXOs by increasing BIM and the cell cycle inhibitors p21Cip1 and p27 Kip1. These results demonstrate that control of FOXOs localization and expression is critical in RAS driven lung cancer cells, suggesting that the dual molecular targeted therapy for MEK and HDACs may be promising as novel therapeutic strategy in NSCLC with specific populations of RAS mutations.
Keywords: RAS, HDAC, MEK, lung cancer, molecular targeted therapy
Introduction
Lung cancer is the leading cause of malignancy-related deaths worldwide (1). Non-small-cell lung cancer (NSCLC) accounts for nearly 85–90% of lung cancers and overall survival is approximately 8–12 months even in good performance status patients in clinical trials with the best conventional chemotherapy (2). NSCLC can be classified by precise molecular subsets based on specific genomic alterations that drive tumorigenesis, such as the epidermal growth factor receptor (EGFR), Kirsten rat sarcoma viral oncogene homolog (KRAS), ALK, HER2, BRAF, RET, ROS1, and NRAS (3). About 15% of non-squamous NSCLC tumors in the United states have known driver alterations that are treated in clinical setting with drugs targeting a specific mutation, such as EGFR tyrosine kinase inhibitors gefitinib, erlotinib, afatinib and, osimertinib, and the ALK inhibitors crizotinib, ceritinib, and alectinib (2), which have improved the quality of life of these patients and increased overall survival when compared to conventional chemotherapy.
The most common of these oncogene mutations is activation of the RAS subfamily (most commonly in KRAS), and is detected in approximately 20% of human cancers (4). In lung adenocarcinoma KRAS is mutated in approximately 30% of cases, but is infrequent in squamous cell carcinoma (5, 6). In addition, KRAS mutations and other driver gene alternations such as EGFR and EML4-ALK are for the most part mutually exclusive (7, 8). KRAS mutations are predominantly found in current or former smokers, mainly in the Caucasian population, and rarely in Asians (9, 10). RAS mutations, including KRAS, NRAS, and HRAS cause constitutive activation of the downstream molecules in the RAS/RAF/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway. This RAS effector-signaling pathway is dysregulated in approximately 20% to 35% of NSCLC (5). Effective drugs targeting KRAS mutant proteins have not been developed, even though RAS mutations were reported more than 30 years ago. Attempts have been made to develop targeted therapies to treat RAS mutated lung cancers, such as farnesyltransferase inhibitors, however, these drugs have not been effective in the clinic (4). Most recently, allele-specific inhibitors against a constitutively active form of mutant KRAS G12C have been developed and are effective against KRAS G12C-driven cancers in vitro, however, high concentrations were needed (IC50; 2.5μM) to inhibit cell growth and the G12C mutation comprises just a portion of the KRAS mutation spectrum (11). Therefore, novel therapeutic strategies are still needed for improving the poor prognosis of patients with KRAS driven lung cancers.
Trametinib (GSK1120212), a MEK1/2 inhibitor, has been approved by the FDA for use in BRAF-mutant melanoma. Several clinical trials using MEK inhibitors have been reported in lung cancer patients with KRAS mutations. A prospective randomized phase II study was performed to assess the efficacy of trametinib as a single agent compared with docetaxel in previously treated patients with advanced KRAS mutant NSCLC. The results in each arm were similar with trametinib providing no better outcome than docetaxel (12). Another prospective randomized phase II study evaluating the efficacy of adding the MEK inhibitor selumetinib to docetaxel in previously treated patients with advanced KRAS mutant NSCLC was conducted based on pre-clinical results. Despite no differences in median overall survival, there were significant improvements in both progression-free survival and objective response rate in patients administered selumetinib (13), albeit with significantly increased toxicity. However, in the recent phase III SELECT-1 trial the addition of selumetinib to docetaxel did not improve progression free survival or overall survival in patients with KRAS mutation-positive, locally-advanced or metastatic NSCLC (14). As these results indicate, the impact of single targeted therapy in combination with a cytotoxic chemotherapy could be insufficient among patients with KRAS mutant cancers. Therefore, combining targeted therapies that hit multiple signaling pathways may be a more promising approach. The goal of this study is to determine a potential therapeutic strategy against RAS mutated lung cancer with agents that affect the FOXO transcription factors; factors known to increase apoptosis through up regulation of apoptotic proteins such as BIM and increase cell cycle inhibitors such as p21Cip1 and p27 Kip1. It is known that the protein stability, localization, and transcriptional activity of the FOXOs are regulated by both phosphorylation and acetylation (15–17).
In this study, we demonstrate the synergistic efficacy of combined targeted therapy for MEK and histone deacetylases (HDACs) through FOXO-mediated transcription of target genes in RAS driven lung cancer cells. To the best of our knowledge, this is the first report to identify the FOXO pathways as critical targetable pathways in RAS driven lung cancer. This suggests that the dual molecular targeted therapy for HDAC and MEK may be promising as novel therapeutic strategy in specific populations of lung cancer patients with mutated RAS.
Material and methods
Cell cultures and reagents
We used 10 human lung cancer cell lines and 2 human lung fibroblast cell lines. The human lung cancer cell lines, Calu-1, Calu-6, H1299, H2009, H2347, and H358 were generously provided by John Minna and Luc Girard (University of Texas, Southwestern, Dallas, TX). H292, H1395, H196, and H1581 were purchased from the American Type Culture Collection (Manassas, VA). The human lung embryonic fibroblast MRC-5 (P30–35) and IMR-90 (P20–25) cell lines were obtained from RIKEN Cell Bank (Ibaraki, Japan). Calu-1 (G12C), Calu-6 (G12C), H2009 (G12A), H358 (G12C), and H292 (G12S) have KRAS mutations, and H1299 (Q61K) and H2347 (Q61R) have NRAS mutations, and H1395 has a BRAF mutation. H196, H1581, MRC-5, and IMR-90 have wild-type KRAS/NRAS/HRAS/BRAF genes. All these cells have wild-type LKB1 genes. Calu-1, Calu-6, H1299, H2009, H2347, H358, H292, H1395, H196, and H1581 were maintained in Roswell Park Memorial Institute 1640 (RPMI1640) medium (GIBCO, Carlsbad, CA), and MRC-5 and IMR-90 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (50 g/mL), in a humidified CO2 incubator at 37°C. All cells were passaged for less than 3 months before renewal from frozen, early-passage stocks. The identity of all cell lines was authenticated by DNA fingerprinting, and all were tested to ensure that they were mycoplasma negative. Belinostat (pan-HDAC inhibitor) and trametinib (MEK1/MEK2 inhibitor) were obtained from Selleckchem (Houston, TX) and ChemieTek (Indianapolis, IN), respectively.
Proliferation assay
The cells were seeded at 2 × 103 per well in 96-well plates, and incubated in antibiotic-containing RPMI 1640 with 10% FBS. After 24 h of incubation, various concentrations of belinostat and/or trametinib were added to each well, and incubation was continued for a further 72 h. These cells were then used for proliferation assay, which was measured using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium) dye reduction method. An aliquot of MTT solution (2 mg/ml; Sigma, St Louis, MO, USA) was added to each well followed by incubation for 2 h at 37 °C as previously described (18). The media were removed and the dark blue crystals in each well were dissolved in 100 μl of dimethyl sulfoxide (DMSO). Absorbance was measured with an MTP-120 microplate reader (Corona Electric, Ibaraki, Japan) at test and reference wavelengths of 550 nm and 630 nm, respectively. The percentage of growth is shown relative to untreated controls. Each sample was assayed in triplicate, with each experiment repeated at least three times independently.
Drug Combination Studies
Characterization of synergistic interactions was quantified by the isobologram and combination-index methods by Chou and Talalay equation (19) using the CalcuSyn software (Biosoft, Ferguson, MO). The combination-index (CI) is a quantitative representation of two-drug pharmacologic interactions. A CI of 1 indicates an additivity between two agents, whereas a CI < 1 or CI > 1 indicates synergism or antagonism, respectively.
Antibodies and western blotting
Protein aliquots of 25 μg each were resolved by SDS polyacrylamide gel (Bio-Rad, Hercules, CA) electrophoresis and transferred to polyvinylidene difluoride membranes (Bio-Rad). After washing 3 times, the membranes were incubated with Blotting-grade blocker (Bio-Rad) for 1 h at room temperature and then incubated overnight at 4°C with primary antibodies to t-FOXO1, t-FOXO3a, p-ERK1/2 (T202/Y204), t-ERK1/2, Acetyl-Histone H3, p21Cip1, p27 Kip1, BIM, cleaved PARP, PARP (1:1000 dilution; Cell Signaling Technology, Danvers, MA, USA), HDAC2, or GAPDH (1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing 3 times, the membranes were incubated for 1 h at room temperature with secondary Ab (horseradish peroxidase-conjugated species-specific Ab). Immunoreactive bands were visualized with SuperSignal West Dura Extended Duration Substrate Enhanced Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL). Each experiment was performed at least three times independently.
Cell apoptosis assay
Cell apoptosis was detected with an Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences Pharmingen, Heidelberg, Germany) in accordance with the manufacturer’s protocols as we described previously (18). The analysis was performed on a FACSCalibur flow cytometer with Cell Quest software (Becton Dickinson, Franklin Lakes, NJ, USA).
RNAi transfection
Silencer® Select siRNAs for FOXO1 (s5258, s5259), FOXO3a (s5260, s5261), and BIM (s195011, s19474) (Invitrogen, Carlsbad, CA) were transfected with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s instructions. Silencer® Select siRNA for Negative Control no.1 (Invitrogen, Carlsbad, CA) was used as scramble control throughout the experiment. FOXO1, FOXO3a, and BIM knockdown were confirmed by western blotting analysis. Each experiment was performed at least in triplicate, and three times independently.
Isolation of nuclear and cytoplasmic fractions
For some experiments nuclear and cytoplasmic extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents from Pierce, and the quality of the preparations was always verified by analysis of proteins differentially enriched in the nucleus (HDAC2) or the cytoplasm (GAPDH).
Dual-luciferase reporter assay
Calu-1, Calu-6, H358, H1299, and H1395 cells were seeded onto 6-well plates at a density of 1.5 × 105 cells per well. After overnight incubation, luciferase reporter FHRE-Luc and pRL-CMV were co-transfected into cells using X-tremeGENE HP DNA transfection reagent (Roche Diagnostics; Indianapolis, IN, USA) according to the manufacturer’s protocol. Calu-1 cells were exposed to RPMI-1640 media with DMSO, 0.2μM trametinib, 2μM orapalib/LY2157299, or 0.2μM trametinib plus 2μM orapalib/LY2157299 following 24 h transfection. Calu-1, Calu-6, H358, H1299, and H1395 cells were exposed to RPMI-1640 media with DMSO, 0.2μM trametinib, 2μM belinostat, or 0.2μM trametinib plus 2μM belinostat following 24 h transfection. Firefly and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system (Promega; Madison, WI, USA) according to the manufacture’s protocol on a GloMax® 96 Microplate Luminometer (Promega; Madison, WI, USA) at 24h after initiation of exposure to each drug.
Subcutaneous xenograft models
Suspensions of H358 cells (5×106) were injected subcutaneously into the flanks of 5-week-old female nude mice (The Jackson Laboratory). After 6 days, the mice were randomized to (a) control group (vehicle treated control), (b) intraperitoneal belinostat (40 mg/kg/daily), (c) oral trametinib (1 mg/kg/daily), and (d) belinostat plus trametinib. Tumor size and mouse body weight were measured twice per week, and tumor volume was calculated, in mm3, as width2 × length/2. The animal protocol was approved by The Ohio State University Institutional Laboratory Animal Care and Use Committee (approval no. #2014A00000116).
Immunohistochemical studies for t-FOXO3a
Paraffin-embedded tissue was cut at 4–5-micron sections and placed on positively charged slides. Slides were baked at 65°C for one hour and immunostaining was performed on the fully automated Bond RX autostaining system (Leica Biosystems, Buffalo Grove, IL). Briefly, heat-induced antigen retrieval was done using ER2 (EDTA buffer) for 20 minutes, slides were stained with a rabbit monoclonal antibody to FOXO3a (clone D19A7 Cat. 12829, Cell Signaling, Danvers, MA) at a 1:800 dilution for 30 minutes and the Bond Polymer Refine (DAB) detection system (Leica Biosystems, Buffalo Grove, IL) was used.
Quantification of Immunohistochemistry for FOXO3a
Five fields containing the highest number of tumors cells were scored for the percent of tumor cells with nuclear staining by light microscopy with a 200-fold magnification. The total number of tumor cells scored in the five fields ranged from 221 to 381 per field. The tumor cells with positive staining nuclei were counted and the percentage of positive cells determined. All results were independently evaluated by two investigators (T.Y. and H.T.).
Statistical analysis
Data from MTT assay and tumor progression of xenograft model are expressed as means of ± S.D. The statistical significance of differences was analyzed by one-way ANOVA and Spearman rank correlations performed with GraphPad Prism Ver. 6.0 (GraphPad Software, Inc., San Diego, CA, USA). For all analyses, a two-sided p-value less than 0.05 was considered statistically significant.
Results
Synergistic effect between belinostat and trametinib in RAS mutated lung cancer cells in vitro.
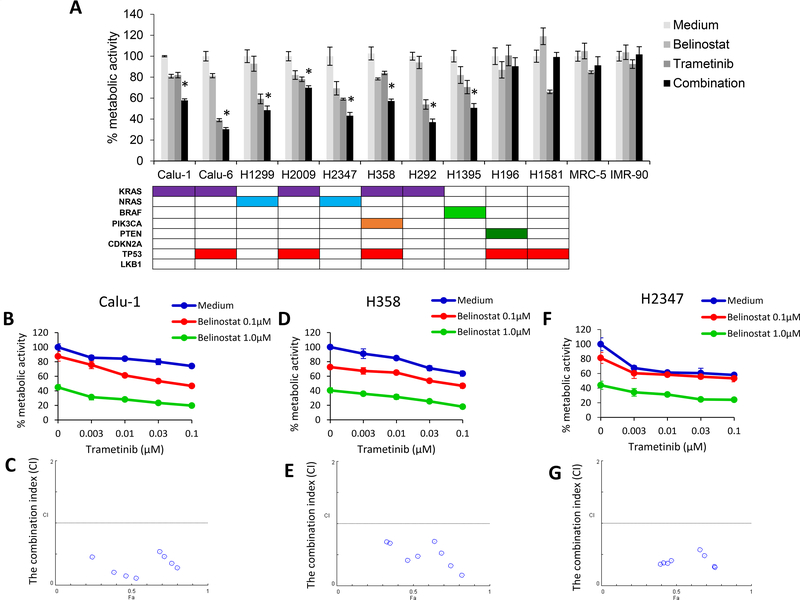
To seek a potential novel treatment for RAS mutated lung cancer patients, we evaluated the effect of trametinib in combination with belinostat on the proliferation of seven RAS mutated and four wild-type RAS cell lines, two lung cancer lines and two fibroblast cell lines. In addition, we included the BRAF mutant cell line H1395, which should also have some dependence on MEK activity. Trametinib in combination with belinostat showed a significant difference in the proliferation of all RAS mutated lung cancer cells and the BRAF mutated cell line when compared with either belinostat or trametinib alone (Fig. 1A). On the other hand, the effects of the combined therapy in the four wild type RAS cell lines were only marginal. Our data suggests that it is KRAS mutated cells that are more likely to be sensitive to the combined therapy with trametinb and belinostat, compared to RAS wild type cells. To assess the synergistic effect, we tested cell metabolic activity with the combination and determined the combination-index (CI) using the method of Chou and Talalay (19). Our data showed that the treatment with belinostat and trametinib resulted in reduced cell metabolic activity and CI values of less than 1.0 indicating synergy for Calu-1, H358, and H2347 (Fig. 1B-G).
Figure 1. Synergistic effect between belinostat and trametinib in RAS mutated lung cancer cells in vitro.
(A) Cell lines with a RAS mutation (Calu-1, Calu-6, H1299, H2009, H2347, H358, and H292), BRAF mutation (H1395), wild-type RAS/BRAF lung cancer cells (H196 and H1581), and human lung embryonic fibroblast cell lines (MRC-5 and IMR-90) (seeded at 2 × 103 per well of a 96-well plate) were incubated with belinostat alone (100 nmol/L), trametinib alone (100 nmol/L), or in combination for 72 h. The MTT assay was used to assess cells for metabolic activity. Data are representative of three independent experiments. *, P < 0.05 for combination when compared with the belinostat alone and trametinib alone. RAS mutated lung cancer cells Calu-1 (B, C), H358 (D, E), and H2347 (F, G) were treated with the indicated doses of belinostat and trametinib for 72 h incubation. Cell metabolic activity was assessed by the MTT assay. Raw proliferation data were expressed as percent of viable cells and CI values were analyzed according to the Chou and Talalay equation using the CalcuSyn software.
Inhibition of HDAC and MEK increases total FOXO1 and FOXO3a expression and regulates apoptosis and cell cycle proteins.
To explore the molecular mechanism of reduced cell numbers with HDAC and MEK inhibition in RAS and BRAF mutated cancer cells we examined the protein expression of FOXO1 and FOXO3a, as well as apoptosis-promoting protein BIM and cell cycle proteins p21Cip1 and p27 Kip1 by Western blotting (Fig. 2A and B). We also examined levels of these proteins in the fibroblast cell line, IMR-90, which was unaffected by single agent tremetinib and belinostat or the combination after 3 days of treatment (Fig. 1A and Supplementary Fig. 1).
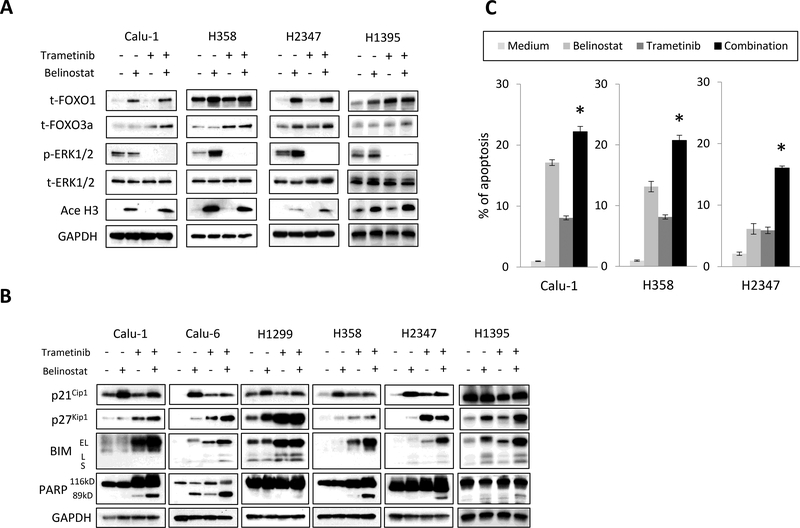
Figure 2. Inhibition of MEK and HDACs increases total FOXO1 and FOXO3a expression and regulates cell apoptosis and cell cycle proteins.
(A) Tumor cells were treated with belinostat (1000 nmol/L) and/or trametinib (100 nmol/L) for 4 h. The cells were lysed and the indicated proteins were detected by immunoblotting. The results shown are representative of 3 independent experiments. (B) Tumor cells were treated with belinostat (1000 nmol/L) and/or trametinib (100 nmol/L) for 48 h. The cells were lysed and the indicated proteins were detected by immunoblotting. The results shown are representative of 3 independent experiments. (C) After 48 h incubation with belinostat (1000 nmol/L) and/or trametinib (100 nmol/L), cell apoptosis was determined with an annexin V-FITC Apoptosis Detection Kit I. *, P < 0.05 for the combination when compared with the belinostat alone and trametinib alone.
All of the cancer cell lines and the fibroblast cell line expressed the FOXO1 and FOXO3a proteins. Belinostat alone and in combination with trametinib clearly increased total FOXO1 in all five of the cell lines and in the fibroblast line (Fig. 2A; Supplementary Fig. 1). Total FOXO3a protein was increased by the combination in all four cancer cell lines, and by trametinib alone in the Calu-1 and H358 cells or belinostat alone in H2347 cells. In the fibroblast cell line, the combination did not increase total FOXO3a protein compared with either single agent, but was still slightly increased above what was seen in untreated cells. Trametinib completely inhibited the phosphorylation of ERK1/2 proteins and belinostat increased the acetylation of histone H3 in all cancer cells. Thus, our findings showed the combination with belinostat and trametinib increases both total FOXO1 and total FOXO3a. We also investigated apoptosis and cell cycle proteins in six cancer cell lines and the IMR-90 fibroblast cell line by Western blotting because FOXO proteins are known to regulate apoptosis and the cell cycle (Fig. 2B; Supplementary Fig. 1). The effect of drug treatments on the p21Cip1 protein were inconsistent, but were always higher in cells treated with either drug, alone or in combination, when compared to untreated cells in five of the six cell lines. The p27 Kip1 protein showed more consistency and in four of the six cell lines examined was highest in the combination treatment. The most dramatic changes occurred with the apoptosis promoting protein BIM, where in five of the six cell lines BIM levels were highest when the combination therapy was used. The resulting increase in cleaved PARP, which is indicative of cell death through apoptosis, was almost always highest in cells treated with the combination. This was the case in five of the six cancer cell lines tested. In contrast to the cancer cell lines, the combination treatment did not increase the expression of p21Cip1 or p27 Kip1, BIM, and cleaved PARP in the fibroblast cell line when compared with either single agent alone (Supplementary Fig. 1). However, combination treatment did marginally increase BIM and PARP levels above those seen in untreated cells, and some toxicity with single agent and combination treatments was seen in cultured fibroblasts five and seven days after the start of treatment (Supplementary Fig. 2).
Furthermore, we also performed the apoptosis assay using Annexin V. Trametinib in combination with belinostat showed a significant increase in apoptosis of Calu-1, H358, and H2347 cells when compared with either belinostat or trametinib alone (Fig. 2C). These results suggested that combined therapy with trametinib and belinostat regulates apoptosis though BIM, and, perhaps to a lesser extent, slows cell growth through the up regulation of the cell cycle inhibitor p27 Kip1.
FOXO1 and FOXO3 accumulate in the nucleus with combination treatment and are responsible for the increased cell death through regulation of BIM.
To gain a better understanding of the mechanism of cell death with combined therapy, we examined localization of FOXO1 and FOXO3a, transcription factors that can shuttle between the nucleus and the cytoplasm. FOXO1 increased in the nuclear fraction of Calu-1 and H358 cells by treatment with either belinostat alone or in cells receiving combination treatment. This is well above the FOXO1 protein level in the nuclear fraction of untreated cells or cells treated with trametinib alone. FOXO3a, on the other hand, appeared to have the highest level of nuclear accumulation when cells were treated with belinostat in combination with trametinib. (Fig. 3A). To assess the FOXO transcriptional activity, we transfected cells with a reporter construct containing a forkhead responsive element (FHRE) that drives luciferase and treated them with trametinib, belinostat, or the combination. The enhancement of FOXO activity was induced by either belinostat alone or in combination with trametinib with a trend toward higher activity in the combination for three of the five cell lines examined (Supplementary Fig. 3).
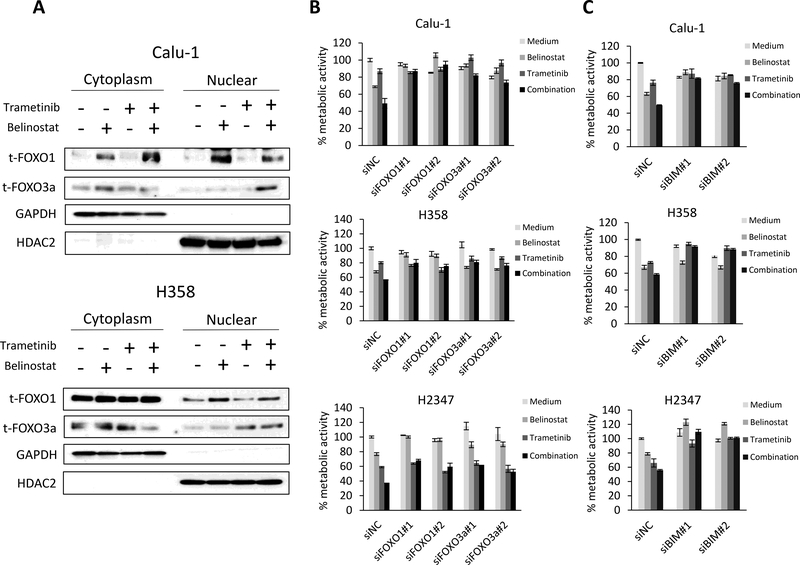
Figure 3. FOXO1 and FOXO3a protein levels increase in the nuclear fraction with combination treatment and are responsible for the increased cell death through regulation of BIM.
(A) Tumor cells were treated with belinostat (1000 nmol/L) and/or trametinib (100 nmol/L) for 4 h. The cells were lysed to extract nuclear and cytoplasmic fractions using the NE-PER Nuclear and Cytoplasmic Extraction Reagents and the indicated proteins were detected by immunoblotting. The results shown are representative of 3 independent experiments. (B) Control or FOXO1- or FOXO3a-specific siRNAs were introduced into Calu-1, H358, and H2347 cells. After 24 h, the cells were incubated with belinostat (100 nmol/L) and/or trametinib (10 nmol/L) for 72 h and metabolic activity was determined by MTT assays. FOXO1 or FOXO3a knockdown was confirmed by immunoblotting (Supplementary Fig. 4). The percentage of metabolic activity is shown relative to untreated controls. Each sample was assayed in triplicate, with each experiment repeated at least 3 times independently. (C) Control or BIM-specific siRNAs were introduced into Calu-1, H358, and H2347 cells. After 24 h, the cells were incubated with belinostat (100 nmol/L) and/or trametinib (10 nmol/L) for 72 h and lung cancer cell metabolic activity was determined by MTT assays. BIM knockdown was confirmed by immunoblotting (Supplementary Fig. 4). The percentage of metabolic activity is shown relative to untreated controls. Each sample was assayed in triplicate, with each experiment repeated at least 3 times independently.
To determine further the potential roles of FOXOs, we performed a knockdown of either FOXO1 or FOXO3a by siRNAs that were transfected into Calu-1, H358, and H2347 cells prior to drug treatment (Fig. 3B; Supplementary Fig. 4). In each of the cell lines examined, the knockdown of FOXO1 and FOXO3a increased drug resistance when compared to a non-targeting control (Figure 3B). We next determined the roles of proapoptotic protein BIM, which is directly activated by FOXO transcription factors, in maintaining cell metabolic activity. To do this, we performed a knockdown of BIM by siRNAs that were transfected into Calu-1, H358, and H2347 cells. BIM siRNAs also increased resistance to the combination of belinostat and trametinib (Fig. 3C; Supplementary Fig. 4). Moreover, the knockdown of either FOXO1 or FOXO3a by siRNAs in Calu-1 cells suppressed the increase of BIM and cleaved PARP compared with control siRNAs when treated with the combination of belinostat and trametinib (Supplementary Fig. 5).
These findings suggest that the FOXO proteins translocate to the nucleus with HDAC and MEK inhibition where they control the expression of the proapoptotic protein BIM, promoting apoptotic death.
The combination of trametinib and belinostat decrease tumor formation in a xenograft model better than either drug alone.
We next examined the antitumor potential of belinostat in combination with trametinib using a xenograft mouse model. The KRAS mutated H358 cell line was implanted into the flanks of immunocompromised nude mice. When tumors reached approximately 100 mm3 mice were treated daily with vehicle, belinostat, trametinib, or the combination. Treatment with either single agent slightly suppressed the growth of H358 tumors. Notably, belinostat in combination with trametinib significantly suppressed the growth of H358 tumors compared with either single agent (P<0.05 by one-way ANOVA) (Fig. 4A; Supplementary Fig. 6A). During treatment with belinostat or trametinib, either alone or in combination, there was no evidence of severe loss in body weight indicating that the combination was well tolerated (Supplementary Fig. 6B). These results suggest that the combination of belinostat and trametinib may provide a potential therapeutic strategy against KRAS mutated lung cancers.
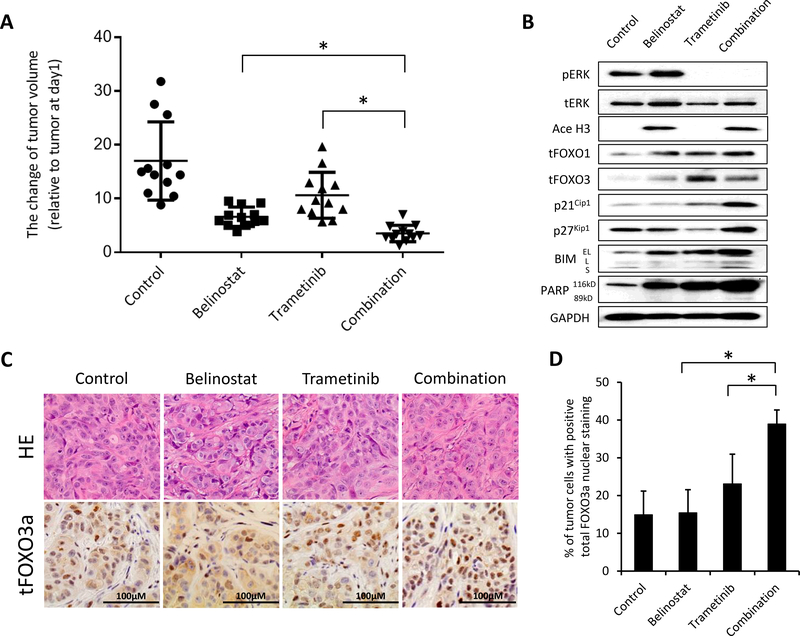
Figure 4. The combination of trametinib and belinostat shows increased efficacy in a xenograft model of KRAS mutated lung cancer.
(A) H358 (5 × 106) cells were inoculated into the flank of nude mice on day −5, and the mice were treated with belinostat 40 mg/kg/mouse i.p. daily, trametinib (1 mg/kg p.o.daily), or in combination from day 1 to 22. The mice were sacrificed on day 22 and xenograft tumors were evaluated as described in Materials and Method. These data show the percent change in the tumor volume on day 22 relative to tumor at day 1. Bars indicating standard error are shown for groups of 12 mice where each symbol represents a single mouse for each of the groups. *, P < 0.05 for the combination compared with either belinostat alone or trametinib alone groups by one-way ANOVA. (B) The tumors were harvested on day 22 and examined for the indicated proteins by western blotting analysis. (C) Tumors harvested on day 22 were histologically examined. Sections were stained with H&E and probed with an anti-human FOXO3a monoclonal antibody. (D) Quantification of total FOXO3a positive nuclei. Columns, mean of five fields; bars, SD. *, P < 0.05 for the combination compared with either belinostat alone or trametinib alone groups by one-way ANOVA.
We checked the protein levels of FOXO1, FOXO3a, p21Cip1, p27 Kip1, BIM, and cleaved PARP in the tumors by western blotting analysis (Fig. 4B). We also confirmed the inhibition of ERK1/2 phosphorylation and the increase in acetylation of Histone H3 to determine that the drugs were working properly. Similar to the in vitro results, we saw increases in total FOXO proteins, cell cycle inhibitors, and the apoptotic protein BIM. These results clearly indicate the therapeutic benefit of combined therapy with belinostat and trametinib against RAS mutated H358 cells. To assess the mechanism by which the combination therapy inhibits tumor growth, we performed FOXO3a immunohistochemical staining of the tumors. We found that the number of cells containing nuclear FOXO3a increased significantly in the tumors of mice treated with the combination of belinostat and trametinib (Fig. 4C and D). These results suggest that HDAC and MEK inhibition promoted the translocation of FOXO3a protein into the nucleus where it induced apoptosis in mouse xenograft tumors through the up regulation of BIM.
Discussion
Many promising drugs have been developed for NSCLC such as molecular targeted therapies for mutated EGFR and ALK translocations and immunotherapy. However, despite many years of research and the development of drugs that target various aspects of RAS biology, an effective treatment for RAS mutant tumors still eludes us. A recent study has shown that KRAS G12C or G12V mutation subgroups tend to have some benefit when compared with other KRAS mutation groups in a phase II trial of the MEK inhibitor selumetinib plus docetaxel in KRAS mutant NSCLC (20). The option of combining targeted therapies hitting different pathways is promising if we can balance toxicity with efficacy for use in the clinic. Combined therapy using PI3K/AKT and MEK inhibitors has activity in pre-clinical studies, but this activity seems relatively limited in clinical trials. For instance, the maximum tolerated dose of both AKT inhibitor MK-2206 and selumetinib could not achieve 70 % inhibition of their targets in colorectal tumors (21).
Recently, we have demonstrated that LKB1 mutant tumors are sensitive to MEK inhibition irrespective of the RAS status. The mechanism appears to be through activation of the FOXO transcription factors, which regulate many cellular processes, including upregulation of BIM and apoptosis (22). When LKB1 is added back to the cells they become resistant to MEK inhibition due to the translocation of the FOXO transcription factors from the nucleus to the cytoplasm where they are sequestered by 14–3-3 proteins (23, 24). It would be beneficial to keep the FOXO proteins in the nucleus where they are active, because they can induce apoptotic proteins BIM, FASL, and TRAIL. In addition, they promote the expression of cell cycle inhibitors, p21Cip1, p27 Kip1, and p15, and induce cell cycle arrest (25). Recently, FOXO proteins were reported to have a critical role in drug resistance. The inhibition of FOXO3a induced resistance to anticancer therapeutics, not only to a MEK inhibitor but also to gefitinib and doxorubicin (26, 27). FOXO3a activity is also frequently attenuated in drug-resistant cancer cells (25). Thus, the control of FOXO activity by increasing nuclear localization is a promising strategy for overcoming drug resistance.
Using the above arguments as a rationale, we focused on using FOXOs as a potential therapeutic target to overcome the resistance to MEK inhibitor trametinib. In our previous LKB1 study, we showed that MEK resistance was due to the presence of LKB1 and relocalization of FOXOs (22). In this study we searched for drug candidates to enhance the activity of transcription factor FOXOs against RAS mutated lung cancer cells with wild type LKB1. Some well-known targeted agents were reported to promote the transcription factor activity of FOXO proteins. HDAC inhibitors were identified to activate E2F1/FOXO transcription and enhanced E2F1-induced apoptosis though FOXO3-dependent pathway in human osteosarcoma cells (28). Another HDAC inhibitor was reported to increase BIM expression though FOXO1 activity, resulting in the increase of apoptosis (15). Besides HDAC inhibitors, the inhibitors of PARP1 or TGF-beta1 were also shown to enhance the translocation of FOXO3a to the nucleus (16, 17). Thus, some agents that target FOXOs for nuclear translocation have already been demonstrated and may be promising drugs to enhance anti-tumor activity via FOXOs. In this study, we assessed the efficacy of inhibitors of PARP1 and TGF-beta1 in combination with trametinib on cell metabolic activity of RAS mutated lung cancer cells. However, in this setting, these drugs did not demonstrate synergistic effects when combined with trametinib (unpublished observations).
HDAC inhibitors have been developed for a broad range of human disorders, such as ischemic stroke (29–31), multiple sclerosis, and Huntington’s disease (32–34). Recently, the FDA has approved multiple HDAC inhibitors, such as vorinostat, romidepsin, belinostat, and panobinostat, for hematopoietic tumors. Belinostat, which inhibits pan-HDAC activities, has been approved for patients with relapsed or refractory peripheral T-cell lymphoma in 2014. However, in the solid tumors, previous clinical trials have failed to show the benefit when using an HDAC inhibitor as a single agent, including belinostat (35). Current clinical studies using HDAC inhibitors have moved toward combined therapy with the other agents (36). It has been reported in several clinical trials that belinostat combined with cytotoxic therapy is active and well tolerated in solid tumors (37, 38). Cell line based pre-clinical studies have shown synergistic inhibitory effects between MEK1/2 and HDAC inhibitor in human leukemia cells and colorectal cancer cells. We now show that in RAS mutated lung cancer cells, and in one BRAF mutated cell line, the MEK inhibitor trametinib in combination with the HDAC inhibitor belinostat induce proteins that promote apoptosis and cell cycle arrest (39, 40). The effect of belinostat in combination with trametinib appears to regulate the expression and activation of both FOXO1 and FOXO3a followed by BIM expression with increased apoptosis of RAS mutated cells. There does appear to be some toxicity in cultured fibroblast cells when they are treated for longer periods of time. However, the combination seemed to be well tolerated in the mice. This caveat suggests that we should pay close attention to the therapeutic window with chronic dosing for clinical development.
Our findings suggest that HDAC and MEK inhibition promotes an increase in FOXO1 and FOXO3a protein levels and higher transcriptional activity through increased nuclear accumulation. The dual molecular targeted therapy for HDAC and MEK may be promising as novel therapeutic strategy in RAS mutated lung cancer.
Supplementary Material
Acknowledgement
We appreciate the gift of the Calu-1, Calu-6, H1299, H2009, H2347, H358, and H441 cells that were provided by John Minna and Luc Girard (University of Texas, Southwestern, Dallas, TX).
Grant support: This study was supported by the DallePezze Thoracic Oncology Fund (David Carbone) and NIH-funding (5U10CA180950 (David Carbone)), and a research grant for developing innovative cancer chemotherapy from the Kobayashi Foundation for Cancer Research (Tadaaki Yamada) and Foundation for Promotion of Cancer Research in Japan (Tadaaki Yamada).
Reference
- 1.Patel JD, Krilov L, Adams S, Aghajanian C, Basch E, Brose MS, et al. Clinical Cancer Advances 2013: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J Clin Oncol 2014;32:129–60. [DOI] [PubMed] [Google Scholar]
- 2.Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 2015;12:511–26. [DOI] [PubMed] [Google Scholar]
- 3.Oxnard GR, Binder A, Jänne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol 2013;31:1097–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov 2007;6:541–55. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Viallet J, Mulshine JL, Linnoila RI, Minna JD, Gazdar AF. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene 1991;6:1353–62. [PubMed] [Google Scholar]
- 6.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002;62:6997–7000. [PubMed] [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339–46. [DOI] [PubMed] [Google Scholar]
- 9.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007;26:3291–310. [DOI] [PubMed] [Google Scholar]
- 10.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016;351:604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumenschein GR Jr, Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann Oncol 2015;26:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14: 38–47. [DOI] [PubMed] [Google Scholar]
- 14.AstraZeneca Provides Update On Phase III Trial Of Selumetinib In Non-Small Cell Lung Cancer. http://www.prnewswire.com/news-releases/astrazeneca-provides-update-on-phase-iii-trial-of-selumetinib-in-non-small-cell-lung-cancer-300310826.html
- 15.Yang Y, Zhao Y, Liao W, Yang J, Wu L, Zheng Z, et al. Acetylation of FoxO1 activates BIM expression to induce apoptosis in response to histone deacetylase inhibitor depsipeptide treatment. Neoplasia 2009;11:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Wang H, Davis BC, Liang J, Cui R, Chen SJ, et al. PARP1 inhibitors attenuate AKT phosphorylation via the upregulation of PHLPP1. Biochem Biophys Res Commun 2011;412:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Q, Zhai Y, Cheng Q, Liu Y, Gao X, Zhang T, et al. The Akt-FoxO3a-manganese superoxide dismutase pathway is involved in the regulation of oxidative stress in diabetic nephropathy. Exp Physiol 2013;98:934–45. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T, Takeuchi S, Fujita N, Nakamura A, Wang W, Li Q, et al. Akt kinase-interacting protein1, a novel therapeutic target for lung cancer with EGFR-activating and gatekeeper mutations. Oncogene 2013;32:4427–35. [DOI] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 20.Jänne PA, Smith I, McWalter G, Mann H, Dougherty B, Walker J, et al. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br J Cancer 2015;113:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Do K, Speranza G, Bishop R, Khin S, Rubinstein L, Kinders RJ, et al. Biomarker-driven phase 2 study of MK-2206 and selumetinib (AZD6244, ARRY-142886) in patients with colorectal cancer. Invest New Drugs 2015;33:720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman JM, Yamada T, Park K, Timmers C, Amann JM, Carbone DP. A transcriptional signature of LKB1 loss identifies LKB1 functional status as a novel determinant of MEK sensitivity. Cancer Res 2017;77:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 1998;391:184–7. [DOI] [PubMed] [Google Scholar]
- 24.Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet 1998;18:38–43. [DOI] [PubMed] [Google Scholar]
- 25.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013;13:482–95. [DOI] [PubMed] [Google Scholar]
- 26.McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther 2009;8:582–91. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Liang C, Xue F, Chen W, Zhi X, Feng X, et al. Salinomycin decreases doxorubicin resistance in hepatocellular carcinoma cells by inhibiting the β-catenin/TCF complex association via FOXO3a activation. Oncotarget 2015;6:10350–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shats I, Gatza ML, Liu B, Angus SP, You L, Nevins JR. FOXO transcription factors control E2F1 transcriptional specificity and apoptotic function. Cancer Res 2013;73:6056–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren M, Leng Y, Jeong M, Leeds PR, Chuang DM. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem 2004;89:1358–67. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Rowe M, Ren M, Hong JS, Chen PS, Chuang DM. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. J Pharmacol Exp Ther 2007;321:892–901. [DOI] [PubMed] [Google Scholar]
- 31.Faraco G, Pancani T, Formentini L, Mascagni P, Fossati G, Leoni F, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol 2006;70:1876–84. [DOI] [PubMed] [Google Scholar]
- 32.Camelo S, Iglesias AH, Hwang D, Due B, Ryu H, Smith K, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol 2005;164:10–21. [DOI] [PubMed] [Google Scholar]
- 33.Jia H, Morris CD, Williams RM, Loring JF, Thomas EA. HDAC inhibition imparts beneficial transgenerational effects in Huntington’s disease mice via altered DNA and histone methylation. Proc Natl Acad Sci U S A 2015;112:E56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 2008;7:854–68. [DOI] [PubMed] [Google Scholar]
- 35.Ramalingam SS, Belani CP, Ruel C, Frankel P, Gitlitz B, Koczywas M, et al. Phase II study of belinostat (PXD101), a histone deacetylase inhibitor, for second line therapy of advanced malignant pleural mesothelioma. J Thorac Oncol 2009;4:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas A, Rajan A, Szabo E, Tomita Y, Carter CA, Scepura B, et al. A phase I/II trial of belinostat in combination with cisplatin, doxorubicin, and cyclophosphamide in thymic epithelial tumors: a clinical and translational study. Clin Cancer Res 2014;20:5392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassen U, Molife LR, Sorensen M, Engelholm SA, Vidal L, Sinha R, et al. A phase I study of the safety and pharmacokinetics of the histone deacetylase inhibitor belinostat administered in combination with carboplatin and/or paclitaxel in patients with solid tumours. Br J Cancer 2010;103:12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu C, Dasmahapatra G, Dent P, Grant S. Synergistic interactions between MEK1/2 and histone deacetylase inhibitors in BCR/ABL+ human leukemia cells. Leukemia 2005;19:1579–89. [DOI] [PubMed] [Google Scholar]
- 40.Morelli MP, Tentler JJ, Kulikowski GN, Tan AC, Bradshaw-Pierce EL, Pitts TM, et al. Preclinical activity of the rational combination of selumetinib (AZD6244) in combination with vorinostat in KRAS-mutant colorectal cancer models. Clin Cancer Res 2012;18:1051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.