Abstract
Suppressor of Variegation 3–9 Homolog 2 (SUV39H2) methylates the lysine 9 residue of histone H3 and induces heterochromatin formation, resulting in transcriptional repression or silencing of target genes. SUV39H1 and SUV39H2 have a role in embryonic development, and SUV39H1 was shown to suppress cell cycle progression associated with Rb. However, the function of human SUV39H2 has not been extensively studied. We observed that forced expression of SUV39H2 decreased cell proliferation by inducing G1 cell cycle arrest. In addition, SUV39H2 was degraded through the ubiquitin-proteasomal pathway. Using yeast two-hybrid screening to address the degradation mechanism and function of SUV39H2, we identified translationally controlled tumor protein (TCTP) as an SUV39H2-interacting molecule. Mapping of the interacting regions indicated that the N-terminal 60 amino acids (aa) of full-length SUV39H2 and the C-terminus of TCTP (120–172 aa) were critical for binding. The interaction of SUV39H2 and TCTP was further confirmed by co-immunoprecipitation and immunofluorescence staining for colocalization. Moreover, depletion of TCTP by RNAi led to up-regulation of SUV39H2 protein, while TCTP overexpression reduced SUV39H2 protein level. The half-life of SUV39H2 protein was significantly extended upon TCTP depletion. These results clearly indicate that TCTP negatively regulates the expression of SUV39H2 post-translationally. Furthermore, SUV39H2 induced apoptotic cell death in TCTP-knockdown cells. Taken together, we identified SUV39H2, as a novel target protein of TCTP and demonstrated that SUV39H2 regulates cell proliferation of lung cancer cells.
Keywords: Histone methyltransferase, SUV39H2, TCTP, Apoptosis, Cell cycle
INTRODUCTION
SUV39H2 is a member of the SUV39H sub-family of histone lysine methyltransferases (Rao et al., 2017). Suppressor of variegation 3–9 homolog 1 (SUV39H1) and suppressor of variegation 3–9 homolog 2 (SUV39H2) trimethylate histone H3 on lysine 9 (H3K9me3), where heterochromatin protein 1 (HP1) recruits and induces heterochromatin formation, resulting in transcriptional repression or silencing of target genes (Rea et al., 2000; Bannister et al., 2001; Lachner et al., 2001). Suv39h1/h2 knockout mouse displayed spermatogenic defects with a delay into meiotic prophase in spermatocytes (Peters et al., 2001). SUV39H1 interacts with Rb, mediates H3K9m3 on E2F target gene promoters including cyclin E and cyclin A, and thereby represses gene expression, indicating a growth-regulatory role of SUV39H1 (Nielsen et al., 2001; Vandel et al., 2001). In addition, Rb-mediated H3K9m3 induction by SUV39H1 and heterochromatin formation was required for repression of S-phase gene promoters in cellular differentiation and senescence (Narita et al., 2003; Ait-Si-Ali et al., 2004). The expression of SUV39H1 and SUV39H2 in mouse embryogenesis is overlapping and single disruption of SUV39H1 or SUV39H2 does not display noticeable phenotype changes (O’Carroll et al., 2000; Peters et al., 2001), suggesting functional redundancy between those two histone methyltransferases.
Previously, it was reported that a single nucleotide polymorphism (SNP) in the 3′-untranslated (3′-UTR) region of SUV39H2 was associated with increased risk of lung cancer (Yoon et al., 2006), suggesting the functional importance of SUV39H2 in lung carcinogenesis. However, the roles and molecular mechanism of SUV39H2 in human cancer are not well established.
In this study, to identify functional partners of SUV39H2 proteins, we performed a yeast two-hybrid screen using SUV39H2 as bait and found that SUV39H2 interacts with translationally controlled tumor protein (TCTP), associated with many important cellular processes.
TCTP is highly conserved and ubiquitously expressed (Fiucci et al., 2003; Bommer and Thiele, 2004), and it was first reported as histamine-releasing factor implying a role in inflammation (MacDonald et al., 1995). TCTP is secreted and found in the exosome associated with TSAP (Amzallag et al., 2004). The expression of TCTP is tightly regulated during early development, and tctp-knockout mice were embryonic lethal displaying small size and higher incidence of apoptosis in the early embryo (Chen et al., 2007; Kubiak et al., 2008). TCTP plays a role in cell growth (Kang et al., 2001; Pickart, 2001) and cell cycle progression, especially in the M phase. The protein binds to the mitotic spindle (Gachet et al., 1999; Burgess et al., 2008), and mutant TCTP at the Plk phosphorylation site inhibited the completion of mitosis (Yarm, 2002). In addition, TCTP is involved in protein synthesis (Cans et al., 2003) and acts as a transcription factor for oct4 and nanog (Koziol et al., 2007). Moreover, phosphorylated TCTP facilitates somatic cell reprogramming (Tani et al., 2007).
TCTP has been reported to regulate several proteins related to apoptosis. TCTP stabilized MDM2 and promoted MDM2-mediated ubiquitination of p53, which targets p53 for proteasomal degradation (Amson et al., 2011). TCTP overexpression led to p53 degradation and thus repressed apoptosis (Rho et al., 2011). Moreover, TCTP stabilized Mcl-1 and Pim-3 by inhibiting the ubiquitin-dependent degradation pathway, resulting in enhanced anti-apoptotic activity (Liu et al., 2005; Zhang et al., 2013). TCTP also has been shown to antagonize bax function (Susini et al., 2008) but activate Bcl-xL (Thebault et al., 2016).
Regulation of the cell cycle and of apoptosis is closely related to tumorigenesis. It was shown that TCTP was highly expressed in tumor cells compared with normal cells, and injection of TCTP-knockdown U937 cells into SCID mice resulted in reduced tumor growth (Tuynder et al., 2002, 2004). TCTP increased Pim-3 kinase stability, which promoted cancer progression in pancreatic cancer (Zhang et al., 2013). More interestingly, TCTP is most frequently down-regulated during tumor reversion (Tuynder et al., 2002, 2004), which is a reprograming of cancer cells into less malignant phenotypes (Telerman and Amson, 2009; Amson et al., 2013).
In this study, we investigated the role of SUV39H2 in lung cancer cells and demonstrated that it regulates cell proliferation inducing G1 cell cycle arrest.
MATERIALS AND METHODS
Cell culture
HEK293, HEK293T, and A549 were purchased from the American Type Culture Collection (Rockville, MD, USA). HEK293 cells were cultured at 37°C in MEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/mL), and streptomycin sulfate (100 μg/mL). To generate stable cell lines expressing SUV39H2 in HEK293, pHM6 or pHM6-SUV39H2 containing a neomycin resistance gene was transfected and selected for stable clones in the presence of 1 mg/mL neomycin. The SUV39H2-overexpressing HEK293 cells were maintained in MEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/mL), streptomycin sulfate (100 μg/mL) and G418 (500 μg/mL). A549 cells, a non-small lung cancer cell line, were cultured at 37°C in RPMI 1,640 supplemented with 10% FBS, penicillin (100 units/mL), and streptomycin sulfate (100 μg/mL) in a humidified atmosphere of 5% CO2. The SUV39H2-overexpressing A549 cells were produced by infection with viral particles produced from infection of 293T cells with pLT-CMV or pLT-CMV-SUV39H2, and selection of stable clones was performed in the presence of 400 μg/mL zeocin.
Cell counting
Control and SUV39H2-overexpressing cells were incubated for the indicated time. Cells were washed twice with PBS and incubated with a trypsin. The cells were harvested with media and counted using a Coulter counter (Beckman Instruments, Fullerton, CA, USA).
Western immunoblot analysis
The cells were washed twice with ice-cold PBS and incubated with RIPA lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nondiet P-40, 0.1% SDS, 1% sodium deoxycholate, 1 mM PMSF, protease inhibitor). After centrifugation at 13,000 rpm for 15 min, the protein concentration of the supernatant was measured. Aliquots of protein were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% non-fat dried milk in TBST buffer for 1 h and incubated overnight with primary antibody in TBST buffer containing 5% non-fat dried milk. The blots were treated with horseradish peroxidase-conjugated secondary antibody in TBST buffer containing 5% non-fat dried milk for 1 h, and immune complex was detected using a Westone ECL detection kit (Intron Biotechnology, Seoul, Korea).
Fluorescence-activated cell sorting (FACS) analysis
The cells were collected and washed twice with PBS. Cells were fixed in 70% ethanol, and stored at −20°C. When ready for use, cells were washed, and incubated with propidium iodide (PI, 50 μg/mL) and RNase (25 μg/mL) for 30 min at room temperature in the dark. PI fluorescence of individual cells were measured by flow cytometry (BD Biosciences, San Jose, CA, USA). The percentage of cells at each phase of the cell cycle was determined using the FlowJo program (BD Biosciences).
Yeast two-hybrid screening
For bait construction with human suppressor of variegation 3–9 homolog 2 (SUV39H2), cDNA encoding full-length human SUV39H2 and N-terminal-truncated SUV39H2 were inserted into the pGilda/LexA yeast shuttle vector at Eco-RI and XhoI enzyme sites. The cDNA encoding full-length and different deletion mutants (Met1-Gly69, Val70-Ala119, Glu120-Cys172) of TCTP were introduced into the pJG4-5/B42 prey vector at EcoRI and XhoI restriction enzyme sites. The bait pGilda/LexA-SUV39H2 and SUV39H2 plasmid were separately transformed into a yeast strain EGY48 [MATa, his3, trp1, ura3-52, leu2::pLeu2-LexAop6/pSH18-34 (LexAop-lacZ reporter)] by a modified lithium acetate method (Rho et al., 1996). The cDNA encoding B42 fusion proteins (pJG4-5 plasmid vector) (Clontech, Palo Alto, CA, USA) were inserted into yeast competent cells containing pGilda/LexA-SUV39H2, and the transformants were selected for tryptophan prototrophy (plasmid marker) on a synthetic medium (Ura−, His−, Trp−) containing 2% (w/v) glucose. All the transformants were pooled and re-spread on the synthetic medium (Ura−, His−, Trp−, Leu−) containing 2% (w/v) galactose to induce cDNA transformation. Cells growing on the selection media were retested on synthetic medium (Ura−, His−, Trp−, Leu−) containing 2% galactose (inducing condition) and 2% glucose (non-inducing condition) to confirm the dependency of their growth on the presence of galactose. Then, selected transformants were confirmed according to the previously described instruction (Rho et al., 1996). The binding activity of the interaction was confirmed by measuring the relative expression level of β-galactosidase, which was determined using an ONPG β-galactosidase system as described previously (Rho et al., 1996).
Co-immunoprecipitation
Cells were lysed with RIPA buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, 1 mM PMSF, protease inhibitor). Solubilized lysates were incubated with specific antibody or preimmune IgG at a final concentration of 1 μg/mL overnight at 4°C. Protein G-Sepharose (GE Healthcare, Waukesha, WI, USA) was then added for 2 h at 4°C. The samples were centrifuged, and washed three times with RIPA buffer and prepared for Western blotting by boiling in sample buffer. The immunoprecipitated proteins were detected by Western blot analysis.
Immunocytochemistry
Cells were fixed with 4% formaldehyde and permeabilized with PBS containing 0.25% Triton X-100. Cells were then incubated with rat anti-HA antibody (Roche, Mannheim, Germany) and mouse anti-TCTP (Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C. After washing, cells were incubated with Alexa 594-conjugated goat anti-rat IgG (Red) and Alexa 488-conjugated goat anti-mouse IgG (Green) for 1 h at room temperature. After washing, the coverslips were mounted using mounting medium (Vector Labs, Burlingame, CA, USA). Cellular localization of HA-SUV39H2 and TCTP staining was visualized using confocal microscopy (Zeiss, Oberkochen, Germany).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNA was synthesized by reverse transcription of equal amounts of total RNA (1 μg) in 20 μL containing 0.5 μg of oligo dT, 2.5 μM of dNTP, 10 mM DTT, 1×first strand buffer and 40 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen). The qRT-PCR was performed in 384-well plates with a LightCycler® 480 real-time PCR instrument (Roche) using the LightCycler® 480 SYBR Green I Master kit (Roche). Single-strand cDNA was amplified by PCR with primers SUV39H2 sense, 5′-TCCCACCTGGTACTCCCATC-3′; SUV39H2 antisense, 5′-CAGCCACGTCCATTGCTAGT-3′; GAPDH sense, 5′-TGATGACATCAAGGTGGTGAAG-3′; GAPDH antisense, 5′-TCCTTGGAGGCCATGTGGGCCAT-3′. For quantification Roche LC480 relative quantification software module LightCycler® 480 software (Roche) was used. All values were normalizaed to the level of the housekeeping gene GAPDH.
Statistical analysis
Data are expressed as the mean ± standard deviation (SD). Data were analyzed using one-way analysis of variance (ANOVA), followed by Student’s t-tests for multiple comparisons. Differences were considered significant if p<0.05. For survival analysis, Kaplan-Meier plotter (Gyorffy et al., 2013) was used to investigate the association of SUV39H2 mRNA or TCTP expression with overall survival of lung cancer patients.
RESULTS
Forced expression of SUV39H2 decreased cell proliferation and induced G1 cell cycle arrest
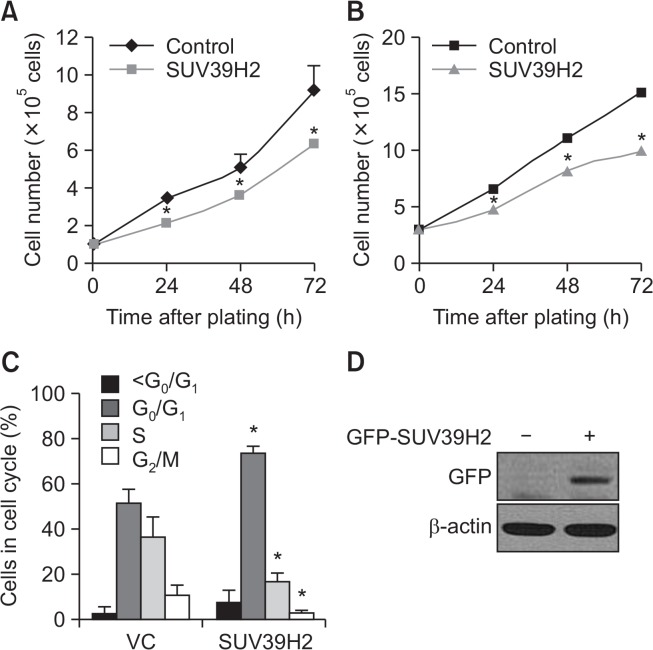
To investigate the effect of SUV39H2 on cell growth, it was stably overexpressed in a non-small cell lung cancer (NSCLC) cell line, A549, and an immortalized cell line, HEK293, and cell number was counted at the indicated time after plating. As shown in Fig. 1A and 1B, cell proliferation was suppressed in SUV39H2-overexpressing cells compared with control cells. Inhibition of cell proliferation can be due to delayed cell cycle progression or increased cell death. Cell cycle analysis was performed in A549 cells transfected with pEGFP or pEGFP-SUV39H2 by measuring the DNA content in GFP-positive cells in each group based on propidium iodide (PI) staining under flow cytometry. Fig. 1C shows that SUV39H2 leads to G1 cell cycle arrest with decreased cell populations in S and G2/M phases. The expression of SUV39H2 in A549 cells is presented in Fig. 1D. These results indicate that SUV39H2 inhibits cell proliferation by inducing G1 cell cycle arrest.
Fig. 1.
Forced expression of SUV39H2 decreased cell proliferation and induced G1 cell cycle arrest. (A) Control or SUV39H2-overexpressing A549 cells were counted at the indicated time after plating using a Coulter counter. Data are presented as mean ± SD of three independent experiments. *p<0.05 indicates significant difference from the control group. (B) Control or SUV39H2-overexpressing HEK293 cells were counted at the indicated time after plating using a Coulter counter. Data are presented as mean ± SD of three independent experiments. *p<0.05 indicates significant difference from the control group. (C) A549 cells were transfected with pEGFP or pEGFP-SUV39H2 and cell cycle analysis was performed by measuring DNA content after propidium iodide (PI) staining. *p<0.05 indicates significant difference from the control group. (D) The level of SUV39H2 protein was detected by Western blotting.
SUV39H2 is poly-ubiquitinated and is degraded through the ubiquitin-proteasome system
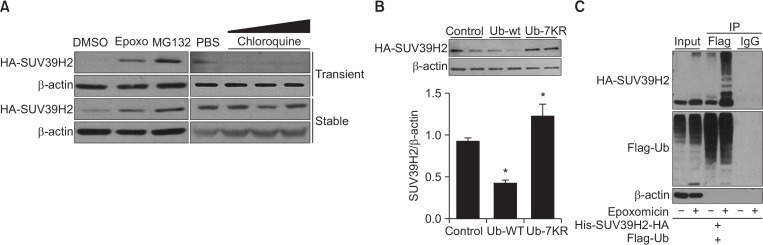
In mammalian cells, the majority of proteins is degraded by lysosomal or proteosomal pathways. Lysosomal degradation mainly occurs under stressed conditions, but ubiquitin-dependent proteosomal degradation occurs in highly selective protein turnover processes (Ciechanover, 1994). The ubiquitinated proteins are targets for degradation by p26 protesome, and the ubiqutin-proteasome system regulates cell cycle-associated proteins including cyclin-dependent kinase inhibitors (Adams, 2004). We examined the effects of epoxomicin and MG132, two inhibitors of proteasomal activity, and chloroquine, an inhibitor of lysosomal activity, on the level of SUV39H2 protein. SUV39H2 protein was stabilized in cells treated with epoxomicin or MG132 (Fig. 2A), indicating that SUV39H2 is degraded by the proteasome. To investigate whether degradation of SUV39H2 is mediated by the ubiquitin-proteasome system, SUV39H2-overexpressing cells were transfected with pcDNA, pcDNA-Ub-wild type, or pcDNA-Ub-7KR in which all seven lysine residues critical for polyubiquitination are replaced with arginine. Fig. 2B shows that SUV39H2 protein level was decreased by wild-type ubiquitin, whereas it was further increased in mutant ubiquitin-expressing cells compared with the control. These results suggest that SUV39H2 is degraded by the ubiquitin-proteasome system. To detect direct ubiquitination of SUV39H2 protein, HEK293 cells were transfected with pHM6-His-SUV39H2-HA and pcDNA-Flag-Ubiquitin. Ubiquitinated SUV39H2 was detected in anti-Flag-ubiquitin immunoprecipitates in the presence of epoxomicin (Fig. 2C). These data indicate that SUV39H2 is poly-ubiquitinated and is degraded through the ubiquitin-proteasome system.
Fig. 2.
SUV39H2 is poly-ubiquitinated and degraded by the ubiquitin-proteasome system. (A) HEK293 cells with transient or stable overexpression of SUV39H2 were treated overnight with proteasome inhibitors, 200 nM of epoxomicin or 20 μM of MG132, or an increasing amount of the lysosome inhibitor chloroquine (10 μM, 30 μM or 100 μM). Cells were harvested, and SUV39H2 protein was detected by Western blotting. (B) The SUV39H2-overexpressing cells were transfected with pcDNA, pcDNA-Ubiquitin-wild type, or pcDNA-Ubiquitin-7KR (lysine residue mutant). Two days after transfection, cells were harvested, and SUV39H2 protein was detected by Western blotting. (C) The HEK293 cells were transiently transfected with pHM6-His-SUV39H2-HA and pcDNA-Flag-Ubiquitin. Cell lysates were immunoprecipitated with anti-Flag antibody and then immunoblotted with anti-HA antibody.
Identification of translationally controlled tumor protein (TCTP) as an SUV39H2-interacting protein
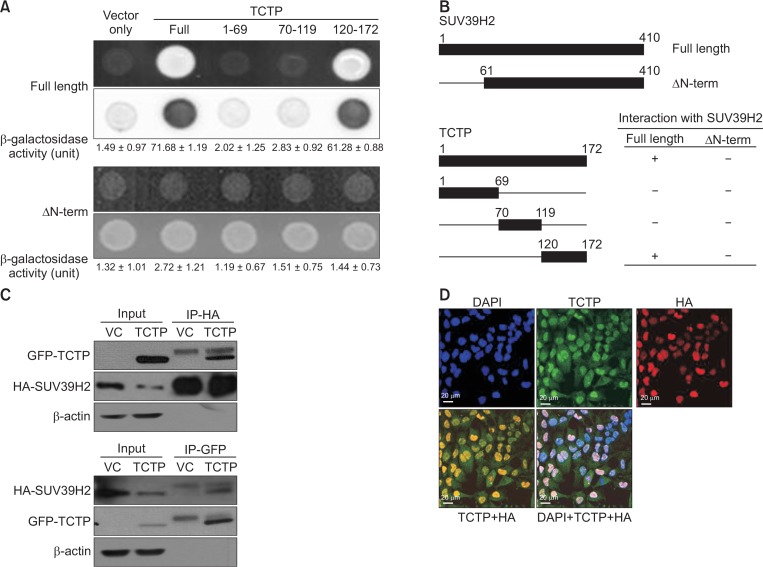
To further address the regulatory mechanism and biological function of SUV39H2 protein, we performed yeast two-hybrid assay to identify potential cellular SUV39H2-interacting partners. The human cDNA library was fused to the transcription activator, pJG4-5/B42 yeast cloning shuttle vector and was introduced into yeast cells containing pGilda/LexA-SUV39H2 (full length) as bait. Approximately 4.4×106 independent transformants were pooled. After re-spreading on selection media (Ura−, His−, Trp−, Leu−), we obtained seven colonies, four of which showed galactose dependency. The plasmids were obtained from the selected yeast cells and transformed into E. coli KC8 to separate the plasmids carrying pJG4-5/B42-cDNA inserts. The plasmids were then segregated by the plasmid marker trp in the E.coli host strain, and the purified plasmids were sequenced. A homology search in GenBank using the BLAST program revealed that all four plasmids encoded human translationally controlled tumor protein (TCTP) (accession number: NM_003295). To confirm this result, positive interaction was measured using cell growth on leucine-deficient and ONPG β-galactosidase activity. As shown in Fig. 3A, β-galactosidase activity was fully activated (71.68 ± 1.19), in the presence of SUV39H2 (full-length) and TCTP, but it was not observed with the empty plasmid (vector only: 1.49 ± 0.97). Subsequently, cDNA constructs containing three deletion mutants were designed to localize the SUV39H2 binding domain of TCTP (Fig. 3B). In the two-hybrid system, the full-length human SUV39H2 cDNA and cDNA with either a plasmid containing a full-length human TCTP or three truncation mutant forms (Fig. 3B, 1–69 aa, 70–119 aa, 120–172 aa) were co-transformed into EGY48 yeast cells. Cells containing full-length SUV39H2 cDNA and one TCTP deletion mutant (120–172 aa) grew on the Ura, His, Trp and Leu deficient plates. Yeast cells transformed with the other deletion mutants (1–69 aa and 70–119 aa) failed to grow. Also, cells containing N-terminal-truncated SUV39H2 failed to grow (Fig. 3A). Quantitation of β-galactosidase activity is shown in Fig. 3A. These results collectively suggest that the N-terminal 60 amino acids of full-length SUV39H2 and the C-terminus of TCTP (120 aa–172 aa) are critical for binding.
Fig. 3.
SUV39H2 interacts with the anti-apoptotic protein TCTP. (A) Direct interaction of TCTP and SUV39H2 was determined in the yeast two-hybrid system. The N-terminal region of SUV39H2 binds to the C-terminal region of TCTP. The binding activity (unit) calculated by adding o-nitrophenyl β-D-galactopyranoside (ONPG) agents, are indicated below their corresponding lanes. An empty plasmid (vector only) was used as the negative control. (B) Schematic representation of cDNA constructs for SUV39H2 (full length and ΔN-term) and for each TCTP truncated mutant and full-length TCTP protein. (C) SUV39H2 overexpressing A549 cells were transfected with pEGFP-TCTP. Cell lysates were immunoprecipitated with anti-HA antibody (upper panel) or anti-GFP (lower panel) and then immunoblotted with anti-GFP (upper panel) or anti-HA (lower panel) antibodies, respectively. (D) Cellular localization of SUV39H2 and endogenous TCTP was determined by immunofluorescence staining in SUV39H2-overexpressing A549 cells. Cell images were captured at a magnification of 200× using confocal microscopy. Scale bar: 20 μm.
Next, we aimed to detect the interaction between SUV39H2 and TCTP in human cancer cells. The SUV39H2-overexpressing A549 cells were transfected with pEGFP-TCTP. Immunoprecipitation was subsequently conducted using anti-HA (SUV39H2) or anti-GFP (TCTP) with whole cell lysates of SUV39H2 stable cells. As shown in Fig. 3C, TCTP was co-immunoprecipitated with SUV39H2. Immunofluorescence staining of SUV39H2 and endogenous TCTP showed co-localization of these two proteins in the nucleus (Fig. 3D). These results demonstrated that SUV39H2 and TCTP interact in lung cancer cells.
TCTP down-regulates SUV39H2 post-translationally
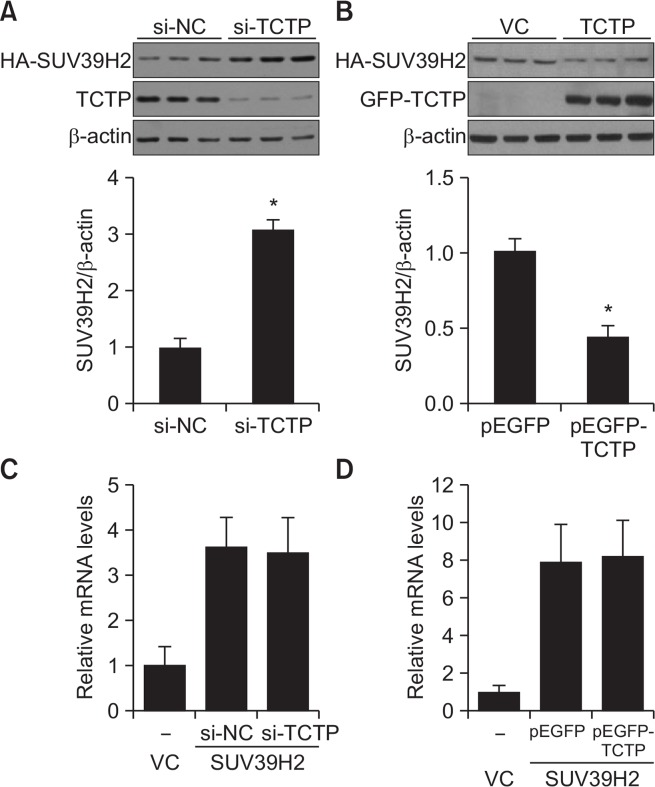
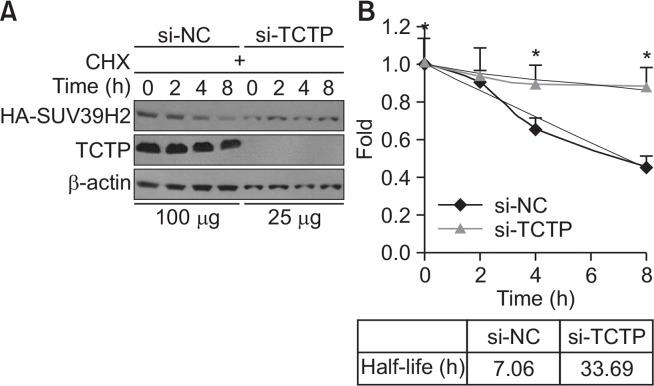
TCTP physically interacts with several proteins including Pim-3, VHL and Mcl-1 and regulates their protein by inhibiting or increasing the ubiquitin-proteasomal degradation pathway (Amson et al., 2011; Chen et al., 2013; Zhang et al., 2013). To determine whether SUV39H2 protein level is changed in the presence or absence of TCTP, SUV39H2-overexpressing A549 cells were transfected with si-negative control (si-NC) or si-TCTP. We demonstrated that the level of SUV39H2 protein in the absence of TCTP was significantly increased by 3-fold (Fig. 4A), while overexpression of TCTP decreased SUV39H2 protein level (Fig. 4B). This led us to investigate the effects of TCTP on SUV39H2 mRNA expression. As shown in Fig. 4C and 4D, qRT-PCR data revealed that the level of SUV39H2 mRNA was not changed with TCTP knockdown or TCTP overexpression. These results indicate that TCTP regulates SUV39H2 at the protein level. To test if regulation of SUV39H2 expression by TCTP is due to changes in protein stability, we measured the half-life of the SUV39H2 protein in the presence or absence of TCTP after treatment with cycloheximide (10 μg/mL) to block further synthesis of SUV39H2 protein. As shown in Fig. 5, SUV39H2 proteins were rapidly degraded with a half-life of 7 h, in the presence of TCTP, whereas the half-life observed in the absence of TCTP (∼33 h) was significantly longer. Taken together, these results indicate that the stability of SUV39H2 is indeed influenced by the cellular level of TCTP.
Fig. 4.
TCTP regulates SUV39H2 at the protein level. (A, C) The SUV39H2-overexpressing A549 cells were transfected with sinegative control (si-NC) or si-TCTP. (B, D) The SUV39H2-overexpressing A549 were transfected with pEGFP or pEGFP-TCTP. The protein or mRNA expression level was analyzed by immunoblotting (A, B) and qRT-PCR (C, D), respectively. Data are presented as mean ± SE of three independent experiments. *p<0.05 indicates significant difference from the control group.
Fig. 5.
TCTP downregulates SUV39H2 post-translationally. (A) The SUV39H2-overexpressing A549 cells were transfected with si-NC or si-TCTP. After transfection, the cells were pre-treated with cycloheximide (CHX, 10 μg/mL) for 30 min. At each time point, whole cell lysates were prepared and analyzed via immunoblot analysis. For si-NC and si-TCTP, 100 μg and 25 μg of protein lysates were loaded, respectively. (B) The SUV39H2 bands were quantified with an Image J analyzer, and the level of SUV39H2 protein at each time point was converted to the fold-change of the level of SUV39H2 protein at time 0 and plotted to determine the turnover rate. Data are presented as mean ± SE of three independent experiments. *p<0.05 indicates significant difference from the control group.
TCTP inhibits apoptotic cell death by negatively regulating SUV39H2
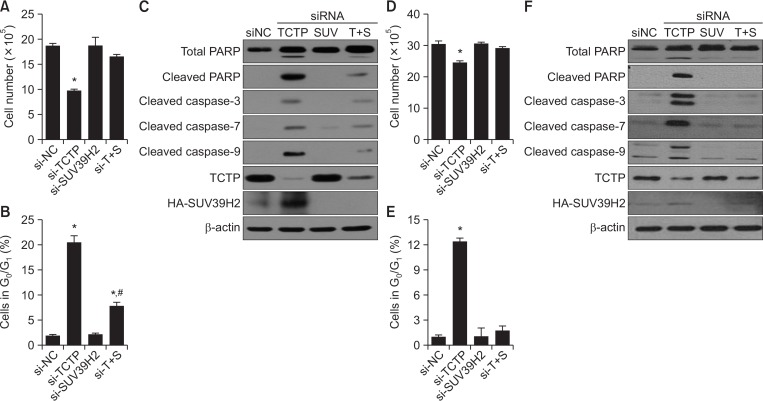
As TCTP is known to play an anti-apoptotic role (Li et al., 2001; Liu et al., 2005; Susini et al., 2008; Amson et al., 2011; Rho et al., 2011; Zhang et al., 2013; Thebault et al., 2016), we tested whether TCTP-knockdown induced apoptosis in A549 lung cancer cells and HEK293 immortalized cells stably transfected with SUV39H2 grown in 0.5% serum containing media. We observed that cell proliferation was decreased (Fig. 6A, 6D) with an increase in subG0/G1 population (Fig. 6B, 6E) in both TCTP-deficient SUV39H2 stable cells compared with control cells. Si-TCTP-transfected, SUV39H2-overexpresing cells displayed increase levels of PARP and cleaved caspase-3, -7, and -9, indicating that TCTP-knockdown induced apoptotic cell death (Fig. 6C, 6F). As TCTP-knockdown induced an increase in SUV39H2 protein (Fig. 4A, 4B, 6C, 6F), we determined if increased SUV39H2 in the absence of TCTP plays a role in inducing apoptosis. Combined treatment of si-TCTP and si-SUV39H2 recovered SUV39H2 stable transfectants from apoptotic cell death with decreased levels of PARP and cleaved caspase-3, -7, and -9, compared with the si-TCTP treated group (Fig. 6). Taken together, our results indicate that TCTP inhibits apoptotic cell death by negatively regulating SUV39H2.
Fig. 6.
TCTP inhibits apoptotic cell death by negatively regulating SUV39H2 protein stability. (A) SUV39H2 stable A549 cells were transfected with indicated siRNAs for 72 h. Twenty-four hours after transfection, cells were cultured in low-dose serum (0.5%) containing media and harvested by trypsinization; cell number was determined using a Coulter counter (Beckman Coulter Life Sciences, Indianapolis, IN, USA). (B) Cell death was assessed as % subG0/G1 population of cells by measuring DNA content after PI staining using flow cytometry. (C) The cell lysates were analyzed by immunoblotting using PARP; cleaved caspase-3, -7, and -9; TCTP; SUV39H2; and β-actin antibodies. Data are mean ± SDs of three independent experiments. *p<0.05 indicates significant difference from the control group. #p<0.05 indicates significant difference from the si-TCTP-treated group. (D) SUV39H2-overxpressing HEK293 cells were transfected with indicated siRNAs for 72 h. Twenty-four hours after transfection, cells were cultured in low-dose serum (0.5%) containing media and harvested by trypsinization; cell number was determined using a Coulter counter. (E) Cell death was assessed as % subG0/G1 population of cells by measuring DNA content after PI staining using flow cytometry. (F) The cell lysates were analyzed by immunoblotting using PARP; cleaved caspase-3, -7, and -9; TCTP; SUV39H2; and β-actin antibodies. Data are presented as mean ± SE of three independent experiments. *p<0.05 indicates significant difference from the control group.
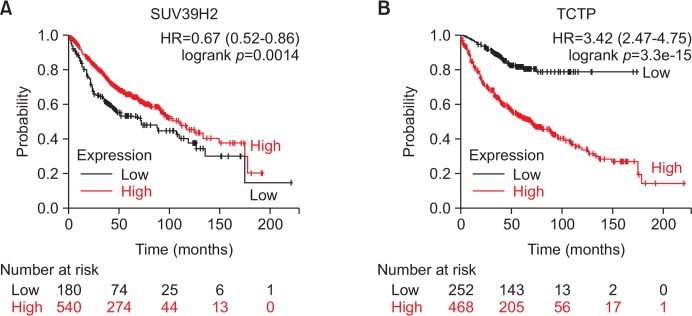
Clinical significance of SUV29H2 and TCTP in lung cancer patients
Associations between clinical outcomes of lung cancer patients and expression of SUV39H2 or TCTP were examined using Kaplan-Meier Plotter (Gyorffy et al., 2013). The results revealed better overall survival of lung cancer patients with higher expression of SUV39H2 compared with patients with lower expression of SUV39H2 (Fig. 7A). However, the level of TCTP mRNA was inversely correlated with overall survival of patients with lung cancer (Fig. 7B).
Fig. 7.
The association of overall survival of human NSCLC patients and the mRNA levels of SUV29H2 or TCTP. (A) The association between the level of SUV39H2 mRNA and overall survival of lung cancer (whole dataset). (B) Correlation of TCTP mRNA expression and overall survival of lung cancer (whole dataset) as analyzed using the Kaplan-Meier Plotter. Hazard ratio (HR) significance was found with log-rank tests.
DISCUSSION
In this study, we showed that overexpression of SUV39H2 decreased cell proliferation and induced cell cycle arrest at the G1 phase in A549 lung cancer cells (Fig. 1). In addition, SUV39H2 induced apoptosis when anti-apoptotic TCTP was depleted, suggesting that SUV39H2 may play a tumor-suppressive role. Notably, lung cancer patients with higher level of SUV39H2 mRNA had favorable clinical outcome with longer overall survival compared with patients with lower SUV39H2 expression, whereas higher TCTP expression led to shorter overall survival (Fig. 7).
Suv39h1 knockout mice display defects in heterochromatin and genome stability and, thereby, an increased risk of late onset lymphomas similar to non-Hodgkin lymphomas in human (Peters et al., 2001). Overexpression of SUV39H1 repressed K-Ras-driven embryonal rhabdomyosarcoma (Albacker et al., 2013), suggesting a tumor suppressive role of SUV39H1.
However, the tumorigenic roles of SUV39H1 and SUV39H2 were described in human cancers recently (Chiba et al., 2015; Shuai et al., 2018; Zheng et al., 2018). These opposing reports imply that the function of SUV39H regarding tumorigenesis depends on the cellular context, although this conclusion remains to be verified.
To further clarify the regulatory mechanism of SUV39H2 degradation via the ubiquitin-proteasomal system (Fig. 2) and to characterize the role of SUV39H2 in cancer cells, we screened for molecules interacting with SUV39H2 using a yeast two-hybrid system and identified TCTP as a binding partner of SUV39H2 (Fig. 3).
TCTP is an oncogenic protein and it has been reported to be increased in several human cancers including breast cancer, colon cancer, pancreatic cancer, prostate cancer, and glioma (Deng et al., 2006; Gnanasekar et al., 2009; Miao et al., 2013; Zhang et al., 2013; Bommer et al., 2017) and has been suggested as a therapeutic target in human cancer (Acunzo et al., 2014). Our results demonstrated that direct binding of TCTP to SUV39H2 protein induced degradation of SUV39H2 and thus shortened the half-life of SUV39H2 protein (Fig. 4, 5). With knockdown of TCTP in lung cancer cells, pro-apoptotic roles of SUV39H2 was uncovered (Fig. 6). Consistent with our data, knock-down of TCTP inhibited cell growth and induced apoptosis in prostate cancer (Gnanasekar et al., 2009). These findings clearly indicate that the oncogenic protein TCTP negatively regulates the expression and function of SUV39H2.
As TCTP exerts its anti-apoptotic function by modulating stability or activity of proteins functioning in apoptosis (Li et al., 2001; Liu et al., 2005; Susini et al., 2008; Amson et al., 2011; Rho et al., 2011; Zhang et al., 2013; Thebault et al., 2016), SUV39H2 may be an additional target of TCTP in regulating apoptosis.
Taken together, our results indicate that SUV39H2 induces G1 cell cycle arrest, and thereby inhibits cell proliferation of lung cancer cells. As a novel binding partner of SUV39H2, TCTP negatively regulates protein stability and the apoptotic role of SUV39H2.
Acknowledgments
This research was supported by National Cancer Center Research Grant (1710310) and the Basic Science Research Program through the National Research Foundation (NRF) funded by the Korean government [NRF-2016R1A2B4016618].
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Acunzo J, Baylot V, So A, Rocchi P. TCTP as therapeutic target in cancers. Cancer Treat. Rev. 2014;40:760–769. doi: 10.1016/j.ctrv.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/S1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albacker CE, Storer NY, Langdon EM, Dibiase A, Zhou Y, Langenau DM, Zon LI. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS ONE. 2013;8:e64969. doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J, Chaloin O, Hoebeke J, Marine JC, Di Fiore PP, Telerman A. Reciprocal repression between P53 and TCTP. Nat. Med. 2011;18:91–99. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- Amson R, Pece S, Marine JC, Di Fiore PP, Telerman A. TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol. 2013;23:37–46. doi: 10.1016/j.tcb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Amzallag N, Passer BJ, Allanic D, Segura E, Thery C, Goud B, Amson R, Telerman A. TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J. Biol. Chem. 2004;279:46104–46112. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int. J. Biochem. Cell Biol. 2004;36:379–385. doi: 10.1016/S1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Bommer UA, Vine KL, Puri P, Engel M, Belfiore L, Fildes K, Batterham M, Lochhead A, Aghmesheh M. Translationally controlled tumour protein TCTP is induced early in human colorectal tumours and contributes to the resistance of HCT116 colon cancer cells to 5-FU and oxaliplatin. Cell Commun. Signal. 2017;15:9. doi: 10.1186/s12964-017-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Labbe JC, Vigneron S, Bonneaud N, Strub JM, Van Dorsselaer A, Lorca T, Castro A. Chfr interacts and colocalizes with TCTP to the mitotic spindle. Oncogene. 2008;27:5554–5566. doi: 10.1038/onc.2008.167. [DOI] [PubMed] [Google Scholar]
- Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, Amzallag N, Allanic D, Tufino R, Argentini M, Moras D, Fiucci G, Goud B, Mirande M, Amson R, Telerman A. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen S, Huang C, Cheng H, Zhou R. TCTP increases stability of hypoxia-inducible factor 1alpha by interaction with and degradation of the tumour suppressor VHL. Biol. Cell. 2013;105:208–218. doi: 10.1111/boc.201200080. [DOI] [PubMed] [Google Scholar]
- Chen SH, Wu PS, Chou CH, Yan YT, Liu H, Weng SY, Yang-Yen HF. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol. Biol. Cell. 2007;18:2525–2532. doi: 10.1091/mbc.e07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Saito T, Yuki K, Zen Y, Koide S, Kanogawa N, Motoyama T, Ogasawara S, Suzuki E, Ooka Y, Tawada A, Otsuka M, Miyazaki M, Iwama A, Yokosuka O. Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int. J. Cancer. 2015;136:289–298. doi: 10.1002/ijc.28985. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Deng SS, Xing TY, Zhou HY, Xiong RH, Lu YG, Wen B, Liu SQ, Yang HJ. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genomics Proteomics Bioinformatics. 2006;4:165–172. doi: 10.1016/S1672-0229(06)60029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiucci G, Lespagnol A, Stumptner-Cuvelette P, Beaucourt S, Duflaut D, Susini L, Amson R, Telerman A. Genomic organization and expression of mouse Tpt1 gene. Genomics. 2003;81:570–578. doi: 10.1016/S0888-7543(03)00047-8. [DOI] [PubMed] [Google Scholar]
- Gachet Y, Tournier S, Lee M, Lazaris-Karatzas A, Poulton T, Bommer UA. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 1999;112:1257–1271. doi: 10.1242/jcs.112.8.1257. [DOI] [PubMed] [Google Scholar]
- Gnanasekar M, Thirugnanam S, Zheng G, Chen A, Ramaswamy K. Gene silencing of translationally controlled tumor protein (TCTP) by siRNA inhibits cell growth and induces apoptosis of human prostate cancer cells. Int. J. Oncol. 2009;34:1241–1246. [PubMed] [Google Scholar]
- Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Lee MJ, Song H, Han SH, Kim YM, Im JY, Choi I. Molecular identification of IgE-dependent histamine-releasing factor as a B cell growth factor. J. Immunol. 2001;166:6545–6554. doi: 10.4049/jimmunol.166.11.6545. [DOI] [PubMed] [Google Scholar]
- Koziol MJ, Garrett N, Gurdon JB. Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr. Biol. 2007;17:801–807. doi: 10.1016/j.cub.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak JZ, Bazile F, Pascal A, Richard-Parpaillon L, Polanski Z, Ciemerych MA, Chesnel F. Temporal regulation of embryonic M-phases. Folia Histochem. Cytobiol. 2008;46:5–9. doi: 10.2478/v10042-008-0001-z. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang D, Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem. 2001;276:47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 2005;25:3117–3126. doi: 10.1128/MCB.25.8.3117-3126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald SM, Rafnar T, Langdon J, Lichtenstein LM. Molecular identification of an IgE-dependent histamine-releasing factor. Science. 1995;269:688–690. doi: 10.1126/science.7542803. [DOI] [PubMed] [Google Scholar]
- Miao X, Chen YB, Xu SL, Zhao T, Liu JY, Li YR, Wang J, Zhang J, Guo GZ. TCTP overexpression is associated with the development and progression of glioma. Tumour Biol. 2013;34:3357–3361. doi: 10.1007/s13277-013-0906-9. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 2000;20:9423–9433. doi: 10.1128/MCB.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Rao VK, Pal A, Taneja R. A drive in SUVs: from development to disease. Epigenetics. 2017;12:177–186. doi: 10.1080/15592294.2017.1281502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, Kim JY, Park SY. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett. 2011;585:29–35. doi: 10.1016/j.febslet.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Rho SB, Lee KH, Kim JW, Shiba K, Jo YJ, Kim S. Interaction between human tRNA synthetases involves repeated sequence elements. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10128–10133. doi: 10.1073/pnas.93.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai W, Wu J, Chen S, Liu R, Ye Z, Kuang C, Fu X, Wang G, Li Y, Peng Q, Shi W, Li Y, Zhou Q, Huang W. SUV39H2 promotes colorectal cancer proliferation and metastasis via tri-methylation of the SLIT1 promoter. Cancer Lett. 2018;422:56–69. doi: 10.1016/j.canlet.2018.02.023. [DOI] [PubMed] [Google Scholar]
- Susini L, Besse S, Duflaut D, Lespagnol A, Beekman C, Fiucci G, Atkinson AR, Busso D, Poussin P, Marine JC, Martinou JC, Cavarelli J, Moras D, Amson R, Telerman A. TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ. 2008;15:1211–1220. doi: 10.1038/cdd.2008.18. [DOI] [PubMed] [Google Scholar]
- Tani T, Shimada H, Kato Y, Tsunoda Y. Bovine oocytes with the potential to reprogram somatic cell nuclei have a unique 23-kDa protein, phosphorylated transcriptionally controlled tumor protein (TCTP) Cloning Stem Cells. 2007;9:267–280. doi: 10.1089/clo.2006.0072. [DOI] [PubMed] [Google Scholar]
- Telerman A, Amson R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat. RevCancer. 2009;9:206–216. doi: 10.1038/nrc2589. [DOI] [PubMed] [Google Scholar]
- Thebault S, Agez M, Chi X, Stojko J, Cura V, Telerman SB, Maillet L, Gautier F, Billas-Massobrio I, Birck C, Troffer-Charlier N, Karafin T, Honore J, Senff-Ribeiro A, Montessuit S, Johnson CM, Juin P, Cianferani S, Martinou JC, Andrews DW, Amson R, Telerman A, Cavarelli J. TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL. Sci. Rep. 2016;6:19725. doi: 10.1038/srep19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, Duflaut D, Besse S, Susini L, Cavarelli J, Moras D, Amson R, Telerman A. Translationally controlled tumor protein is a target of tumor reversion. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15364–15369. doi: 10.1073/pnas.0406776101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, Telerman A. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14976–14981. doi: 10.1073/pnas.222470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarm FR. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ, Lee JE, Jang YK, Park JG, Lee JS. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]
- Zhang F, Liu B, Wang Z, Yu XJ, Ni QX, Yang WT, Mukaida N, Li YY. A novel regulatory mechanism of Pim-3 kinase stability and its involvement in pancreatic cancer progression. Mol. Cancer Res. 2013;11:1508–1520. doi: 10.1158/1541-7786.MCR-13-0389. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Li B, Wang J, Xiong Y, Wang K, Qi Y, Sun H, Wu L, Yang L. Identification of SUV39H2 as a potential oncogene in lung adenocarcinoma. Clin. Epigenetics. 2018;10:129. doi: 10.1186/s13148-018-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]