Abstract
Mixed lineage leukemia proteins (MLL) are the key histone lysine methyltransferases that regulate expression of diverse genes. Aberrant activation of MLL promotes leukemia as well as solid tumors in humans, highlighting the urgent need for the development of an MLL inhibitor. We screened and isolated MLL1-binding ssRNAs using SELEX (Systemic Evolution of Ligands by Exponential enrichment) technology. When sequences in sub-libraries were obtained using next-generation sequencing (NGS), the most enriched aptamers—APT1 and APT2—represented about 30% and 26% of sub-library populations, respectively. Motif analysis of the top 50 sequences provided a highly conserved sequence: 5′-A[A/C][C/G][G/U][U/A]ACAGAGGG[U/A]GG[A/C] GAGUGGGU-3′. APT1, APT2, and APT5 embracing this motif generated secondary structures with similar topological characteristics. We found that APT1 and APT2 have a good binding activity and the analysis using mutated aptamer variants showed that the site information in the central region was critical for binding. In vitro enzyme activity assay showed that APT1 and APT2 had MLL1 inhibitory activity. Three-dimensional structure prediction of APT1-MLL1 complex indicates multiple weak interactions formed between MLL1 SET domain and APT1. Our study confirmed that NGS-assisted SELEX is an efficient tool for aptamer screening and that aptamers could be useful in diagnosis and treatment of MLL1-mediated diseases.
Keywords: Aptamers, SELEX, MLL1, Next-generation sequencing, ssRNA
INTRODUCTION
The myeloid/lymphoid or mixed lineage leukemia proteins (MLLs) are the key histone lysine methyltransferases (HKMTs) that regulate the expression of a number of genes, including homeobox (HOX) gene, thereby affecting hematopoiesis and development in mammals (Yu et al., 1995; Milne et al., 2002, 2005; Terranova et al., 2006). Of the six MLL members, MLL1 catalyzes mono-, di-, or trimethylation of histone H3 on Lys4 via the SET domain. The full-length MLL1 is activated after cleavage by taspase 1 into N- and C-terminal fragments (320 and 180 kDa respectively) which reassemble to form a stable complex (Yokoyama et al., 2002; Hsieh et al., 2003). The SET is located at the c-terminal end of the MLL-C fragment and responsible for the methyltransferase activity. MLL1 SET core domain is composed of distinctive subdomains of SET-N, SET- I, and SET-C (Fig. 7) (Cosgrove and Patel, 2010). MLL1 alone exhibits a low methyltransferase activity but becomes highly active by forming a complex with cofactors such as WDR5, RbBP5, and ASH2L (Dou et al., 2006; Patel et al., 2009). The crystallographic structure of the MLL1 complex containing S-adenosylmethionine, its cofactor, and a short histone peptide substrate showed that the cofactor and substrate are located in a well-defined pocket which is made up of residues from the SET-I, SET-C, and post-SET regions (Southall et al., 2009).
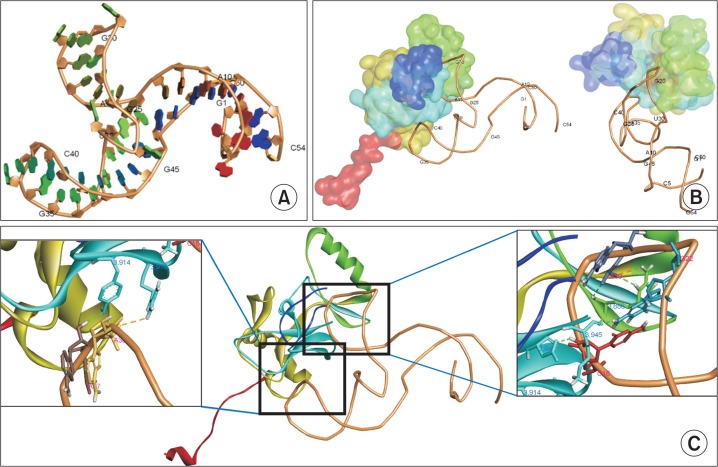
Fig. 7.
3D-structure predictions for APT1-MLL1 interactions. (A) Predicted 3D-structure of APT1 was constructed. The numbers represent the order of nucleotide residues. Some nucleotide residues are indicated. (B) 3D-structure of the complex was calculated using NPDock web server and illustrated in two different views. MLL1 SET domain is represented as subdomains with different colors such as the N-flanking region in red, SET-N in dark yellow, SET-I in green, SET-C in cyan, and post-SET in blue. (C) Hydrogen bonds formed between APT1 and MLL1 protein in two regions (dotted lines). The bases interacting with MLL1 amino acids are illustrated.
The protein MLL1 has been studied extensively because its dysfunction is involved in acute lymphoblastic or myelogenous leukemia (Ziemin-van der Poel et al., 1991; Hess, 2004; Morillon et al., 2005; Marschalek, 2010). In leukemogenesis, one allele of MLL1 is truncated, resulting in loss of the SET domain and production of oncogenic fusion proteins with over 70 partners (Dou and Hess, 2008). Of note, the other wild-type MLL1 allele is required for most MLL1 fusion protein-mediated leukemogenesis, indicating the importance of the activity of the intact SET domain in pathology (Thiel et al., 2010; Cao et al., 2014). In addition, intact MLL1 activity has been reported to be essential for growth of solid tumors (Ansari et al., 2013) or estrogen-dependent gene expression (Jeong et al., 2014). Therefore, elaborate regulation of its activity by developing inhibitors has been considered crucial for cancer therapy (Cosgrove and Patel, 2010). In spite of several efforts conducted by some research groups, no inhibitors have been in clinical use (Bolshan et al., 2013; Karatas et al., 2013; Borkin et al., 2015).
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technique was developed by Ellington and Szostak (1990) and Tuerk and Gold (1990) for in vitro selection and cyclic enrichment of single stranded (ss)RNA/ssDNA molecules (aptamers) that bind specific ligands with high selectivity and sensitivity. Aptamers are good replacement for antibodies, owing to their several advantages over antibodies. For instance, aptamers are less expensive, easy to synthesize, accessible to chemical modification without loss of function, and easy to immobilize (Nieuwlandt, 2000). Following the SELEX process, the aptamer sequences can be obtained from the canonical cloning work and Sanger sequencing. Cloning work is laborious and time consuming with low yield. These disadvantages are overcome with the recent introduction of high-throughput sequencing (HTS) technology (Cho et al., 2010; Hoon et al., 2011). By producing more than millions of sequences at once, this technique can provide more quantitative and comprehensive results in less time. Furthermore, sequences with low frequency can be effectively analyzed using HTS technology.
To our knowledge, there are no reports on MLL1-specific aptamers. In this study, we performed 16 rounds of SELEX and obtained sequences of MLL1-binding RNA aptamers using next-generation sequencing (NGS) technology. We also present evidences for the specific binding of aptamers to MLL1 and in vitro inhibition of MLL1. We suggest that MLL1-specific aptamers could be further developed for useful applications in various areas, including as therapeutic or diagnostic agents in the biomedical field.
MATERIALS AND METHODS
Materials
ssRNA aptamers were synthesized with in vitro transcription (IVT) using T7 Quick High Yield RNA Synthesis kit (New England Biolabs, MA, USA) and purified using PAGE. Mutated derivatives for aptamers were synthesized from Bioneer (Daejeon, Korea). Tumor necrosis factor-α was purchased from PeproTech (Cat No:300-01A, NJ, USA) The overall process is illustrated in Fig. 1.
Fig. 1.
A graphical overview of the SELEX process applied in this study.
Purification of MLL1 protein
The expression plasmid for MLL1 (SET) was a kind gift from Yali Dou (University of Michigan). His-tagged MLL1 (SET) (amino acids 3762-3970) proteins were expressed in Escherichia coli BL21(DE3) strain and purified with Ni-NTA agarose beads (Qiagen, GmbH, Hilden, Germany). Bound proteins were washed with phosphate-buffered saline (PBS) containing 25 mM imidazole and eluted twice with PBS containing 250 mM imidazole. The concentration of unbound protein was determined by Bradford assay.
Preparation of initial ssDNA library and ssRNA library
The initial ssDNA library was constructed with a centrally randomized region sequence (25 nt) and flanking regions (Macrogen, Korea). Briefly, the sequence of the starting ss-DNA library was 5′-CGACTCACTATAGGCTCGAGG-(N25)-GGAAGCTTACGGTACCTAGC-3′ (average MW=21374.7). The dsDNA library was constructed using 5′-GCTAATACGACTCACTATAGGCTCGAGG-3′ and 5′-GCTAGGTACCGTAAGCTTCC-3′ as the forward and reverse primers, respectively using 100 ng of ssDNA (4.75 pmol, 2.82E+12 molecules). The forward primer contained T7 promoter sequence for in vitro transcription IVT (underlined) to generate the initial ssRNA library (S0). ssRNA aptamers were synthesized with IVT using T7 Quick High Yield RNA Synthesis kit (New England Biolabs, USA) and purified using 8% denaturing PAGE. The same primer set was also used for reverse transcription and PCR amplification during SELEX rounds.
SELEX rounds
Before use, ssRNA oligonucleotides were heated at 95°C for 5 min and immediately cooled on ice for 5 min, followed by their equilibration at room temperature (RT). One μg of the initial RNA library (S0) was incubated with Ni-NTA agarose beads five times to remove non-specific binding. Ten microgram of initial ssRNA library (12.1 μM) was treated with 1 μg of MLL1 (0.524 μM) bound Ni-NTA agarose beads in 50 μl of NETN buffer media (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) at 37°C for 1 h. Unbound RNA sequences were washed with NETN buffer and MLL1-bound RNA reverse transcribed to obtain complementary DNA (cDNA) pool containing MLL1-binding sequences using SuperScript® II Reverse Transcriptase kit (Invitrogen, CA, USA). The cDNA pool was amplified 20 cycles with PCR using Enzynomics kit (Enzynomics, Daejeon, Korea) to generate dsDNA library. The dsDNA pool was used to obtain ssRNA sub-library (S1) with IVT. These processes were iterated for the next cycles of SELEX and the ssRNA sub-library dissolved in NETN buffer. The overall process is illustrated in Fig. 1. We increased the stringency for interaction by reducing the protein to RNA ratio from 1:10 to 1:20 (0.5 μg (0.262 μM) of protein: 10 μg (12.1 μM) of library) and increasing incubation time (from 30 min up to 1 h) during SELEX.
Quantitative real-time PCR (qRT-PCR)
The ssRNA was used as a template for cDNA synthesis using SuperScript® II Reverse Transcriptase kit (Invitrogen). The same primer set as library construction was used for PCR. Aliquots (1/100) of cDNA reaction mixtures were analyzed by qRT-PCR on Stratagene Mx3005P (Agilent Technologies, CA, USA) according to the manufacturer’s protocol.
Acquisition and processing of aptamer sequences by NGS
Suitable forms of S1, S3, and S16 libraries for sequencing were prepared with PCR using the following primer sets; forward 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCT ACACGACGCTCTTCCGATCTCGACTCACTATAGGC-3′ and reverse 5′-CAAGCAGAAGACGGCATACGAGAT-(index)-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGCTAGGTACCGTAAG-3′. These primers contained adapter sequences (italic) and sequencing primer sequences (underlined) and were sequenced using MiSeq sequencer (Illumina, CA, USA). Different indices such as CGATGT, TGACCA, and CAGATC were used to identify libraries. We isolated PCR products of 150–200 bp with gel extraction using Pippin prep (Sage Science, MA, USA) and 3% agarose gel cassette. The quality of purified PCR products was evaluated using Agilent Bioanalyzer 2100 (Agilent Technologies). The Quant-iTTM Picogreen® dsDNA Reagent (Invitrogen) was used to quantify the purified products using Synergy H1 hybrid reader (BioTek Instruments, VT, USA). Each library (7 pM) was sequenced and the sequences obtained from NGS were refined with the removal of byproducts that lacked adaptor sequences and flanking regions. Adaptor sequences and flanking regions were filtered out to obtain the exact sequences of aptamers. Data parsing was performed using Perl script software (version 5.18, comprehensive Perl archive network, https://www.perl.org/).
Consensus motif analysis
Consensus motif of RNA aptamers was determined using Gapped Local Alignment of Motifs (GLAM2) (Frith et al., 2008) (http://meme-suite.org/tools/glam2/) (Ver. 4.10.2) and Multiple Em for Motif Elicitation (MEME) (Bailey et al., 2009) (http://meme-suite.org/tools/meme/) (Ver. 4.10.2). MEME discovers novel, un-gapped motifs (recurring, fixed-length patterns), while GLAM2 discovers gapped motifs (recurring, variable-length patterns) in sequences. We uploaded 50 top sequences of library S16 to MEME or GLAM2 under default conditions for consensus motif analysis.
Binding assay
Percentage of binding of RNAs for MLL1 were determined by a qRT-PCR (Fig. 2A, 5A). For binding assay, 1 μg of RNAs (1.21 μM), unless otherwise noted, were incubated with 1 μg of MLL1 (0.524 μM) in 50 μl of NETN buffer. All aptamers were incubated with MLL1 after heating and cooling as described in Materials. Ni-NTA agarose bead-bound MLL1 protein were incubated with aptamers for 30 min. The beads were washed three times with NETN buffer to remove the unbound aptamers. Remained aptamers were reverse transcribed and amplified by qRT-PCR. To draw standard curve, qRT-PCR was also run for standard amount (0, 1, 10 and 100 ng) of RNA without binding. Amount of bound RNA was calculated by applying CT values in the standard curve.
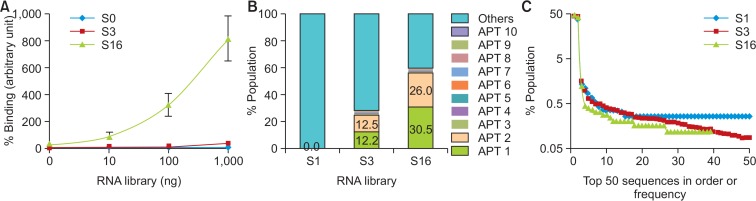
Fig. 2.
Evaluation of enrichment of MLL1-binding aptamers during SELEX. (A) Binding activity was measured using various amounts of S0, S3, and S16 libraries and 1 μg of MLL1 protein. The binding activity was calculated by qRT-PCR as described in “Materials and Methods”. Data are presented as means ± SEM. (B) Sequencing data of S0, S3, and S16 libraries were obtained using NGS. The percentage population of the five most popular abundant aptamers was revealed. (C) Sequence abundance in the sub-population consisting of the top 50 sequences. The percentage in the population of top 50 unique sequences was determined from S1, S3, and S16 libraries.
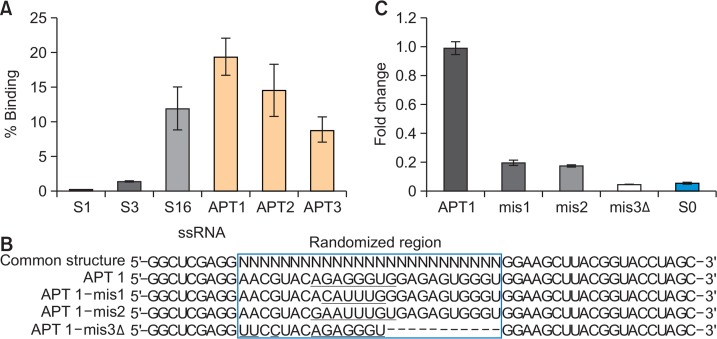
Fig. 5.
Evaluation of binding affinity of aptamers. (A) 1 μg of APT1, APT2, APT3, and three libraries were incubated with 1 μg of MLL1 protein and the binding activity was determined by qRT-PCR analysis. The percentage of bound RNA was expressed. (B) Mutation or deletion was introduced in APT1 as illustrated. The randomized region of APT1 are enclosed in a box, mutated nucleotides are underlined, and the deleted nucleotides are marked with hyphen. (C) The binding activity of each variant (1 μg) was measured by qRT-PCR analysis and the amount of bound RNA was calculated. Relative amount of bound RNAs was expressed as a fold change using wild-type APT1 was used as a control.
In vitro MLL1 activity assay
The MLL1 activity was measured using MLL1 Complex Chemiluminescent Assay Kit (BPS Bioscience, SD, USA) following the manufacturer’s protocol. Briefly, 0.25 μg MLL1 and its complex proteins (WDR5, Ash2, RbBP5, and DPY30) were incubated with aptamers in assay buffer with S-adenosylmethionine for 1 h at RT in a 96-well plate precoated with histone H3 peptide substrate. Following incubation, wells were washed with TBST buffer (50 mM Tris, 150 mM NaCl, and 0.05% Tween 20), dried with paper towel, and incubated with 100 μL blocking buffer for 10 min. The wells were further treated with 100 μL of 800-fold primary antibody 2 (diluted with blocking buffer) for 1 h, followed by incubation with horseradish peroxidase (HRP)-labeled secondary antibody 1 (1,000-fold diluted with blocking buffer). Chemiluminescence substrate (50 μL substrate A+50 μL substrate B mixed well on ice) was added to each well and chemiluminescence measured using fluorescent microplate reader (BioTek Instruments). The experiments were repeated three times, and the samples were analyzed in triplicate.
3D-structure prediction of MLL1-APT1 complex
The 3D-structure of APT1 was generated using RNAComposer (http://rnacomposer.ibch.poznan.pl/) (Popenda et al., 2012). The secondary structure of APT1 was predicted by Mfold. In silico docking between APT1 and MLL1 was conducted using NPDock (Ver. 3.0.2) (http://iimcb.genesilico.pl/NPDock). The best scored structure by the server was chosen. All 3D-stuructres are displayed using Discovery Studio (Ver. 16.1.015350) (BIOVIA, CA, USA).
Statistical analysis
Data are expressed as means ± SEM. Comparison of the means was performed using unpaired Student’s t-test, and null hypotheses of no difference were rejected if p-values were less than designated values denoted in the figure legends.
RESULTS
Enrichment and sequence identification of MLL1-specific ssRNAs
We performed 16 rounds of SELEX to achieve high enrichment of MLL1-specific aptamers. Enrichment was confirmed with the measurement of the binding capabilities of the libraries at various amounts using quantitative real-time (qRT)-PCR analysis. The S16 library was efficiently enriched with its high MLL1 binding capabilities compared to S0 and S3 (Fig. 2A).
In comparison to the classical sequencing process embracing canonical cloning steps, HTS can save time required for cloning and provide massive sequence information (Cho et al., 2010; Hoon et al., 2011; Schutze et al., 2011). With the help of NGS technology, we obtained approximately 1.5×106 sequences from each library of S1, S3, and S16. Aptamers were named after the frequency order in S16 library (APT1, APT2, and so on); the top ten sequences are reported in Table 1. APT1 and APT2 were predominant in S3 and S16 libraries (Fig. 2B). Some sequences among the top ten aptamers, such as APT1 and APT2, were enriched more than hundred times at early rounds of SELEX (S1 to S3) but at a slower rate at later stages (S3 to S16). The result shows that the evolution of target-specific sequences was sufficiently established at very early rounds of SELEX in our study. Analysis using subgroups consisting of 50 top aptamers revealed that >40% of the population comprised one species of aptamer in S0, S3, S16 libraries (Fig. 2C). Besides, the trend line of population was steeper as SELEX progressed (Fig. 2C). These data indicate that the HTS technology can identify strong binders at early rounds of SELEX and further identify aptamers with low frequencies in extended rounds.
Table 1.
The most enriched aptamer sequences obtained by NGS
| Aptamer | Sequences 5′-GGCUCGAGG-(N25)-GGAAGCUUACGGUACCUAGC-3′ | Population (%) | Enrichment (fold) | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| S16 | S3 | S1 | S3/S1 | S16/S3 | ||
| APT1 | AACGUACAGAGGGUGGAGAGUGGGU | 3.0×101 | 1.2×101 | 3.6×10−2 | 338 | 2.5 |
| APT2 | ACGUAACAGAGGGAGGCGAGUGGGU | 2.6×101 | 1.3×101 | 3.1×10−2 | 399 | 2.1 |
| APT3 | ACCGAAGUCGAGGGGGACGUGAGGG | 7.9×10−1 | 4.3×10−1 | 1.0×10−3 | 419 | 1.8 |
| APT4 | ACCUAAGUGGGAAGGUGAGCGGGUG | 2.8×10−1 | 1.0×10−1 | 6.9×10−4 | 149 | 2.7 |
| APT5 | AACGUACAGAGGGCGGAGAGUGGGU | 2.5×10−1 | 1.2×10−1 | 6.9×10−5 | 1676 | 2.2 |
| APT6 | AAAUGCAAGGAGGUCGGAGGUCGGA | 2.3×10−1 | 1.4×10−1 | 5.5×10−4 | 255 | 1.6 |
| APT7 | ACCGACGACGAGGGGGACGUGAGGG | 2.0×10−1 | 7.3×10−2 | 1.1×10−3 | 66 | 2.8 |
| APT8 | AUAAGCAUGGGGGUCGAGAGGGGAC | 2.0×10−1 | 1.9×10−1 | 1.2×10−2 | 15 | 1.1 |
| APT9 | AACGUACAGAGGGAGGCGAGUGGGU | 1.8×10−1 | 2.8×10−1 | 1.0×10−3 | 274 | 0.6 |
| APT10 | AACGUACAGAGGGUGGAGAGUGGGU | 1.8×10−1 | 7.0×10−2 | 1.0×10−3 | 67 | 2.6 |
Sequence information and percentage of ten aptamers were addressed from libraries S1, S3, and S16. Ratios of frequency of S3/S1 and S16/S3 were calculated, showing large increases in their populations at early rounds of SELEX.
Analysis of consensus motif
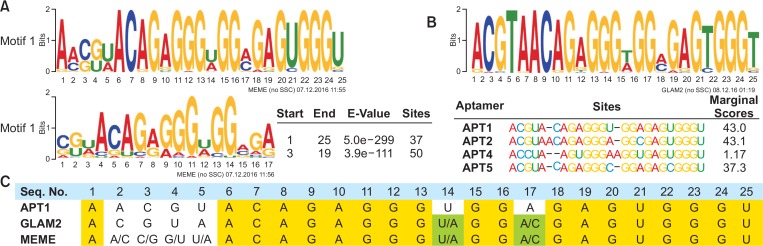
MEME discovers novel, un-gapped motifs (recurring, fixed-length patterns), while GLAM2 discovers gapped motifs (recurring, variable-length patterns) in sequences. Fifty aptamers from S16 were analyzed for the consensus motif using Multiple Em for Motif Elicitation (MEME) (Bailey et al., 2009). The analysis revealed two significantly conserved motifs, Motif 1 and Motif 2 (Fig. 3A), wherein Motif 1 represented all sites of randomized region (25 nt); 5′-A[A/C][C/G][G/U][U/A] ACAGAGGG[U/A]GG[A/C]GAGUGGGU-3′ (Motif 1, E-value= 5.0e–299). Motif 2 represented 17 nt (starting at +3 of random region) having same base compositions as Motif 1. The sites were well conserved over the sequence, except at +2, +3, +4, +5, +14, and +17 positions. Both APT1 and APT2 were highly homologous to Motif 1 (75%), with differences recorded only at the less conserved sites in Motif 1 (noted above). Meanwhile, Gapped Local Alignment of Motifs (GLAM2) algorithm (Frith et al., 2008) produced the consensus motif 5′-ACGUAACAGAGGG[U/A]GG[A/C]GAGUGGGU-3′ with the allowance of gap insertion (Fig. 3B). GLAM2 algorithm showed that APT1, APT2, and APT5 were highly homologous to each other but not to APT3. Moreover, the conserved sites in two algorithms were perfectly matched in 84% of residues (Fig. 3C), indicating that these conserved sites are components of the core region critical for aptamer-MLL1 binding.
Fig. 3.
Consensus motifs analysis. (A, B) Consensus motifs in the random region were analyzed from 50 most popular aptamers of the S16 library using MEME (A) or GLAM2 (B) software. Sequence logos consist of stacks of letter for each position in the motif sequence. The height of the individual letter indicates the frequency of each nucleotide, and the positions where the motif starts were presented. Sites show the number of sequences contributing to the construction of the motif. The marginal scores of GLAM2 reflect the degree of matches between segments. (C) APT1 sequence was aligned with the most conserved motifs from MEME and GLAM2. Yellow color shows the sequences common in all motifs, whereas green color shows variable sequences in MEME and GLAM2 motifs.
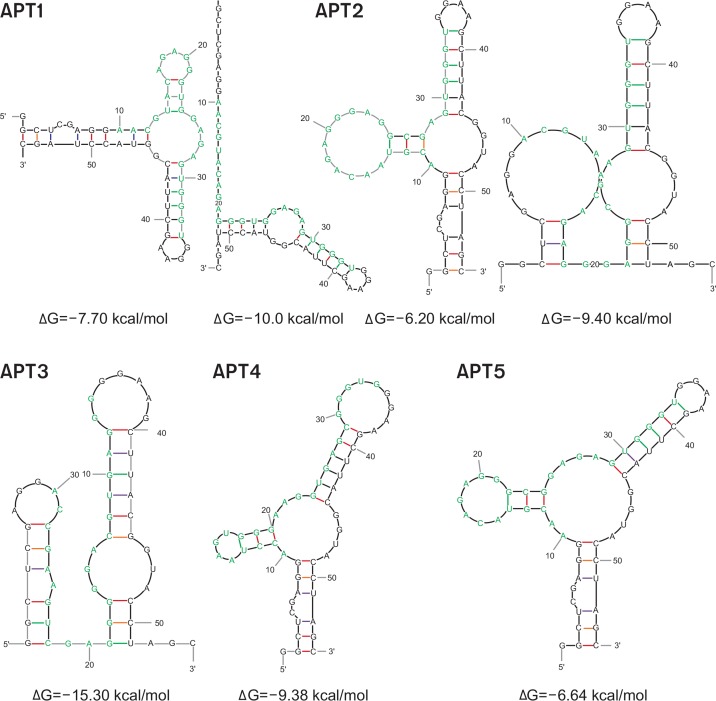
Secondary structure of aptamers
Secondary structures of five aptamers were predicted using the Mfold software (Fig. 4) (Zuker, 2003). All aptamers had significant secondary structure, including protruding loops and stems, with comparatively low free energy loops present in the structure of RNA molecules. APT1, APT2, and APT5 showed similar structure bearing one central multi-branched loop surrounded by two hairpin loops and one terminal open branch. Consensus strings from rG18 to rG22 of those aptamers were located on the outermost location from the branching point of the loop. On the contrary, APT3 failed to generate this characteristic structural pattern. These findings suggest that the consensus motif from APT1, APT2, and APT5 may produce a certain characteristic secondary structure. In addition, we evaluated the possibility of formation of G-quadruplex structure using the web-based Quadruplex forming G-Rich Sequences (QGRS) Mapper software, owing to the presence of multiple guanine bases in aptamers. Analysis of five most popular aptamers showed that all exhibit some potential to form quadruplex structures (Supplementary Table 1).
Fig. 4.
Determination of secondary structure by Mfold. The secondary structure of five most popular sequences of RNA-aptamers from S16 was predicted using Mfold software. Bases from randomized region are colored in blue.
Measurement of binding affinity of MLL1-binding aptamer
We verified the interaction between aptamers (APT1, APT2, and APT3) and MLL1 protein using qRT-PCR analysis as described in Materials and Methods. Results showed that higher fraction of APT1 bound with MLL1 (19%) compared to APT2 (14%) and APT3 (9%) (Fig. 5A). An increase in the band intensity for APT-MLL1 complex was observed with increase in the concentration of MLL1. Moreover, an increase in the binding capability for RNA sub-library pools was observed during the progress of SELEX cycles (0%, 1%, and 12% for S1, S3, and S16, respectively) (Fig. 5A), suggesting that multiple rounds of SELEX significantly enriched strong binding aptamers.
To verify the binding affinity and the role of internal region, we performed regional mutations in aptamer sequences and tested the binding affinity of the mutants. We first mutated the sites within the randomized region of APT1 (Fig. 5B); 5′-A17G18A19G20G21G22U23G24-3′ was mutated to 5′-A17C18A19 U20U21U22G23G24-3′ (APT1-mis1) or 5′-G17A18A19U20U21U22G23U24-3′ (APT1-mis2). We observed that both mutated aptamers showed a significant loss as low as 0.2-fold in their binding activity (Fig. 5C). When deletion was applied along with a site mutation (APT1-mis3Δ), the activity decreased further by 0.05-fold. Thus, our results indicate that the internal randomized region has important information that determines the binding affinity of the aptamer with MLL1 protein.
Evaluation of the interaction between MLL1 and aptamers in silico
We evaluated the interaction between aptamers and MLL1 protein with catRAPID algorithm package (Agostini et al., 2013). First, catRAPID signature algorithm showed that MLL1 SET domain may have an intrinsic RNA-binding ability (over-all interaction score=0.6) (Supplementary Fig. 1A). The binding mode was predicted to be the non-classical RNA-binding protein (RBP) class (0.62) rather than the classical (0.37) or putative class (0.26). Furthermore, the binding profile showed the distribution of moderate RNA-binding ability along the internal region (Supplementary Fig. 1B). Interaction propensities calculated from catRAPID fragments showed that the region of SET domain spanning amino acid residues 101–152 displayed highest interacting probability for APT1, APT2, and APT3 (Supplementary Table 2). The interaction strengths of aptamers with MLL1 were 96% (APT1), 95% (APT2), and 99% (APT3), as evaluated from catRAPID strength (Supplementary Fig. 1C). These data show that selected aptamers may have strong interaction abilities to bind MLL1.
Aptamers inhibit MLL1 activity
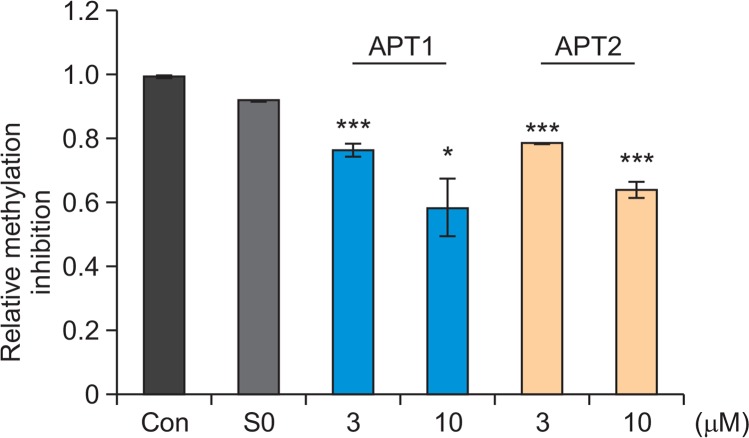
We tested if aptamers can directly modulate MLL1 activity. MLL1 methyltransferase activity is highly enhanced by complexing with cofactors such as WDR5, RbBP5, and ASH2L while marginal without those cofactors (Dou et al., 2006; Patel et al., 2009). We measured the methyltransferase activity of MLL1 complex containing the cofactors using antibody recognizing methylated Lys4 residue of Histone H3 substrate. The presence of enzyme MLL1 showed significant elevation in the methylation levels of histone H3 peptide substrate. However, in presence of APT1 or APT2 (3 μM), the MLL1 activity was significantly lowered by 23% and 22%, respectively (Fig. 6). Increase in the concentration of APT1 and APT2 (10 μM) resulted in decrease in MLL1 activity by about 40% for both aptamers.
Fig. 6.
Effect of aptamers on MLL1 activity. MLL1 activity was measured using MLL1 Complex Chemiluminescent Assay Kit. S-adenosylmethionine was added as a methyl-group donor. MLL1 (2 ng/μl) was incubated with or without aptamers. S0 library was used as a control. *p<0.05, ***p<0.005 versus control. Data are presented as means ± SEM. Statistical significance was determined with unpaired Student’s t-test.
In silico prediction of 3D-structure of APT1 and aptamer-MLL1 complex
MLL1 is a large protein of 3969 amino acid residues and the SET domain, which has methyltransferase activity, occupies the carboxyl terminus of the protein. MLL1 SET domain contains two conserved SET-N and SET-C regions which is separated by a less conserved region, SET-I. To understand the mechanism for inhibitory activity of APT1, we predicted the 3D-structure of the APT1-MLL1 complex using NPDock software (Tuszynska et al., 2015). Fig. 7A and 7B illustrates the 3D-sturcture of APT1 and its complex with MLL 1 SET domain (PDB-2W5Y), respectively. The prediction showed that the region containing 5′-C16A17G18A19G20G21G22-3′ might penetrate the histone H3-binding pocket. Multiple hydrogen bonds, electrostatic, or hydrophobic interactions were predicted to form between two molecules. Among weak interactions, hydrogen bonds are illustrated in Fig. 7C (boxes). The magnified structure shows two major regions of interaction. The motif region of APT1 formed hydrogen bonds with residues at the active site of MLL1 (right box) through interactions between C16 HO2’, A19 OP1, and G22 H21 with Asp3943 O, Arg3886 HE/HH21, and Lys3945 O respectively. Additional interactions formed between A37 HO2’ and Try3914 OH, between A38 OP1 and Arg3916 HH21 fortified their interaction. This prediction indicates that the motif sequence may inhibit enzyme activity by blocking the active site of MLL1.
Taken together, our study shows that SELEX process was effectively replaced the multiple processes following the HTS technology and that selected MLL1-binding aptamers with a notable inhibitory activity were efficiently enriched.
DISCUSSION
In this report, we describe the development of ssRNA aptamers for MLL1 methyltransferase. The methyltransferase activity of MLL1 is associated with a variety of disease types, and therefore, MLL1 has been considered as an attractive target for treatment of MLL1-related diseases. However, only a limited number of MLL1 inhibitors are currently under use. For the development of a novel MLL1-specific inhibitor, we selected ssRNA aptamers targeting the catalytic SET domain. Our study provides evidence of the inhibitory effects of aptamers on MLL-1 activity. The aptamers inhibited MLL1 activity effectively at micromolar concentration (Fig. 6) indicating that a half-maximum inhibitory concentration (IC50) would be in a micromolar range and dissociation constant (Kd) less the IC50. The in vitro inhibitory effect of the aptamers indicates that these might regulate the MLL1-mediated functions in cells although the cellular effects were not further explored here. We expect that APT1 and ATP2 might be promising candidates for therapeutic applications for MLL1-related pathological conditions.
With HTS technology, we acquired massive number of sequences. The NGS technology helped us eliminate the time-consuming canonical cloning process and Sanger sequencing processes reducing the time for overall steps. Our study showed that only three rounds of SELEX can discriminate good binding aptamers from library pool with even low copies (Fig. 2C). By 16 rounds of SELEX process, sequences with higher capability for MLL1 became dominant; population of APT1, APT2, APT3, APT4, and APT5 increased significantly from S1 to S16.
In this study, we obtained a highly conserved motif (Motif 1) (Fig. 3). Our results demonstrated that well-conserved consensus sites play a key role in binding to MLL1. First, Motif 1 was highly homologous (76%) to APT1 and APT2, which were the major populations in S16 (30% and 26%, respectively). Second, mutations in this conserved region disabled aptamers from binding (Fig. 5C). Third, aptamers homologous to Motif 1 shared similar characteristic secondary structure (Fig. 4), which may be crucial to generate proper conformations for binding. Forth, in 3D complex structure prediction, the loop structure containing consensus motif (rG19, rA20, rG21, rG22, and rG23) was expected to pass through and make multiple noncovalent interactions with the active site of MLL1 SET domain. Especially, rC16, rA19, and rG22 were expected to interact with the residues of MLL1’s active site forming hydrogen bonds (Fig. 7). Fifth, the computational evaluation of their binding by catRAPID signature and strength algorithms showed high possibility (Supplementary Fig. 1A). All evidences suggest that the motif sequence contains the core sequence information for its effective binding to MLL1.
Deregulation of MLL1 is involved in 5%–10% of acute myeloid leukemia (AML) in adults and around 70% of acute lymphoblastic leukemia (ALL) in infants. The translocation in one MLL1 allele causes aberrant expression of hematopoietic transcription factors leading abnormal cell proliferation (Cao et al., 2014). Besides, the targeting of wild-type MLL1 methyltransferase activity was proven to be effective to reduce leukemogenesis-related genes (Patel et al., 2009). Our ssRNA aptamers are efficient binders to MLL1 and act as activity modulators. APT1 and APT2 showed inhibitory effect against MLL1 methyltransferase activity in vitro (Fig. 6). At present, only a few inhibitors have been reported to inhibit MLL1, and to our knowledge, no reports on the development of aptamers with MLL1 inhibitory activity have been published. We suggest that APT1 and APT2 are prominent candidates for development of diagnosis or therapy in MLL1-related diseases. First, by developing these aptamers as chemical probes, the function and biological properties can be examined. Second, these aptamers can be tested whether these are used as a therapy related to aberrant regulation of methylase activity of MLL1. In the future, the study of these aptamers will provide deeper insights and more therapeutic potential.
Acknowledgments
This study was supported with grants from the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (2016R1D1A1B01012515), Republic Korea.
REFERENCES
- Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 2013;29:2928–2930. doi: 10.1093/bioinformatics/btt495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2013;32:3359–3370. doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshan Y, Getlik M, Kuznetsova E, Wasney GA, Hajian T, Poda G, Nguyen KT, Wu H, Dombrovski L, Dong A, Senisterra G, Schapira M, Arrowsmith CH, Brown PJ, Al-Awar R, Vedadi M, Smil D. Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction. ACS Med. Chem. Lett. 2013;4:353–357. doi: 10.1021/ml300467n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, Wen B, Zong H, Jones M, Danet-Desnoyers G, Guzman ML, Talpaz M, Bixby DL, Sun D, Hess JL, Muntean AG, Maillard I, Cierpicki T, Grembecka J. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, Hess JL, Atadja P, Lei M, Qin ZS, Malek S, Wang S, Dou Y. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. MolCell. 2014;53:247–261. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Patel A. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J. 2010;277:1832–1842. doi: 10.1111/j.1742-4658.2010.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int. J. Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol. Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Hoon S, Zhou B, Janda KD, Brenner S, Scolnick J. Aptamer selection by high-throughput sequencing and informatic analysis. Biotechniques. 2011;51:413–416. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KW, Andreu-Vieyra C, You JS, Jones PA, Stallcup MR. Establishment of active chromatin structure at enhancer elements by mixed-lineage leukemia 1 to initiate estrogen-dependent gene expression. Nucleic Acids Res. 2014;42:2245–2256. doi: 10.1093/nar/gkt1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, Lei M, Dou Y, Wang S. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 2013;135:669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 2010;277:1822–1831. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. MolCell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. MolCell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt D. In vitro selection of functional nucleic acid sequences. Curr. Issues Mol. Biol. 2000;2:9–16. [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze T, Wilhelm B, Greiner N, Braun H, Peter F, Morl M, Erdmann VA, Lehrach H, Konthur Z, Menger M, Arndt PF, Glokler J. Probing the SELEX process with next-generation sequencing. PLoS ONE. 2011;6:e29604. doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. MolCell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, Hua X. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Tuszynska I, Magnus M, Jonak K, Dawson W, Bujnicki JM. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res. 2015;43:W425–430. doi: 10.1093/nar/gkv493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mllmutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, 3rd, Patel Y, Harden A, Rubinelli P, Smith SD, LeBeau MM, Rowley JD, Diaz MO. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.