Summary.
Working memory (WM) was one of the first cognitive processes studied with functional magnetic resonance imaging (fMRI). With now over 20 years of studies on WM, each study with tiny sample sizes, there is a need for meta-analysis to identify the brain regions consistently activated by WM tasks, and to understand the inter-study variation in those activations. However, current methods in the field cannot fully account for the spatial nature of neuroimaging meta-analysis data or the heterogeneity observed among WM studies. In this work, we propose a fully Bayesian random-effects meta-regression model based on log-Gaussian Cox processes, which can be used for meta-analysis of neuroimaging studies. An efficient MCMC scheme for posterior simulations is presented which makes use of some recent advances in parallel computing using graphics processing units (GPUs). Application of the proposed model to a real dataset provides valuable insights regarding the function of the WM.
Keywords: functional magnetic resonance imaging, working memory, random effects meta-analysis, meta-regression
1. Introduction
1.1. The working memory
Humans depend on working memory (WM) for many behaviours and cognitive tasks. WM includes both the retention of information (aka short term memory), as well as the manipulation of information over a short duration. An example of the former is remembering a phone number until you dial it, while an example of the latter is building a ‘mental map’ while receiving directions. WM is impaired in a number of neurological and psychiatric diseases, most notably in all forms of dementia.
With its central role in everyday behaviour and implication in disease, WM has been frequently studied with functional brain imaging techniques like functional magnetic resonance imaging (fMRI). fMRI is sensitive to changes in blood flow, volume and oxygenation level in the brain, and provides a noninvasive way to identify regions of the brain associated with a given task or behaviour. However, each fMRI study has traditionally had very small samples, rarely exceeding 20. Thus, there is a need for meta-analysis methods to pool information over studies, separating consistent findings from those occurring by chance, as well as meta-regression methods (Greenland, 1994) to understand heterogeneity in terms of study-specific characteristics.
1.2. Neuroimaging meta-analyses
In fMRI there are two broad approaches for meta-analysis. When the full statistical images from each study are available, that is effect sizes and associated standard errors for all voxels in the brain, an intensity-based meta-analysis (IBMA) can proceed by means of standard meta-analytic methods (see Hartung et al. (2008) for an overview). However, these statistic images (200,000+ voxels) traditionally have not been shared by authors. Instead, researchers only publish the x,y,z brain atlas coordinates of the local maxima in significant regions of the statistic image. We call these coordinates the foci (singular focus). When only foci are available then a coordinate-based meta-analysis (CBMA) is conducted. As can be expected, the transition from full images to the lists of foci involves a heavy loss of information (Salimi-Khorshidi et al., 2009). However, since the vast majority of researchers rarely provide the full images, CBMA constitutes the main approach for fMRI meta-analysis.
Most work in the field is focused on the so-called kernel-based methods such as activation likelihood estimation (Turkeltaub et al., 2002; Eickhoff et al., 2012, ALE), multilevel kernel density analysis (Wager et al., 2004, 2007, MKDA) and signed differential mapping (Radua and Mataix-Cols, 2009; Radua et al., 2012, SDM). Roughly, these methods construct a statistic map as the convolution of the foci† with 3D spatial kernels, but not exactly correspond to traditional kernel density estimation. In particular, these methods give special treatment to foci that appear close in one study, decreasing their influence relative to dispersed points.
Areas of the map with large values suggest brain regions of consistent activation across studies. For statistical inference, the map is thresholded by reference to a Monte Carlo distribution under the null hypothesis of no consistent activation across studies. Kernel-based methods are not based on an explicit probabilistic model and hence often lack interpretability. Moreover, for some methods it is difficult to obtain standard errors and hence only p-values are reported for each voxel. Some of these approaches cannot accept study-level covariates, and thus can’t conduct meta-regression, and all are massively univariate in that they have no model of spatial dependence and can make only limited probabilistic statements about sets of voxels.
Recently, some model-based methods were proposed to address the limitations of kernel-based methods, such as the Bayesian hierarchical independent Cox cluster process model of Kang et al. (2011), the Bayesian nonparametric binary regression model of Yue et al. (2012), the hierarchical Poisson/Gamma random field model of Kang et al. (2014) and the spatial Bayesian latent factor model of Montagna et al. (2017). However, most of these methods do not allow for meta-regression. Further, current model-based approaches do not account for dependence induced when a single publication reports the results of multiple studies using the same cohort of participants. (In this work, we refer to ‘study’ as the result of one statistical map; typically a publication will report results from several maps).
1.3. Contribution and outline
The contributions of this work are twofold. The first contribution is methodological. In particular, we propose a Bayesian spatial point process model, extension of the log-Gaussian Cox process model (Møller et al., 1998) that can account for study specific characteristics as explanatory variables thus allowing for metaregression. Compared to the model of Montagna et al. (2017), which is the only existing coordinate-based meta-regression method, our model has two advantages. Firstly, it is less mathematically complex and therefore easier to communicate to practitioners and elicit prior distributions for its parameters. Secondly, by introducing random-effect terms, our model can capture heterogeneities that cannot be captured by the covariates and also reduce biases caused by the assumption that studies in the meta-analysis are independent one of another.
The second contribution of this paper is to conduct a meta-analysis of working memory fMRI studies using the proposed model. Even though previous metaanalyses of working memory studies exist (Wager and Smith, 2003; Owen et al., 2005; Rottschy et al., 2012), none of these studies uses some of the available model-based methods and hence the inferences they provide are limited. Further, our analyses quantifies the effect of some important covariates and thus provides new insights regarding the function of working memory.
The remainder of this manuscript is structured as follows. In Section 2 we present the data under investigation and state the questions that our metaanalysis wishes to answer. Motivated by the data in 2, we introduce our LGCP model in Section 3. The algorithm used for posterior inference is presented in Section 4. The results of the real-data analysis can be found in Section 5. Finally, Section 6 summarises our findings and sets some possible directions for future research.
2. Motivating dataset
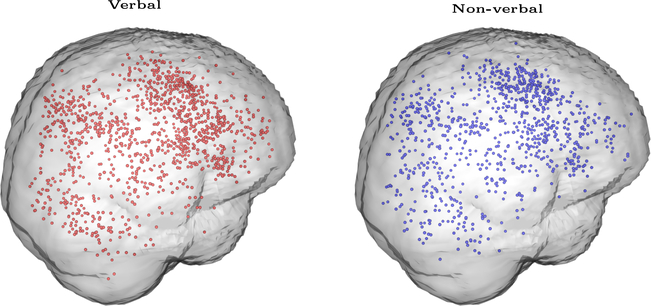
Our investigations are motivated by data from Rottschy et al. (2012). The data have been retrieved from 89 publications on working memory but some of these publications conduct multiple studies (experiments). The average number of studies per publication is 1.76 (ranging 1–7). Overall, we include 157 studies in the meta-analysis and the total number of foci is 2107. As well as the foci, for each study we observe the stimulus type (where 102 studies used verbal stimuli and 55 studies used non-verbal), the sample size (mean 14.94, SD 5.64) and the average age of the participants (mean 32, SD 10.99). See Table 1 for more descriptives, whereas a graphical representation of the data can be found in Figure 1. Note that, the dataset that we use is a subset of the dataset of Rottschy et al. (2012); this is due to missing values for the covariate age.
Table 1:
Data summaries
| Dataset composition | ||||
|---|---|---|---|---|
| Min. | Median | Mean | Max. | |
| Studies per publication | 1 | 1 | 1.76 | 7 |
| Foci per study | 1 | 11 | 13.42 | 55 |
| Participants per study | 6 | 14 | 14.94 | 41 |
| Mean participant age | 21.25 | 29.20 | 32.00 | 75.11 |
| Verbal | ||||
| Min. | Median | Mean | Max. | |
| Foci per study | 1 | 10 | 11.83 | 39 |
| Participants per study | 7 | 14 | 14.91 | 41 |
| Mean participant age | 21.80 | 30.12 | 33.80 | 75.11 |
| Non-verbal | ||||
| Min. | Median | Mean | Max. | |
| Foci per study | 2 | 15 | 16.36 | 55 |
| Participants per study | 6 | 13 | 14.98 | 33 |
| Mean participant age | 21.25 | 28.00 | 28.64 | 61 |
Fig. 1:
Graphical representation of the meta-analysis dataset. Data consist of 2,107 foci from 157 studies on working memory. Of these, 1,207 are obtained from studies using verbal stimuli (shown in red) whereas the remaining 900 are obtained from studies using non-verbal stimuli (shown in blue). The code used to generate this figure is courtesy of Jian Kang.
Our meta-analysis aims to address the following questions related to the function of working memory: I) what the regions of the brain that are consistently engaged by working memory across studies? II) do these regions differ depending on the type of stimulus presented to the participants? III) is the organisation of working memory affected by age? IV) does sample size affect the total number of activations reported? In order to ensure that the answers to these questions are not driven by influential publications conducting multiple studies, our investigations should account for such dependencies.
3. A model for CBMA meta-regression
To address the questions raised in Section 2, we propose a model for CBMA meta-regression. First, we set notation. Suppose that there are a total I studies in the meta analysis and that each study i comes with a point pattern xi, a set of foci, where B is the support of the analysis, usually set from a standard atlas of the brain, and j = 1,...,ni, where ni is the number of foci in a study. Additionally, suppose that for each point pattern there is a set of K study specific characteristics,. Henceforth, we will occasionally refer to these characteristics as covariates.
We assume that each point pattern xi is the realisation of a Cox point process Xi defined on B, driven by a random intensity λi(·). We can then model the intensity function at each point ξ ∈ B as
| (1) |
where αi is the random effect of study i, βk(·) are the regression coefficients for the covariates that have a local effect (k = 0,...,K∗), zik are covariate values where k = 0 is for the intercept (zi0 = 1), and are βk the regression coefficients for the covariates that have a global (homogenous) effect (k = K∗ + 1,...,K).
Equation (1) defines a spatial log-linear model over the brain. Foci are more likely to occur in regions of the brain with high intensity values whereas we expect almost no foci in regions as the intensity approaches zero. The exact rates are given by the properties of a Cox process. In particular, given λi(·), the expected number of foci in any bounded B ⊆ B is a Poisson random variable with mean ∫Bλi(ξ)dξ (Møller and Waagepetersen, 2004).
The inclusion of the random effect terms is an important feature of our model. Firstly, by assuming that αi = αj for studies i and j retrieved from the same publication, we relax the assumption of independence between their reported activations. This assumption is taken by all existing CBMA approaches but is unlikely to hold for studies from the same publication. For example, a multi-study publication will typically engage the same participants in all of its experiments. By using a common random effect for studies from the same publication, our model prevents publications with several studies to drive the estimates of the regression coefficients. Secondly, the random effects can allow for additional variability in the total number of foci that cannot be captured by the Poisson log-linear model. In a recent study, Samartsidis et al. (2017) found that CBMA data do show such overdispersion and thus inclusion of the random effect terms can potentially improve the fit to the data.
Separation of the covariates into those with a localised and those with a global effect should be done with caution. If one is interested in investigating whether the effect of a covariate varies from one region of the brain to another, such as age in our application, a spatially varying regression coefficient is needed. However, the total number of parameters associated with a spatially varying effect is large and therefore assigning a spatially varying coefficient to a covariate with a global effect may substantially increase the uncertainty associated to the other model parameters. In order to determine if a spatially varying coefficient for a covariate is required, one can fit two models, one that assumes that the covariate has a global effect and one that assumes a local effect. If the more complex model improves the fit to the data substantially‡, then it should be preferred for inference instead of the simple model. Sometimes, it plausible to assume a global effect solely based on prior expectation. For instance, a covariate for multiple testing correction can be assumed to have a global effect; for studies not applying any corrections, we expect false positives to appear uniformly across the brain.
A Bayesian model is defined with prior distributions on model parameters, which here include the functional parameters βk(·) (k = 0,...,K∗), and scalar parameters βk, (k = K* + 1,...,K). A natural way to proceed is to assume that βk(·) are realisations of Gaussian processes and that the βk have normal distributions. That way, when αi = 1, the right hand side of Equation (1) is also a Gaussian process, and each point process is a log-Gaussian Cox process (LGCP) (Møller et al., 1998). The log-Gaussian Cox process is a flexible model for spatial point data that can account for aggregation (Møller et al., 1998; Møller and Waagepetersen, 2007) or even repulsion between points (Illian et al., 2012a) and has therefore found applications in several fields such as disease mapping (Benes et al., 2002; Liang et al., 2009) and ecology (Møller and Waagepetersen, 2003; Illian et al., 2012b).
By the definition of a Cox process, Xi is a Poisson point process on B conditional on λi(·) (Møller and Waagepetersen, 2004). The density (Radon-Nikodym derivative) of this point process with respect to the unit rate Poisson process is
| (2) |
for i = 1,...,I, with |B| denoting the volume of the brain. We can view π (xi | λi) as the density of the sampling distribution of the data. If we further assume independent studies, then we posterior distribution of the model parameters conditional on the foci is given, up to a normalising constant by
| (3) |
where π(αi), π(βk(·)) and π(βk) are the priors on the random effects, functional and scalar parameters, respectively, which we discuss the priors below in Section 3.2.
3.1. Choice of correlation function
We will assume an isotropic, Gaussian correlation structure, that is for points ξ,ξ0 ∈ B we have
| (4) |
where ρk > 0 are the correlation decay parameters and δk = 2 for all k = 1,...,K∗. Note that for numerical stability with the discrete Fourier transform (see Section 4) we set δ = 1.9 in our implementations. The same correlation structure was used by Møller et al. (1998) and Møller and Waagepetersen (2003) in the context of LGCPs.
A Gaussian correlation function is used instead of alternative correlation structures (see e.g. Rasmussen and Williams (2005)) because it allows us to calculate the gradient of the posterior with respect to the correlation parameters ρk, which we use to design an efficient algorithm for posterior simulations (see Section 4 for details). Further, in exploratory work using other correlation structures, our neuroscientist colleagues preferred the appearance of results from Gaussian correlation, perhaps because of the pervasive use of Gaussian kernel smoothing in fMRI. Finally, it is well known that estimating the correlation parameters for more flexible correlation structures can be extremely challenging in practice, see e.g. discussions by Zhang (2004) and Diggle et al. (2013) for the Matérn correlation function.
3.2. Posterior approximation
Calculation of the posterior in Equation (3) requires the evaluation of the infinite dimensional Gaussian processes βk (·), k = 0,...,K∗, which we approximate with a finite dimensional distribution. Following Møller et al. (1998) and Benes et al. (2002), we consider the discretisation of the 3D volume with a regular rectangular grid W ⊃ B. We use V cubic cells (i.e. voxels) in W with volume A = a3, where a is the length of the side. In neuroimaging, analysis with 2mm3 cubic voxels is typical, leading to a box-shaper grid of about 1 million voxels, of which about 200,000 are in the brain or cerebellum. Note that for simplicity, we consider both grey matter and white matter voxels in our implementations. Voxels are indexed v = 1,...,V , and the coordinate of v is the location of the center .
For any k = 0,...,K∗, the Gaussian process βk(·) can be now approximated with a step function which is constant within each voxel v and equal to the value of βk(·) at the location of the center, i.e. βk(νv). Waagepetersen (2004) shows that the accuracy of this approximation improves as a goes to zero. By definition, βk = [βk(ν1),...,βk(νV )] are multivariate Gaussian vectors. We parametrise βk as
| (5) |
where μk are the overall (scalar) means, 1V is a V -vector of ones, σk are the marginal standard deviations, Rk are the V × V correlation matrices with elements , and γk are the a priori vectors, k = 0,...,K∗. The same parametrisation is used by Møller et al. (1998), Christensen and Waagepetersen (2002) and is advocated by Christensen et al. (2006) because it allows for computationally efficient posterior simulations.
Priors for the V -vectors γk are induced by the parametrisation of Equation (5). The priors for the remaining model parameters are set as follows. We assign weakly informative to the scalar parameters μk, σk and βk. Further, we assume that , which we found corresponded to smoothness ranges found in single-study fMRI statistic maps. Finally, in order to ensure identifiability, we a priori let . In our analyses, we set κ = 10 since we expect 90% of the multiplicative random effects to be within the interval [0.5,1.5].
Once the latent Gaussian processes are approximated, one can also approximate λi with a step function as before. The intensities at the center of each voxel are given by
| (6) |
where λi is the V -vector, the discretised intensity. We will write λiv = (λi)v for the v-element of study i’s intensity. The approximated posterior is
| (7) |
where , Av takes on the value A when νv ∈ B and 0 otherwise, v(xij) is the index of the voxel containing xij, and π(θ) is the joint prior distribution of the parameters. The posterior distribution in Equation (7) is still analytically intractable due to the presence of an unknown normalising constant and thus we need to resort to Monte Carlo simulation or approximation techniques to obtain samples from it. The method that we use is described in Section 4.
4. Sampling algorithm details
Bayesian methodology for inference on LGCPs can be broadly divided into two main categories: simulation based approximations of the posterior such as Markov chain Monte Carlo (Møller et al., 1998, MCMC) and elliptical slice sampling (Murray et al., 2010), and deterministic approximations to the posterior such as integrated nested Laplace approximations (Illian et al., 2012a; Simpson et al., 2016, INLA) and variational Bayes (Jaakkola and Jordan, 2000). In a recent study, Taylor and Diggle (2014) compare the Metropolis-adjusted Langevin (MALA) algorithm with INLA and find that both methods give similar results. In our application, we choose to use simulation based methods because application on our 3D problem is more straightforward.
We propose a hybrid MCMC algorithm to sample from the posterior (7), where parameters are updated in two blocks. The first block includes the random effect terms whereas the second block includes the remaining model parameters The gamma prior is conjugate for the elements of α; hence, they are simulated from their full conditional distributions given the remaining model parameters, see web-based supplementary materials for details. Even though it is possible, we choose not to update α jointly with θ∗ because that would increase computation time of our algorithm.
Sampling from the full conditional of θ∗ given α is challenging due to its dimensionality. Girolami and Calderhead (2011) showed that of all possible strategies, their Riemann manifold Hamiltonian Monte Carlo (RMHMC) sampler is the computationally most efficient for LGCPs in a 2D setting. Unfortunately, application in this problem (3D setting) is prohibitive as it would require the inversion of a huge V × V tensor matrix. Alternatives to RMHMC include the MALA and the standard Hamiltonian Monte Carlo (Duane et al., 1987; Neal, 2011, HMC) algorithms. We choose to use HMC because Girolami and Calderhead (2011) found that it is more efficient compared MALA in a 2D setting. This finding was confirmed in our preliminary 2D simulation studies with synthetic CBMA data, where HMC outperformed MALA in terms of computational efficiency (mixing/running-time tradeoff).
HMC initially appeared in the physics literature by Duane et al. (1987) under the name hybrid Monte Carlo, and later emerged into statistics literature by Neal (2011). HMC emulates the evolution of a particle system which is characterised by its position (q) and momentum (p) over time. In our case, q will be the parameter vector of interest θ∗, and p will be introduced artificially from a distribution, with d being the dimensionality of the problem and M the mass matrix. The dynamics of the system are described by a set of differential equations, known as Hamilton’s equations.
HMC alternates between moves for the position vector θ∗ and the momentum vector p based on Hamilton’s equations. If the solutions of the equations can be found analytically then moves will be deterministic; if not, numerical integration is required and an acceptance/rejection step must be performed to account for integration error. Integration is done in fictitious time ϵL, w here is the stepsize and L is the number of steps. Typically the leapfrog integrator is employed, which for L = 1 and starting at time t is performed as (Neal, 2011)
| (8) |
Overall, if the method is applied correctly, it will produce samples from the desired full conditional distribution Gradient expressions for the elements of θ∗, including correlation parameters ρk, can be found in webbased supplementary materials Section 1. Since it is well known that grouping of variables can lead to samplers with faster convergence properties (Park and van Dyk, 2009), we choose to update all elements of θ∗ jointly using the HMC. The solutions to Hamilton’s equations are not available analytically so we need to use the Leapfrog integrator and include an accept/reject step at the end of it.
Our sampler requires the specification of a stepsize and a total number of leapfrog steps L for the HMC step. Hoffman and Gelman (2014) show how tuning can be achieved automatically but when we applied this method to our problem running time was increased substantially. Therefore we use an alternative approach to tune these parameters. The stepsize is automatically adjusted during the burn-in phase of the HMC to give an overall acceptance rate close to the 65% suggested by Neal (2011). In particular, if t is the stepsize at iteration t and qt1 is the acceptance rate over the past t1 iterations, then every t2 iterations we calculate the new stepsize as
| (9) |
Specifically we use t1 = 100 and t2 = 10. A similar approach is employed by Marshall and Roberts (2012) for MALA. The latter (number of leapfrog steps), is always fixed to L = 50. We took this approach because we found that, for our LGCP application, the mixing properties of the algorithm scale linearly with L but also with the total number of HMC iterations. Hence one can use a relatively large L and few iterations or relatively smaller L and more iterations, the total computation time staying relatively constant.
The last tuning parameter in the HMC algorithm is the variance-covariance matrix of the zero mean normal momentum parameters, M. To our knowledge, there is only limited off the shelf methodology on how to adjust M. As a starting place we set M = I. Neal (1996) suggests that if an estimate of the posterior variance is available then a good practice is to set . In principle, can be estimated during the burn-in phase of HMC but in practice this is not possible due to the dimensionality of the problem. In our simulations, we found that the mean posterior variance of the elements of the γk was higher compared to the scalar parameters, followed by βk or σk and then ρk. Especially for the ρk the scale is typically much smaller compared to the other parameters in our applications and so we use 100 × ρk instead of ρk. After the reparametrisation we found that setting the mass for parameters of γk, βk, σk and ρk equal to 1, 9, 16 and 25 respectively worked well in most of our implementations on simulated and real data. However, users might need to adjust these parameters if mixing of the chains is slow. For example, estimates of the posterior variance of the scalar parameters can be obtained based on preliminary runs of the algorithm for a few iterations. In Section 2 of the web-based supplementary materials, we perform a series of simulations studies which demonstrate that the proposed HMC algorithm can efficiently sample from the posterior distribution of the highdimensional parameter vector .
The most computationally demanding part of the algorithm is the the calculation of the large matrix-vector products appearing in the intensity functions of Equation (6). Luckily, an elegant solution to this problem is given by Møller et al. (1998) based on circulant embedding that was first proposed by Dietrich and Newsam (1993) and Wood and Chan (1994). The key to the approach is the linear algebra result that a circulant matrix has the discrete Fourier basis as its eigenvectors. Rk is not circulant but is block Toeplitz and can be embedded in a (2V )×(2V ) matrix that is circulant. Thus the matrix square root, inversion and multiplication can be accelerated by using (the highly efficient) discrete Fourier transform (DFT) of the embedded matrix and manipulating Fourier coefficients, followed by inverse DFT and extracting the appropriate sub-matrix/sub-vector. See Rue and Held (2005, Section 2.6.2) for more details.
We close this section by stressing that despite the massive dimensionality of the parameter vector, the problem has a very high degree of parallelisation. Intensities can be evaluated in blocks of thousands of voxels simultaneously making the algorithm suitable for implementation in a graphics processing unit (GPU). The most computationally intensive part of our model, namely operations with DFTs, is also amenable to parallelisation and there exist libraries such as NVIDIA’s cuFFT library that are designed for this specific task. Overall, we believe that implementation of the log-Gaussian Cox process model described above will soon become a routine task for any moderately powerful GPU device.
5. Analysis of the WM dataset
5.1. Model, algorithm details and convergence diagnostics
For i = 1,...,157 we fit the model
| (10) |
where di0 and di1 are indicator variables of verbal and non-verbal stimuli, respectively, and ni is the total number of participants in study i. Continuous parameters were standardised before implementation.
We run the MCMC algorithm described in Section 4 for 22,000 iterations, discarding the first 7,000 as a burn-in. The algorithm run for approximately 30 hours on an NVIDIA Tesla K20c GPU card. We then apply a thinning factor of 15 to the chains and therefore end up with 1,000 draws from the posterior distribution of the model parameters. The total number of leapfrog steps is set to L = 50 and the stepsize is initialised at ϵ=0.00001. We use a diagonal mass matrix with units specified in Section 4. A preliminary run of the algorithm revealed that the posterior variance of the scalar parameters ρ2 and σ2 of β2 was higher compared to the corresponding parameters of β0 and β1. Therefore, in order to improve mixing of the algorithm, we set the mass parameters to 1 and 4 for ρ2 and σ2, respectively.
Convergence of the MCMC chain is assessed visually by inspection of posterior traceplots for the model parameters. We run a total of 2 MCMC chains in order to examine if they all converge to the same values. Posterior traceplots are shown in web-based supplementary materials Section 3. Due to the large number of parameters we mainly focus on the scalar parameters of the model and some summary statistics, see Section 3 in the web-based supplementary materials for more details. Results indicate that our chains have converged to their stationary distribution. This is verified by the fact that posterior values from the 2 different runs overlap one with another for all the quantities that we examine.
5.2. Results
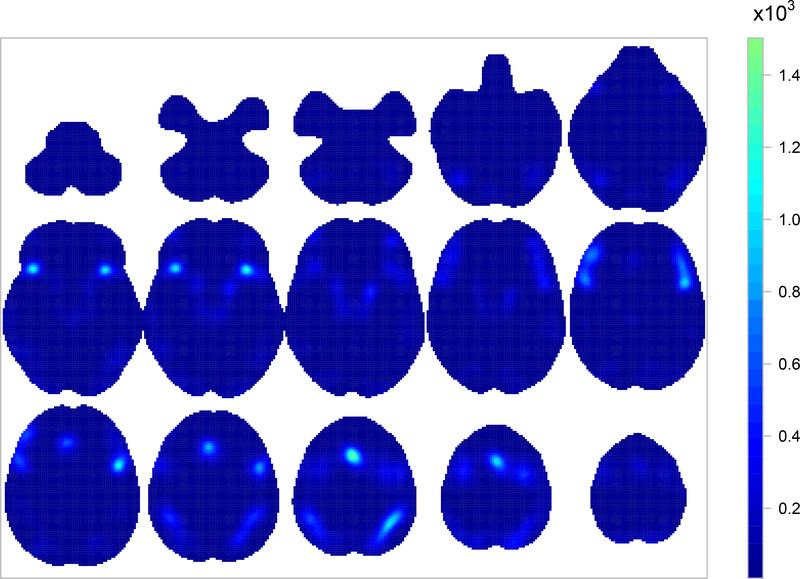
Figure 2 shows the mean posterior of λ, the average intensity of a working memory study, where λ = (λv + λnv)/2, λv is the intensity for verbal WM studies and λnv is for non-verbal WM studies (mean age and number of participants are set equal equal to the average values in our dataset). We can see that working memory engages several regions of the brain. The regions mostly activated are the frontal orbital cortex (axial slice z = −10, left), the insular cortex (z = −10, right and z = −2, left and right), the precentral gyrus (z = +30, left), Broca’s areas (z = +22 & z = +30, bilateral), the angular gyrus (z = +46, left), the superior parietal lobule (z = +46, right) and the paracingulate gyrus (z = +46, middle).
Fig. 2:
Voxelwise mean posterior of λ, the average intensity of a working memory study. Top row shows (from left to right) axial slices z = −50,−42,−34,−26 and −18, respectively. Middle row shows axial slices z = −10,−2,+6,+14 and +22, respectively. Bottom row shows axial slices z = +30,+38,+46,+54 and +62, respectively.
Our results are qualitatively similar to results obtained by Rottschy et al. (2012) who used the ALE method. However, our model-based approach allows us to derive several quantities of interest along with credible intervals, that cannot be obtained by any of the kernel-based methods. For example, one may calculate the probability of observing at least one focus in a set of voxels, e.g. an ROI or the entire brain. Table 2 summarises the posterior distribution of , the probability of observing at least one focus in B, for several ROIs B. A full brain analysis can be found in web-based supplementary materials Section 4. The division of the brain in ROIs is done according to the Harvard-Oxford atlas (Desikan et al., 2006).
Table 2:
Posterior % probabilities of observing at least once focus for several ROIs. All quantities have been calculated based on 1,000 MCMC samples.
| ROI | Mean | 95% CI | Verbal | Non-verbal |
|---|---|---|---|---|
| Frontal orbital cortex | 36.94 | [27.27,43.06] | 37.26 | 36.48 |
| Insular cortex | 33.39 | [26.68,39.36] | 32.79 | 33.86 |
| Precentral gyrus | 68.47 | [59.96,73.72] | 64.10 | 72.09 |
| Inferior frontal gyrus, PO | 39.88 | [31.06,45.96] | 43.66 | 35.69 |
| Angular gyrus | 21.69 | [14.39,26.34] | 24.30 | 18.91 |
| Superior parietal lobule | 36.16 | [26.16,42.31] | 38.81 | 33.24 |
| Paracingulate gyrus | 46.22 | [35.94,52.89] | 42.91 | 49.14 |
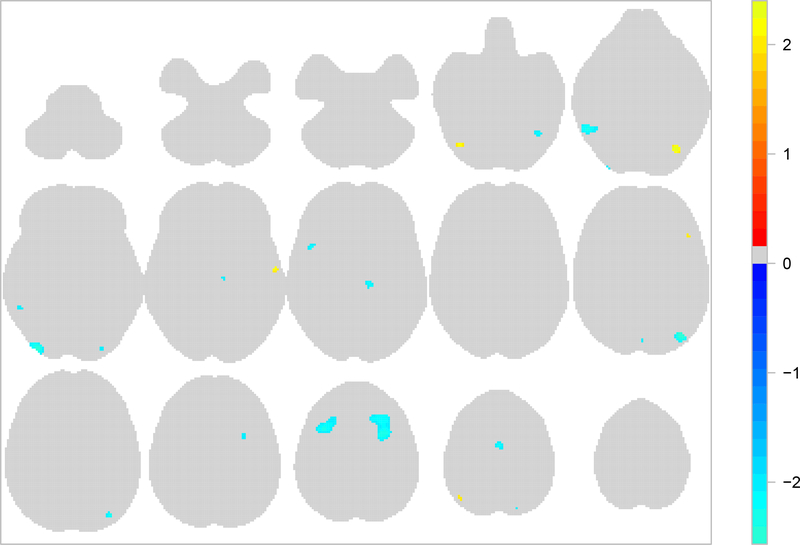
We use posterior intensities λv and λnv to compare activation between the two types of studies in our sample, namely studies using verbal and studies non-verbal stimuli. We start with an ROI analysis. In particular, for each type and ROI we calculate the probability of at least one focus observed as explained above. These are shown in Table 2 for a few ROIs, whereas a full brain analysis of the two types can be found in Section 4 in the web-based supplementary materials. We see that even though the two types show similar patterns of activation, there several ROIs where the probabilities of at least one focus have credible intervals with little overlap. The main differences are found in the superior frontal gyrus, the middle frontal gyrus, the lateral occipital cortex, superior division and the inferior frontal gyrus, pars opercularis. A voxel-by-voxel comparison is also feasible. To answer this, we use the mean standardised posterior difference . This is shown in Figure 3. Large positive values indicate regions that are activated by verbal stimuli more than non-verbal stimuli. Such regions appear the occipital fusiform gyrus (z = −18, right). Based on the mean standardised posterior difference, regions mostly activated in studies using non-verbal are located in the middle frontal gyrus (z = +46).
Fig. 3:
Voxelwise mean standardised posterior difference between β1 and β2, the intensities of studies using verbal and nonverbal stimuli, respectively. Top row shows (from left to right) axial slices z = −50,−42,−34,−26 and −18, respectively. Middle row shows axial slices z = −10,−2,+6,+14 and +22, respectively. Bottom row shows axial slices z = +30,+38,+46,+54 and +62, respectively. Voxels for which the mean posterior λ is low (below the 75% quantile over the brain) or the absolute mean standardised posterior difference is less than two have been set to zero.
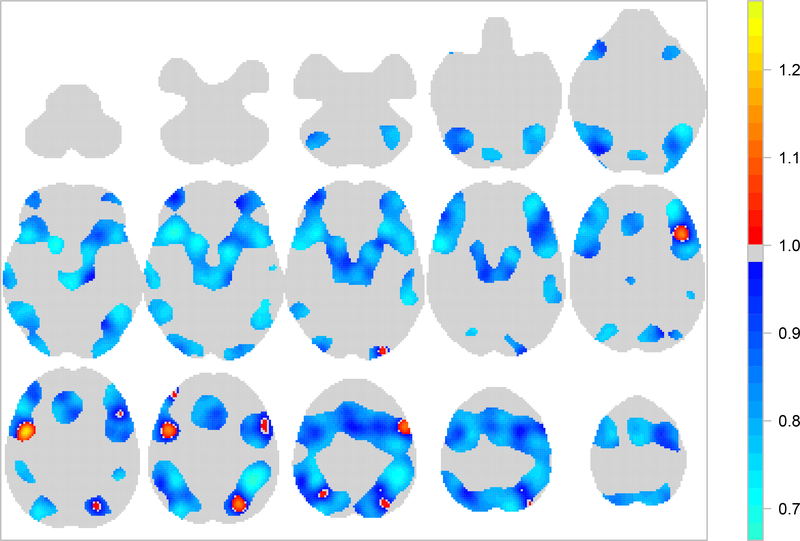
Our results provide evidence that age has an important effect on the function of working memory. The point estimate for the overall age effect μ2 is −0.22 (95% CI [−0.337,−0.120]) thus suggesting that we expect a decrease of 20% in the total number of reported activations per study, each time the average age of the participants increases by 10.99 years. Localised age effects can be identified through the posterior distribution of exp{β2}, the mean of which is shown in Figure 4. The map represents the multiplicative effect that an increase of the average participant age by 10.99 years has on the intensity of both verbal and non-verbal studies. Large negative age effects can be found near the left putamen (z = −2 and z = −10, middle), the insular cortex (z = −2, left) and near the superior parietal lobule (z = +38 and z = +46, right). A positive age effect is found near the precentral gyrus (z = +30, left). However, due to the limited number of studies, the posterior variance of these estimates is large in some regions of the brain, see Figure 9 in web-based supplementary materials Section 3.
Fig. 4:
Mean posterior of exp{β2}, the multiplicative age effect on the intensity of both verbal and non-verbal studies. Top row shows (from left to right) axial slices z = −50,−42,−34,−26 and −18, respectively. Middle row shows axial slices z = −10,−2,+6,+14 and +22, respectively. Bottom row shows axial slices z = +30,+38,+46,+54 and +62, respectively. Voxels for which the mean posterior λ is low (below the 75% quantile over the brain) have been set to one.
The 95% CI for the sample size covariate is [−0.088,0.064] thus indicating that there is no significant effect on the total number of reported activations. The result is counter-intuitive as one would expect that studies with few participants would be underpowered and thus detect fewer activations. Thus, further investigation is required.
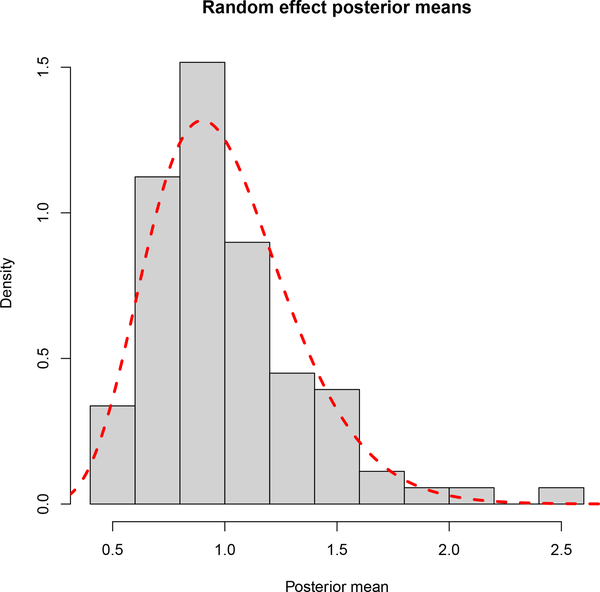
Figure 5 shows the mean posterior of the 89 unique random effect terms αi, one for each publication considered. We see that despite most of the mass being near 1, there are publications whose mean posterior random effect is different than 1, thus suggesting that observed variability of the foci counts is larger compared to what can be explained by the Poisson log-linear model. The importance of allowing for this additional variability can be seen by comparing the proposed random effects model to the standard LGCP model, which we also fit to the data. We use posterior predictive checks (Gelman et al., 1996) to assess how well the two models fit the data. For each study and MCMC draw, we simulate from the posterior predictive distribution of NXi(B), the total number of foci, given the covariates. Based on these draws, we calculate the 95% predictive intervals of NXi(B) and check if they contain the observed values. For our model, the coverage of the intervals is 90% compared to 66% obtained using the standard LGCP model, which implies that our model provides a better fit to the data compared to the standard LGCP. A comparison of the predictive intervals that takes into account the length of these intervals can be based on the mean interval score (Gneiting and Raftery, 2007). This is 22.45 and 76.93 for the random effects and standard LGCP models, respectively, thus suggesting that the inclusion of αi leads to improved prediction of the study counts.
Fig. 5:
Histogram of the mean posterior random effect terms, αi. We only plot the 89 unique random effects, one for each publication considered in the metaanalysis. Means are based on a sample of 1,000 MCMC draws from the posterior. The dashed red line represents the density of the Gamma prior.
Some of the estimated effects are affected by inclusion of the random effect terms. For instance, the expected number of foci for verbal studies is estimated as 12.80 (95% CI [11.57,14.14]) by the random effects LGCP as opposed to 11.67 (95% CI [10.97,12.36]) by the fixed effects LGCP model. One possible explanation for this is that our model is assigning a low random effect to publications systematically reporting only a few foci. Such a behaviour is desired since, e.g. this underreporting could be solely due to author preference. Further, the random effects model provides credible intervals that fully account for the uncertainty in the regression coefficients. For example, the 95% CI for the overall age effect μ2 provided by the fixed effects LGCP is [−0.309,−0.151], shorter than the CI provided by our model.
6. Discussion
In this work, we have presented a new CBMA model, extension of the logGaussian Cox process model. To our knowledge, this is the first application of the random effects LGCP with covariates in a 3D problem with multiple realisations. The model has an appealing interpretation being a spatial GLM and several interesting inferences can be obtained based on the properties of the spatial Poisson process that cannot be obtained with the commonly used kernelbased approaches. An advantage of our model compared to most of the existing methods is the inclusion of covariates in the analysis thus allowing for metaregression. Finally, a novel feature introduced in our work is the inclusion of random-effect terms which can account for additional heterogeneity in the total number of activations, compared to the standard Poisson model.
Application of our model on a meta-analysis of working memory studies have given valuable insights regarding the data. While our maps for the overall pattern of WM activations (Fig. 2) and the differential effect of verbal vs. non-verbal WM tasks (Fig. 3) reflect previous findings found by Rottschy et al. (2012), our fully Bayesian approach allowed us to make direct inference on probability of any foci and expected number of foci. Our model found no regions with evidence of different rates of foci between verbal and non-verbal WM tasks (web-based supplementary materials Section 4, Table 3). Importantly, our model allows a meta-regression, and we examined the effect of age and found no strong effects but generally negative effects of age on the number of foci.
There are few limitations to our work. Firstly, even though we found that the proposed MCMC algorithm performed well in most of the applications considered, we believe that there is room for further for improvement. For example, one can consider adaptive schemes in order to automatically adjust the mass matrix M of the HMC which we found that is crucial for the mixing properties of the algorithm. Secondly, we are currently not considering the problem of learning the hyperparameter κ that controls the posterior variability of the random effect terms, but rather make use of our prior expectations to tune it. However, since we found that results are sensitive to the specification of κ, it is plausible to consider estimating it along with remaining model parameters.
Our work can be extended in several ways. One possible direction for future research is to perform a head-to-head comparison of existing methodologies that can be used for posterior inference with the proposed LGCP model in the context of CBMA. However, given the computation time required to apply these methods to a 3D problem, such a comparison might be too long. Another potential future direction is to study the conditions, such as sample size or minimum number of foci, under which it is possible to estimate several global or spatially varying effects using the LGCP. Such work can be of importance for practical implementations since it will provide some guidance regarding the complexity of meta-regression models that can be fit to a given dataset.
Another open problem is how to use some additional information about the foci such as p-values or T-scores. These values can be attached as marks to the existing point patterns. Such an approach can enrich the inferences obtained from a CBMA by characterising the magnitude of activation in each region as opposed to the localisation of activations, which is the question that current methods address. Finally, it is worth considering a zero-truncated LGCP model. The reason is that several CBMAs use data from databases such as BrainMap (Laird et al., 2005), where only studies with at least one focus are registered. For such applications, a model that does not account for the zero-truncation can provide biased intensity estimates, especially when the expected number of foci per study is low. Currently, very few of the existing approaches propose adjustments for this potential problem.
Dataset composition
Supplementary Material
Acknowledgements
This work was largely completed while PS and TEN were at the University of Warwick, Department of Statistics. PS, TDJ and TEN were supported by NIH grant 5-R01-NS-075066; TEN was supported by a Wellcome Trust fellowship 100309/Z/12/Z and NIH grant R01 2R01EB015611–04. The work presented in this paper represents the views of the authors and not necessarily those of the NIH or the Wellcome Trust Foundation.
Footnotes
Precisely, this is a convolution of a Dirac delta function located at each focus with a given kernel.
As determined by a goodness-of-fit measure, e.g. posterior predictive checks (Gelman et al., 1996; Leininger and Gelfand, 2017).
Contributor Information
Pantelis Samartsidis, University of Cambridge.
Claudia R. Eickhoff, Heinrich-Heine University Düsseldorf
Simon B. Eickhoff, Heinrich-Heine University Düsseldorf
Tor D. Wager, University of Colorado at Boulder
Lisa Feldman Barrett, Northeastern University.
Shir Atzil, Massachusetts General Hospital.
Timothy D. Johnson, University of Michigan
Thomas E. Nichols, University of Oxford
References
- Benes V, Bodlák K, Møller J and Waagepetersen RP (2002) Bayesian analysis of log Gaussian Cox processes for disease mapping. Tech. rep, Depertment of Mathematical Sciences, Aalborg University. [Google Scholar]
- Christensen OF, Roberts GO and Sköld M (2006) Robust Markov chain Monte Carlo methods for spatial generalized linear mixed models. Journal of Computational and Graphical Statistics, 15, 1–17. [Google Scholar]
- Christensen OF and Waagepetersen RP (2002) Bayesian prediction of spatial count data using generalized linear mixed models. Biometrics, 58, 280–286. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Sgonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS and Killiany RJ (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31, 968–980. [DOI] [PubMed] [Google Scholar]
- Dietrich CR and Newsam GN (1993) A fast and exact method for multidimensional Gaussian stochastic simulations. Water Resources Research, 29, 2861–2869. [Google Scholar]
- Diggle PJ, Moraga P, Rowlingson B and Taylor BM (2013) Spatial and spatio-temporal log-gaussian cox processes: Extending the geostatistical paradigm. Statist. Sci, 28, 542–563. [Google Scholar]
- Duane S, Kennedy AD, Pendleton BJ and Roweth D (1987) Hybrid Monte Carlo. Physics Letters B, 195, 216–222. [Google Scholar]
- Eickhoff SB, Bzdok D, Laird AR, Kurth F and Fox PT (2012) Activation likelihood estimation meta-analysis revisited. NeuroImage, 59, 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Meng X-L and Stern H (1996) Posterior predictive assessment of model fitness via realized discrepancies. Statistica Sinica, 6, 733–807. [Google Scholar]
- Girolami M and Calderhead B (2011) Riemann manifold Langevin and Hamiltonian Monte Carlo methods. Journal of the Royal Statistical Society, Series B: Statistical Methodology, 73, 123–214. [Google Scholar]
- Gneiting T and Raftery AE (2007) Strictly proper scoring rules, prediction, and estimation. Journal of the American Statistical Association, 102, 359–378. [Google Scholar]
- Greenland S (1994) Invited commentary: a critical look at some popular metaanalytic methods. American Journal of Epidemiology, 140, 290–296. [DOI] [PubMed] [Google Scholar]
- Hartung J, Knapp G and Sinha BK (2008) Statistical Meta-Analysis with Applications. Hoboken: John Wiley & Sons. [Google Scholar]
- Hoffman M and Gelman A (2014) The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. Journal of Machine Learning Research, 15, 1593–1623. [Google Scholar]
- Illian JB, Sørbye SH and Rue H (2012a) A toolbox for fitting complex spatial point process models using integrated nested Laplace approximation (INLA). The Annals of Applied Statistics, 6, 1499–1530. [Google Scholar]
- Illian JB, Sørbye SH, Rue H and Hendrichsen DK (2012b) Using INLA to fit a complex point process model with temporally varying effects – A case study. Journal of Environmentl Statistics, 3, 1–25. [Google Scholar]
- Jaakkola T and Jordan M (2000) Bayesian parameter estimation via variational methods. Statistics and Computing, 10, 25–37. [Google Scholar]
- Kang J, Johnson TD, Nichols TE and Wager TD (2011) Meta analysis of functional neuroimaging data via Bayesian spatial point processes. Journal of the American Statistical Association, 106, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Nichols TE, Wager TD and Johnson TD (2014) A Bayesian hierarchical spatial point process model for multi-type neuroimaging metaanalysis. The Annals of Applied Statistics, 8, 1561–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Lancaster JJ and Fox PT (2005) Brainmap: the social evolution of a human brain mapping database. Neuroinformatics, 3, 65–77. [DOI] [PubMed] [Google Scholar]
- Leininger TJ and Gelfand AE (2017) Bayesian inference and model assessment for spatial point patterns using posterior predictive samples. Bayesian Analysis, 12, 1–30. [Google Scholar]
- Liang S, Carlin BP and Gelfand AE (2009) Analysis of Minnesota colon and rectum cancer point patterns with spatial and nonspatial covariate information. The Annals of Applied Statistics, 3, 943–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall T and Roberts G (2012) An adaptive approach to Langevin MCMC. Statistics and Computing, 22, 1041–1057. [Google Scholar]
- Møller J, Syversveen AR and Waagepetersen RP (1998) Log Gaussian Cox processes. Scandinavian Journal of Statistics, 25, 451–482. [Google Scholar]
- Møller J and Waagepetersen RP (2003) An introduction to simulation-based inference for spatial point processes In Spatial Statistics and Computational Methods (ed. Møller J), chap. 4, 143–198. Springer-Verlag. [Google Scholar]
- Møller J (2004) Statistical Inference and Simulation for Spatial Point Processes. Boca Raton: Chapman and Hall/CRC. [Google Scholar]
- Møller J (2007) Modern statistics for spatial point processes. Scandinavian Journal of Statistics, 34, 643–684. [Google Scholar]
- Montagna S, Wager T, Barrett LF, Johnson TD and Nichols TE (2017) Spatial bayesian latent factor regression modeling of coordinate-based metaanalysis data. Biometrics. URL: 10.1111/biom.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray I, Adams RP and MacKay DJ (2010) Elliptical slice sampling. Journal of Machine Learning Research: Workshop and Conference Proceeding, 9, 541–548. [Google Scholar]
- Neal RM (1996) Bayesian Learning for Neural Networks. Secaucus, NJ, USA: Springer-Verlag New York, Inc. [Google Scholar]
- Neal RM (2011) MCMC using Hamiltonian dynamics In Handbook of Markov Chain Mote Carlo (eds. Brooks S, Gelman A, Jones GL and Meng X), chap. 5, 113–162. Chapman & Hall/CRC. [Google Scholar]
- Owen AM, McMillan KM, Laird AR and Bullmore E (2005) N-back working memory paradigm: A meta-analysis of normative functional neuroimaging studies. Human brain mapping, 25, 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T and van Dyk DA (2009) Partially collapsed gibbs samplers: Illustrations and applications. Journal of Computational and Graphical Statistics, 18, 283–305. [Google Scholar]
- Radua J and Mataix-Cols D (2009) Voxel-wise meta-analysis of grey matter changes in obsessive-compulsive disorder. The British Journal of Psychiatry : the Journal of Mental Science, 195, 393–402. [DOI] [PubMed] [Google Scholar]
- Radua J, Mataix-Cols D, Phillips ML, El-Hage W, Kronhaus DM, Cardoner N and Surguladze S (2012) A new meta-analytic method for neuroimaging studies that combines reported peak coordinates and statistical parametric maps. European Psychiatry, 27, 605–611. [DOI] [PubMed] [Google Scholar]
- Rasmussen CE and Williams CKI (2005) Gaussian Processes for Machine Learning (Adaptive Computation and Machine Learning). The MIT Press. [Google Scholar]
- Rottschy C, Langner R, Dogan I, Reetz K, Laird AR, Schulz JB, Fox PT and Eickhoff SB (2012) Modelling neural correlates of working memory: a coordinate-based meta-analysis. NeuroImage, 60, 830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue H and Held L (2005) Gaussian Markov Random Fields: Theory and Applications Chapman & Hall/CRC Monographs on Statistics & Applied Probability. Taylor & Francis. [Google Scholar]
- Salimi-Khorshidi G, Smith SM, Keltner JR, Wager TD and Nichols TE (2009) Meta-analysis of neuroimaging data: a comparison of image-based and coordinate-based pooling of studies. NeuroImage, 45, 810–823. [DOI] [PubMed] [Google Scholar]
- Samartsidis P, Montagna S, Laird AR, Fox PT, Johnson TD and Nichols TE (2017) Estimating the number of missing experiments in a neuroimaging meta-analysis. bioRxiv, 225425. [DOI] [PMC free article] [PubMed]
- Simpson D, Illian J, Lindgren F, Sørbye S and Rue H (2016) Going off grid: computationally efficient inference for log-Gaussian Cox processes. Biometrika, 103, 49–70. [Google Scholar]
- Taylor BM and Diggle PJ (2014) INLA or MCMC? A tutorial and comparative evaluation for spatial prediction in log-Gaussian Cox processes. Journal of Statistical Computation and Simulation, 84, 2266–2284. [Google Scholar]
- Turkeltaub PE, Eden GF, Jones KM and Zeffiro TA (2002) Metaanalysis of the functional neuroanatomy of single-word reading: method and validation. NeuroImage, 16, 765–780. [DOI] [PubMed] [Google Scholar]
- Waagepetersen RP (2004) Convergence of posteriors for discretized log Gaussian Cox processes. Statistics and Probability Letters, 66, 229–235. [Google Scholar]
- Wager TD, Jonides J and Reading S (2004) Neuroimaging studies of shifting attention: a meta-analysis. NeuroImage, 22, 1679–1693. [DOI] [PubMed] [Google Scholar]
- Wager TD, Lindquist M and Kaplan L (2007) Meta-analysis of functional neuroimaging data: current and future directions. Social Cognitive and Affective Neuroscience, 2, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD and Smith EE (2003) Neuroimaging studies of working memory. Cognitive, Affective, & Behavioral Neuroscience, 3, 255–274. [DOI] [PubMed] [Google Scholar]
- Wood ATA and Chan G (1994) Simulation of stationary Gaussian processes in [0,1]d. Journal of Computational and Graphical Statistics, 3, 409–432. [Google Scholar]
- Yue YR, Lindquist MA and Loh JM (2012) Meta-analysis of functional neuroimaging data using Bayesian nonparametric binary regression. The Annals of Applied Statistics, 6, 697–718. [Google Scholar]
- Zhang H (2004) Inconsistent estimation and asymptotically equal interpolations in model-based geostatistics. Journal of the American Statistical Association, 99, 250–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.