Abstract
Backgrounds
Rapid discrimination between Mycobacterium tuberculosis (MTB) and nontuberculous mycobacteria (NTM) is critical for patient treatment and to avoid unnecessary expenditure on infection control. Because real‐time PCR assays distinguish MTB from NTM, we evaluated the performance of two real‐time PCR assays (AdvanSure and PowerChek).
Methods
This study used 143 DNA samples from respiratory specimens which were collected based on routine PCR results using Anyplex kit. A total of 87 positive samples (65 MTB and 22 NTM) and 56 negative samples were collected consecutively during 6 months and 1 month, respectively. The diagnostic performance of PCR assays (AdvanSure and PowerChek) was retrospectively analyzed based on the results of conventional mycobacterial tests and routine PCR assay.
Results
Based on culture results, the sensitivities/specificities of AdvanSure and PowerChek were 90.7%/87.6% and 92.6%/85.4%, respectively, for MTB detection. For PCR‐positive specimens, the quantification cycle (Cq) values of smear‐negative specimens were higher than those of the smear‐positive specimens (P < 0.001). As expected, the two PCR assays had the same sensitivities for NTM detection, viz. 90.0%, and their specificities were 99.2% and 98.4%, respectively. The overall agreement rate between the three PCR assays was 96.5% for MTB and 97.9% for NTM.
Conclusion
The sensitivities of PCR assays in our study might be overestimated, because this study enrolled relatively lower number of PCR‐negative samples which potentially missed PCR‐negative but culture‐positive specimens. However, the two real‐time PCR assays for detecting MTB and NTM perform equally well in relative performance evaluation and their Cq values can be considered suitable for predicting smear‐positive specimens.
Keywords: Mycobacterium tuberculosis, nontuberculous mycobacteria, NTM, real‐time PCR, tuberculosis
1. INTRODUCTION
Tuberculosis (TB) is one of the top 10 causes of death worldwide.1 Although the TB burden in Korea is decreasing every year,2 Korea still ranks highest for TB incidence rate and mortality among Organization for Economic Cooperation and Development countries.3
Rapid detection of Mycobacterium tuberculosis (MTB) is essential for controlling TB transmission in hospitals and in the community. Acid‐fast bacilli (AFB) culture is the gold standard for TB diagnosis, but several weeks are required for growth of Mycobacterium.4 The AFB smear can provide early diagnosis, but the method has low sensitivity and cannot distinguish MTB from nontuberculous mycobacteria (NTM).4
Nontuberculous mycobacteria infections are increasing worldwide. Until the early 1990s, 97%‐98% of AFB culture isolates from sputum samples in Korea contained MTB; however, by the early 2010s, the proportion of NTM had increased to 50%‐70%.5 Because molecular testing can discriminate MTB from NTM and provide high sensitivity with a short turn‐around time, real‐time PCR assays targeting MTB and NTM have been widely used in Korea. The AdvanSure TB/NTM Real‐time PCR Kit (LG Life Science, Korea) is the most widely used kit, followed by the Real‐Q M. tuberculosis Kit (Biosewoom, Korea) and Anyplex MTB/NTM Real‐time Detection (Seegene, Korea) in Korea (Korean Association of External Quality Assessment Service, 2017, unpublished data). The PowerChek MTB/NTM Real‐time PCR Kit (Kogene Biotech, Korea) designed for simultaneous detection of MTB and NTM was introduced in 2013. Among these kits, Anyplex and PowerChek are available worldwide.
In this study, we evaluated and compared the diagnostic performance of two real‐time PCR kits: the widely used AdvanSure TB/NTM Real‐time PCR kit and the newly developed PowerChek MTB/NTM Real‐time PCR.
2. MATERIALS AND METHODS
2.1. Specimens
A total of 143 DNA samples extracted from respiratory specimens of 118 patients were included in this study. The DNA samples were collected based on routine PCR results using Anyplex. PCR‐positive samples were collected consecutively between March and September 2016, and PCR‐negative samples were collected consecutively in the last month of this study. Sixty‐five samples were positive for MTB, 22 were positive for NTM, and 56 samples were negative results in routine PCR assay (Table 1). The DNA samples have been stored frozen at −70°C after routine PCR and retrospectively tested using AdvanSure and PowerChek. This study was approved by the Institutional Review Board of Hanyang University Guri Hospital (2017‐08‐017‐002).
Table 1.
Baseline mycobacterial characteristics of specimens
| Specimen type | No. | Anyplex PCR | AFB culture | AFB smear | |||||
|---|---|---|---|---|---|---|---|---|---|
| MTB | NTM | N | MTB | NTM | NG | + | − | ||
| Sputum | 106 | 43 (40.6)a | 19 (17.9) | 44 | 39 (36.8) | 16 (15.1) | 51 | 41 (38.7) | 65 |
| Bronchial aspirate | 37 | 22 (59.5) | 3 (8.1) | 12 | 15 (40.5) | 4 (10.8) | 18 | 10 (27.0) | 27 |
| Total | 143 | 65 (45.5) | 22 (15.4) | 56 | 54 (37.8) | 20 (14.0) | 69 | 51 (35.7) | 92 |
AFB, acid‐fast bacilli; MTB, Mycobacterium tuberculosis; N, negative; NG, no growth; NTM, nontuberculous mycobacteria.
Percentage of positives for each test modality are written in parentheses.
2.2. Conventional mycobacterial tests
Each patient provided three respiratory specimens simultaneously; one was used for the PCR assays and the others for conventional mycobacterial tests. The specimens were processed with 2% N‐acetyl‐L‐cysteine‐sodium hydroxide (NALC‐NaOH) and subsequently neutralized with phosphate buffer. They were then centrifuged at 3000 g for 18 minutes, and the pellets were resuspended in 1 mL of phosphate buffer (PBS) and used in the AFB smear test. The smears were stained with the auramine‐rhodamine fluorescence stain using an AT‐2000F AFB Fluorescence Stainer (Dagatron, Korea) and confirmed by Ziehl‐Neelsen staining. The smear results were reported using the guidelines of the American Thoracic Society/Centers for Disease Control and Prevention.4
For AFB culture, pretreated specimens were inoculated in 3% Ogawa medium (Korean Institute of Tuberculosis, Korea) for 8 weeks and in a BACTEC MGIT 960 system (Becton Dickinson, USA) for 6 weeks at the Green Cross Reference Laboratory (Korea). Positive culture isolates were confirmed by Ziehl‐Neelsen staining and MPT64 antigen detection (Standard Diagnostics, Korea). When isolates gave discrepant results in any tests, they were assessed by PCR using the AccuPower MTB & NTM real‐time PCR assay (Bioneer, Korea).
2.3. DNA extraction for PCR assays
For sputum specimens, an equal volume of 4% NaOH was added to each specimen with 1 minute of vortexing, and 1 mL of the mixture was transferred to a sterile tube after 15 minutes. The mixture was centrifuged, and the pellet was washed three times with 1 mL of PBS. For bronchial washings, 1 mL of specimen was transferred to a sterile tube and centrifuged for 5 minutes at 15 000 g (13 000 rpm), and the pellet was washed once with 1 mL of PBS. The sputum and bronchial pellets were mixed with 100 μL of the DNA extraction solution supplied with the Anyplex kit, with 1 minute of vortexing. After heating at 100°C for 20 minutes, the mixtures were incubated in room temperature for 5 minutes and centrifuged for 5 minutes at 15 000 g, and the supernatants were transferred to sterile tubes and used for PCR amplification. When the Anyplex PCR assays had been completed, the remaining DNA samples were stored at −70°C for subsequent AdvanSure and PowerChek PCR assays.
2.4. Anyplex MTB/NTM real‐time PCR
Primers for IS6110 and mpb64 of MTB and the 16S rRNA gene of NTM were used, and PCR was performed with the CFX96 real‐time PCR detection system (Bio‐Rad Laboratories, Inc., USA).6 Separate channels for FAM, HEX and Quasar 670 fluorophore signals were used to detect MTB, mycobacteria and the internal control (IC), respectively. In cases where the quantification cycle (Cq) values of MTB and IC were both below 45, the result was interpreted as “MTB positive” regardless of mycobacteria positivity; where the Cq value of mycobacteria was below 40 and the IC was positive, the result was interpreted as “NTM positive” and where the Cq values of all three channels were below their limits and the Cq value of mycobacteria was lower than that of MTB, the result was interpreted as “codetection with MTB and NTM.”
2.5. AdvanSure TB/NTM real‐time PCR
The PCR assays were performed on the frozen DNA samples after thawing. This kit detects the IS6110 region of MTB and the rpoB gene of NTM. PCR was performed with a SLAN real‐time PCR detection system (LG Life Sciences). FAM, HEX, and Cy5 fluorophore signals were used to detect MTB, mycobacteria, and IC, respectively, according to the instructions of the manufacturer. The data interpretation strategy was the same as for the Anyplex assay, except for the Cq cutoff values to consider a sample positive, which was 35 for all three channels.
2.6. PowerChek MTB/NTM real‐time PCR
Frozen DNA samples were used after thawing. The PowerChek kit targets the MTB complex‐specific IS6110 gene and the ITS region of Mycobacterium. PCR was performed with a CFX96 real‐time PCR detection system (Bio‐Rad Laboratories). FAM, HEX and Cy5 fluorophore signals were again used, and the data interpretation strategy was the same as for the AdvanSure assay.
2.7. Statistical analysis
Statistical analyses were performed using Statistical Package for the Social Sciences (version 18.0; USA) and GraphPad Prism (version 7; USA). The chi‐squared test was used to analyze tabular data, the Mann‐Whitney U test was used to analyze differences between groups, and a correlation study was performed with Spearman's rank correlation coefficient (r s). All tests were two‐sided, and a P value <0.05 was considered statistically significant.
3. RESULTS
A total of 143 DNA samples were tested in this study. Percentage of positives for MTB and NTM were 45.5% and 15.4% for Anyplex PCR and 37.8% and 14.0% for AFB culture (Table 1). AFB smears were positive in 35.7% of specimens. The DNA samples were extracted from 106 (74.1%) sputum samples and 37 (25.9%) bronchial aspirate samples. Based on Anyplex PCR results, MTB positivity of bronchial aspirates was 59.5% (22/37) which was higher than sputum samples; 40.6% (43/106) (P = 0.047). There was no significant difference between sample types in analyses of positivity for NTM by PCR, NTM or MTB by culture and AFB smear.
Based on culture results, the overall sensitivities and specificities of the two assays for detecting MTB were 90.7% (95% CI, 79.7‐96.9) and 87.6%% (95% CI, 79.0‐93.7), respectively, for AdvanSure, and 92.6% (95% CI, 82.1‐97.9) and 85.4% (95% CI, 76.3‐92.0), respectively, for PowerChek (Table 2). The two methods gave identical results for the smear‐positive samples, with sensitivities and specificities of 97.1% (95% CI, 85.1‐99.9) and 87.5% (95% CI, 61.7‐98.5), respectively. The sensitivities and specificities of the smear‐negative specimens were 78.9% (95% CI, 54.4‐94.0) and 87.7% (95% CI, 77.9‐94.2), respectively, for AdvanSure, and 84.2% (95% CI, 60.4‐96.6) and 84.9% (95% CI, 76.6‐92.2), respectively, for PowerChek.
Table 2.
Performances of real‐time PCR assays for detection of Mycobacterium tuberculosis depending on culture results
| PCR Assay | MTB growth | MTB no growth | Sensitivity%, (95% CI) | Specificity %, (95% CI) | |||
|---|---|---|---|---|---|---|---|
| PCR + | PCR − | PCR + | PCR − | ||||
| All | AdvanSure | 49 | 5 | 11 | 78 | 90.7 (79.7‐96.9) | 87.6 (79.0‐93.7) |
| PowerChek | 50 | 4 | 13 | 76 | 92.6 (82.1‐97.9) | 85.4 (76.3‐92.0) | |
| Smear + | AdvanSure | 34 | 1 | 2 | 14 | 97.1 (85.1‐99.9) | 87.5 (61.7‐98.5) |
| PowerChek | 34 | 1 | 2 | 14 | 97.1 (85.1‐99.9) | 87.5 (61.7‐98.5) | |
| Smear − | AdvanSure | 15 | 4 | 9 | 64 | 78.9 (54.4‐94.0) | 87.7 (77.9‐94.2) |
| PowerChek | 16 | 3 | 11 | 62 | 84.2 (60.4‐96.6) | 84.9 (76.6‐92.2) | |
CI, confidence interval; MTB, Mycobacterium tuberculosis.
We examined the relationship between grade of AFB smear gradation and Cq value of MTB (Figure 1). There were moderate inverse correlations between the AFB smear grades and the Cq values of AdvanSure and PowerChek (r s = −0.65 and −0.64, respectively). The Cq values of the smear‐negative specimens were higher than those of the smear‐positive samples (P < 0.001) and also higher than those of the 1 + smears (P = 0.019 and 0.010), while the Cq values of the latter were higher than those of the other smear‐positive samples (P = 0.005 and 0.007). However, the Cq values of the AFB 2‐4 + samples did not differ significantly. There was no difference between Cq values of sputum and bronchial aspirates. A Cq of 25.73 was 95.8% sensitive and 66.7% specific for smear‐positive status in AdvanSure assay, and a Cq of 25.77 was 96.3% sensitive and 61.1% specific for smear‐positive status in PowerChek assay.
Figure 1.
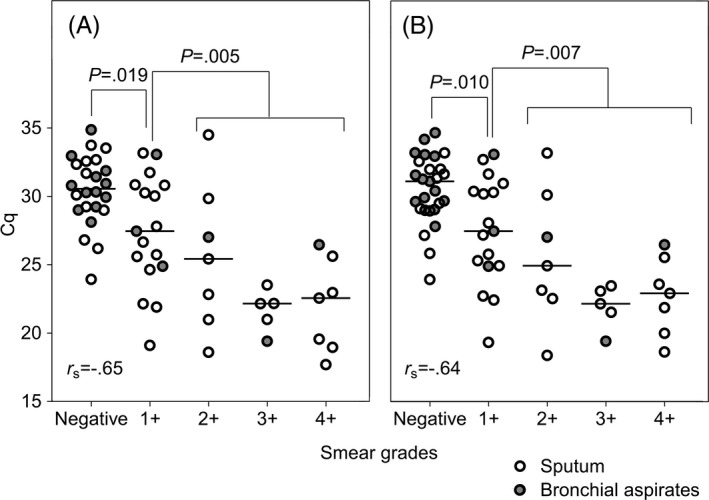
Comparison of acid‐fast bacilli smear grades and quantification cycle (Cq) values obtained in two real‐time PCR assays for Mycobacterium tuberculosis in respiratory specimens. There were moderate inverse correlations (r s = −0.65 and −0.64) between the smear grades and Cq values determined by AdvanSure (A) and PowerChek (B), respectively. The Cq values of the smear‐negative specimens were higher than those of the 1 + smears (P = 0.019 and 0.010), and the Cq values of the 1 + smears were higher than those of the other smear‐positive specimens in all three PCR assays (P = 0.005 and 0.007)
The two PCR assays had the same sensitivities for detecting NTM, namely 90.0% (95% CI, 68.3‐98.8) (Table 3). The specificities of the assays were 99.2% (95% CI, 95.6‐100) for AdvanSure and 98.4% (95% CI, 94.3‐99.8) for PowerChek. The results of the PCR assays were identical to those for the smear‐positive samples. The sensitivities and specificities were all 100% (95% CI, 76.8‐100 and 90.5‐100, respectively). For the smear‐negative specimens, sensitivities were 66.7% (95% CI, 22.3‐95.7), and specificities were 98.8% (95% CI, 93.7‐100) and 97.7% (95% CI, 91.9‐99.7) for AdvanSure, and PowerChek, respectively.
Table 3.
Performances of real‐time PCR assays for detection of nontuberculous mycobacteria depending on culture results
| PCR Assay | NTM growth | NTM no growth | Sensitivity %, (95% CI) | Specificity %, (95% CI) | |||
|---|---|---|---|---|---|---|---|
| PCR + | PCR − | PCR + | PCR − | ||||
| All | AdvanSure | 18 | 2 | 1 | 122 | 90.0 (68.3‐98.8) | 99.2 (95.6‐100) |
| PowerChek | 18 | 2 | 2 | 121 | 90.0 (68.3‐98.8) | 98.4 (94.3‐99.8) | |
| Smear + | AdvanSure | 14 | 0 | 0 | 37 | 100 (76.8‐100) | 100 (90.5‐100) |
| PowerChek | 14 | 0 | 0 | 37 | 100 (76.8‐100) | 100 (90.5‐100) | |
| Smear − | AdvanSure | 4 | 2 | 1 | 85 | 66.7 (22.3‐95.7) | 98.8 (93.7‐100) |
| PowerChek | 4 | 2 | 2 | 84 | 66.7 (22.3‐95.7) | 97.7 (91.9‐99.7) | |
CI, confidence interval; NTM, nontuberculous mycobacteria.
The overall agreement of the three PCR assays (Anyplex, AdvanSure, and PowerChek) was 96.5% for MTB (138/143) and 97.9% for NTM (140/143). There were discrepancies in the case of eight samples (5.6%) (Table 4). All of the assays yielding discordant results were positive by Anyplex and negative by AdvanSure PCR. PowerChek detected four of these samples as positive and 4 as negative.
Table 4.
Discordant results in the three real‐time PCR assays
| No. | Specimen | Anyplex | AdvanSure | PowerChek | Culture | AFB stain | Diagnosis |
|---|---|---|---|---|---|---|---|
| 1 | Br. asp | MTB | ‐ | MTB | No growth | ‐ | TB pleurisy |
| 2 | Br. asp | MTB | ‐ | ‐ | MTB | 1+ | Pulmonary TB |
| 3 | Br. asp | MTB | ‐ | MTB | No growth | ‐ | COPD, pneumonia |
| 4 | Br. asp | MTB | ‐ | MTB | MTB | ‐ | Pulmonary TB |
| 5 | Sputum | MTB | ‐ | ‐ | MTB | ‐ | Pulmonary TB |
| 6 | Sputum | NTM | ‐ | NTM | No growth | ‐ | COPD |
| 7 | Sputum | NTM | ‐ | ‐ | No growth | ‐ | Pneumonia |
| 8 | Sputum | NTM | ‐ | ‐ | No growth | ‐ | Pneumonia |
AFB, acid‐fast bacilli; Br. asp, bronchial aspirate; COPD, chronic obstructive pulmonary disease; MTB, Mycobacterium tuberculosis; NTM, nontuberculous mycobacteria; TB, tuberculosis.
4. DISCUSSION
Many laboratories have adopted the use of nucleic acid amplification tests to detect viruses or fastidious microorganisms to achieve early diagnosis and proper infection control. For MTB and NTM, real‐time PCR assays are commonly used to overcome the limitations of conventional tests, and newer assays are being actively developed to improve the accuracy of TB diagnosis and these need to be tested. Therefore, we evaluated and compared the performances of two real‐time PCR assays for MTB and NTM, one of which is a new assay.
The sensitivities of the PCR assays analyzed in this study were all over 90% for detecting MTB in respiratory specimens, comparable to or higher than the values previously reported (86.4% ‐86.8% by Anyplex; 70.9%‐88.7% by AdvanSure).6, 7, 8 The specificities in our study were below 90%, comparable to or lower than in previous studies (94.9%‐99.0% by Anyplex; 93.9%‐98.1% by AdvanSure).6, 7, 8 High sensitivity combined with low specificity of our study could be affected by relatively lower number of PCR‐negative samples which potentially involved the exclusion of PCR‐negative but culture‐positive specimens. Among the 13 PCR‐positive but culture‐negative specimens in our study, two yielded MTB in culture along with other specimens, and nine were from patients under treatment for TB or who had a recent history of treatment. AFB culture can be negative in patients undergoing tuberculosis treatment because of low numbers of viable bacilli; however, the DNA of MTB can persist for a long time.9 If we take into account the presence of these patients, the specificities in our study appear to be comparable to those in previous studies.
The AFB smear is widely used to assess infectivity in pulmonary TB patients at initial diagnosis. Because smear classification has a highly variable operational sensitivity, there have been several studies of the use of molecular assays for checking smear‐positive specimens.10, 11, 12 Previous research on the Xpert MTB/RIF assay (Cepheid, USA) showed that the Cq results correlated well with smear microscopic grades (r s = −0.71) and suggested a Cq cutoff value of 25 could predict smear‐positive status with 95% sensitivity and 65% specificity.10 We found that the Cq values of MTB were moderately correlated with smear grades and that smear‐positive specimens could be predicted from Cq values. We suggest that Cq values of 25.73 and 25.77 which were determined by AdvanSure or PowerChek could provide consistent results with smear microscopy. This molecular method could be useful especially in laboratory settings where personnel is insufficient or lack the required experience in AFB smear method.
Because the incidences of MTB and NTM are both high in Korea, rapid discrimination between these two organisms is critical for patient treatment and to avoid unnecessary expenditure for infection control. However, there have been few reports evaluating the performance of molecular tests for detecting NTM. Reported sensitivities for detecting NTM in respiratory specimens are 45.5%‐73.1% by Anyplex and 68.2%‐75.0% by AdvanSure.6, 8, 13 These researchers reported 98.4%‐100% specificity for both methods. Detection of NTM has not been important in the past in Korea, and the sensitivity for detection of NTM is sometimes adjusted downwards to avoid false positive results from NTM contamination. However, with the increasing incidence of NTM infections, molecular discrimination of NTM from MTB has become crucial, especially in smear‐positive cases, which can confuse clinicians. The sensitivity for detecting NTM in our study was considerably higher than in previous studies; 90.0% overall and 100% for smear‐positive specimens. The sensitivities of our study might be overestimated by unintended exclusion of PCR‐negative but culture‐positive specimens in the step of sample recruitment. However, we note that the sensitivity for detection of NTM in all the previous studies was lower than that for MTB, whereas the sensitivity for NTM in our study was comparable to that for MTB, and the specificity in our case (98.4%‐99.2%) was as high as in the previous reports. We therefore conclude that real‐time PCR assays simultaneously targeting MTB and NTM are useful for distinguishing between the two types of infection, although this needs to be confirmed.
In the cases where the results of the real‐time PCR assays were discordant, the detection rate of Anyplex seemed to be higher than the detection rates of AdvanSure and PowerChek. This result might have been influenced by the sample inclusion criteria which were based on Anyplex data. Although all the specimens were collected consecutively, the sample collection period was different for positive samples and negative samples which were determined by Anyplex assay. In addition, the DNA samples used were extracted with Anyplex extraction kit according to a protocol that had been optimized for the Anyplex assay. AdvanSure and PowerChek recommended the same sample preparation method for DNA extraction which was different from Anyplex. The method of AdvanSure and PowerChek used 5‐minute incubation with NaOH and two times of PBS washing for sputum specimens. However, Anyplex used 1‐minute incubation with NaOH and three times of PBS washing. For bronchial aspirates specimens, PBS washing step was different between the methods (AdvanSure and PowerChek, two times; Anyplex, one time). Because three assays were recommended to use different extraction solutions for each assay, use of Anyplex kit to other assays could affect the performance evaluated. In retrospective study with AdvanSure and PowerChek, samples stored at −70°C for up to 7 months were used, but the effects of sample storage on these assays (AdvanSure and PowerChek) could not be estimated because we were unable to assess sample stability. However, the diagnostic performances of the three real‐time PCR were not significantly different, and this limitation should not affect the conclusion that the performances of PowerChek and AdvanSure were comparable and acceptable.
In conclusion, the two real‐time PCR assays, namely AdvanSure and PowerChek, performed comparably for detecting Mycobacterium species. The Cq values of these assays are suitable for predicting smear‐positive specimens.
AUTHOR'S CONTRIBUTION
M.H.B. and C.‐K.K. contributed equally to this work as corresponding authors.
ACKNOWLEDGMENTS
AdvanSure TB/NTM Real‐time PCR Kits and PowerChek MTB/NTM Real‐time PCR Kits were donated by LG Life Sciences and Kogene Biotech, respectively.
Lim J‐H, Kim C‐K, Bae MH. Evaluation of the performance of two real‐time PCR assays for detecting Mycobacterium species. J Clin Lab Anal. 2019;33:e22645 10.1002/jcla.22645
Mi Hyun Bae and Chang‐Ki Kim contributed equally to this work as corresponding authors.
Contributor Information
Chang‐Ki Kim, Email: psoas95@gmail.com.
Mi Hyun Bae, Email: mhbae@hanyang.ac.kr.
REFERENCES
- 1. World Health Organization . Global Tuberculosis Report 2017. 2018. http://www.who.int/tb/publications/global_report/en/. Accessed May 1, 2018.
- 2. Jeon JS, Kim JK, Choi Q, Kim JW. Distribution of Mycobacterium tuberculosis in Korea in the preceding decade. J Clin Lab Anal. 2018;32:e22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kang H, Cha J, Park O. Global burden of tuberculosis, 2015. Public health weekly report. 2016;9:862‐866.
- 4. American Thoracic Society . Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med. 2000;161(4):1376‐1395. [DOI] [PubMed] [Google Scholar]
- 5. Kwon YS, Koh WJ. Diagnosis and treatment of nontuberculous mycobacterial lung disease. J Korean Med Sci. 2016;31(5):649‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choe W, Kim E, Park SY, Chae JD. Performance evaluation of anyplex plus MTB/NTM and AdvanSure TB/NTM for the detection of Mycobacterium tuberculosis and nontuberculous mycobacteria. Ann Clin Microbiol. 2015;18(2):44‐51. [Google Scholar]
- 7. Cho WH, Won EJ, Choi HJ, et al. Comparison of AdvanSure TB/NTM PCR and COBAS TaqMan MTB PCR for detection of Mycobacterium tuberculosis complex in routine clinical practice. Ann Lab Med. 2015;35(3):356‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sali M, De Maio F, Caccuri F, et al. Multicenter evaluation of anyplex plus MTB/NTM MDR‐TB assay for rapid detection of Mycobacterium tuberculosis complex and multidrug‐resistant isolates in pulmonary and extrapulmonary specimens. J Clin Microbiol. 2016;54(1):59‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedrich SO, Rachow A, Saathoff E, et al. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 2013;1(6):462‐470. [DOI] [PubMed] [Google Scholar]
- 10. Blakemore R, Nabeta P, Davidow AL, et al. A multisite assessment of the quantitative capabilities of the Xpert MTB/RIF assay. Am J Respir Crit Care Med. 2011;184(9):1076‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huh HJ, Koh WJ, Song DJ, Ki CS, Lee NY. Evaluation of the Cobas TaqMan MTB test for the detection of Mycobacterium tuberculosis complex according to acid‐fast‐bacillus smear grades in respiratory specimens. J Clin Microbiol. 2015;53(2):696‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee H, Park KG, Lee G, Park J, Park YG, Park YJ. Assessment of the quantitative ability of AdvanSure TB/NTM real‐time PCR in respiratory specimens by comparison with phenotypic methods. Ann Lab Med. 2014;34(1):51‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang S, Oh KJ, Jang IH, et al. Evaluation of the diagnostic performance of the AdvanSure TB/NTM Real‐Time PCR kit for detection of mycobacteria. Ann Clin Microbiol. 2011;14(2):55‐59. [Google Scholar]


