Water from the Hickey Run Tributary of the Anacostia River is being collected quarterly (beginning August 2018) and analyzed to create high-resolution baseline taxonomic profiles of microbiota associated with this important aquatic ecosystem, which has a long history of exposure to residential and commercial effluents from Washington, DC. These United States National Arboretum Microbial Observatory data are available under NCBI BioProject number PRJNA498951.
ABSTRACT
Water from the Hickey Run Tributary of the Anacostia River is being collected quarterly (beginning August 2018) and analyzed to create high-resolution baseline taxonomic profiles of microbiota associated with this important aquatic ecosystem, which has a long history of exposure to residential and commercial effluents from Washington, DC. These United States National Arboretum Microbial Observatory data are available under NCBI BioProject number PRJNA498951.
ANNOUNCEMENT
When the National Arboretum Act became law in 1927, a national garden long envisioned by U.S. presidents was established as a research center to advance the scientific and economic prosperity of American agriculture (1). The mission of the United States National Arboretum (USNA) includes study and management of plants for long-term contributions to environmental sustainability and agricultural prosperity (1), a goal inextricably linked with water health. The 444 acres that comprise the USNA are situated between Mount Hamilton and the Anacostia River and are bisected by Hickey Run. Thus, study of microbiota from Hickey Run will provide valuable information about this aquatic ecosystem and contribute to our understanding of its intersection with the more than 16,000 cultivars, natural landscapes, and soils of the USNA. These data will also support a broader range of scientific inquiry, spanning objectives for sustainable stewardship of natural resources to public health surveillance of eukaryotic and bacterial pathogens.
Collection of 100 liters of water from each of 2 sites along Hickey Run (38.912341, −76.96536, and 38.9147, −76.966873) is being carried out quarterly using ultrafiltration according to previously described methods (2). To date, collections occurred on 28 August 2018, 26 November 2018, and 27 February 2019, with future collections planned for May and August of 2019. Metadata, including dates, times, location (latitude and longitude), temperature, additional site details, and photos, have been stored in the EpiCollect5 (3) project at the USNA Microbial Observatory, which will be made public upon completion of one full year of sample collection. Following ultrafiltration with Hemodialyzer Rexeed 25S filters (AsahiKasei, Chiyoda, Tokyo, Japan), using a Geopump peristaltic pump (Geotech, Denver, CO), filters are capped, bagged and stored at 4°C, until backflushing, centrifugation (using an Eppendorf 5810 instrument at 4,000 rpm for 10 min), and homogenization of filtrate. Aliquots of homogenate are then used for culture-independent DNA extraction (DNeasy Power soil kit; Qiagen, Germantown, MD) and enrichment of specific taxa, such as Salmonella enterica and Escherichia coli, according to methods described in the Bacteriological Analytical Manual (BAM) of the Food and Drug Administration (FDA) (4, 5) and for quasimetagenomic profiling (shotgun sequencing) of the 24-h point of enrichment (6–8). DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Germantown, MD), and the library was prepared using the Nextera DNA Flex library prep kit (Illumina, San Diego, CA) according to the manufacturers’ specifications. Libraries were sequenced on an Illumina NextSeq 550 system using a NextSeq 500/550 high-output kit v2 (300 cycles, 2 × 150 bp).
Reads were trimmed with Trimmomatic (9), and preliminary taxonomic annotation was achieved using the CosmosID analytic pipeline (December 2018 database; Rockville, MD) (8, 10, 11). Phylotypes of E. coli were described using Center for Food Safety and Applied Nutrition (CFSAN) (FDA) in-house k-mer- and single-nucleotide polymorphism (SNP)-based pipelines (12). Additional boutique pipelines for description of protist species were used to annotate relevant eukaryotic species (13). Data from the 28 August 2018 sampling time point averaged 4.1 Gb (∼27 million reads) per replicate.
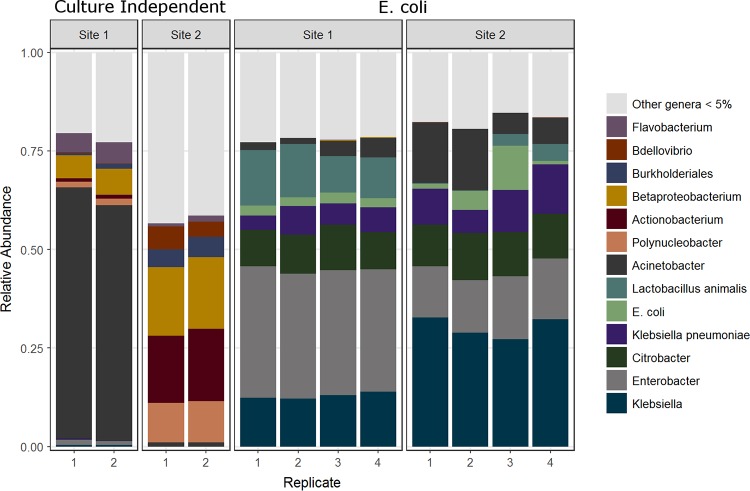
The following bacterial taxa were observed (in order of relative abundance): Acinetobacter spp. (including Acinetobacter junii), Bdellovibrio, Flavobacterium, Betaproteobacterium, Polynucleobacter, Burkholderidales, Enterobacter cloacae, Klebsiella spp., Lactobacillus animalis, Aeromonas caviae, Aeromonas hydrophilia, and Clostridium spp (Fig. 1). As many as 34 official H and 112 official O antigens of Escherichia coli were observed, in addition to Shigella sonnei wzx and wzy genes. The following protists were described: Dictyostelium citrinum, Thalassiosira, Paramecium biaurelia, Balantidium coli, and Balantidium hominis, Naegleria, Toxoplasma gondii, Saprolegnia parasitica, Chatonella, and Pseudo-nitzschia.
FIG 1.
Microbial taxa from water collected at two sites along Hickey Run from DNA extractions of culture-independent and of 24-h E. coli enrichments.
Data availability.
The USNA Microbial Observatory data from the 28 August 2018 collection time point are available under National Center for Biological Information (NCBI) BioProject number PRJNA498951 and SRA accession numbers SRR8126707 to SRR8126738. Data from subsequent time points will be added to the BioProject as completed.
REFERENCES
- 1.USNA Strategic Planning Team. 2013. U.S. National Arboretum strategic plan 2013–2017. Office of National Programs, USDA, Agricultural Research Service, Beltsville, MD. [Google Scholar]
- 2.Mull B, Hill VR. 2012. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J Microbiol Methods 91:429–433. doi: 10.1016/j.mimet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aanensen DM, Huntley DM, Feil EJ, al-Own F, Spratt BG. 2009. EpiCollect: linking smartphones to Web applications for epidemiology, ecology and community data collection. PLoS One 4:e6968. doi: 10.1371/journal.pone.0006968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng P, Weagant SD, Grant MA, Burkhardt W. 2017. Enumeration of Escherichia coli and the coliform bacteria, chapter 4. In Bacteriological analytical manual. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 5.Andrews WH, Wang H, Jacobson A, Hammack T. 2018. Salmonella, chapter 5. In Bacteriological analytical manual. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 6.Ottesen A, Ramachandran P. 2019. Food microbiomes, a new paradigm of food ecology In Doyle MP, Buchanan RL (ed), Food microbiology: fundamentals and frontiers. ASM Press, Washington, DC. [Google Scholar]
- 7.Allard MW, Bell R, Ferreira CM, Gonzalez-Escalona N, Hoffmann M, Muruvanda T, Ottesen A, Ramachandran P, Reed E, Sharma S, Stevens E, Timme R, Zheng J, Brown EW. 2018. Genomics of foodborne pathogens for microbial food safety. Curr Opin Biotechnol 49:224–229. doi: 10.1016/j.copbio.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, Subramanian P, Ryan G, Jarvis K, Grim C, Daquiqan N, Hanes D, Allard M, Colwell R, Brown E, Chen Y. 2016. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol 16:275. doi: 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasan NA, Young BA, Minard-Smith AT, Saeed K, Li H, Heizer EM, McMillan NJ, Isom R, Abdullah AS, Bornman DM, Faith SA, Choi SY, Dickens ML, Cebula TA, Colwell RR. 2014. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One 9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vázquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayford AE, Mammel MK, Lacher DW, Brown EW. 2011. Single nucleotide polymorphism (SNP)-based differentiation of Shigella isolates by pyrosequencing. Infect Genet Evol 11:1761–1768. doi: 10.1016/j.meegid.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Commichaux S, Ramachandran P, Reed E, Ottesen A, Strain E, Pop M. 2018. Using mitochondrial and plastid genomes to characterize protists in shotgun metagenomic data, poster 424A. Int Soc Microb Ecol. Leipzig, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The USNA Microbial Observatory data from the 28 August 2018 collection time point are available under National Center for Biological Information (NCBI) BioProject number PRJNA498951 and SRA accession numbers SRR8126707 to SRR8126738. Data from subsequent time points will be added to the BioProject as completed.