Abstract
The technological advancements of the modern era have enabled the collection of huge amounts of data in science and beyond. Extracting useful information from such massive datasets is an ongoing challenge as traditional data visualization tools typically do not scale well in high-dimensional settings. An existing visualization technique that is particularly well suited to visualizing large datasets is the heatmap. Although heatmaps are extremely popular in fields such as bioinformatics, they remain a severely underutilized visualization tool in modern data analysis. This paper introduces superheat, a new R package that provides an extremely flexible and customizable platform for visualizing complex datasets. Superheat produces attractive and extendable heatmaps to which the user can add a response variable as a scatterplot, model results as boxplots, correlation information as barplots, and more. The goal of this paper is two-fold: (1) to demonstrate the potential of the heatmap as a core visualization method for a range of data types, and (2) to highlight the customizability and ease of implementation of the superheat R package for creating beautiful and extendable heatmaps. The capabilities and fundamental applicability of the superheat package will be explored via three reproducible case studies, each based on publicly available data sources.
Keywords: Data Visualization, Exploratory Data Analysis, Heatmap, Multivariate Data
1. Introduction
The rapid technological advancements of the past few decades have enabled us to collect vast amounts of data with the goal of finding answers to increasingly complex questions both in science and beyond. Although visualization has the capacity to be a powerful tool in the information extraction process of large multivariate datasets, the majority of commonly used graphical exploratory techniques such as traditional scatterplots, boxplots, and histograms are embedded in spaces of 2 dimensions and rarely extend satisfactorily into higher dimensions. Basic extensions of these traditional techniques into 3 dimensions are not uncommon, but tend to be inadequately represented when compressed to a 2-dimensional format. Even graphical techniques designed for 2-dimensional visualization of multivariate data, such as the scatterplot matrix (Cleveland, 1993; Andrews, 1972) and parallel coordinates (Inselberg, 1985, 1998; Inselberg and Dimsdale, 1987) become incomprehensible in the presence of too many data points or variables. These techniques suffer from a lack of scalability. Effective approaches to the visualization of high-dimensional data must subsequently satisfy a tradeoff between simplicity and complexity. A graph that is overly complex impedes comprehension, while a graph that is too simple conceals important information.
1.1. The heatmap for matrix visualization
An existing visualization technique that is particularly well suited to the visualization of highdimensional multivariate data is the heatmap. Today, heatmaps are widely used in areas such as bioinformatics (often to visualize large gene expression datasets, for example in Trakhtenberg et al. (2016) and Tardu et al. (2017)), yet are significantly underemployed in other domains. There exist a wide range of standard heatmap software available, including inbuilt R functions such as image and heatmap, as well as functions from R packages such as heatmap.2 from the gplots package, heatmap.3 from the GMD package, the pheatmap package (Kolde, 2012) and its extension aheatmap (Gaujoux and Seoighe, 2010) from the NMF package.
A heatmap can be used to visualize a data matrix by representing each matrix entry by a color corresponding to its magnitude, enabling the user to visually process large datasets with thousands of rows and/or columns. While the computational power of the 21st century has enabled researchers to produce increasingly rich and complex heatmaps, the earliest sources of the heatmap date back to at least the 1800s, where Loua (1873) used color to represent the numerical values of various social statistics in Paris.
Even the modern practice of emphasizing structure in the data by clustering together similar rows and columns of the heatmap is not new. Authors such as Brinton (1914) used such techniques over 100 years ago to highlight relationships in educational data. A more recent development is the practice of appending a dendrogram to the rows and/or columns of a heatmap to present the hierarchy of clusters in the data. Authors such as Ling (1973); Gower and Digby (1981) and Chen (2002) originally developed heatmaps that displayed both the reordered/clustered data matrix as well as adjacent diagonal similarity matrices with dendrograms attached. These more complex (but perhaps more informative) versions of the clustered heatmap later morphed into the more common version we see today which appends the dendrograms directly to the clustered data matrix (Wilkinson, 1994; Eisen et al., 1998). For more details on the history of the heatmap, thereader is referred to Wilkinson and Friendly (2009).
While augmentation by a cluster dendrogram has been fairly common practice for the past two decades, it remains fairly uncommon to augment heatmaps by other types of information. Recently, interest has arisen in combining heatmaps with other traditional plot types such as barplots, scatterplots, and histograms, and several authors have produced software for producing such visualizations such as the ComplexHeatmap (Gu et al., 2016). Another recent avenue for expanding the traditional heatmap toolbox is the incorporation of hover and click interactivity such as in heatmaply (Galili et al., 2017). Both interactivity and additional subplots have been combined in the iheatmapr R package by Schep and Kummerfeld (2017). Our R package, superheat, was one of the early packages (originally developed in 2015) to incorporate additional information in the form of adjacent subplots such as barplots, boxplots, line plots, scatterplots and more. The greater ease of implementation, flexibility of customization, and visual attractiveness of superheat, as we will show throughout this paper, sets our software apart from its static competitors.
1.2. Choosing row/column ordering and color mapping in heatmaps
While heatmaps can be incredibly useful for visualizing large matrices, they can also be misinterpreted if designed improperly (Cook et al., 2007). Two features of the heatmap most likely to lead to misrepresentation of the data are (1) the choice of row/column ordering in generating clustered heatmaps, and (2) the choice of color mapping. In this section, we will provide some brief advice on the use of for row/column ordering, and discuss how a quantile color mapping helps alleviate issues that can arise when manually the choosing the scale for a heatmap color map.
Row and column ordering
Interpretation of a heatmap can vary based on the ordering of the rows/columns, so it is always a good idea to ensure that any patterns highlighted by a clustered or re-ordered heatmap are stable. Our recommended approach to assess the stability of the patterns identified in such heatmaps is to re-generate the heatmap on various random subsets of the data to ensure that the patterns identified are consistent (Yu, 2013).
Color maps
A color map consists of two components: (1) the choice of color space/scheme, and (2) the functional mapping that dictates which color (within the specified color space) each data point is mapped to. Care must be taken when defining the color scheme and mapping (Wong, 2011).
It is important that the selected heatmap color space is perceptually uniform, i.e. the difference between two colors, as perceived by the human eye, is proportional to the Euclidean distance between the two colors in the color space. The default color scheme for our superheat package, viridis, is perceptually uniform. It has been shown, however, that many of the popular color schemes, such as the “rainbow” color scheme, are not (Tajima, 1983; Bujack et al., 2018).
Having selected an appropriate color scheme, the next decision is how to map data into the corresponding color space. For the majority of heatmap software, the default functional mapping from data to color is linear: equal distances in data space are represented as equal distances in color space. In most datasets, however, the data is not spread uniformly throughout the range from the smallest to the largest value. Instead, the data might be more dense in the middle of the range, or be skewed towards larger or smaller values. In this case, a linear mapping from data space to color space will highlight outliers by emphasizing the data points that have largest distances from the bulk of the data. As a result, it is common in practice for users to manually adjust the color transition positions until they feel that they have highlighted as many trends in the center of the data as possible. Manual selection of the data-to-color mapping can potentially lead to a scenario in which the researcher is simply highlighting noise or is unintentionally hiding information. For example, a color map that represents all negative values as black and all positive values as ranging from dark blue to light blue will hide the information contained within the negative data space.
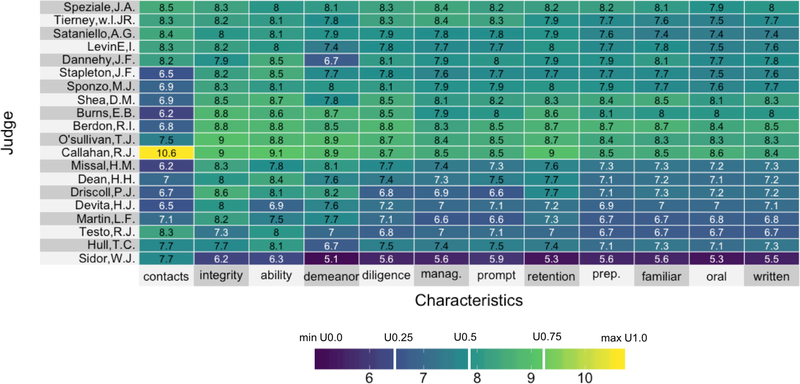
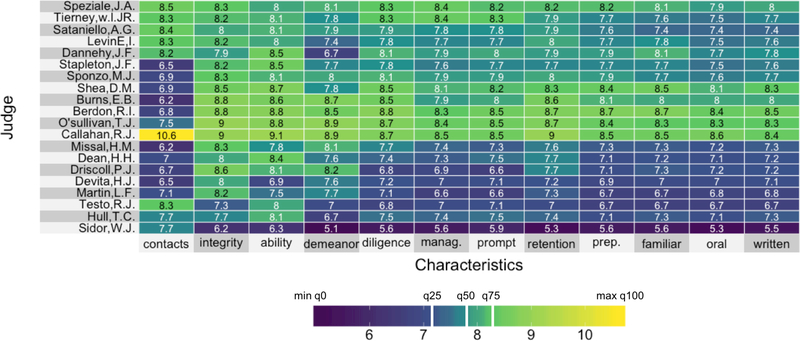
Figures 1 and 2 present two heatmaps, each using the same viridis color space but different color mappings: Figure 1 implements a linear color map, while Figure 2 implements a quantile color map (described below). The data underlying the heatmaps comes from lawyers’ ratings of a subset of 20 state judges in the US Superior Court from the New Haven Register in 1977. These ratings were collected on 12 characteristics: contacts (the number of contacts of the lawyer with the judge), judicial integrity, physical ability, demeanor, diligence, case flow managing, prompt decisions, worthy of retention, preparation for trial, familiarity with law, sound oral rulings, and sound written rulings. The data can be found as a part of the inbuilt datasets package in R.
Figure 1:
A heatmap with a viridis color space and linear color map of the lawyers’ ratings of 20 state Judges in the US Superior Court. The white vertical bars in the legend represent the positions of three central (equidistant) colors in color space.
Figure 2:
A heatmap with a viridis color space and quantile color map of the Lawyers’ ratings of 20 state Judges in the US Superior Court. The numbers in the cells show the actual ratings. The white vertical bars in the legend represent the same three colors from Figure 1, and in this example, their positions are mapped from the 25th, 50th, and 75th quantiles in the data.
With a linear mapping (Figure 1), color is distributed uniformly throughout the range of the data-to-color map (as represented by the equidistant vertical white bars in the legend representing three equidistant colors in color space). As a result of the linear mapping, there is a notable lack of contrast among the bulk of the data in Figure 1. Since the data are far from uniformly distributed (80% of the data lies between the values 7 and 9), most of the ratings are presented as being very close together, which, relative to the range of the data, they are. Unfortunately, this feature of the linear mapping makes it very difficult to tease out the patterns in the data when the majority of the data values do not uniformly span the range of the data. Linear color maps would be appropriate if the user wanted to highlight the outliers and subdue patterns within the region of typical data values, however, this is usually not the goal of a heatmap.
Figure 2 shows the same heatmap with an alternative quantile color mapping which allows for quicker transitions between the colors in regions where the bulk of the data lie. The quantile color map uses the quantiles of the data to dictate where the color transitions should take place within the heatmap. In a color space that is defined by a set of five sequential colors, the first color is centered at the minimum value in the data (or the 0th quantile), the second color is centered at the 25th quantile, the third color is centered at the median of the data (the 50th quantile), the fourth color is centered at the 75th quantile, and the fifth color is centered at the maximum value of the data (the 100th quantile). The transitions from one color to the next happen in between these quantiles. The positions of three central colors that are equidistant in color space (but whose mapping is defined by the 25th, 50th and 75th quantiles in the data) are presented as white bars in the legend of Figure 2. Compare the positions of the same central colors in Figure 1.
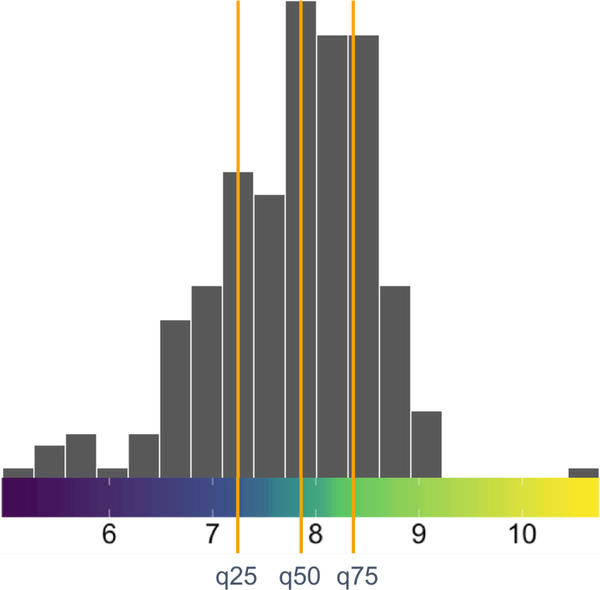
Notice that are more distinct groupings visible in Figure 2 as compared to Figure 1. Quantile color maps are appropriate for users who want to highlight the typical data values and reduce the influence of outliers on the color transitions. Figure 3 shows the distribution of the data with the quantile color map.
Figure 3:
The distribution as a histogram of the lawyer’s ratings on US superior court judges placed on top of the quantile color map (from Figure 2). The quantiles are highlighted by vertical orange lines.
While alternatives to heatmaps such as scatterplot matrices and parallel coordinate plots are less sensitive to choices of color and order, they quickly become intractable in the presence of even tens of variables. Heatmaps are able to display substantially more information in less space than either of these popular counterparts. A detailed comparison of the heatmap with scatterplot matrices and parallel coordinates will feature in our first case study in Section 3.2.
2. Superheat
Inspired by a desire to visualize a design matrix in a manner that is supervised by some response variable, we developed an R package superheat (short for “supervised heatmap”) for producing “supervised” heatmaps that extend the traditional heatmap via the incorporation of additional information. Superheatmaps are flexible, customizable and very useful for presenting a global view of complex datasets. Such plots would be difficult and time-consuming to produce without the existence of software that can automatically generate the plots given the user’s preferences. Superheat, builds upon the infrastructure provided by the ggplot2 (Wickham, 2016) R package to develop an intuitive heatmap function that possesses the aesthetics of ggplot2 with the simple implementation of the inbuilt heatmap functions. While ggplot2 itself contains functions for producing visually appealing heatmaps, it requires the user to convert the data matrix to a long-form data frame consisting of three columns: the row index, the column index, and the corresponding fill value. Although this data structure is intuitive for other types of plots, it can be somewhat cumbersome for producing heatmaps. For this reason, superheat accepts matrix inputs directly and does not make use of the ggplot2 grammar of graphics (Wickham, 2010).
Below we highlight some key features and usage of superheat. Readers looking for more details can find the wide variety of features as well as extensive instructions on usage of superheat in the online Vignette (see Supplementary Materials for the URL). The superheat package contains a single function: the self-named superheat function. The data matrix to be plotted is to be provided as the first argument, X. All other arguments of the superheat function are optional, and some are described below.
2.1. Adding additional information
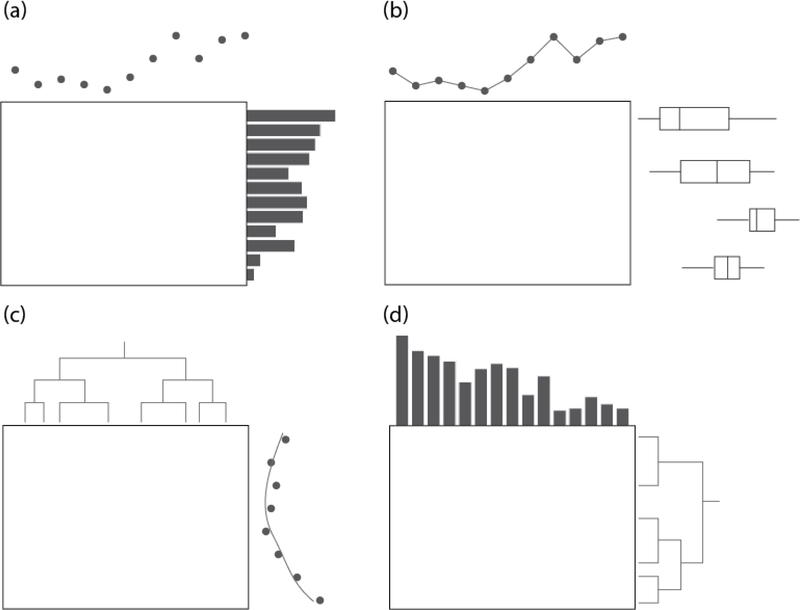
One of the primary components of superheat is the ability to add additional sources of information in the form of scatterplots, barplots, boxplots, line plots, and dendrograms adjacent to the rows and columns of the heatmap. These adjacent plots allow the user to explore their data to greater depths, and to take advantage of the heterogeneity present in the data to inform analysis decisions. Some examples of the basic structure of a superheatmap are presented in Figure 4.
Figure 4:
Four examples of superheat layouts. Panel (a) shows a scatterplot added to the columns, and a bar plot added to the rows. Panel (b) shows a scatter-line plot added to the columns and grouped boxplots added to the rows. Panel (c) shows a dendrogram added to the columns and a scatter-smooth plot (a scatterplot with a smoothed curve) added to the rows. Panel (d) shows a bar plot added to the columns and a dendrogram added to the rows.
To add a plot above the heatmap, the user provides a vector to the yt (“y top”) argument, where the length of the vector is equal to the number of columns in the heatmap. Similarly, to add a plot to the right of the heatmap, the user provides a vector to the yr (“y right”) argument. The type of plot can be specified by setting the yt.plot.type or yr.plot.type argument to ‘scatter’, ‘bar’, ‘boxplot’, ‘scattersmooth’, ‘smooth’, ‘scatterline’, or ‘line’. Note that boxplots can only be added when the rows or columns are grouped (see Section 2.2). Overlaid text such as the data itself can be added to the heatmap using the X.text argument. Row or column dendrograms can be added by setting the row.dendrogram or col.dendrogram to be TRUE.
2.2. Specifying row/column ordering and grouping
By default, superheat does not reorder the rows or columns of the matrix provided. The order of the rows and columns (and simultaneously the data in the adjacent plots) can be changed by providing the order.rows and order.cols arguments with an index vector specifying the position of the columns/rows. For users that would like superheat to automatically rearrange the rows/columns in order to highlight structure, setting the arguments pretty.order.rows = TRUE and pretty.order.cols = TRUE will apply a hierarchical clustering algorithm and rearrange the rows/columns accordingly.
Superheat has inbuilt clustering capabilities wherein the user can specify the number of row or column clusters they would like using the n.clusters.rows and n.clusters.cols arguments. Superheat will then run a k-means (the default clustering algorithm) on the data matrix and will group together the rows or columns that are in the same cluster (while respecting the order of the rows/columns specified by order.rows and order.cols within each cluster). To select the number of clusters, it is recommended that the user does so prior to the implementation of the superheatmaps using standard methods such as Silhouette plots (Rousseeuw, 1987). Users can also provide their own cluster membership vectors using the membership.rows and membership.cols arguments.
When using clustering within superheat, the resulting heatmap is a “grouped” heatmap, to which boxplots and aggregate bar plots can be added as an adjacent plot for each group of rows or columns. Grouped heatmaps with a large number of rows/columns can be smoothed so that each row/column group is presented by a single color corresponding to the median entry, rather than to show each matrix entry individually. An example of a grouped heatmap with smoothing can be seen in our second case study in Figure 9.
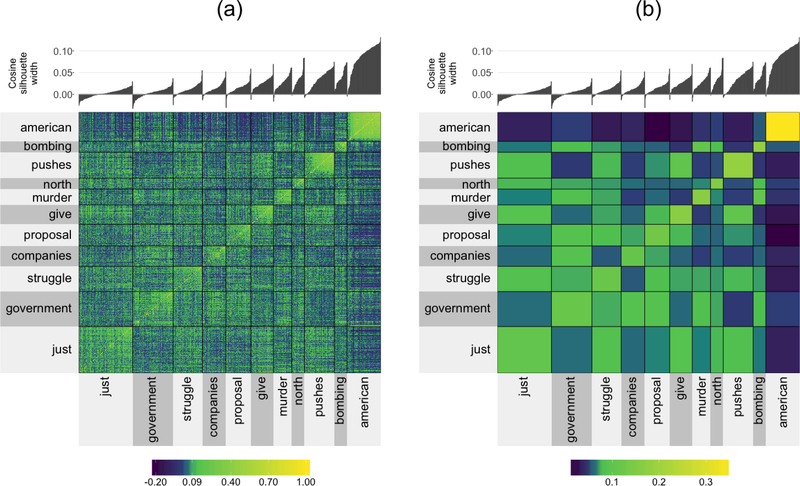
Figure 9:
A clustered cosine similarity matrix for the 855 most common words from the NY Times headlines that also appear in the Google News corpus. The clusters were generated using PAM and the cluster label is given by the medoid word of the cluster. Panel (a) displays the raw clustered 855×855 cosine similarity matrix, while panel (b) displays a “smoothed” version where the cells in the cluster are aggregated by taking the median of the values within the cluster.
2.3. Specifying the color map and color scheme
By default, superheat uses a quantile color map (see Section 1.2) with the perceptually uniform viridis color scheme. Users who wish to deviate from the default viridis quantile color map can specify their own color palette using the heat.pal argument, or users can choose alternative color schemes from among the sequential color brewer (Harrower and Brewer, 2003) schemes (setting heat.col.scheme to one of ‘red’, ‘purple’, ‘blue’, ‘grey’, ‘green’). Users can manually specify a data-to-color map using the heat.pal.values argument, which expects a vector whose length equals heat.pal and which specifies the center position of each color specified in heat.pal. For example, if we have a dataset whose minimum value is 0 and whose maximum value is 10, if we set heat.pal = c(‘white’, ‘blue’, ‘black’) and heat.pal.values = c(0, 0.2, 1), then the data value of 0 will map to “white”, the data value of 2 will map to “blue”, and the value of 10 will map to “black”, with linear transitions between each of these colors.
2.4. Further implementation information
The development page for superheat is hosted on GitHub (see Supplementary Materials), where the user can also find a detailed Vignette describing further information on the specific usage of superheat as well as a host of options for functional and aesthetic customizability. Details of the analytic pipeline and code for the case studies presented in this paper can be found in the Supplementary Materials.
The remainder of this paper will present three case studies that highlight the ability of superheat to (1) combine multiple sources of data together, (2) uncover correlational structure in data, and (3) evaluate heterogeneity in the performance of data models.
3. Case study I: combining data sources to explore global organ transplantation trends
The worldwide demand for organ transplantation has drastically increased over the past decade, leading to a gross imbalance of supply and demand. In the United States, there are currently over 100,000 people waiting on the national transplant lists but there simply aren’t enough donors to meet this demand (Abouna, 2008). This imbalance is worse in some countries than others as organ donation rates vary hugely from country to country, and it has been suggested that organ donation and transplantation rates are correlated with country development (Garcia et al., 2012).
This case study will explore combining multiple sources of data in order to examine the recent trends in organ donation worldwide as well as the relationship between organ donation and the Human Development Index (HDI).
The organ donation data was collected from the WHO-ONT Global Observatory on Donation and Transplantation, which represents the most comprehensive source to date of worldwide data concerning activities in organ donation and transplantation derived from official sources. The database (available from GODT (2016)) contains information from a questionnaire annually distributed to health authorities from the 194 Member States in the six World Health Organization (WHO) regions: Africa, The Americas, Eastern Mediterranean, Europe, South-East Asia and Western Pacific.
The HDI was created to emphasize that people and their capabilities (rather than economic growth) should be the ultimate criteria for assessing the development of a country. The HDI is calculated based on life expectancy, education and per capita indicators and is hosted by the United Nations Development Program’s Human Development Reports (available from UNDP (2015)).
3.1. Exploration
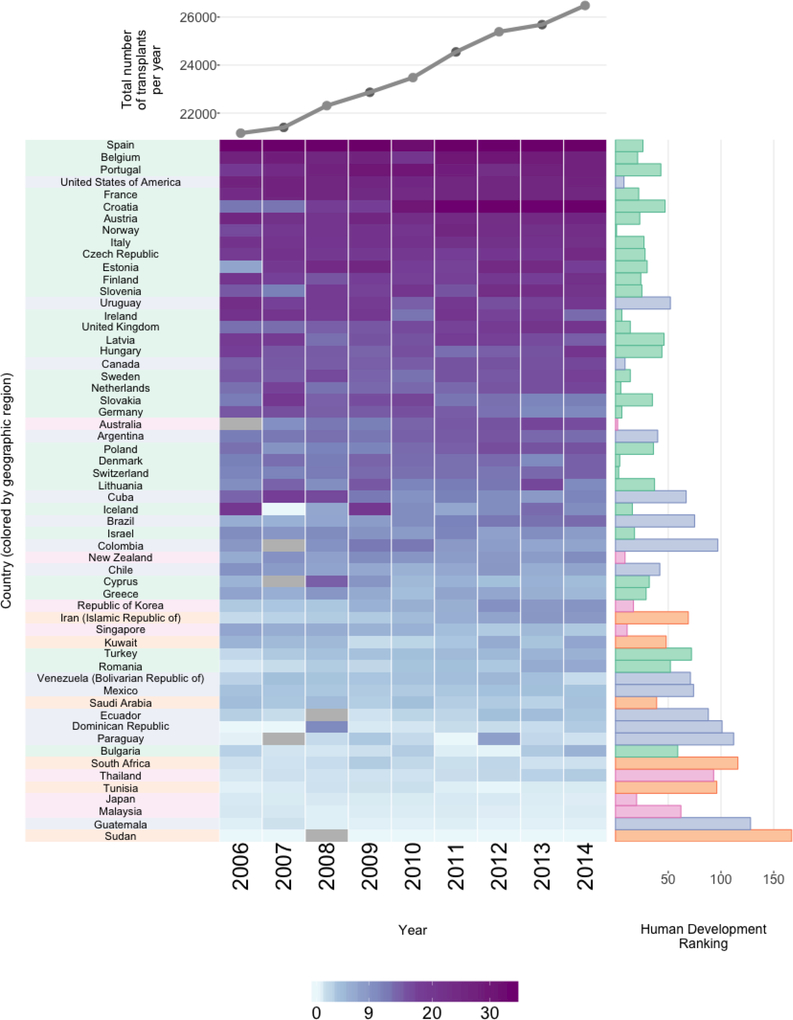
In the superheatmap presented in Figure 5, the central heatmap presents the total number of donated organs from deceased donors per 100,000 individuals between 2006 to 2014 for each country, restricting to countries for which data was collected for at least 8 of the 9 years.
Figure 5:
Organ donations and HDI by country. The right-hand bar plot displays the HDI ranking (lower is better). Each heatmap cell shows the number of organ donations from deceased donors per 100K. Grey cells correspond to missing values. The rows (countries) are ordered by average transplants per 100K. The country labels and HDI bar plot are colored based on region: Europe (green), Eastern Mediterranean (purple), Western Pacific (yellow), America (orange), South East Asia (pink) and Africa (light green). The upper line plot shows total organs donated per year.
Note that relaxing the country inclusion requirement to available data for 7 of the 9 years would include some additional countries (Bhutan, Costa Rica, Kenya, Luxembourg, Myanmar, Nigeria, Oman, Panama and the Syrian Arab Republic), however, in the interests of space, we do not include these. Further note that there are several countries (China and India included) for which there is no total deceased organ donor data available.
Above the heatmap, a line plot displays the overall number of donated organs over time, aggregated across all 58 countries represented in the figure. We see that overall, the organ donation rate is increasing, with approximately 5,000 more recorded organ donations occurring in 2014 relative to 2006. To the right of the heatmap, next to each row, a bar displays the country’s HDI ranking (a lower HDI ranking is better). Each country is colored based on which global region it belongs to: Europe (green), Eastern Mediterranean (purple), Western Pacific (yellow), America (orange), South East Asia (pink) and Africa (light green).
From Figure 5, we see that Spain is the clear leader in global organ donation, however there has been a rapid increase in donation rates in Croatia, which had one of the lower rates of organ donation in 2006 but has a rate equaling that of Spain in 2014. However, in contrast to the growth experienced by Croatia, the rate of organ donation appears to be slowing in several countries including as Germany, Slovakia and Cuba. For some unexplained reason, Iceland reported zero organ donations recorded from deceased donors in 2007.
The countries with the most organ donations are predominantly European and American. In addition, there appears to be a general correlation between organ donations and HDI ranking: countries with lower (better) HDI rankings tend to have higher organ donation rates. Subsequently, countries with higher (worse) HDI rankings tend to have lower organ donation rates, with the exception of a few Western Pacific countries such as Japan, Singapore and Korea, which have fairly good HDI rankings but relatively low organ donation rates.
In this case study, superheat allowed us to visualize multiple trends simultaneously without resorting to mass over-plotting. In particular, we were able to examine the organ donation over time and for each country and compare these trends to the country’s HDI ranking while visually grouping countries from the same region together. No other 2-dimensional graph would be able to provide such an in-depth, yet uncluttered, summary of the trends contained in these data. In the next section, we will compare superheat to alternative graphs: parallel coordinates and scatterplot matrices.
The code used to produce Figure 5 is provided in the supplementary materials.
3.2. A comparison with scatterplot matrices and parallel coordinates
The superheatmap in Figure 5 provides a clear, uncluttered view of this multivariate dataset (treating the primary variables in the data as the number of organ donations per year and the HDI ranking for each country). In this section, we will examine alternative views of the same data as presented by the two other popular multivariate plots: scatterplot matrices and parallel coordinate plots.
Figure 6 displays the organ donation data (originally presented in Figure 5) as a scatterplot matrix created using the ggpairs function from the GGally R package. Each row/column of the matrix corresponds to the number of organ donations for a given year and the final row/column corresponds to the HDI ranking. The scatterplot matrix presents each pair of variables as a scatterplot.
Figure 6:
A scatterplot matrix of the organ donation data created using the ggpairs function from the GGally R package. The matrix contains of pairwise scatterplots for the following variables: the number of organ donations for each country each year from 2006 to 2014 and the country’s HDI ranking. Each point is colored by region as in Figure 5.
While the scatterplot matrix highlights the correlation between donation counts from one year to the next, unfortunately since we cannot follow a single country through time using these pairwise plots, it does not allow us to explore a time trend in the same way that the heatmap does.
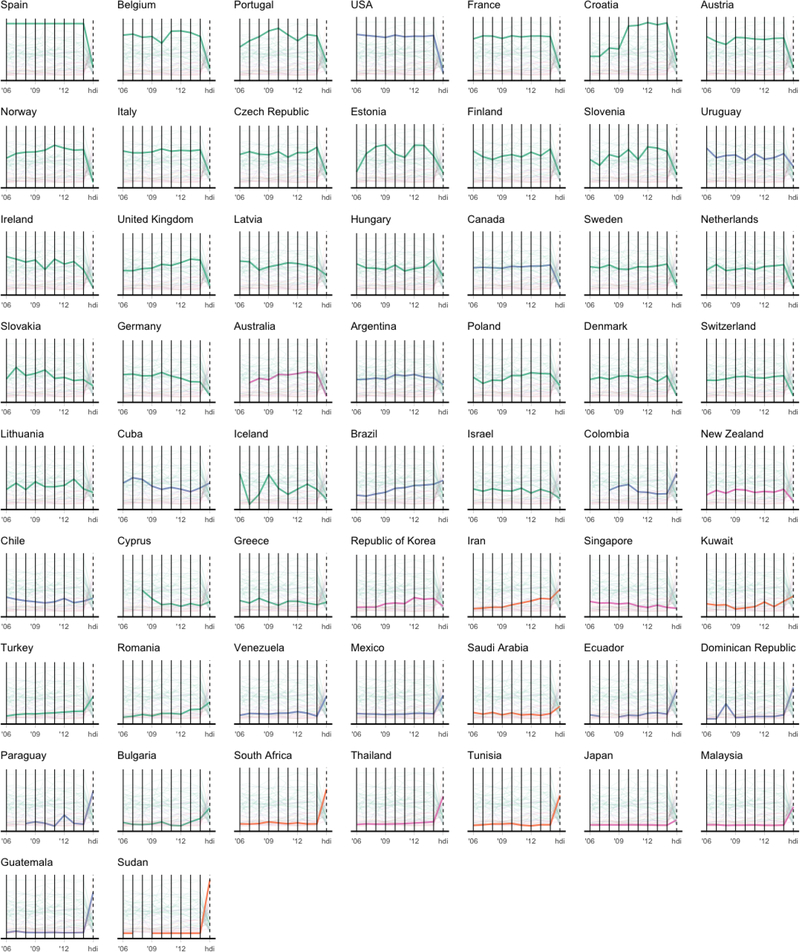
Another alternative presentation of this data is in the form of a parallel coordinates plot. Instead of showing all countries on a single parallel coordinates plot, we show a tiling of parallel coordinate plots wherein each panel highlights a single country in Figure 7. Each country is represented by a line and each vertical axis represents a variable (the first 9 variables are the donations per year from 2006 to 2014, and the final variable is the HDI ranking). Each variable is scaled so that the bottom of each vertical axis line represents the smallest observed value for that variable and the top corresponds to the largest observed value. The parallel coordinates plot allows us to follow each country’s donations over time and provides quite an effective representation of the donation trends over time for each country, as well as the country’s performance relative to the other countries.
Figure 7:
A series of parallel coordinates plots of the organ donation data built using the ggplot2 R package. Each country corresponds to a line that traverses a path from one variable to another. Each variable has been scaled so that the bottom of the vertical line representing the variable corresponds to the smallest observed value and the top corresponds to the largest observed value. Each country is colored based on region as in Figure 5.
In both Figure 6 and Figure 7, the HDI ranking is presented as the same type of variable as the donations per year. As a result, the comparison of each country’s organ donation trends with HDI is much less obvious than in the superheatmap from Figure 5. Moreover, the overall trend over time (the line plot above the heatmap in Figure 5) is absent from the parallel coordinate and scatterplot matrix versions.
In higher dimensional datasets, such as in our third case study in Section 5, neither a scatterplot matrix nor a parallel coordinates plot are appropriate due to unavoidable mass over-plotting. Thus, while all three of superheatmaps, scatterplot matrices, and parallel coordinates can be effective visualizations for data that reaches up to at most 50 dimensions, only the heatmap is able to handle datasets with hundreds or even thousands of rows or columns.
4. Case study II: uncovering clusters in language using Word2Vec
Word2Vec is an extremely popular group of algorithms for embedding words into high-dimensional spaces such that their relative distances to one another convey semantic meaning (Mikolov et al., 2013). The canonical example highlighting the impressiveness of these word embeddings is
That is, that if you take the word vector for “man”, subtract the word vector for “king” and add the word vector for “woman”, you approximately arrive at the word vector for “queen”. These algorithms are quite remarkable and represent an exciting step towards teaching machines to understand language.
In 2013, Google published pre-trained vectors trained on part of the Google News corpus, which consists of around 100 billion words. Their algorithm produced 300-dimensional vectors for 3 million words and phrases (Google (2013)).
The majority of existing visualization methods for word vectors focus on projecting the 300dimensional space to a low-dimensional representation using methods such as t-distributed stochas-tic neighbor embedding (t-SNE) (Maaten and Hinton, 2008).
4.1 Visualizing cosine similarity
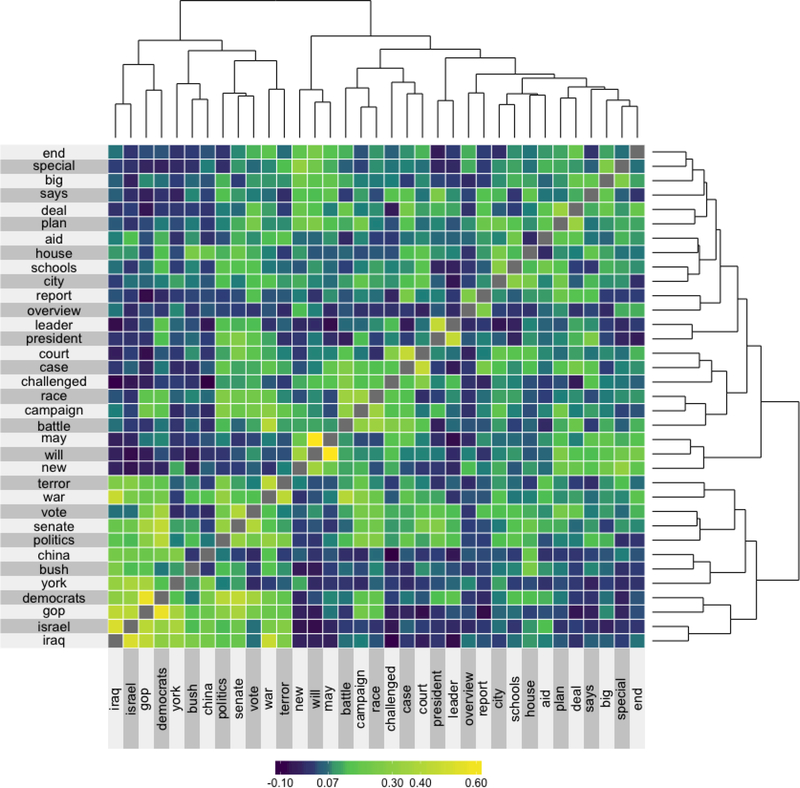
In this superheat case study we present an alternative approach to visualizing word vectors, which highlights contextual similarity. Figure 8 presents the cosine similarity matrix for the Google-News word vectors of the 35 most common words from the NY Times headlines dataset (from the RTextTools package). The rows and columns are ordered based on a hierarchical clustering and are accompanied by dendrograms describing this hierarchical cluster structure. From this super-heatmap we observe that words appearing in global conflict contexts such as “terror” and “war” have high cosine similarity (implying that these words appear in similar contexts). Words that are used in legal contexts such as “court” and “case” as well as words with political context such as “Democrats” and “GOP” also have high pairwise cosine similarity. The code used to prepare Figure 8 is provided in the Supplementary Materials.
Figure 8:
The cosine similarity matrix for the 35 most common words from the NY Times headlines that also appear in the Google News corpus. The rows and columns are ordered based on hierarchical clustering. This hierarchical clustering is displayed via dendrograms.
Although the example presented in Figure 8 displays relatively few words (we are presenting only the 35 most frequent words) and we have reached our capacity to be able to visualize each word individually on a single page, it is possible to use superheat to represent hundreds or thousands of words simultaneously by aggregating over word clusters.
4.2. Visualizing word clusters
Figure 9(a) displays the cosine similarity matrix for the Google News word vectors of the 855 most common words from the NY Times headlines dataset where the words are grouped into 11 clusters generated using the Partitioning Around Medoids (PAM) algorithm (Kaufman and Rousseeuw, 1990; Reynolds et al., 2006) applied to the rows/columns of the cosine similarity matrix. As PAM forces the cluster centroids to be data points, we represent each cluster by the word that corresponds to its center (these are the row and column labels that appear in Figure 9(a)). A silhouette plot is placed above the columns of the superheatmap in Figure 9(a), and the clusters are ordered in increasing average silhouette width.
The silhouette width is a traditional measure of cluster quality based on how well each object lies within its cluster, however we adapted its definition to suit cosine-based distance so that the cosine-silhouette width for data point i is defined to be:
where is the average cosine-dissimilarity of i with all other data within the same cluster (Ci is the index set of the cluster to which i belongs), and b(i) = minC≠Ci dcosine(xi,C) is the lowest average dissimilarity of i to any other cluster of which i is not a member. dcosine(x,y) is a measure of cosine “distance”, which is equal to (where scosine is standard cosine similarity).
The number of clusters (k = 11) was chosen based on the value of k that was optimal based on two types of criteria: (1) performance-based (Rousseeuw, 1987): the maximal average cosinesilhouette width, and (2) stability-based (Yu, 2013): the average pairwise Jaccard similarity based on 100 membership vectors each generated by a 90% subsample of the data. Plots of k versus average silhouette width and average Jaccard similarity are presented in the Appendix.
Word clouds displaying the words that are members of each of the 11 word clusters are presented in the Appendix. For example, the “government” cluster contains words that typically appear in political contexts such as “president”, “leader”, and “senate”, whereas the “murder” cluster contains words such as “case”, “drugs”, and “crime”.
Figure 9(b) presents a “smoothed” version of the cosine similarity matrix in panel (a), wherein the smoothed cluster-aggregated value corresponds to the median of the original values in the original “un-smoothed” matrix. The smoothing provides an aggregated representation of Figure 9(a) that allows the viewer to focus on the overall differences between the clusters. Note that the color range is slightly different between panels (a) and (b) due to the extreme values present in panel (a) being removed when we take the median in panel (b).
What we find is that the words in the “American” cluster have high silhouette widths, and thus is a “tight” cluster. This is reflected in the high cosine similarity within the cluster and low similarity between the words in the “American” cluster and words from other clusters. However, the words in the “murder” cluster have relatively high cosine similarity with words in the “government”, “struggle”, and “bombing” clusters. The clusters whose centers are not topic-specific such as “just” and “pushes” tend to consist of common words that are context agnostic (see their word clouds in the Appendix), and these clusters have fairly high average similarity with one another.
The information presented by Figure 9 far surpasses that of a standard silhouette plot: it allows the quality of the clusters to be evaluated relative to one another. For example, when a cluster exhibits low between-cluster separability, we can clearly see which clusters it is close to.
A brief comparison of how the clusters change when the number of clusters generated, k is increased from 11 to 12 is shown in the Appendix. We see that a new cluster is generated whose center is the word “children”, and which consists of words that originally came from a range of clusters (“just”, “struggle”, “government”, “murder”, “bombing”, and “companies”). Notice that one might have expected when k is increased from 11 to 12 that an existing cluster would split apart into two distinctive clusters. Instead, an entirely new cluster is formed from words from a combination of clusters and all of the previous clusters remained more or less in-tact. This observation is consistent over many different implementations of the clustering algorithm.
The code used to produce Figure 9 is provided in the supplementary materials.
5. Case study III: evaluation of heterogeneity in the performance of predictive models for fMRI brain signals from image inputs
Our final case study evaluates the performance of a number of models of the brain’s response to visual stimuli. This study is based on data collected from a functional Magnetic Resonance Imaging (fMRI) experiment performed on a single individual by the Gallant neuroscience lab at UC Berkeley (Vu et al., 2009, 2011).
fMRI measures oxygenated blood flow in the brain, which can be considered as an indirect measure of neural activity (the two processes are highly correlated). The measurements obtained from an fMRI experiment correspond to the aggregated response of hundreds of thousands of neurons within cube-like voxels of the brain, where the segmentation of the brain into 3D voxels is analogous to the segmentation of an image into 2D pixels.
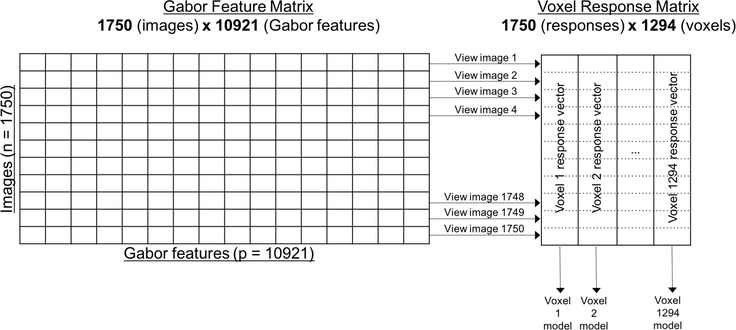
The data contains the fMRI measurements (averaged over 10 runs of the experiment) for each of 1,294 voxels located in the V1 region of the visual cortex of a single individual in response to viewings of 1,750 different images (such as a picture of a baby, a house or a horse). Each image is a 128 × 128 pixel grayscale image, which is represented as a vector of length 10,921 through a Gabor wavelet transformation (Lee, 1996). Figure 10 displays a graphical representation of the data structure.
Figure 10:
A diagram describing the fMRI data: a design matrix with 1,750 observations (images) and 10,921 features (Gabor wavelets) for each image, and a voxel response matrix consisting of 1,294 distinct voxel response vectors, where, for each voxel, the responses to each of the 1,750 images were collected. We fit a predictive model for each voxel using the Gabor feature matrix (1,294 models). The heatmap in Figure 11 corresponds to the voxel response matrix.
The data are hosted on the Collaborative Research in Computational Neuroscience repository (Kay et al. (2011)). However, unfortunately, only the voxel responses and raw images are available. The Gabor wavelet features are not provided.
5.1. Modeling brain activity
We developed a model for each voxel that predicts its response to visual stimuli in the form of greyscale images. Since each voxel responds quite differently to the image stimuli, instead of fitting a single multi-response model, we fit 1,294 independent Lasso models as in Vu et al. (2011).
The models are then evaluated based on how well they predict the voxel responses to a set of 120 withheld validation images.
5.2. Simultaneous performance evaluation of all 1,294 voxel-models
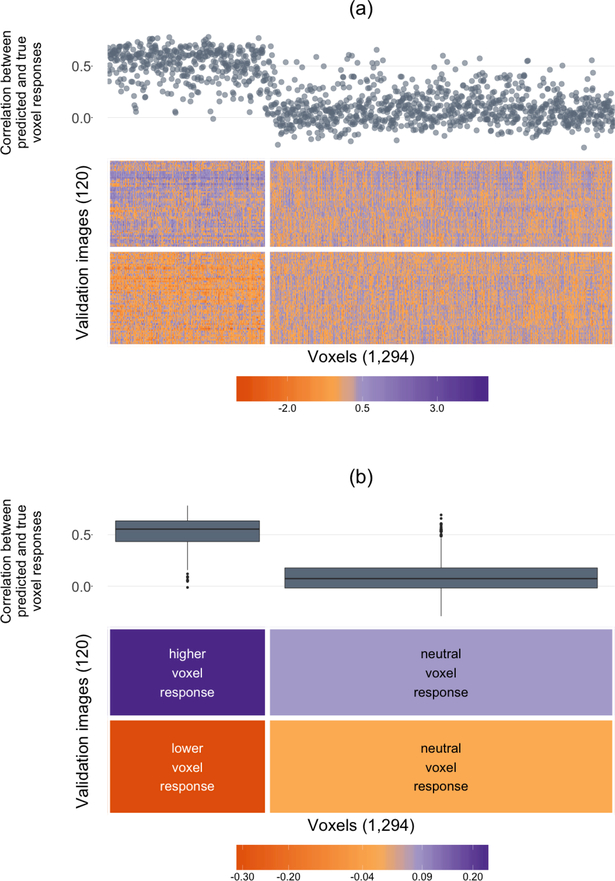
The voxel response matrix is displayed in Figure 11(a). The rows of the heatmap correspond to the 120 images from the validation set, while the columns correspond to the 1,294 voxels. Each cell displays the voxel’s response to the image. The rows and columns are clustered into two groups using K-means. As in Figure 9(a), the heatmap is extremely grainy. Figure 11(b) displays the same heatmap with the cell values smoothed within each cluster (by taking the median value).
Figure 11:
A superheatmap displaying the validation set voxel response matrix (Panel (a) displays the raw matrix, while Panel (b) displays a smoothed version). The images (rows) and voxels (columns) are each clustered into two groups (using K-means). The left cluster of voxels are more “sensitive” wherein their response is different for each group of images (higher than the average response for top cluster images, and lower than the average response for bottom cluster images), while the right cluster of voxels are more “neutral” wherein their response is similar for both image clusters. Voxel-specific Lasso model performance is plotted as correlations above the columns of the heatmap (as a scatterplot in (a) and cluster-aggregated boxplots in (b)).
The Appendix displays four randomly selected images from each of the two image clusters. We find that the bottom image cluster consists of images for which the subject is easily identifiable (e.g. Princess Diana and Prince Charles riding in a carriage, a bird, or an insect), whereas the contents of images from the top cluster of images are less easy to identify (e.g. rocks, a bunch of apples, or an abstract painting). Further, from Figure 11, it is clear that the brain is much more active in response to the images from the top cluster (whose contents were less easily identifiable) than to images from the bottom cluster.
Furthermore, there are two distinct groups of voxels:
Sensitive voxels that respond very differently to the two groups of images (for the top image cluster, their response is significantly lower than the average response, while for the bottom image cluster, their response is significantly higher than the average response).
Neutral voxels that respond similarly to both clusters of images.
In addition, above each voxel (column) in the heatmap, the correlation of that voxel-model’s predicted responses with the voxel’s true response is presented (as a scatterplot in Panel (a) and as aggregate boxplots in Panel (b)).
It is clear that the models for the voxels in the first (sensitive) cluster perform significantly better than the models for the voxels in the second (neutral) cluster. That is, the responses of the voxels that are sensitive to the image stimuli are much easier to predict (the average correlation between the predicted and true responses was greater than 0.5) than the responses of the voxels whose responses are neutral (the average correlation between the predicted and true responses was close to zero).
Further examination revealed that the neutral voxels were primarily located on the periphery of the V1 region of the visual cortex, whereas the sensitive voxels tended to be more centrally located.
Although a standard histogram of the predicted and observed response correlations would have revealed that there were two groups of voxels (those whose responses we can predict well, and those whose responses we cannot), superheat allowed us to examine this finding in context. In particular, it allowed us to take advantage of the heterogeneity present in the data: we were able to identify that the voxels whose response we were able to predict well were exactly the voxels whose response was sensitive to the two clusters of images.
Note that we also ran Random Forest models for predicting the voxel responses and found the same results, however, the overall correlation was approximately 0.05 higher on average.
The code used to produce Figure 11 is provided in the supplementary materials.
6. Conclusion
In this paper, we have proposed the superheatmap that augments traditional heatmaps via the inclusion of extra information such as a response variable as a scatterplot, model results as boxplots, correlation information as barplots, text information, and more. These augmentations provide the user with an additional avenue for information extraction, and allow for exploration of heterogeneity within the data. The superheatmap, as implemented by the superheat package written by the authors, is highly customizable and can be used effectively in a wide range of situations in exploratory data analysis and model assessment. The usefulness of the superheatmap was high-lighted in three case studies. The first combined multiple sources of data to assess the relationship between organ donation and country development worldwide. The second explored the structure of the English language by visualizing word clusters from Word2Vec data, while highlighting the hierarchical nature of these word groupings. Finally, the third case study evaluated heterogeneity in the performance of Lasso models designed to predict fMRI brain signals in response to visual stimuli in the form of image viewings. We hope that we have demonstrated clearly that the heatmap is an extremely useful data visualization tool, particularly for high-dimensional datasets.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the Gallant Lab at UC Berkeley for providing the fMRI data. This research is partially supported by NSF grants DMS-1107000, CDS&E-MSS 1228246, DMS-1160319 (FRG), NHGRI grant 1U01HG007031–01 (ENCODE), AFOSR grant FA9550–14-1–0016, and the Center for Science of Information (CSoI), an USNSF Science and Technology Center, under grant agreement CCF-0939370.
Footnotes
SUPPLEMENTARY MATERIAL
Appendices: appendix word clusters.pdf, appendix word clouds.pdf, and appendix image examples.pdf: pdfs showing additional information for the Word2Vec and fMRI analyses.
knitr documents: organ.rmd, word.rmd, fMRI.rmd: R markdown documents that provide the code for cleaning the data and producing the heatmaps for each of our case studies.
Package GitHub repository: https://github.com/rlbarter/superheat
Online vignette: https://rlbarter.github.io/superheat/
Contributor Information
Rebecca L. Barter, Department of Statistics, University of California, Berkeley
Bin Yu, Department of Statistics, University of California, Berkeley.
References
- Abouna GM (2008). Organ Shortage Crisis: Problems and Possible Solutions. Transplantation Proceedings 40(1), 34–38. [DOI] [PubMed] [Google Scholar]
- Andrews DF (1972). Plots of High-Dimensional Data. Biometrics 28(1), 125–136. [Google Scholar]
- Brinton W (1914). Graphic Methods for Presenting Facts,. New York: The Engineering Magazine Company. [Google Scholar]
- Bujack R, Turton TL, Samsel F, Ware C, Rogers DH, and Ahrens J (2018, January). The Good, the Bad, and the Ugly: A Theoretical Framework for the Assessment of Continuous Colormaps. IEEE Transactions on Visualization and Computer Graphics 24(1), 923–933. [DOI] [PubMed] [Google Scholar]
- Chen C (2002). Generalized Association Plots: Information Visualization via Iteratively Gener-ated Correlation Matrices. Statistica Sinica (12), 7–29. [Google Scholar]
- Cleveland WS (1993). Visualizing Data. At&T Bell Laboratories. [Google Scholar]
- Cook D, Hofmann H, Lee E-K, Yang H, Nikolau B, and Wurtele E (2007). Exploring gene expression data, using plots. Journal of Data Science 5(2), 151. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, and Botstein D (1998). Cluster analysis and display of genome-wide expression patterns. PNAS 95(25), 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili T, OCallaghan A, Sidi J, and Sievert C (2017, October). heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia GG, Harden PN, and Chapman JR (2012). The global role of kidney transplantation. Kidney International 81(5), 425–427. [DOI] [PubMed] [Google Scholar]
- Gaujoux R and Seoighe C (2010, July). A flexible R package for nonnegative matrix factorization. BMC bioinformatics 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GODT (2016). Global Observatory on Donation and Transplantation Database.
- Google (2013). Google Code Archive.
- Gower J and Digby P (1981). Expressing Complex Relationships in Two Dimensions Interpreting Multivariate Data, ed. Barnett V, Chichester, U.K.: Wiley, 83–118. [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016, September). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18), 2847–2849. [DOI] [PubMed] [Google Scholar]
- Harrower M and Brewer CA (2003, June). ColorBrewer.org: An Online Tool for Selecting Colour Schemes for Maps. The Cartographic Journal 40(1), 27–37. [Google Scholar]
- Inselberg A (1985). The plane with parallel coordinates. The Visual Computer 1(2), 69–91. [Google Scholar]
- Inselberg A (1998). Visual Data Mining with Parallel Coordinates SSRN Scholarly Paper ID 85868, Social Science Research Network, Rochester, NY. [Google Scholar]
- Inselberg A and Dimsdale B (1987). Parallel Coordinates for Visualizing Multi-Dimensional Geometry. In Kunii DTL (Ed.), Computer Graphics 1987, pp. 25–44. Springer; Japan. [Google Scholar]
- Kaufman L and Rousseeuw PJ (1990). Partitioning Around Medoids (Program PAM) In Finding Groups in Data, pp. 68–125. John Wiley & Sons, Inc. [Google Scholar]
- Kay K, Naselaris T, and Gallant J (2011). fMRI of human visual areas in response to natural images. CRCNS.org.
- Kolde R (2012). Pheatmap: pretty heatmaps. R package version 61. [Google Scholar]
- Lee TS (1996). Image representation using 2d Gabor wavelets. IEE Transactions on Pattern Analysis and Machine Intelligence 18(10), 959–971. [Google Scholar]
- Ling R (1973). A Computer Generated Aid for Cluster Analysis. Communications of the ACM (16), 355–361. [Google Scholar]
- Loua T (1873). Atlas statistique de la population de Paris.
- Maaten L. v. d. and Hinton G (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9(Nov), 2579–2605. [Google Scholar]
- Mikolov T, Chen K, Corrado G, and Dean J (2013). Efficient Estimation of Word Representa-tions in Vector Space. arXiv:1301.3781 [cs]. arXiv: 1301.3781.
- Reynolds AP, Richards G, Iglesia B. d. l., and Rayward-Smith VJ (2006). Clustering Rules: A Comparison of Partitioning and Hierarchical Clustering Algorithms. Journal of Mathematical Modelling and Algorithms 5(4), 475–504. [Google Scholar]
- Rousseeuw PJ (1987). Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics 20, 53–65. [Google Scholar]
- Schep AN and Kummerfeld SK (2017, August). iheatmapr: Interactive complex heatmaps in R
- Tajima J (1983, March). Uniform color scale applications to computer graphics. Computer Vision, Graphics, and Image Processing 21(3), 305–325. [Google Scholar]
- Tardu M, Bulut S, and Kavakli IH (2017, January). MerR and ChrR mediate blue light induced photo-oxidative stress response at the transcriptional level in Vibrio cholerae. Scientific Reports 7, 40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakhtenberg EF, Pho N, Holton KM, Chittenden TW, Goldberg JL, and Dong L (2016, August). Cell types differ in global coordination of splicing and proportion of highly expressed genes. Scientific Reports 6, 32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNDP (2015). Human Development Data. [Google Scholar]
- Vu VQ, Ravikumar P, Naselaris T, Kay KN, Gallant JL, and Yu B (2011). Encoding and decoding V1 fMRI responses to natural images with sparse nonparametric models. Annals of Applied Statistics 5(2), 1159–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu VQ, Yu B, Naselaris T, Kay K, Gallant J, and Ravikumar PK (2009). Nonparametric sparse hierarchical models describe V1 fMRI responses to natural images In Koller D, Schu-urmans D, Bengio Y, and Bottou L (Eds.), Advances in Neural Information Processing Systems 21, pp. 1337–1344. Curran Associates, Inc. [Google Scholar]
- Wickham H (2010). A Layered Grammar of Graphics. Journal of Computational and Graphical Statistics 19(1). [Google Scholar]
- Wickham H (2016). ggplot2: elegant graphics for data analysis Springer. [Google Scholar]
- Wilkinson L (1994). SYSTAT for DOS: Advanced Applications, Version 6. Evanston, IL: SYSTAT Inc.. [Google Scholar]
- Wilkinson L and Friendly M (2009). The History of the Cluster Heat Map. The American Statistician 63(2), 179–184. [Google Scholar]
- Wong B (2011, June). Points of view: Avoiding color. [DOI] [PubMed]
- Yu B (2013). Stability. Bernoulli 19(4), 1484–1500. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.