Abstract
4D printing represents one of the most advanced fabrication techniques for prospective applications in tissue engineering, biomedical devices, and soft robotics, among others. In this study, a novel multiresponsive architecture is developed through stereolithography-based 4D printing, where a universal concept of stress-induced shape transformation is applied to achieve the 4D reprogramming. The light-induced graded internal stress followed by a subsequent solvent-induced relaxation, driving an autonomous and reversible change of the programmed configuration after printing, is employed and investigated in depth and details. Moreover, the fabricated construct possesses shape memory property, offering a characteristic of multiple shape change. Using this novel multiple responsive 4D technique, a proof-of-concept smart nerve guidance conduit is demonstrated on a graphene hybrid 4D construct providing outstanding multifunctional characteristics for nerve regeneration including physical guidance, chemical cues, dynamic self-entubulation, and seamless integration. By employing this fabrication technique, creating multiresponsive smart architectures, as well as demonstrating application potential, this work paves the way for truly initiation of 4D printing in various high-value research fields.
Keywords: 4D bioprinting, multiresponsive, shape memory, smart biomaterials, tissue engineering
1. Introduction
4D printing was first unveiled in 2013, and immediately caught great attention in various research areas including but not limited to autonomous robots, smart materials, and biomedical research.[1–3] Based on 3D printing which has demonstrated versatile capacity in constructing complex structures, customizing mass distribution and minimizing weigh/volume while achieving requisite strength properties, 4D printing further integrates time-dependent transforming information directly into the printed structure which performs shape changes when actuated by external stimuli.[1–5]
Various ink materials and printing techniques have been explored for achieving advanced 4D printing.[1,6–10] Water absorption structures, printed with two different materials having different porosities and water swelling capacities, initiated the concept of 4D printing.[6] Hinges were designed using this two-layer-material structure; the hinged constructs were programmed to deform into predesigned 3D configurations when immersed in water. Besides folding, other transformations such as curling, twisting, and linear expansion were achieved using highly specific joint designs.[7] Similar to water stimuli structures, hygroscopic and ethyl acetate active architectures were fabricated with bilayered structural designs using multimaterial printing techniques.[8,11] In a recent study, a novel acrylamide ink, containing nanofibrillated cellulose, was printed, enabling a swelling-induced 4D shape change using a single material patterned in a onestep process;[1] by controlling the swelling anisotropies of the partially aligned cellulose component, different folding results upon swelling can be precisely predetermined.[8] Additionally, digital light exposure on light-curable monomers of multidimensional responsive polymers, including hydrogels and wax-based shape memory polymers, is reported as an ultrafast 4D printing process via gel swelling/wax uptake as well.[9] Different from water/wax-induced anisotropicity, a heat-induced 4D printing architecture was developed using shape memory polymer very recently, enabling direct and rapid shape change upon heating without any thermomechanical loading.[12] Although these examples of 4D printing research successfully realize self-morphing structures, the shape change triggered by swelling or temperature results in intrinsic configuration changes for the printed materials, thereby affecting mechanical or other physicochemical properties. Moreover, 4D architectures are highly desirable, but their real applications are still scarce.
The 4D process may elicit proper biological responses that not only mimic the dynamic growth process of native tissues/ organs, but also achieve a more complicated, dynamic architecture for tissue implants that meet the criteria of multiple variation with precisely controlled stimuli processes.[13] Among various active materials with different physicochemical properties, shape memory polymers have attracted particular attention owing to their thermomechanical programming characteristics. The reversible “temporary-permanent” transformation has shown great potential for facile, minimally invasive, surgical delivery with in situ shape activation for considerable reduction of trauma and significant improvement of patient comfort.[14–23] Seamless integration between the scaffold and tissue defect can also be better facilitated and addressed through the inherent shape memory effect.[14] We expect that a 4D-printed architecture, which additionally possesses shape memory characteristic (multiple responsive 4D feature), will significantly increase the value and application potential of 4D techniques and materials in biomedical applications.
Herein, we developed a novel multiresponsive architecture through stereolithography (SL)-based 4D bioprinting, using a naturally derived, photocrosslinkable monomer (soybean oil epoxidized acrylate, SOEA). Differing from other 4D printing techniques, an integrated design of stress-induced shape transformation and shape memory effesazct was proposed to achieve a multiple responsive 4D concept. The unique, light-induced, graded internal stress, followed by a subsequent solvent-induced relaxation was employed to drive an autonomous, reversible change of the programmed configuration after printing. Moreover, the naturally derived shape memory construct is able to trigger an additional “thermomechanical programming” shape transformation which is over the usual 4D effect. A typical nanomaterial amplification on 4D effect was also observed, not only verifying the mechanism of light-induced graded internal stress, but also advancing the specific needs of various applications. Using this novel multiple responsive 4D technique, a proof-of-concept, reprogrammable nerve guidance conduit was demonstrated with human mesenchymal stem cells (hMSCs) which high-potentially differentiate into neural cell types on the SL-induced, aligned, surficial microstructures of graphene hybrid 4D constructs providing multifunctional characteristics for nerve regeneration.
2. Results and Discussion
2.1. 4D Printing and Internal-Stress-Induced 4D Transformation
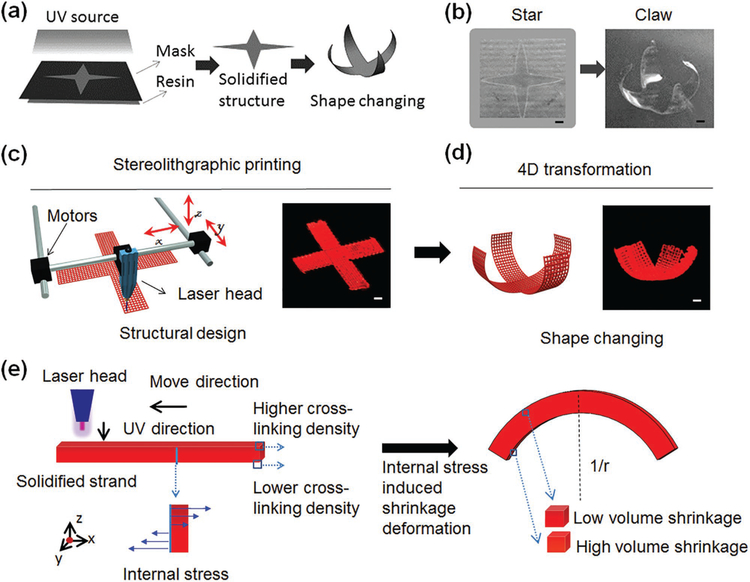
The crucial characteristic for 4D transformation is curvature changing under external stimuli.[3,9,24] Inspired by the “stressrelaxation” phenomenon in material manufacturing, we propose a universal manufacturing-induced 4D printing technique. We observed the shape-changing of UV cross-linked SOEA, as shown in Figure 1a,b, when the solidified flat star structure was immersed in ethanol to remove the uncured ink and the star dynamically transformed into a claw structure. Inspired by this shape-changing process, stereolithographic printing of SOEA was performed at a printing speed of 40 mm s−1 and laser frequency of 20 000 Hz. Shape changing, evident by the bending of the printed structure upon immersion, brought this study into the realm of 4D printing (Figure 1c,d).[6] We assume the laser-induced graded internal stress, introduced through the printing process, is the major driving force of this 4D dynamic variation (Figure 1e). Internal-stress-induced volume shrinkage has been observed and studied in frontal photopolymerization where a polymer film is continuously cured by light from one side in a thick layer of liquid resin.[25,26] By controlling light penetration, light intensity decreased along the sample thickness, inducing an intensity gradient in the resin.[25] As a result, a nonuniform stress field was developed across sample thickness, driving the sample to bend.[25] Different from this reported frontal polymerization which shots light from one side, our 4D SL printing draws desired structural patterns with a laser head on the surface of SOEA resin, which is versatile in pattern design and has advantages in achieving more complex shape changing such as torsion (see next section). But the attenuation of laser light in the resin shares the same mechanism with the generation of light gradient intensity in the frontal polymerization, which leads to the internal stress to drive shape change. In the frontal polymerization, internal stress is released by peeling off the sample from substrate, resulting in the shape change immediately.[25] In the present study, the sample is printed directly on the resin, but shape change is not observed even after taking the sample out of the resin. The high sticky feature of the SOEA resin may prevent the internal stress release and shape change. After immersing the sample in ethanol, the sticky SOEA on the surface of the sample will be dissolved, triggering the release of internal stresses and driving the dynamic shape change process (Figure S1, Supporting Information).
Figure 1.
Novel 4D printing using stress-induced shape transformation. a) Illustration of shape changing of UV-cured SOEA by stress relaxation after ethanol immersion. b) The photos of UV-cured “star” architecture (left) and reprogrammed “claw” architecture after ethanol immersion (right). c) Illustration of the stereolithographic (SL) printing process. A cross architecture (structural design, left; sample image, right) is printed by SL printer. d) Illustration of 4D transformation that the claw architecture (structural design, left; sample image, right) is formed after stress relaxation. e) Mechanism of the internal-stress-induced shape changing process. Uneven cross-linking density generates the difference of modulus between upper and lower surfaces due to laser energy attenuation, leading to different volume shrinkage after stress relaxation. Scale bar, 2 mm.
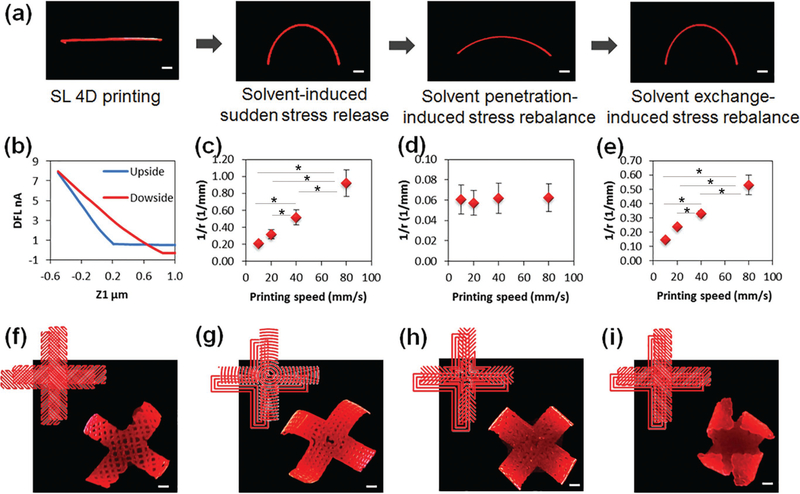
Another interesting phenomenon is that the bending architecture gradually flatted after remaining in ethanol for more than 30 min. In order to further investigate this process, a printed single strip of SOEA was thoroughly studied further developing the concept of a reversible, solvent-induced 4D transformation. This 4D reprogrammable process is concisely illustrated in Figure 2a through a reversible, dynamic variation of the SL-printed strip under ethanol stimuli; that is, ethanol lets the sample swell which enables the shape change. As the straight strip was immersed in ethanol, bending (curvature = 0.5 mm−1) was clearly observed in several seconds; with increasing time, the curved strip decreased its curvature gradually in ethanol (curvature smaller than 0.07 mm−1); interestingly, the strip curved again, with a curvature over 0.35 mm−1, after it was transferred from ethanol to water. Most importantly, when repeating this process, we can observe a reversible, cyclical variation. The entire 4D transformation process of the printed strip is also presented in Movie S1 (Supporting Information). When the freshly printed strip is immersed in ethanol, the immediate shape change is driven by internal stress as discussed earlier. The following “flat–curving–flat” recycle process is attributed to a resolvation/desolvation process, which will be discussed next.
Figure 2.
4D-printed single strand and flower structures. a) Stimulation process of a printed strand containing 0.01% Nile red. b) DFLvs z1 analyzed with AFM. c) Curvature versus printing speed when the printed strand was immediately immersed in ethanol, the strand was printed at 40 mm s−1 with laser intensity of 20 kHz. d) Curvature versus printing speed when the strand was thereafter immersed in ethanol for 30 min. e) Curvature versus printing speed when the strand was then immersed in water for 30 min. f–i) Various 4D transformations were performed by different pattern designs (see the details in Figure S4, Supporting Information). The 4D-printed flower samples, which were printed at 40 mm s−1 with laser intensity of 20 kHz and contained 0.01% Nile red. Scale bar, 2 mm. The error bar indicates the standard error of the mean (n = 6). * indicates p < 0.05 from ANOVA with Tukey’s multiple pairwise comparison. Data shown are from a representative set of experiments.
When printing architectures using SL, a higher crosslinking density is expected in the printed strip’s surface facing the laser head (upside) while a lower cross-linking density is expected in the opposite surface (downside) largely due to laser energy attenuation (Figure 1e). As confirmed in Figure 2b, the cross-linking characteristics of the upside, determined by atomic force microscopy (AFM), are significantly different from that of the downside; accordingly, the surficial modulus on the upside, 75.1 ± 9.3 MPa, is significantly higher than that on the downside, 37.3 ± 5.4 MPa, which supports the lower cross-linking density on downside. When the freshly printed strip is placed in ethanol, the 4D shape change is achieved immediately driven by the internal stress. However, if the sample is remained in ethanol, the gradual permeation of ethanol into the gradient cross-linked structure of the printed hydrophobic resin will thereafter stretch the strip, subsequently decreasing the curvature; this is a typical resolvation process. As the strip is shifted into water, the permeated ethanol in the strip is extracted but is not replaced by external water because the hydrophilicity and high surface tension of water cannot permeate the hydrophobic resin; this process is the intrinsic mechanism to achieve the recovery of curvature, and a typical desolvation process. Desolvation/resolvation process has been used to develop self-assembly of photopatterned polymers.[27–29] Herein, the desolvation/resolvation process is responsible for the reversible sample shape change in ethanol/ water after 4D printing. In particular, both internal-stressinduced shape change and desolvation/resolvation process are observed in our dynamic shape change process. The remarkable difference between these two processes is that the former is exerted very rapidly in several seconds and not reversible while the latter is relatively slow, about 30 min, and reversible. Strictly, the internal-stress-induced shape change has brought this research into the field of 4D printing; the reversible desolvation/resolvation process is a multiple responsive 4D feature.[9,11]
In order to further understand the laser-induced graded internal stress, the printing speed was changed to investigate its effect on the curvature of the printed strip, as shown in Figure 2c–e. With increasing printing speeds, the curvature of the strip increased significantly as it was initially immersed in ethanol (Figure 2c). In contrast, after remaining in ethanol for 30 min, all strips exhibited similar curvature in the rebalance phase (Figure 2d). The strips printed at higher speeds still showed higher curvature after moving from ethanol to water (Figure 2e). We deduce that the thickness of the printed strip plays an important role in the 4D curvature. Our results (Figure S2, Supporting Information) also exhibit that thinner strips are obtained at higher printing speeds due to the decrease in time of laser exposure. At lower printing speed, bending stiffness may increase because of increased strip thickness, which makes the strip more difficult to bend.[10,25]
2.2. A Mathematical Model for 4D Reprogrammable Change by Anisotropic Stress Relaxation
Harnessing the anisotropic stress relaxation allows precision control over the curvature of our 4D-printed architectures. To mathematically characterize this phenomenon (Figure S3, Supporting Information) we constructed a mathematical model for the anisotropic mechanics to elaborate on the relationship among all parameters, including pore size, pore volume fraction, diffusion rate, and dissolution rate/solubility of the resins, among others. Considering the complexity of multiple factors in 4D reprogramming process involving the degree of photopolymerization and flows in porous media with mass transfer, we simplified mechanical model with certain linear assumptions to effectively model the phenomenon and direct our CAD design. As we discussed above, the thickness of the printed strip depends on the printing speed, which is related to the energy offered by the laser. The higher the printing speed, the less the exposure time of the ink and thereby the less polymerization, leading to a thinner strip with lower cross-linking density. In order to model this phenomenon, we may simply assume the validity of the following relation[30]
| (1) |
where Q with a unit of length is a physical constant, α is another constant with units of velocity, and ν is the printing speed. Using the above equation and fitting to the data in Figure S2 (Supporting Information), we calculate Q = 61.86 × 10−6 m, and α = 0.23 m s−1.
The relationship between the curvature and printing speed can be modeled using the following equation (see detailed derivations in Figure S3, Supporting Information)
| (2) |
where r is the radius of the curvature and χ0 is the shrinkage which is dependent on both the material and the immersed medium. Here, χ0 = 0.012 s−1 for water, and χ0 = 0.019 s−1 for ethanol. The above simple equation correlates the curvature of the strip to other controllable parameters, which could help print objects with desirable curvatures or shapes. More interestingly, the Young’s modulus does not contribute to the final curvature directly. It is their difference and ratio that make a difference. This is mainly due to the linear assumption on the Young’s modulus. According to our theoretical model, through varying printing speed, we are able to control the 4D curvature of the programmed architectures, which provides a significantly convenient and universal approach to achieve 4D transformation for photocuring materials.
The ability to program and optimize the bending primitive on topographical designs suggests complicated 4D architectures are highly promising and expected. As shown in Figure 2f–i, the architectures show the different shape changes under stimuli rising from various topographical designs (Figure S4, Supporting Information). In our design, the structure in Figure 2f has a cross orientation for the two layers, thus the packed architecture shows a unidirectional bending (vertical plane) due to the counterbalance effect. The architectures in Figure 2g,h have the same pattern on the first layer; when changing the patterns on the second layer, we observed different 4D transformation behaviors. Compared to architectures in Figure 2f–h, the architecture in Figure 2i shows a complicated bending structure (a beautiful flower shape); here, the horizontal direction had an additional torsion of 45° attributed to the anisotropic orientation on the different layers. Additionally, the curvature of pattern also has a direct effect on the 4D shape changes of the printed architectures. The curvatures of architectures in Figure 2f,g have a noticeable deviation while architectures in Figure 2h,i perfectly agree with the expected pattern designs and printing paths on the second layer. Therefore, these reprogrammable and tunable processes through a precise topographic design make our 4D study versatile for different forms.
2.3. Multiple Responsive 4D Programming: Reversible Shape Change and Shape Memory Property
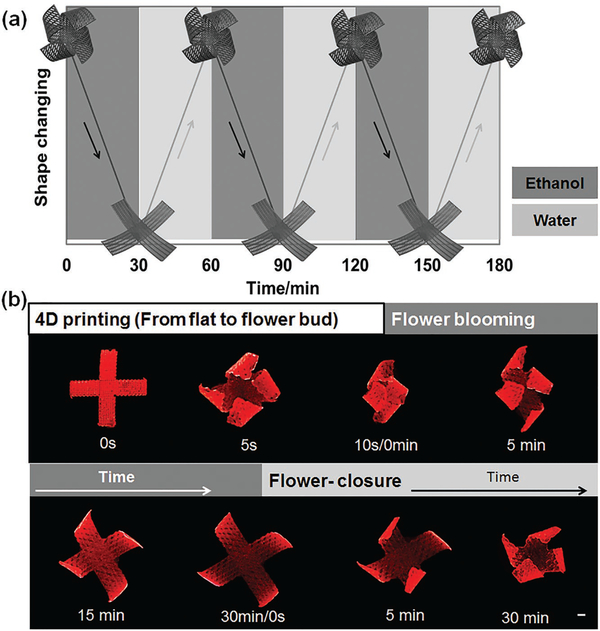
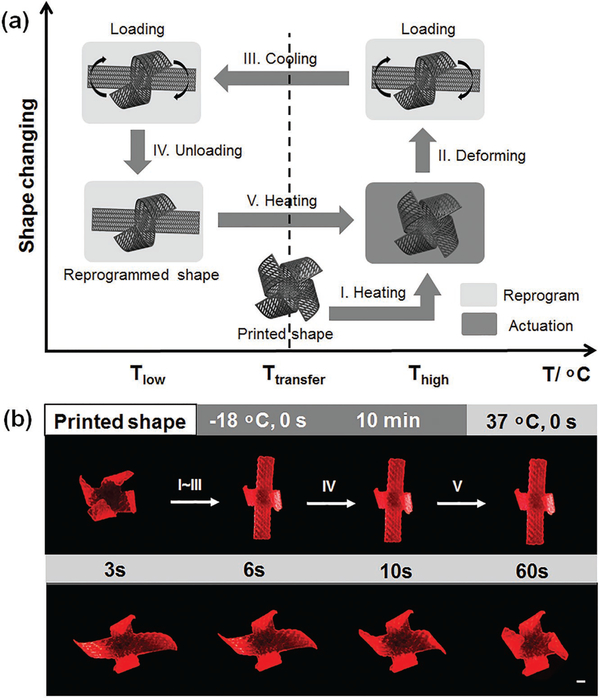
Based on aforementioned simulation and theory analysis, we designed an anisotropic architecture, achieving a maximum, multidirectional 4D transformation. As observed in the recycling bending of the printed single strip (Figure 2), the final 4D flower architecture also demonstrates a reversible shape change between ethanol and water immersion processes, as shown in Figure 3. After internal stress relaxation by ethanol, this 4D flower reprogrammable architecture is permanent and stable in water/air. However, this flower architecture gradually flattens again after reimmersion in ethanol for long periods of time (Movie S2, Supporting Information). This dynamic shape change process is driven by the resolvation of ethanol, which repenetrates to swell the resin, as discussed in earlier section. After reimmersed in water, the flat architecture is slowly closed to form a cage-like or flower bud architecture in a revolving fashion as the ethanol is extracted from the resin while water cannot penetrate (Movie S3, Supporting Information). The creation of reversibly actuating components that alter their shapes in a controllable manner in response to environmental stimuli is highly attractive in active material design, which is even beyond the 4D realm and has a wide array of application potentials.[31] Moreover, the 4D flower possesses excellent shape memory effect, as exhibited in Figure 4, achieving an additional thermomechanical-programmed 4D transformation. Two petals of the flower are fully flattened by external force, and fix a temporary shape at −18 °C; the original flower shape is recovered after exposure to a 37 °C environment (Movie S4, Supporting Information). In the printed samples, the chemical cross-linking networks from SOEA are utilized to set the permanent shape while a glass transition temperature (Tg) is used to control the molecular switching segments that fix the temporary shape. The Tg of solidified SOEA is 20 °C (Figure S5, Supporting Information). As the sample is heated above Tg (37 °C), the molecular segments between cross-linking points are “soft” and a deformation can be exerted to set the temporary shape; when the temperature is decreased below the Tg (−18 °C), the molecular segments will “freeze” to immobilize the predesigned temporary shape. The sample will recover their permanent shape upon returning to a temperature over Tg since the molecular segments are soft again allowing the cross-link networks to revert the structure back to the original shape. The temperature diagram is also indicated in Figure 4. The “thermomechanical programming” shape transformation enables additional functions to exceed the original 4D effect, facilitating a partial or surplus change for more complicated requirements on real applications. Therefore, the multiple responsive 4D feature, which is the combination of reversible shape changing and shape memory properties, may greatly inspire further development of multitransformative smart architectures, a highly active research area in recent years.[32] In this study, only shape memory property is further utilized in the nerve conduit construct; thus, the 4D feature is restricted to shape memory property in the next context.
Figure 3.
Beyond 4D printing—reversible dynamic shape change. a) The illustration of the reversible dynamic process of opening in ethanol and closing in water of the 4D-printed flower structure, which can be performed repeatedly. b) Photo of the reversible shape change process with 4D-printed flower structure; 0 s, the printed structure before 4D shape change; 5 s, the printed structure after 4D shape change as it was immersed in ethanol immediately; 0–30 min, the flower structure gradually flatted as it was soaked in ethanol; flower closure (0–30 min), the flatted structure gradually recovered its flower shape after it was immersed in water. Scale bar, 2 mm.
Figure 4.
Beyond 4D printing—shape memory effect. a) Illustration of the shape memory process. (I) Increase temperature over transferring temperature; (II) exert a shape change with enforced restriction; (III) fix a temporary shape at lower temperature; (IV) remove external enforced restriction; (V) increase temperature to recover the original shape. b) Demonstration of the shape memory process with 4D-printed flower structure. The samples were printed at 40 mm s−1 with laser intensity of 20 kHz. (I)–(IV) Two petals of the flower were fixed at a flat temporary structure as demonstrated in (a). (V) (0–60 s), the petals of the flower gradually recovered their original shape as the structure was immersed in 37 °C water. Scale bar, 2 mm.
2.4. Graphene Nanohybrid for 4D Effect Enhancement
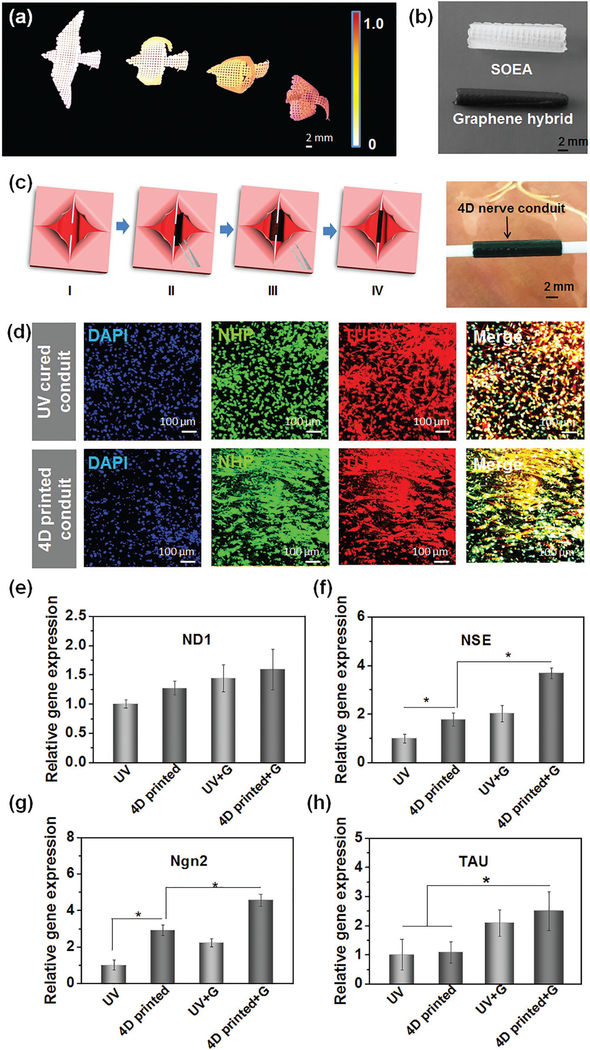
Owing to the laser-induced graded internal stress mechanism, we expect nanohybrids can further affect the attenuation of laser energy, contributing to the regulation of the curvature in 4D transformation. In this study, we chosen typical functional nanoparticles to prepare the composite nanoinks, including iron oxide magnetic nanopowder, carbon nanofiber, hydroxyapatite, and graphene. We observed that all of the nanohybrids significantly increase the curvature of the 4D-printed architectures compared to the pure resin, as presented in Figure S6 (Supporting Information). In fabricating origami by frontal polymerization, photoabsorbers were added to control light penetration to induce an intensity gradient in the resin.[25] Herein, these nanoparticles have a similar role with photoabsorbers in control of laser penetration as the printing ink exhibits significantly decreased light penetration when the nanoparticles are added (data not shown here). Interestingly, by varying the content of nanoparticles (graphene) in the inks, a bird with a serial of flying actions was easily generated after the 4D printing (Figure 5a and Figure S6, Supporting Information), indicating the 4D curvature can be tuned by varying the nanoparticle concentrations. This phenomenon can be explained that the addition of graphene hinders the penetration of laser through the ink, leading to a weak cross-linking effect. As shown in Figure S6 (Supporting Information), the thickness of the architectures decreases significantly with the increasing graphene content. When the strip with 0.8% graphene has same thickness as the strip without graphene, the former exhibits a greater curvature as shown in Figure S6 (Supporting Information). Additionally, we also observed nanomaterial-induced surficial porous structures in the 4D-printed architectures (Figure S6, Supporting Information). The porous surficial structures on the downside could result in greater difference from the surface facing the laser head, as interpreted in Figure S1 (Supporting Information), playing a greater role in increasing the curvature. Thus, either nanoparticle types or concentration is able to manipulate or amplify our 4D effect, not only verifying the mechanism of light-induced graded internal stress, but also advancing the specific needs of various applications.
Figure 5.
Graphene nanohybrid for enhancing 4D curvature, and 4D reprogrammable nerve guidance conduit. a) A serial of bird flying architectures fabricated with different graphene contents ranging from 0 to 0.8%. b) 4D reprogrammable nerve guidance conduit fabricated without graphene (white) and with 0.8% graphene. A rectangular architecture in 0.8 × 1.5 cm was printed, and then a conduit was formed through an interior stress-induced 4D transformation. c) Illustration of the full entubulation of 4D nerve guidance conduit via a “thermomechanical programming” shape transformation. (I) The model of a damaged nerve which is present as two stumps of a severed nerve; (II) place the 4D reprogrammable nanohybrid construct (black) which is fixed at a flat temporary structure under the injured nerve; (III) the 4D reprogrammable nanohybrid construct will autonomously recover its original rolling structure at body temperature; (IV) the nerve is fully wrapped by the 4D reprogrammable nanohybrid conduit. The photo illustrates the 4D-printed nanohydrid conduit integrated well with the two assumed stumps of a severed nerve, suggesting the 4D transformation potentially facilitates the surgical operation on conduit implantation. d) Immunofluorescence images of neurogenic differentiation of hMSCs on 4D-printed nanohydrid conduits and their ultraviolet-cured counterpart. Although both of them showed excellent neurogenic ability, the 4D-printed nanohydrid showed a significant aligned topography due to the printing-induced microfeature. Gene expression of neurogenic differentiation of hMSCs by RT-PCR technique, including e) ND1, f) NSE, g) Ngn2, and h) TAU. The results indicate a 4D-printed graphene hybrid conduit can significantly improve neurogenic differentiation of hMSCs. The error bar indicates the standard error of the mean (n = 6). * indicates p < 0.05 from ANOVA with Tukey’s multiple pairwise comparison. Data shown are from a representative set of experiments.
2.5. 4D Reprogrammable Nerve Guidance Conduit
To explore the potential application of our 4D-printed architectures, we design and fabricate a reprogrammable nerve guidance conduit as demonstrated in Figure 5b,c for potentially repairing peripheral nerve injuries. 4D nerve guidance conduit was easily fabricated with high curvature and flexibility without any supporting materials, achieving dynamic self-entubulation and seamless integration for damaged nerve. A tightscrolled, tubular architecture was obtained by adding only 0.8% graphene compared to the counterpart without graphene (Figure 5b) and is capable of improved integration with the two assumed stumps of a damaged or severed nerve providing tension necessary for guiding axonal regrowth (Figure 5c). Additionally, the “thermomechanical programming” shape memory characteristic enables the closed conduit to be temporarily opened and fixed, facilitating the surgical operation on conduit implantation, as demonstrated in Figure 5c. Moreover, graphene has been used to enhance conduit conductivity and neuronal differentiation,[33] thus we expect the 4D-printed graphene nanohybrid may provide excellent physical and chemical signals for nerve regeneration.
For this proof-of-concept study, neural differentiation of hMSCs by addition of neural differentiation medium on the 4D-printed architectures was performed to fully investigate their potential for neural engineering. As shown in Figure 5d, although neural differentiation of hMSCs was observed on all the samples, alignment of hMSCs was only noticed on the 4D-printed architectures. It is evident that the cell alignment originates from the microstrip orientation of the printed architectures (Figure 5b). A crucial feature of neural tissues is highly organized and structurally anisotropic components. The aligned topographical (physical) cues on the 4D conduits may better recapitulate native cellular microenvironments, and have an impact on improving neural alignment.[34] However, in the immunostaining study, we did not observe significant difference between the graphene conduits and their counterparts. Therefore, we further quantitatively investigated the neural differentiation of hMSCs on various samples by a real-time quantitative polymerase chain reaction (rt-qPCR) analysis. The typical neurogenic genes including neurogenin 2 (Ngn2), neurogenic differentiation 1 (ND1), neuron specific enolase (NSE), and Tau proteins (TAU) were analyzed. As shown in Figure 5d, compared to control samples, the 4D-printed samples showed greater gene expression of ND1, NSE, and Ngn2. Moreover, all neurogenic gene expression was upregulated in the graphene conduits when compared with those conduits without graphene. Among these samples, the 4D-printed graphene conduit exhibited the best results on neural differentiation confirmed by rt-qPCR. Although there are few evidences on axon formation, our studies demonstrate that mesenchymal stem cells have high potential on differentiating into neural cell types on the 4D aligned graphene hybrid conduits. The 4D printing technique offers outstanding, multi functional characteristics for developing novel nerve conduit including physical guidance and chemical cues, as well as the potential ability of self-entubulation for dynamic and seamless integration.
3. Conclusion
In summary, a novel 4D dynamic process was realized using laser-induced graded internal stress; beyond 4D bioprinting, the fabricated construct showed multiple, value-added, additional functions including reversible shape transformation and shape memory effect, which provide great inspiration for not only biomedical studies but also for other 4D reprogramming research such as smart robot design. A reprogrammable nerve guidance conduit was demonstrated with this novel 4D bioprinting, significantly inspiring the advancement of this 4D printing technique in neural tissue engineering.
4. Experimental Section
Chemicals and Materials:
Alpha minimum essential medium (MEM alpha (1×)+Gluta MAXTM-1), l-glutamine, and penicillin/streptomycin solution were obtained from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) was purchased from Gemini Bioproducts (West Sacramento, CA). DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) was purchased from Anaspec Inc. (Fremont, CA). Phosphate buffered saline (PBS), without calcium and magnesium, and trypsin/ ethylenediaminetetraacetic acid (EDTA) (1× 0.25% trypsin/2.21 × 10−3 M EDTA in Hank’s balanced salt solution without sodium bicarbonate, calcium, and magnesium) were supplied by Mediatech Inc. (Manassas, VA). Primary hMSCs were obtained from healthy consenting donors at the Texas A&M Health Science Center, Institute for Regenerative Medicine. Mouse anti-tubulin β 3 (TUBB3) antibody was purchased from BioLegend (San Diego, CA). Rabbit anti-neurofilament heavy polypeptide (NHP) antibody, goat anti-mouse IgG-H&L (Alexa Fluor 594) antibody, and goat anti-rabbit IgG-H&L (Alexa Fluor 488) antibody were purchased from Abcam (Cambridge, UK). Unless otherwise listed, all solvents and reagents were purchased from Aldrich Chemical Co. (St. Louis, MO) and used as received.
Preparation of 4D Bioprinting Ink:
Briefly, 100 g soybean oil epoxidized acrylate was mixed with 100 mL acetone in a 500 mL brown glass bottle wrapped with two layers of aluminum foil to protect the mixture from visible light. Then 1.26 g bis(2,4,6-trimethylbenzoyl)-phenylphosphineoxide was added. The mixture was shaken mildly with hands and a 150 rpm Standard Analog Shaker (VWR International, PA, USA) alternatively to reach a homogenous yellow solution that was subsequently put into a vacuumed container overnight to remove acetone. The resulting yellow, sticky liquid was utilized directly as ink for bioprinting.
Design of Scaffolds:
The expected structure was drawn with the software Autodesk123D (Autodesk Inc., CA, USA), and saved as a stl format file which was then loaded into the software Slic3r which is licensed under the GNU Affero General Public License, version 3. The infill density and the printing speed were assigned in Slic3r. Other typical parameters assigned in Slic3r include: 0.2 mm layer height, 0.2 mm first layer height; vertical shells—perimeters 0; horizontal shells—solid layers, top 0, bottom 0; 90° infill angle, 10 mm2 solid infill threshold area; skirt, loop 0; extrusion width, first layer 0%.
3D Laser Bioprinting:
The designed micropatterns were printed via stereolithography with a table top SL system developed in a lab based on the existing Solidoodle 3D printer platform.[35] Open source software (Prontrface) was employed to control the three stepper motors with an effective resolution of 100 μm in the x, y, and z-axis. The major modification to the existing platform is the incorporation of a 110 μm fiber optic-coupled solid-state UV (355 nm) laser (MarketTech, Scotts Valley, CA). Per the manufacturer’s specifications, the effective spot size of the emitted light is 190 ± 50 μm with an energy output of 20 μJ at 15 kHz. A glass petri dish fixed on the print bed acted as a minivat for the addition of liquid photocurable resin. The ability to alter the frequency of the pulsed signal facilitates power control at the material’s surface ranging from ≈40 to 110 mW. Activating the laser and drawing lines at various print speeds cured the ink. After polymerization, the micropattern was lifted off the petri dish and was soaked overnight in ethanol to remove unpolymerized ink. Finally, the micropattern was sterilized with 70% ethanol for 30 min and soaked in PBS overnight prior to cell culturing.
Video Shooting:
Shape transformation was recoded in a self-made black box. The inks for the recorded samples contained 0.01% of Nile red giving the samples a red color appearance during the recording process. A black paper was used as background. A petri dish containing PBS buffer or ethanol was placed on the black paper. Samples were placed into the solvent-containing petri dish when recording the reversible shape change. An empty petri dish was applied when recording the shape recovery process. The dynamic shape change was recorded with a PowerShot ELPH 360HS Cannon camera.
Neurogenic Differentiation and Immunofluorescence:
hMSCs (passage nos. 3–6) were cultured in complete media composed of alpha minimum essential medium supplemented with 16.5% FBS, 1% (v/v) l-glutamine, and 1% penicillin:streptomycin solution, under standard cell culture conditions (37 °C, a humidified, 5% CO2/95% air environment). hMSCs were seeded at a cell density of 100 000 cells/specimen, and cultured under standard cell culture conditions for 24 h. After discarding the complete medium, hMSC neurogenic differentiation medium (PromoCell) was added to the culture, and the medium was changed every 3 d. After two weeks of culture, the samples were fixed with 10% formalin for 15 min, permeabilized in 0.1% Triton X-100 for 10 min, and blocked in 2% BSA for 30 min. The cells were then incubated overnight with primary antibodies at 4 °C. The following primary antibodies were used for staining: mouse TUBB3 antibody (BioLegend) and rabbit anti-NHP antibody (Abcam). After incubation with primary antibodies, goat anti-mouse IgG-H&L (Alexa Fluor 594) (Abcam) and goat anti-rabbit IgG-H&L (Alexa Fluor 488) (Abcam) as secondary antibodies were added and incubated for 1 h each, respectively. Finally, the nuclei were stained with DAPI. Fluorescence images were observed, and taken using a Zeiss LSM 710 confocal microscope.
Real-Time Quantitative Polymerase Chain Reaction:
The neurogenic gene expression of all samples including Ngn2, ND1, NSE, and TAU were analyzed by rt-qPCR assay. Briefly, the total RNA contents were extracted from the samples by using Trizol reagent. The RNA quality and concentration were determined from the absorbance at 260 and 280 nm with a microplate reader. RNA samples were reverse-transcribed to cDNA using a Prime ScriptRT reagent Kit (TaKaRa). RT-PCR was then performed on a CFX384 Real-Time System (BIORAD) by using SYBR Premix Ex Taq (TaKaRa) according to the manufacturer’s protocol. The gene expression level of target genes was normalized against the house-keeping gene beta-actin (β-actin). The relative gene expression of the 4D-printed scaffold was normalized against the control group (UV-cured sample) to obtain relative gene expression fold values, and calculated via the 2-delta delta (2−ΔΔCt) cycle-threshold method. Primer sequences are as follows: β-actin (NM_001101), forward primer 5′-CCCTTGCCATCCTAAAAGCC-3′ and reverse primer 5′-TGCTATCACCTCCCCTGTGT-3′; Ngn2 (NM_024019.2), forward primer 5′-CCTGGAAACCATCTCACTTCA-3′ and reverse primer 5′-TACCCAAAGCCAAGAAATGC-3′; ND1 (NM_002500.2), forward primer 5′-CCACGGATCAATCTTCTCAG-3′ and reverse primer 5′-CATGATGTGAATGGCTATCG-3′; NSE (NM_001975.2), forward primer 5′-TAACTTCCGTAATCCCAGTGT-3′ and reverse primer 5′-AAGAGGTCAGGTAAGCCAA-3′; TAU (NM_016835.3), forward primer 5′-TACAGACCTGCGGCTTCATAA-3′ and reverse primer 5′-CCAGAAATAGTCCTGCTCAACA-3′.
Statistical Analysis:
The curvatures of printed strips versus printing speeds were plotted using Excel (Figure 2c–e). Relative gene expression data were plotted using Origin Pro 8.5 (Figure 5e–h). The mean and standard deviation was plotted for each sample group (n = 6). Then, a one-way analysis of variance (ANOVA) (p < 0.05) with Tukey’s multiple pairwise comparison was performed on each set of data using GraphPad Prism (GraphPad Software Inc., San Diego, CA). The statistical significance was indicated with an asterisk. That is, samples marked with an asterisk were significantly different.
Supplementary Material
Acknowledgements
S.M. and H.C. contributed equally to this work. This work was supported by NSF MME program Grant No. 1642186, NIH Director’s New Innovator Award 1DP2EB020549–01, and March of Dimes Foundation’s Gene Discovery and Translational Research Grant.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adbi.201800101
Contributor Information
Dr. Shida Miao, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Dr. Haitao Cui, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Dr. Margaret Nowicki, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Dr. Lang Xia, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Dr. Xuan Zhou, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Se-Jun Lee, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu.
Dr. Wei Zhu, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Prof. Kausik Sarkar, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu
Prof. Zhiyong Zhang, Translational Research Centre of Regenerative Medicine and 3D Printing Technologies of Guangzhou Medical University, The Third Affiliated Hospital of Guangzhou Medical University, No. 63 Duobao Road, Liwan District, Guangzhou City, Guangdong, Province 510150, P. R. China
Prof. Lijie Grace Zhang, Department of Aerospace and Mechanical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA, lgzhang@gwu.edu Department of Biomedical Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA; Department of Electrical and Computer Engineering, The George Washington University, 800 22nd St NW, Washington, DC 20052, USA; Department of Medicine, The George Washington University, 2150 Pennsylvania Ave NW, Washington, DC 20052, USA.
References
- [1].Gladman AS, Matsumoto EA, Nuzzo RG, Mahadevan L, Lewis JA, Nat. Mater 2016, 15, 413. [DOI] [PubMed] [Google Scholar]
- [2].Truby RL, Lewis JA, Nature 2016, 540, 371. [DOI] [PubMed] [Google Scholar]
- [3].Miao S, Castro N, Nowicki M, Xia L, Cui H, Zhou X, Zhu W, Lee S-J, Sarkar K, Vozzi G, Mater. Today 2017, 20, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ge Q, Qi HJ, Dunn ML, Appl. Phys. Lett 2013, 103, 131901. [Google Scholar]
- [5].Ge Q, Dunn CK, Qi HJ, Dunn ML, Smart Mater. Struct 2014, 23, 094007. [Google Scholar]
- [6].Tibbits S, Archit. Des 2014, 84, 116. [Google Scholar]
- [7].Raviv D, Zhao W, McKnelly C, Papadopoulou A, Kadambi A, Shi B, Hirsch S, Dikovsky D, Zyracki M, Olguin C, Sci. Rep 2014, 4, 7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Correa D, Papadopoulou A, Guberan C, Jhaveri N, Reichert S, Menges A, Tibbits S, 3D Print. 2015, 2, 106. [Google Scholar]
- [9].Huang L, Jiang R, Wu J, Song J, Bai H, Li B, Zhao Q, Xie T, Adv. Mater 2016, 29, 1605390. [DOI] [PubMed] [Google Scholar]
- [10].Su J-W, Tao X, Deng H, Zhang C, Jiang S, Lin Y, Lin J, Soft Matter 2018, 14, 765. [DOI] [PubMed] [Google Scholar]
- [11].Kokkinis D, Schaffner M, Studart AR, Nat. Commun 2015, 6, 8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ding Z, Yuan C, Peng X, Wang T, Qi HJ, Dunn ML, Sci. Adv 2017, 3, e1602890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Morrison RJ, Hollister SJ, Niedner MF, Mahani MG, Park AH, Mehta DK, Ohye RG, Green GE, Sci. Transl. Med 2015, 7, 285ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang D, George OJ, Petersen KM, Jimenez-Vergara AC, Hahn MS, Grunlan MA, Acta Biomater. 2014, 10, 4597. [DOI] [PubMed] [Google Scholar]
- [15].Miao S, Zhu W, Castro NJ, Leng J, Zhang LG, Tissue Eng., Part C 2016, 22, 952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yakacki CM, Shandas R, Lanning C, Rech B, Eckstein A, Gall K, Biomaterials 2007, 28, 2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lendlein A, Kelch S, Angew. Chem., Int. Ed 2002, 41, 2034. [PubMed] [Google Scholar]
- [18].Hearon K, Wierzbicki MA, Nash LD, Landsman TL, Laramy C, Lonnecker AT, Gibbons MC, Ur S, Cardinal KO, Wilson TS, Adv. Healthcare Mater 2015, 4, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Small IW, Singhal P, Wilson TS, Maitland DJ, J. Mater. Chem. 2010, 20, 3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Miao S, Sun L, Wang P, Liu R, Su Z, Zhang S, Eur. J. Lipid Sci. Technol. 2012, 114, 1165. [Google Scholar]
- [21].Miao S, Wang P, Su Z, Zhang S, Acta Biomater. 2014, 10, 1692. [DOI] [PubMed] [Google Scholar]
- [22].Miao S, Callow N, Wang P, Liu Y, Su Z, Zhang S, J. Am. Oil Chem. Soc 2013, 90, 1415. [Google Scholar]
- [23].Miao S, Wang P, Su Z, Liu Y, Zhang S, Eur. J. Lipid Sci. Technol. 2012, 114, 1345. [Google Scholar]
- [24].Momeni F, Liu X, Ni J, Mater. Des 2017, 122, 42. [Google Scholar]
- [25].Zhao Z, Wu J, Mu X, Chen H, Qi HJ, Fang D, Sci. Adv 2017, 3, e1602326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Robertson ID, Yourdkhani M, Centellas PJ, Aw JE, Ivanoff DG, Goli E, Lloyd EM, Dean LM, Sottos NR, Geubelle PH, Nature 2018, 557, 223. [DOI] [PubMed] [Google Scholar]
- [27].Jamal M, Zarafshar AM, Gracias DH, Nat. Commun 2011, 2, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao Z, Wu J, Mu X, Chen H, Qi HJ, Fang D, Macromol. Rapid Commun 2017, 38. [DOI] [PubMed] [Google Scholar]
- [29].Yoon C, Xiao R, Park J, Cha J, Nguyen TD, Gracias DH, Smart Mater. Struct 2014, 23, 094008. [Google Scholar]
- [30].Fuh J, Choo Y, Nee A, Lu L, Lee K, Mater. Des 1995, 16, 23. [Google Scholar]
- [31].Ionov L, Stoychev G, Jehnichen D, Sommer JU, ACS Appl. Mater. Interfaces 2016, 9, 4873. [DOI] [PubMed] [Google Scholar]
- [32].Thérien-Aubin HL, Wu ZL, Nie Z, Kumacheva E, J. Am. Chem. Soc 2013, 135, 4834. [DOI] [PubMed] [Google Scholar]
- [33].Gardin C, Piattelli A, Zavan B, Trends Biotechnol. 2016, 34, 435. [DOI] [PubMed] [Google Scholar]
- [34].Wang ZY, Teoh SH, Johana NB, Chong MSK, Teo EY, Hong M-H, Chan JKY, San Thian E, J. Mater. Chem. B 2014, 2, 5898. [DOI] [PubMed] [Google Scholar]
- [35].Castro NJ, O’Brien J, Zhang LG, Nanoscale 2015, 7, 14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.