Abstract
Background
The prevalence of carbapenem‐resistant Enterobacteriaceae (CRE) is alarming worldwide causing serious infections. Rapid and accurate identification of CRE is crucial to reduce the mortality and morbidity. In this study, we tried to develop an in‐house Carba NP test for detection of CRE and evaluate its performance with others.
Methods
A prospective study was conducted with 40 nonrepeating Enterobacteriaceae isolates over a period of 3 months. All the isolates were screened for carbapenem resistance as per CLSI 2016 guidelines followed by PCR for blaNDM‐1, blaOXA‐48, blaKPC, blaVIM, and blaIMP genes. All the isolates were subjected to five phenotypic tests, that is, in‐house Carba NP (iCarba NP), commercial Carba NP (cCarba NP), Blue‐Carba, modified Hodge test (MHT), and CHROMagar.
Results
Among the 40 isolates, 87.5% were identified as Escherichia coli, 7.5% were Klebsiella pneumoniae, 2.5% were Enterobacter cloacae, and 2.5% were Citrobacter freundii. Thirty‐three of 40 (82.5%) isolates were found to harbor one or more resistant genes. Considering PCR to be the gold standard test, sensitivity of the phenotypic methods for CRE detection ranged from 63.6% (MHT) to 96.9% (CHROMagar). Both cCarba NP and iCarba NP observed to have highest specificity. The performance of iCarba NP was found comparable with cCarba NP by kappa score 1 and found approximately 10 times less expensive than cCarba NP.
Conclusion
CHROMagar was observed most sensitive assay for detection of CRE followed by both Carba NP tests. iCarba NP was proved cheaper and equally good as cCarba NP for detection of CRE.
Keywords: Carba NP test, carbapenem‐resistant Enterobacteriaceae, CHROMagar, modified Hodge test
Abbreviation
- BCarba
Blue‐Carba
- B‐PERII
Bacterial Protein Extraction Reagent
- cCarba NP
commercial Carba NP
- CDC
Centers for Disease Control and Prevention
- CLSI
Clinical and Laboratory Standards Institute
- CP‐CRE
carbapenamase‐producing carbapenem‐resistant Enterobacteriaceae
- CRE
carbapenem‐resistant Enterobacteriaceae
- iCarba NP
In‐house Carba NP
- mCIM
modified carbapenemase inhibition method
- MHA
Mueller‐Hinton agar
- MHT
modified Hodge test
- NPV
negative predictive value
- PPV
positive predictive value
1. INTRODUCTION
Carbapenem‐resistant Enterobacteriaceae (CRE) has become a major clinical challenge globally. Centers for Disease Control and Prevention (CDC), the United States, estimated 9300 patients infected with CRE causing 610 deaths annually.1 Carbapenem resistance is manifested either due to production of carbapenemase enzymes or due to noncarbapenemase mechanisms, for example, alterations in porin channels, mutation in the expression of efflux pumps, or AmpC production.2 Carbapenamase‐producing carbapenem‐resistant Enterobacteriaceae (CP‐CRE) is observed as a major threat to healthcare system due to its ability to disseminate rapidly among various bacterial clones through plasmids. The most prevalent carbapenemase enzymes include blaNDM‐1, blaOXA‐48, blaOXA‐23, blaKPC, blaVIM, and blaIMP.3 In addition, these isolates often carry other non–β‐lactam resistance determinants that give rise to multi–drug‐resistant and pan–drug‐resistant isolates.4 Rapid and accurate identification of CRE is crucial not only to reduce the mortality and morbidity, but also to implement infection control measures. The presence of multiple resistance mechanisms among CREs makes their identification difficult.5 Currently, several phenotypic tests (e.g, Carba NP test, MHT, CHROMagar, modified carbapenemase inhibition method [mCIM], and Blue‐Carba [BCarba]) and molecular methods are available for the detection of CRE. However, all these methods have their own limitations in terms of sensitivity, specificity, rapidity, cost‐effectiveness, and availability of laboratory facility.
Here, in this study we tried to develop an iCarba NP test for rapid and accurate identification of CP‐CRE. We also tried to evaluate its performance with other available phenotypic methods cCarba NP (RAPIDECw CARBA NP), BCarba, MHT, and CHROMagar.
2. MATERIALS AND METHODS
A prospective study was conducted with 40 nonrepeating Enterobacteriaceae isolates over a period of 3 months (from November 2016 to January 2017). All the isolates were phenotypically identified by the MALDI‐TOF (BioMerieux, Durham, NC, USA).
2.1. Screening test for CRE
All the 40 isolates were screened for carbapenem resistance as per Clinical and Laboratory Standards Institute (CLSI) 2016 guidelines using four carbapenem disks (imipenem 10 μg, meropenem 10 μg, ertapenem 10 μg, and doripenem 10 μg).6 Isolates were further confirmed by PCR for five carbapenamase genes (blaNDM‐1, blaOXA‐48, blaKPC, blaVIM, and blaIMP) using published primers.7 PCR was considered as gold standard test to evaluate the performance of all the assays.
2.2. In‐house Carba NP
The iCarba NP test was performed using the following method. Two solutions were prepared.
2.2.1. Solution A
One microliter solution (made up of 3 mg of imipenem monohydrate + 0.5% phenol red solution + 0.1 mmol/L ZnSO4 at pH 7.8).
2.2.2. Solution B
One microliter solution (made up of 0.5% phenol red solution + 0.1 mmol/L ZnSO4 at pH 7.8).
Two calibrated loops (10 μL) of the tested isolates from Mueller‐Hinton agar (MHA) plate were suspended in 200 μL of Bacterial Protein Extraction Reagent (B‐PERII). The mixture was vortexed for 1 minute and incubated at room temperature for 30 minutes. One hundred microliters of the bacterial suspension was put in a 96‐well U‐bottom microtiter plate in two adjacent wells as duplicate. One hundred microliters of solution A and 100 μL of solution B were added in the first column and in adjacent column, respectively. The plate was incubated at 37°C for 2 hours. Klebsiella pneumoniae ATCC‐BAA 1705 and ATC C‐BAA 1706 were used as positive control and negative control, respectively. Any color change from red to yellow or red to orange of the test solution was considered as positive test result, whereas red‐orange was considered as indeterminate. No color change was considered as negative. Any color change in solution B was considered as invalid result6 (Figure 1).
Figure 1.
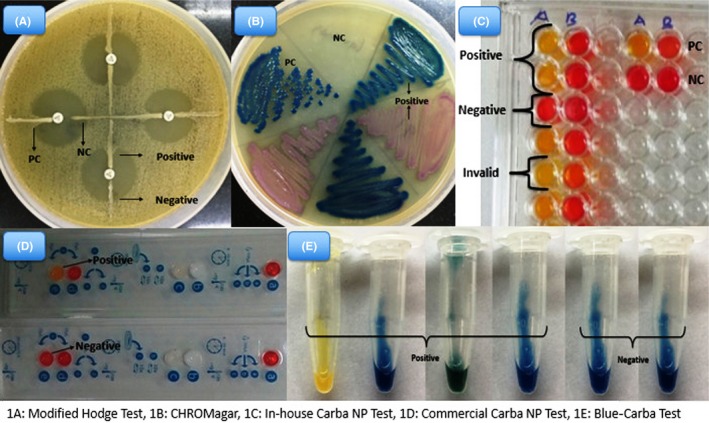
Figure showing various phenotypic tests for detection of carbapenem resistance
2.3. Commercial Carba NP (cCarba NP)
The RAPIDEC CARBA NP test was performed and interpreted according to the manufacturer's instructions.
2.4. BCarba
The BCarba test was performed as follows.8
2.4.1. Test solution
Test solution was prepared by mixing 3 mg of imipenem monohydrate in 1‐mL of 0.04% bromothymol blue solution containing 0.1 mmol/L ZnSO4 and adjusted to pH 7.
2.4.2. Control solution
Control solution was prepared by mixing 1 mL of 0.04% bromothymol blue solution containing 0.1 mmol/L ZnSO4. pH was adjusted to 7.
Two calibrated loops (10 μL) of the test isolate from Mueller‐Hinton agar were suspended in 100 μL of test and 100 μL of control solutions in two different Eppendorf tubes. The mixture was homogenized by vortexing for 15‐30 seconds and incubated at 37°C for 2 hours with agitation (150 rpm). Klebsiella pneumoniae (ATCC‐BAA 1705 and ATCC‐BAA 1706) were used as positive and negative control strains, respectively. Change in color either from blue to green, blue to yellow, or green to yellow was considered positive. No change in initial green or blue was considered negative, whereas any color change in control solution was treated as invalid.
2.5. Modified Hodge test
Modified Hodge test was performed as per CLSI 2016 guidelines.6
2.6. CHROMagar KPC
All the isolates were inoculated on chromogenic medium, that is, CHROMagar KPC (CHROMagar Company, Paris, France). The result was interpreted after 18‐24 hours of aerobic incubation as per the manufacturer's instruction.
2.7. Statistical analysis
The performance of all the assays was calculated in terms of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Kappa value was calculated to check the inter‐rater agreement between all the assays.
3. RESULTS
Among 40 Enterobacteriaceae isolates, 87.5% (35/40) were Escherichia coli, 7.5% (3/40) were Klebsiella pneumoniae, 2.5% (1/40) were Enterobacter cloacae, and 2.5% (1/40) were Citrobacter freundii. Using carbapenem disk diffusion test, 80%(32/40) of the total isolates were resistant, 7.5% (3/40) were intermediate, and 12.5% (5/40) were sensitive. Thirty‐three of 40 isolates were found to harbor one or more resistant genes by PCR. Nineteen of 33 (57.6%) contain blaNDM‐1, 6 (18.2%) contain blaOXA‐48, and only 1 (3%) contain blaIMP gene. Seven isolates (21.2%) observed to contain both blaNDM‐1 and blaOXA‐48 genes. All the isolates were found negative for blaKPC and blaVIM genes. The bacterial isolates of this study were isolated from rectal swabs of hematological malignancy patients receiving chemotherapy. Thirty‐six of 40 patients (90%) had history of antibiotic intake (either single or multiple including β‐lactams) within previous 30 days as a course of their treatment or prophylaxis. As the study was planned with one‐time screening of patients during the first visit, the outcome could not be observed. Thirty of 36 (83.3%) patients who were on antibiotics observed to harbor one or more carbapenamase genes. Three of rest 4 (75%) patients without previous antibiotic history also showed the presence of carbapenamase gene, that is, only blaNDM‐1.
Considering PCR as gold standard, the sensitivity and specificity of all the phenotypic assays were calculated (Table 1). Our methodology for iCarba NP is a modification of the original test process described in CLSI in terms of increasing the inoculum size and additional incubation step after the addition of protein extraction reagent. The sensitivity of the phenotypic methods in this study ranged from 63.6% to 96.9%. CHROMagar was observed to have the highest sensitivity followed by the cCarba NP. Both cCarba NP and iCarba NP had highest specificity in comparison with other assays. The performance of iCarba NP was comparable to cCarba NP. Overall, iCarba NP and cCarba NP performed well for the detection of metallo‐β‐lactamases (blaNDM‐1, blaIMP); however, only one isolate positive for blaOXA‐48 could not be detected. Among all the assays, MHT observed to have lowest sensitivity (63.6%) and specificity (71.4%), respectively. iCarba NP and cCarba NP had observed highest PPV followed by BCarba. NPV was ranged from 83.3% for CHROMagar to 29.4% in case of MHT. PPV and NPV of iCarba NP and cCarba NP were same. Considering CHROMagar as standard test, kappa value for iCarba NP and cCarba NP was observed same, that is, 0.7, suggesting good agreement between the CHROMagar assay and the two Carba NP tests. Moreover, considering cCarba NP as the standard, kappa value of iCarba NP was calculated to be 1. We also compared time to positivity of the result for both iCarba NP and cCarba NP assays at different time interval, that is, 15 minutes, 30 minutes, 1 hour, and 2 hours of putting up of the test (Table 2). More than 60% of isolates were detected positive within 15 minutes of putting up of the test. On further analysis, it was found that 65% of these isolates which gave positive result within 15 minutes of the test were carrying metallo‐β‐lactamase (blaNDM‐1, blaIMP) gene with or without blaOXA‐48, whereas only 16.7% isolates carrying blaOXA‐48 gene alone were positive by 15 minutes. Comparing the price of both the Carba NP tests, iCarba NP test was found 10 times cheaper than cCarba NP.
Table 1.
Comparison of the iCarba NP with other phenotypic methods for detection of CP‐CRE
| Phenotypic tests | Positive (%) | Negative (%) | Indeterminate (%) | Sensitivity (95% CI) | Specificity (95% CI) | Positive predictive value (95% CI) | Negative predictive value (95% CI) | Unweighted kappa value |
|---|---|---|---|---|---|---|---|---|
| iCarba NP (n = 40) | 33 (82.5%) | 4 (10%) | 3 (7.5%) | 93.9% (78.3‐98.9) | 71.4% (30.2‐94.8) | 93.9% (78.3‐98.9) | 71.4% (30.2‐94.8) | 0.653 |
| cCarba NP (n = 40) | 33 (82.5%) | 3 (7.5%) | 4 (10%) | 93.9% (78.3‐98.9) | 71.4% (30.2‐94.8) | 93.9% (78.3‐98.9) | 71.4% (30.2‐94.8) | 0.653 |
| MHT (n = 40) | 23 (57.5%) | 16 (40%) | 1 (2.5%) | 63.6% (45.1‐79) | 71.4% (30.2‐94.9) | 91.3% (70.5‐98.5) | 29.4% (11.4‐56) | 0.724 |
| CHROMagar (n = 40) | 34 (85%) | 6 (15%) | 0 (0%) | 96.9% (82.4‐99.8) | 71.4% (30.2‐94.8) | 94.1% (78.9‐98.9) | 83.3% (36.4‐99.1) | 0.224 |
| BCarba (n = 40) | 35 (87.5%) | 5 (12.5%) | 0 (0%) | 96.9% (82.5‐94.8) | 57.1% (20.2‐88.2) | 91.4% (75.8‐97.7) | 80% (29.8‐98.9) | 0.609 |
Kappa value ≥0.6 is significant.
MHT, modified Hodge test; iCarba NP, in‐house Carba NP test; cCarba NP, commercial Carba NP test; BCarba, Blue‐Carba.
Table 2.
Comparison of time to positivity and cost between iCarba NP and cCarba NP
| Phenotypic tests | After 15 min | After 30 min | After 1 h | After 2 h | Cost per test in ₹ |
|---|---|---|---|---|---|
| iCarba NP (n = 33) | 21 | 5 | 2 | 5 | 35 |
| cCarba NP (n = 33) | 22 | 6 | 3 | 2 | 300 |
BCarba, Blue‐Carba; cCarba NP, commercial Carba NP test; iCarba NP, in‐house Carba NP test.
4. DISCUSSION
The prevalence of CRE is increasing worldwide causing serious community‐acquired and nosocomial infections. Carbapenem resistance is exhibited by two main mechanisms, firstly due to the presence of carbapenemase enzyme, and secondly due to the loss of porin function or expression in the efflux pumps.2 Identification of CP‐CRE can be made by several phenotypic methods. This includes rapid colorimetric‐based assays (Carba NP test); growth‐based assays (MHT), carbapenem hydrolysis assays; and immunochromatogenic assays. Molecular methods such as PCR and sequencing can also be used for the detection of carbapenemase genes, but less commonly practiced being costly and need good laboratory facility.9 Among the carbapenemases, blaNDM‐1 gene was observed to be present in both the groups of patients with or without history of intake of antibiotics. Hence, the presence of blaNDM‐1 gene may not be directly related to previous antibiotic intake. Carba NP is a colorimetric assay based on enzymatic hydrolysis of the β‐lactam ring of carbapenem group of drugs. Sensitivity and specificity of this assay have been reported to be very high, that is, 90%‐100% and 100%, respectively.10, 11, 12, 13, 14, 15, 16 However, less optimal results were observed for the detection of carbapenemases enzymes with low hydrolytic activity, particularly for blaOXA‐48 and blaGES‐5.12, 14 Increased sensitivity for detection of blaOXA‐48 had been already established in literatures by increasing the inoculum size.9 It was also proven that there might be buffer inhibition effect by B‐PER II, which reduces the sensitivity for detection of low hydrolyzing enzymes such as blaOXA‐48. Alkaline agent such as NaHCO3 can be added to neutralize this effect. The decreased sensitivity of iCarba NP test might be due to the poor detection of few isolates with blaOXA‐48. As per CLSI 2016 guidelines, MHT and Carba NP test can be used as confirmatory test for detection of CP‐CRE.6 However, the sensitivity and specificity Carba NP test are reported to be >90% for isolates carrying blaKPC gene, blaNDM‐1, and other metallo β‐lactamases, but on the other hand, it can be as low as 11% for blaOXA‐48 gene.17 Contrarily, the performance of MHT is poor for the detection of CP‐CRE harboring blaNDM‐1.18, 19, 20
In this study, we tried to compare in–house‐developed Carba NP test with four other phenotypic tests, that is, cCarba NP, BCarba, CHROMagar, and MHT. Among the phenotypic tests, CHROMagar and BCarba tests showed excellent performances in terms of sensitivity and PPV for detection of CP‐CRE isolates.9, 21, 22 The performance of iCarba NP showed concordant result with the gold standard test, that is, PCR and other phenotypic tests. The sensitivity of iCarba NP and cCarba NP was observed 93.9%, which is in concordance with the published reports.23 Although reports showed specificity of 98%‐100% for Carba NP tests, specificity in the current study was found to be lower. Our result was concordant with the results by Thomson et al.24 Specificity of all the phenotypic tests was found less in comparison with published reports.9, 22 It might be due to the presence of other carbapenemase genes apart from the five genes used in the study. Among all the phenotypic tests, CHROMagar and both the Carba NP tests showed high sensitivity and specificity and hence can be used as screening tool to detect production of CP‐CRE. Sensitivity of MHT observed least among all the assays. Our study result showed the presence of NDM‐1 and IMP‐like carbapenemases in higher amount gives faster positive result than low hydrolyzing enzymes such as OXA‐48. The result of time to positivity of various Carba NP in our study was in concordance with the published literature.25 Comparing the cost, the iCarba NP test was observed 10 times cost‐effective than cCarba NP test.
5. CONCLUSION
Among the available phenotypic tests, CHROMagar was observed the most sensitive and specific followed by Carba NP test for detection of CP‐CRE. The performance of iCarba NP was comparable with cCarba NP test and was able to differentiate CP‐CRE from other carbapenem‐resistant isolates. Both the Carba NP tests were found rapid, accurate, and easy to perform and interpret the result. Hence, it can be used as screening test for the rapid detection of CP‐CRE in clinical settings.
ETHICAL APPROVAL
This study was approved by the Ethical Committee of All India Institute of Medical Sciences, New Delhi, India.
Bir R, Mohapatra S, Kumar A, et al. Comparative evaluation of in‐house Carba NP test with other phenotypic tests for rapid detection of carbapenem‐resistant Enterobacteriaceae. J Clin Lab Anal. 2019;33:e22652 10.1002/jcla.22652
REFERENCES
- 1. Centers for Disease Control and Prevention (CDC) . Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: CDC; 2013. [Google Scholar]
- 2. Viau R, Frank KM, Jacobs MR, et al. Intestinal carriage of carbapenemase‐producing organisms: current status of surveillance methods. Clin Microbiol Rev. 2016;29:1‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordmann P, Poirel L. The difficult‐to‐control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821‐830. [DOI] [PubMed] [Google Scholar]
- 4. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597‐602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller S, Humphries RM. Clinical laboratory detection of carbapenem‐resistant and carbapenemase‐producing Enterobacteriaceae. Expert Rev Anti Infect Ther. 2016;14:705‐717. [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; Twenty‐Third Informational Supplement, M100‐S26. Wanye, PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 7. Asthana S, Mathur P, Tak V. Detection of carbapenemase production in gram‐negative bacteria. J Lab Physicians. 2014;6:69‐75. 10.4103/0974-2727.141497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pires J, Novais Â, Peixe L. Blue‐Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013;51:4281‐4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aguirre‐Quiñonero A, Martínez‐Martínez L. Non‐molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother. 2017;23:1‐11. [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase‐producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503‐1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dortet L, Brechard L, Poirel L, Nordmann P. Rapid detection of carbaoenemase‐producing Enterobacteriaceae from blood cultures. Clin Microbiol Infect. 2014;20:340‐344. [DOI] [PubMed] [Google Scholar]
- 12. Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. Evaluation of the Carba NP test for rapid detection of carbapenemase‐producing Enterobacteriaceae and Pseudomonas aeruginosa . Antimicrob Agents Chemother. 2013;57:4578‐4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dortet L, Poirel L, Nordmann P. Further proofs of concept for the Carba NP test. Antimicrob Agents Chemother. 2014;58:1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. Reply to “Further proofs of concept for the Carba NP test”. Antimicrob Agents Chemother. 2014;58:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garg A, Garg J, Upadhyay GC, Agarwal A, Bhattacharjee A. Evaluation of the Rapidec Carba NP test kit for detection of carbapenemase‐producing Gram negative bacteria. Antimicrob Agents Chemother. 2015;59:7870‐7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pantel A, Souzy D, Sotto A, Lavigne JP. Evaluation of two phenotypic screening tests for carbapenemase‐producing Enterobacteriaceae. J Clin Microbiol. 2015;53:3359‐3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasoo S, Cunningham SA, Kohner PC, et al. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase‐producing Gram‐negative bacilli. J Clin Microbiol. 2013. Sep;51(9):3097‐3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50:3877‐3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50:477‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saito R, Koyano S, Dorin M, et al. Evaluation of a simple phenotypic method for the detection of carbapenemase‐producing Enterobacteriaceae. J Microbiol Methods. 2015;108:45‐48. [DOI] [PubMed] [Google Scholar]
- 21. Samra Z, Bahar J, Madar‐Shapiro L, Aziz N, Israel S, Bishara J. Evaluation of CHROMagar KPC for rapid detection of carbapenem‐resistant Enterobacteriaceae. J Clin Microbiol. 2008;46:3110‐3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bayramoğlu G, Uluçam G, Gençoğlu Özgür Ç, Kılıç AO, Aydın F. Comparison of the modified Hodge test and the Carba NP test for detection of carbapenemases in Enterobacteriaceae isolates. Mikrobiyol Bul. 2016;50:1‐10. [DOI] [PubMed] [Google Scholar]
- 23. Tamma PD, Opene BN, Gluck A, Chambers KK, Carroll KC, Simner PJ. Comparison of 11 phenotypic assays for accurate detection of carbapenemase‐producing Enterobacteriaceae. J Clin Microbiol. 2017;55:1046‐1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomson G, Turner D, Brasso W, Kircher S, Guillet T, Thomson K. High‐stringency evaluation of the automated BD phoenix CPO Detect and Rapidec Carba NP tests for detection and classification of carbapenemases. J Clin Microbiol. 2017;55:3437‐3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. García‐Fernandez S, Morosini MI, Gijon D, et al. Detection of carbapenemase production in a collection of Enterobacteriaceae with characterized resistance mechanisms from clinical and environmental origins by use of both Carba NP and Blue‐Carba Tests. J Clin Microbiol. 2015;54:464‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]


