Summary
Phytophthora sojae is a destructive pathogen of soybean [Glycine max (L.) Merr.] which causes stem and root rot on soybean plants worldwide. However, the pathogenesis and molecular mechanism of plant defence responses against P. sojae are largely unclear. Herein, we document the underlying mechanisms and function of a novel BTB/POZ protein, GmBTB/POZ, which contains a BTB/POZ domain found in certain animal transcriptional regulators, in host soybean plants in response to P. sojae. It is located in the cell nucleus and is transcriptionally up‐regulated by P. sojae. Overexpression of GmBTB/POZ in soybean resulted in enhanced resistance to P. sojae. The activities and expression levels of enzymatic superoxide dismutase (SOD) and peroxidase (POD) antioxidants were significantly higher in GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) transgenic soybean plants than in wild‐type (WT) plants treated with sterile water or infected with P. sojae. The transcript levels of defence‐associated genes were also higher in overexpressing plants than in WT on infection. Moreover, salicylic acid (SA) levels and the transcript levels of SA biosynthesis‐related genes were markedly higher in GmBTB/POZ‐OE transgenic soybean than in WT, but there were almost no differences in jasmonic acid (JA) levels or JA biosynthesis‐related gene expression between GmBTB/POZ‐OE and WT soybean lines. Furthermore, exogenous SA application induced the expression of GmBTB/POZ and inhibited the increase in P. sojae biomass in both WT and GmBTB/POZ‐OE transgenic soybean plants. Taken together, these results suggest that GmBTB/POZ plays a positive role in P. sojae resistance and the defence response in soybean via a process that might be dependent on SA.
Keywords: BTB/POZ domain, enzymatic antioxidants, Phytophthora sojae, salicylic acid
Introduction
Plants have evolved to defend themselves against microbial pathogens using multiple defence mechanisms, including chemical or physical barriers, such as preformed antimicrobial compounds or a waxy cuticle, as well as harbouring innate plant immunity systems (Schneider, 2002). These immunity systems have great potential to protect plants from pathogen attack and to enable plants to defend themselves against widespread disease (Vidhyasekaran, 2015). This form of immune defence can be divided into two branches of response that are interconnected: pathogen/microbe‐associated molecular patterns (PAMPs or MAMPs)‐triggered immunity (PTI), which is based on the perception of PAMPs and effectors by cell surface‐localized pattern recognition receptors (PRRs), and effector‐triggered immunity (ETI), which is based on host disease resistance (R) proteins (Dangl et al., 2013; Jones and Dangl, 2006). PTI and ETI are associated with the activation of common defence networks mediated by kinases, transcription factors, pathogenesis‐related (PR) proteins, reactive oxygen species (ROS) and phytohormones (Baxter et al., 2014; Chandran et al., 2014; Cui et al., 2015). Plant hormones, including salicylic acid (SA) and jasmonic acid (JA), perform a key role in plant immunity systems and major roles in the regulation of plant basal defence responses (Koornneef and Pieterse, 2008; Thaler et al., 2012). SA acts as a crucial signalling element in systemic acquired resistance (SAR) signalling pathways; the SA‐mediated pathway is typically activated by biotrophic pathogens (Glazebrook, 2005). Plant defence responses that depend on JA are usually induced by necrotrophic pathogens (Glazebrook, 2005). In general, the SA pathway is prioritized over the JA pathway, and crosstalk between the two signalling pathways is usually antagonistic and might represent an adaptive response in plants (Thaler et al., 2012).
The BTB/POZ domain (Broad Complex, Tramtrack, Bric‐a‐brac⁄Pox virus and Zinc finger) was originally identified as a conserved motif present in Drosophila melanogaster bric‐à‐brac, tramtrack and broad complex transcriptional regulators and in many pox virus zinc finger proteins (Bardwell and Treisman, 1994; Koonin et al., 1992; Zollman et al., 1994). The BTB/POZ domain is an NH3‐terminal motif of approximately 120 amino acids that is highly conserved and hydrophobic (Oyake et al., 1996). BTB/POZ proteins can also contain other types of domain in addition to the evolutionarily conserved BTB/POZ domain (Csankovszki et al., 2001; David et al., 1998; Stogios et al., 2005). Specifically, many BTB/POZ proteins also contain one or more BTB‐BACK‐kelch (BBK) motifs (Stogios et al., 2005) and may also contain a secondary protein domain, such as C2H2, ANKYRIN, NPH3 or MATH [meprin and TRAF (tumour necrosis factor receptor‐associated factor) homology] motifs (Csankovszki et al., 2001; David et al., 1998; Dong et al., 1996). The BTB/POZ domain is usually found as a single‐copy protein domain in proteins containing only one or two other types of domain, thereby classifying them as members of the BTB‐zinc finger (BTB‐ZF), BTB‐NPH3, MATH‐BTB and BTB‐BACK‐PHR (BBP) families of BTB/POZ proteins (Stogios et al., 2005). By contrast, the Skp1 and ElonginC BTB/POZ proteins almost exclusively consist of only the core BTB/POZ domain (Salas‐Vidal et al., 2005; Weber et al., 2005).
The BTB/POZ domain is a widely distributed structural motif found in an array of proteins involved in various biological processes, including transcriptional and cytoskeleton regulation and the formation of voltage‐gated channels (Ahmad et al., 2003; Bomont et al., 2000; Collins et al., 2001; Ziegelbauer et al., 2001). BTB/POZ proteins have been discovered in organisms from yeasts to humans (Furukawa et al., 2003; Pintard et al., 2004). NPR1 was the first BTB/POZ protein identified in plants: it was discovered in Arabidopsis thaliana and Arabidopsis npr1 mutants were unable to respond to SA or to express PR genes induced by SA (Cao et al., 1997; Delaney et al., 1995). PR‐1 gene expression may also be regulated by the interaction of NPR1 and bZIP transcription factors, thereby enhancing the resistance of host plants to pathogenic microorganisms (Zhang et al., 1999). Furthermore, NPR1 improves the resistance of several plant species to pathogen attack (Lin et al., 2004; Makandar et al., 2006; Meur et al., 2008). It has been shown that a subfamily of nucleus‐localized BTB/POZ proteins encoded by LIGHT‐RESPONSE BTB1 (LRB1) and LRB2 in Arabidopsis strongly influences photomorphogenesis (Christians et al., 2012). In barley aleurone cells, GMPOZ, which contains a BTB/POZ domain, is implicated in the regulation of hormone‐responsive gene expression (Woodger et al., 2004). To date, over 100 plant BTB/POZ proteins have been identified (Gingerich et al., 2005, 2007). Excluding NPR1, a systematic study of the roles played by BTB/POZ proteins in disease resistance in soybean has not yet been reported.
Soybean BTB/POZ protein interacts with GmLHP1, as revealed in a bimolecular fluorescence complementation (BiFC) assay (Fig. S1, see Supporting Information), and GmLHP1 may play a crucial role in the response to P. sojae (Cheng et al., unpublished data). In this study, we isolated the soybean BTB/POZ gene, encoding a protein containing a BTB/POZ domain that is found in certain animal transcriptional regulators, which was designated as GmBTB/POZ [GenBank accession no. XM_006578889; National Center for Biotechnology Information (NCBI) protein no. XP_006578952]. GmBTB/POZ is localized to the nucleus and fails to activate transcription in yeast cells. To further elucidate the role of GmBTB/POZ in plant defence, we investigated the effect of GmBTB/POZ overexpression on Phytophthora root rot resistance in soybean. Several physiological and biochemical analyses showed that GmBTB/POZ plays an important role in the pathogenic process of P. sojae in soybean by enhancing SA levels, providing evidence that GmBTB/POZ is directly involved in the defence response of soybean to P. sojae attack.
Results
Expression of GmBTB/POZ is induced by P. sojae
We obtained the full‐length cDNA sequence of GmBTB/POZ (GenBank accession no. XM_006578889) from total RNA isolated from soybean cultivar ‘Suinong 10’. Sequence analysis suggested that the full‐length GmBTB/POZ cDNA is 1108 bp, containing a 777‐bp open reading frame (ORF), encoding a 258‐amino‐acid peptide (Fig. S2, see Supporting Information) with a predicted molecular mass of 29.099 kDa. Phylogenetic analysis and multiple sequence alignment demonstrated that GmBTB/POZ shares 48.99%–81.78% identity in overall amino acid sequence with its other plant species orthologues, including Capsella rubella CrBTB/POZ (XP_006288349.1), Populus euphratica PeBTB/POZ (XP_011027431.1), Populus trichocarpa PtBTB/POZ (XP_002300959.1), Brassica rapa BrBTB/POZ (XP_009134091.1), Pyrus × bretschneideri PbBTB/POZ (XP_009354024.1), Cucumis sativus CsBTB/POZ (XP_004139801.1), Cucumis melo CmBTB/POZ (XP_008451217.1), Ziziphus jujuba ZjBTB/POZ (XP_015885781.1), Jatropha curcas JcBTB/POZ (XP_012078336.1), Morus notabilis MnBTB/POZ (XP_010105442.1), Lupinus angustifolius LaBTB/POZ (XP_019464120.1), Cicer arietinum CaBTB/POZ (XP_004502546.1), Citrus clementina CcBTB/POZ (XP_020212618.1), Phaseolus vulgaris PvBTB/POZ (XP_007137484.1), Vigna angularis VaBTB/POZ (XP_017420040.1) and Vigna radiata VrBTB/POZ (XP_014498765.1) (Figs [Link], [Link], see Supporting Information).
To evaluate the expression profiles of GmBTB/POZ, we performed quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) to examine GmBTB/POZ transcript levels in the soybean resistant cultivar ‘Suinong 10’ and the soybean susceptible cultivar ‘Dongnong 50’. As shown in Fig. 1A, the expression levels of GmBTB/POZ in the plant tissues of Suinong 10 were much higher than those in the plant tissues of Dongnong 50. We further explored the gene expression pattern of GmBTB/POZ during infection with zoospores of P. sojae, which revealed that GmBTB/POZ expression was induced by P. sojae in the plant tissues of Suinong 10, reaching a peak at 24 h, followed by a steep decline (Fig. 1B). By contrast, in the plant tissues of Dongnong 50, there was almost no change in GmBTB/POZ transcript abundance after treatment (Fig. 1B), demonstrating the differential expression pattern of GmBTB/POZ between the resistant and susceptible cultivars.
Figure 1.
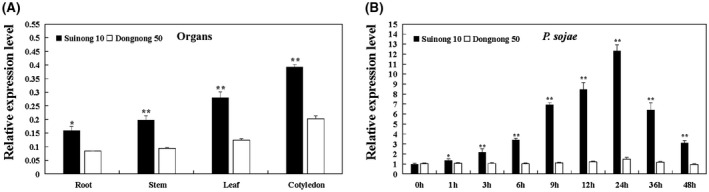
Expression patterns of GmBTB/POZ in Phytophthora sojae‐resistant and P. sojae‐susceptible soybean cultivars. (A) The tissue‐specific expression patterns of GmBTB/POZ in resistant cultivar ‘Suinong 10’ and susceptible cultivar ‘Dongnong 50’ under normal conditions. (B) Relative expression of GmBTB/POZ in soybean cultivars ‘Suinong 10’ and ‘Dongnong 50’ on P. sojae infection. The infected samples were collected at 0, 1, 3, 6, 9, 12, 24, 36 and 48 h after P. sojae infection. The relative expression levels of GmBTB/POZ were compared with those of mock‐treated plants (plants treated with sterile water) at the same time point. Fourteen‐day‐old soybean plants were used for analysis. The housekeeping gene of soybean GmEF1 (NM_001248778) was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean.
Subcellular localization and transcriptional activation ability of GmBTB/POZ
To examine the subcellular localization of GmBTB/POZ, the expression of the GmBTB/POZ‐GFP fusion protein was analysed in Arabidopsis protoplasts (Fig. 2A). The green fluorescent protein (GFP) signal was observed in the entire cell of protoplasts harbouring the 35S:GFP vector alone, whereas the GmBTB/POZ‐GFP green fluorescent signal was strongly displayed in the nucleus in transformed cells, similar to the signal produced by GmWRKY31‐GFP, reported by Fan et al. (2017), clearly indicating that GmBTB/POZ is a nucleus‐localized protein (Fig. 2A).
Figure 2.
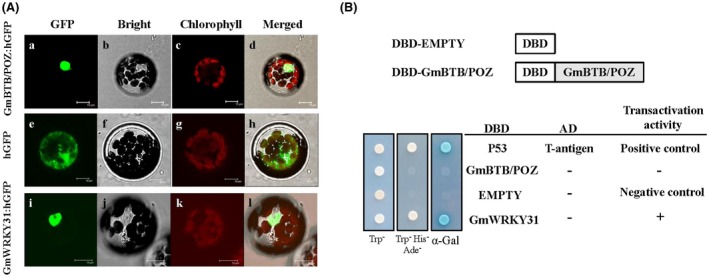
Subcellular localization and transcriptional activation analysis of GmBTB/POZ. (A) Subcellular localization was investigated in Arabidopsis protoplasts via confocal microscopy. Images of bright‐field (b, f, j), green fluorescent protein (GFP) fluorescence (green) only (a, e, i), chlorophyll autofluorescence (red) only (c, g, k) and their combination (d, h, l) are shown. Scale bars indicate 10 µm. (B) The open reading frame (ORF) of GmBTB/POZ was amplified into the pGBKT7 (GAL4 DBD) vector to generate the DBD‐GmBTB/POZ constructs. The yeast strain AH109 was transformed with pGBKT7‐53+pGADT7‐T, pGBKT7‐GmBTB/POZ, pGBKT7‐GmWRKY31 and pGBKT7. The transformed cells were grown on synthetic dropout medium without tryptophan [SD (‐Trp)], SD medium without Trp, histidine and adenine [SD (‐Trp/‐His/‐Ade)] and SD medium without Trp, His and Ade but with α‐galactosidase [SD (‐Trp/‐His/‐Ade/α‐gal)] for 3 days at 30 °C. Transcriptional activation was monitored by the detection of yeast growth and performance of an α‐Gal assay. [Color figure can be viewed at wileyonlinelibrary.com]
The BTB/POZ domain is a highly conserved structural motif involved in transcriptional regulation (Aravind and Koonin, 1999; Collins et al., 2001; Hu et al., 2010). To test the activation of the transcription function of GmBTB/POZ, we performed a transient expression assay in yeast cells using a GAL4‐responsive reporter system. As shown in Fig. 2B, transformed yeast cells harbouring DBD‐P53+T‐antigen (pGBKT7‐53+pGADT7‐T, positive control) and DBD‐GmWRKY31 (pGBKT7‐GmWRKY31), which exhibited transcriptional activation ability in our previous studies, grew well in synthetic dropout medium without tryptophan, histidine and adenine [SD (‐Trp/‐His/‐Ade)] and showed α‐galactosidase (α‐gal) activity, whereas yeast cells containing DBD‐GmBTB/POZ (pGBKT7‐GmBTB/POZ) or empty (pGBKT7, negative control) exhibited no α‐gal activity. These data confirm that GmBTB/POZ has no transcriptional activation activity.
GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) transgenic soybean plants exhibit enhanced resistance to P. sojae
To determine the role of GmBTB/POZ in the response to P. sojae, we transformed soybean plants with a construct harbouring GmBTB/POZ‐Myc driven by the 35S promoter via Agrobacterium‐mediated transformation. The expression of recombinant GmBTB/POZ‐Myc protein was confirmed by immunoblotting using an anti‐Myc antibody (Fig. 3B). At 96 h post‐inoculation (hpi), wild‐type (WT) soybean roots inoculated with P. sojae zoospores exhibited more serious symptoms, with watery and rotting lesions, than those of GmBTB/POZ‐OE soybean lines (Fig. 3A). Moreover, we analysed the relative biomass of P. sojae in soybean roots based on the presence of the transcript level of the P. sojae TEF1 gene (EU079791) (Fig. 3C); the results indicated that overexpression of GmBTB/POZ enhances the resistance to P. sojae after root infection. Furthermore, the cotyledons of WT inoculated with P. sojae zoospores exhibited more serious symptoms than those of GmBTB/POZ‐OE soybean lines (Fig. 4A), and the lesion area of GmBTB/POZ‐transformed soybean plants was much smaller than that of WT lines (Fig. 4B). In addition, the relative biomass of P. sojae was significantly lower in GmBTB/POZ‐transformed plants than in WT in infected living cotyledons after 96 h. The results indicate that the overexpression of GmBTB/POZ in soybean plants could enhance the resistance to P. sojae in soybean.
Figure 3.
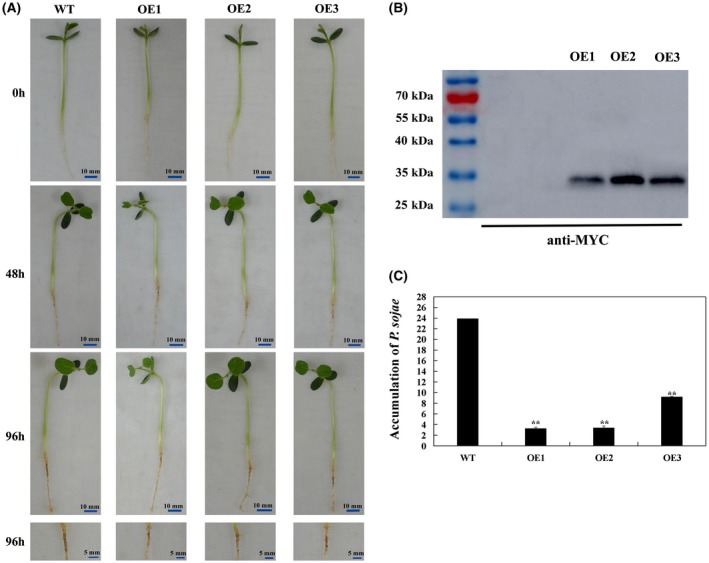
Resistance analysis of GmBTB/POZ transgenic soybean plants. (A) Disease symptoms on the roots of GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) and wild‐type (WT) plants at 96 h after inoculation with Phytophthora sojae. (B) Immunoblot analysis of the expression of GmBTB/POZ in three positive overexpressing transgenic soybean lines (OE1, OE2 and OE3). The corresponding protein bands of GmBTB/POZ‐Myc (total molecular mass of 34 kDa) were detected. (C) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the P. sojae relative biomass in three GmBTB/POZ‐OE soybean plants and WT soybean plants based on the transcript level of the P. sojae TEF1 gene (EU079791). The experiment was performed on three biological replicates, each with three technical replicates, and statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
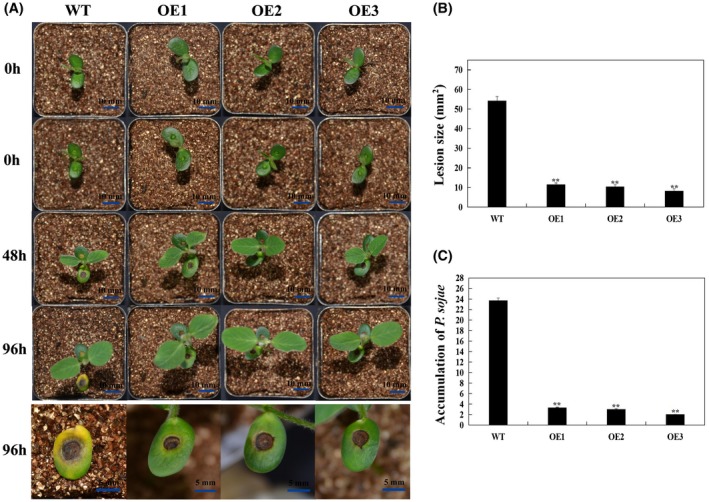
GmBTB/POZ enhances resistance to Phytophthora sojae in transgenic soybean cotyledons. (A) Disease symptoms on living cotyledons of GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) and wild‐type (WT) plants at 96 h after inoculation with P. sojae. (B) Lesion size measured from photographed cotyledons of GmBTB/POZ‐OE transgenic and WT plants at 96 h post‐inoculation (hpi). The lesion size of each independent soybean line (n = 3) was calculated, and the lesion sizes are shown in the columns based on a comparison with the average lesion area on WT soybean. (C) Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the relative biomass of P. sojae in GmBTB/POZ transgenic lines and WT soybean based on P. sojae TEF1 transcript levels. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean. [Color figure can be viewed at wileyonlinelibrary.com]
Soybean GmBTB/POZ‐regulated defence response to P. sojae involves the antioxidant defence system and oxidative stress damage
Plants experience various types of environmental stress which can lead to the production of ROS. Excess ROS concentrations induce oxidative damage or apoptotic cell death (Hückelhoven and Kogel, 2003; Yu and Liu, 2003). Plant responses to pathogen attack are closely related to the formation of ROS (Nandinip et al., 2008; Soosaar et al., 2005). We therefore analysed the relative ROS levels in WT and GmBTB/POZ‐OE soybean plants at 0, 12, 24 and 48 hpi. The changes in ROS levels displayed a similar tendency in WT and GmBTB/POZ‐OE lines, whereas the relative ROS levels in GmBTB/POZ‐OE lines were significantly lower than in WT at all time points during the incubation period (Fig. 5A). We also analysed the hydrogen peroxide (H2O2) contents at 24 hpi via 3,3′‐diaminobenzidine (DAB) staining in soybean leaves (Fig. 5B). The amount of DAB staining was clearly lower in GmBTB/POZ‐OE plants relative to WT. The results show that GmBTB/POZ might play a crucial role as an antioxidant in protecting soybean from P. sojae infection.
Figure 5.
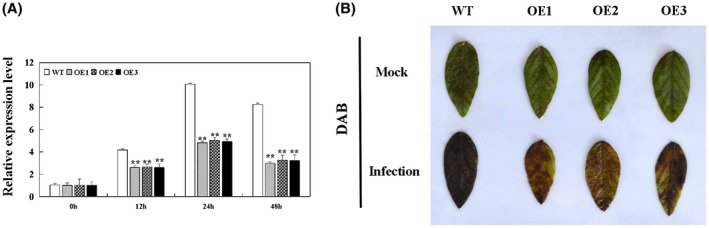
Reactive oxygen species (ROS) levels and 3,3′‐diaminobenzidine (DAB) staining analysis. (A) Relative expression levels of ROS in GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) and wild‐type (WT) lines at 0, 12, 24 and 48 h post‐inoculation (hpi). (B) H2O2 levels were detected by DAB staining in plants infected with P. sojae at 24 hpi. Values are relative to the value of mock‐treated (treated with sterile water) plants at the same time point. The results from three biological replicates, each with three technical replicates, were averaged and statistically analysed using Student’s t‐test (*P < 0.05; **P < 0.01). Bars indicate the standard error of the mean. [Color figure can be viewed at wileyonlinelibrary.com]
Plants have well‐developed antioxidant defence systems that efficiently scavenge ROS, involving the antioxidant enzymes superoxide dismutase (SOD) and peroxidase (POD) (Du et al., 2001). We therefore analysed SOD and POD activity, as well as the expression of GmSOD1 (NM_001248369) and GmPOD (XM_006575142), in infected soybean plants. SOD and POD activity and the expression levels of related genes were much higher in GmBTB/POZ‐OE transgenic soybean plants than in WT under both mock treatment and at 24 hpi (Fig. 6). In general, the SOD and POD activity are influenced to a certain extent by the expression levels of the corresponding enzymes (Badawi et al., 2004; McKersie and Jones, 1999; Melchiorre et al., 2009; Wang et al., 2015). Accordingly, we deduced that the antioxidant enzymatic activities were increased because of the higher expression level of the corresponding enzymatic genes in GmBTB/POZ‐OE soybean plants, and thus may eliminate ROS as a plant defence response to P. sojae infection.
Figure 6.
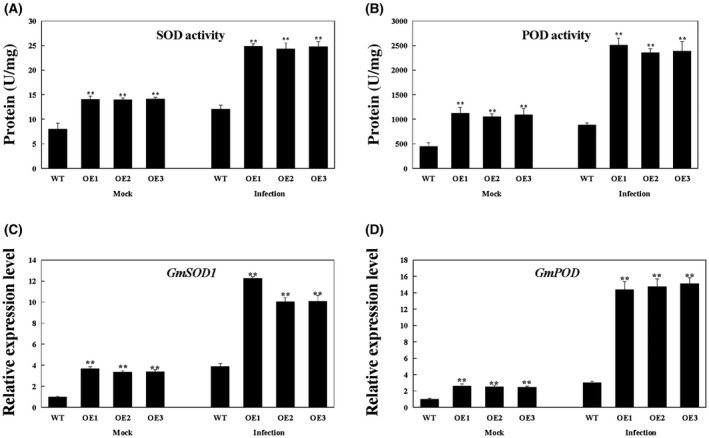
Analysis of antioxidant enzyme activity (A, B) and the relative expression of genes (C, D) under mock treatment and infected by Phytophthora sojae at 24 h post‐inoculation (hpi). The activity of the control sample [mock‐treated wild‐type (WT) plants] was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean. POD, peroxidase; SOD, superoxide dismutase.
GmBTB/POZ regulates defence‐associated gene expression in response to P. sojae infection
To evaluate the gene expression efficiency of GmBTB/POZ as an indicator of the defence response to P. sojae infection, we monitored GmBTB/POZ transcript levels by quantitative RT‐PCR. GmBTB/POZ transcript levels were significantly higher in GmBTB/POZ‐OE plants compared with WT plants under both mock treatment and after P. sojae inoculation at 24 hpi (Fig. 7).
Figure 7.
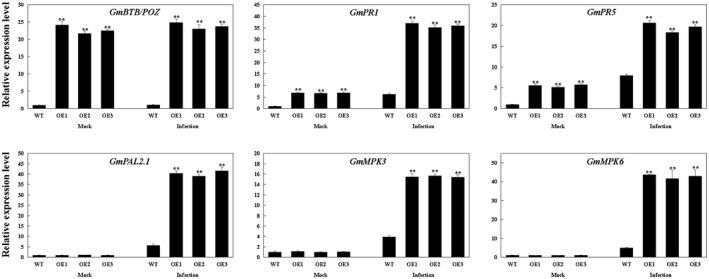
Relative expression levels of defence‐associated genes in soybean plants under mock treatment and infected by Phytophthora sojae at 24 h post‐inoculation (hpi). The housekeeping gene of soybean GmEF1 was used as an internal control to normalize the data. The expression level of the control sample [mock‐treated wild‐type (WT) plants] was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean.
PR proteins are crucial components involved in the plant defence response to invading pathogens and can affect the defence reactions of plants (Lcvan and Eavan, 1999; Loon et al., 2006). For example, MPK3 and MPK6 are two PR proteins that have been implicated to play critical roles in plant defence responses against pathogens (Colcombet and Hirt, 2008; Rasmussen et al., 2012). It has been demonstrated previously that phenylalanine ammonia‐lyase (PAL) positively regulates the defence response to pathogen infection in soybean (Zhang et al., 2017). To investigate the potential defence mechanism of the GmBTB/POZ‐regulated resistance to P. sojae, we examined the expression of some candidate defence‐related genes, such as the PR genes GmPR1 (AF136636) and GmPR5 (M21297), pathogen‐responsive mitogen‐activated protein kinase genes GmMPK3 (Glyma11g15700) and GmMPK6 (Glyma02g15690), and, finally, the PAL gene GmPAL2.1 (NM_001250027). The transcript abundances of these resistance genes were significantly higher in GmBTB/POZ‐OE plants than in WT at 24 hpi (Fig. 7). The results suggest that GmBTB/POZ improves defence against P. sojae by affecting defence‐related gene expression.
GmBTB/POZ is a positive regulator of SA‐dependent signalling during the defence response to P. sojae
Notably, GmPR1 and GmPR5 were constitutively induced in GmBTB/POZ‐OE soybean plants under mock treatment, indicating that the defence response of these plants was activated (Fig. 7). Increased production of SA has been associated with constitutively activated defence responses (Chen and Klessig, 1991). Furthermore, research has shown that SA and JA are functional signalling molecules for disease resistance in plants (Dong, 1998). To dissect whether the GmBTB/POZ‐regulated defence response is dependent on these phytohormones, we measured both SA and JA content in GmBTB/POZ transgenic and WT plants. In addition, the transcript levels of GmICS1 (XM_003522145) and GmAOS (NM_001249516) were measured, which play crucial roles in SA and JA biosynthesis, respectively. As shown in Fig. 8A,B, SA levels were significantly higher in GmBTB/POZ‐OE leaves than in WT leaves, whereas there was no significant difference in JA levels between the two groups. Consistent with the SA and JA measurements, GmICS1 transcript levels were markedly higher in the GmBTB/POZ‐OE group than in the WT group, whereas GmAOS expression differed little between the GmBTB/POZ‐OE plant group and the WT plant group (Fig. 8C,D). It is important to note that two different pathways of SA biosynthesis have been proposed in plants (Chen et al., 2009): the first is the chorismate pathway controlled by isochorismate synthase (ICS) (Catinot et al., 2008; Chen et al., 2009; Wildermuth et al., 2001), whereas the second is the PAL‐controlled phenylpropanoid pathway (Huang et al., 2010). The results revealed that the expression levels of GmPAL2.1 were not constitutively induced in non‐infected GmBTB/POZ‐OE soybean plants (Fig. 7). The soybean genome contains eight GmPAL members (Rawal et al., 2013; Schlueter, 2010). To determine whether there were gene expression level changes for any of the other GmPAL genes in the GmBTB/POZ‐OE group, the expression levels of GmPAL1.1 (Glyma.19G182300), GmPAL1.2 (Glyma.03G181700), GmPAL1.3 (Glyma.03G181600), GmPAL2.2 (Glyma.20G180800), GmPAL2.3 (Glyma.13G145000), GmPAL2.4 (Glyma.10G209800) and GmPAL3.1 (Glyma.02G309300) were also analysed. The results showed that the transcripts of GmPAL1.1, GmPAL1.2 and GmPAL1.3 changed significantly in each of the GmBTB/POZ‐OE lines compared with WT soybean plants, whereas there were no significant differences in the expression levels for each of GmPAL2.2, GmPAL2.3, GmPAL2.4 and GmPAL3.1 (Fig. S5, see Supporting Information). These results indicate that GmBTB/POZ may play a role in regulating both the ICS and PAL pathways of SA accumulation.
Figure 8.
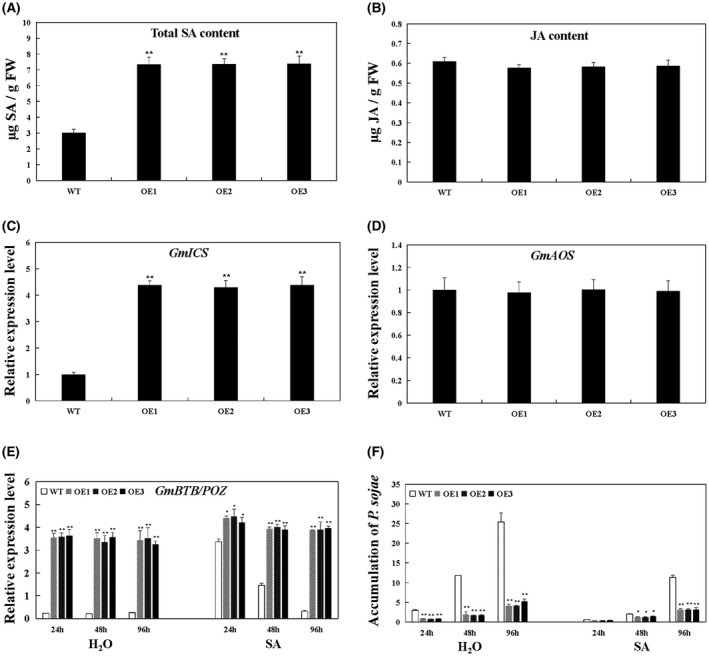
Investigation of the relationship between GmBTB/POZ and the salicylic acid (SA) pathway in soybean. (A) SA contents in leaves of transgenic and wild‐type (WT) soybean. FW, fresh weight. (B) Jasmonic acid (JA) contents in leaves of transgenic and WT soybean. (C) Relative transcript level of GmICS in GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) transgenic and WT soybean. The level of the control sample (WT plants) was set to unity. (D) Relative transcript level of GmAOS in GmBTB/POZ‐OE transgenic and WT soybean. The level in the control sample (WT plants) was set to unity. (E) Expression patterns of GmBTB/POZ in WT and GmBTB/POZ transgenic soybean with treatment of H2O or SA (0.5 mm). (F) Relative biomass of Phytophthora sojae in infected cotyledons after 24, 48 and 96 h of H2O or SA (0.5 mm) treatment based on P. sojae TEF1 transcript levels. The housekeeping gene of soybean GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean.
Pre‐treatment with SA increases resistance to P. sojae infection in GmBTB/POZ transgenic soybean plants
To further confirm the relationship between GmBTB/POZ and the SA pathway in soybean, we analysed the expression of GmBTB/POZ in WT and GmBTB/POZ transgenic soybean lines treated with H2O (control) and SA (0.5 mm). As shown in Fig. 8E, GmBTB/POZ transcript abundance was much higher in GmBTB/POZ transgenic soybean than in WT plants on treatment with H2O at 24, 48 and 96 h. In plants on SA treatment, GmBTB/POZ expression was significantly up‐regulated at 48 and 96 h in GmBTB/POZ‐OE plants vs. WT. To further determine whether the GmBTB/POZ‐regulated resistance response was related to SA accumulation, we tested the relative biomass of P. sojae in GmBTB/POZ‐OE and WT plants with application of exogenous SA. As expected, SA‐treated plants displayed clearly reduced pathogen biomass compared with H2O‐treated plants (Fig. 8F). More interestingly, the differences in the relative biomass of P. sojae between GmBTB/POZ‐OE and WT lines were not obvious at 24 hpi, after which the difference became increasingly noticeable (Fig. 8F). Therefore, it is possible to speculate that GmBTB/POZ improves the defence resistance to P. sojae probably by affecting SA accumulation. Moreover, NPR1 is the key node of the SA signalling pathway whose expression is affected by the binding of SA and can mediate the SA‐dependent activation of PR genes (Cao et al., 1997; Delaney et al., 1995; Rochon et al., 2006). In the present study, the transcript levels of GmNPR1 (NM_001251745) in GmBTB/POZ‐OE were significantly higher than those in WT plants (Fig. S5). In summary, these data suggest that GmBTB/POZ plays an integral role in the resistance of soybean to P. sojae via a mechanism that is mainly linked to the SA signalling pathway.
Discussion
BTB/POZ proteins are involved in the modulation of various biological processes, including transcriptional regulation (Ahmad et al., 2003; Melnick et al., 2000), cytoskeleton regulation (Bomont et al., 2000; Ziegelbauer et al., 2001) and protein ubiquitination/degradation (Kobayashi et al., 2004; Xu et al., 2003). However, except for NPR1, the potential functions of BTB/POZ proteins in soybean in the plant response to pathogen infection have remained largely uncharacterized. In this study, we identified GmBTB/POZ, a novel BTB/POZ gene in soybean, and determined that overexpression of GmBTB/POZ serves to increase resistance to P. sojae.
Changes in gene expression induced by pathogen infection play important roles in the responses of plants to pathogens (Buscaill and Rivas, 2014; Koh et al., 2005; Tsuda and Somssich, 2015). In this work, we analysed the effects of GmBTB/POZ expression in the leaves of P. sojae‐resistant and P. sojae‐susceptible soybean cultivars. We detected significant induction of GmBTB/POZ in ‘Suinong 10’, with significant differences in expression in the resistant vs. susceptible cultivar, suggesting that GmBTB/POZ plays an important role in the response of soybean to P. sojae. Transgenic soybean overexpressing GmBTB/POZ displayed significantly higher resistance to P. sojae compared with WT.
ROS function as signalling molecules and are involved in the regulation of many growth and developmental processes in plants and in responses to environmental cues (Hirt, 2006; Mittler, 2002). Furthermore, ROS are involved in host–pathogen interactions, including interactions with fungal and oomycete pathogens, and the scavenging of excess ROS can improve plant resistance to many pathogens (Shetty et al., 2003, 2008). Thus, we tested the relative ROS levels in GmBTB/POZ‐OE and WT lines during infection with P. sojae, and these levels were significantly lower in GmBTB/POZ‐OE plants than in WT. Moreover, DAB staining showed that H2O2 levels in leaves when quantified at 24 hpi were clearly lower in the GmBTB/POZ‐OE group relative to the WT group. Further analysis indicated that GmBTB/POZ positively regulates the activities and expression of the enzymatic antioxidants SOD and POD. Within a cell, SOD provides the first line of defence against ROS (Alscher et al., 2002). Conversely, POD serves as an efficient scavenger of ROS, preventing cellular damage (Tewari et al., 2006). Therefore, these findings suggest that GmBTB/POZ improves antioxidant enzymatic activity in soybean, thereby eliminating ROS in response to P. sojae, and thus providing sufficient protection against oxidative damage.
Phytohormones, such as SA and JA, mediate the activation of sophisticated plant defence mechanisms to ward off pathogen attack (Alazem and Lin, 2015; Robert‐Seilaniantz et al., 2011; Vlot et al., 2009). PR1 and PR5 usually act as effector genes for SAR, a process mediated by SA; high expression levels of these genes indicate that SA signalling has been activated (He et al., 2007; Ward et al., 1991). Herein, we determined that the SA marker genes, such as GmNPR1, GmPR1 and GmPR5, were constitutively induced in the group of GmBTB/POZ‐OE soybean plants. Moreover, SA accumulation and the transcript abundance of SA biosynthesis genes were much higher in GmBTB/POZ‐OE leaves than in WT. By contrast, JA levels and GmAOS expression differed little between GmBTB/POZ transgenic and WT plants. These results suggest that GmBTB/POZ plays an important role in the regulation of SA‐dependent defence signalling and downstream defence gene expression, but not JA signalling. Interestingly, exogenous application of SA also demonstrated that SA positively regulated GmBTB/POZ expression. Combining the results of SA accumulation and the transcript abundance of SA synthesis genes in GmBTB/POZ‐OE soybean plants, these data suggest that GmBTB/POZ up‐regulates the related genes for SA synthesis, and SA enhances the expression of GmBTB/POZ, creating a positive feedback loop of the SA signal. These results are similar to previous reports indicating that EDS1 and PAD4 are located upstream of SA synthesis genes and may promote the synthesis of SA, and also suggesting that there is a positive feedback mechanism involving SA that enhances the expression of EDS1 and PAD4 (Brodersen et al., 2006; Shah, 2003). Furthermore, exogenous application of SA limited the increase in biomass of P. sojae. These findings suggest that GmBTB/POZ plays a positive role in P. sojae resistance and that this defence response might be dependent on SA.
Taken together, we speculate that GmBTB/POZ plays a crucial role in soybean resistance to P. sojae, which depends mainly on the SA signalling pathway. In addition, some reports have suggested that many BTB/POZ proteins serve as transcription factors, repressors and transcriptional activators (Ahmad et al., 2003; Cao et al., 1997; Hu et al., 2010; Melnick et al., 2000). Our results showed that GmBTB/POZ is a nuclear‐localized protein with no transcriptional activator activity. However, it is by no means clear whether, like some BTB/POZ domain proteins, GmBTB/POZ functions as a transcriptional repressor. The BTB/POZ domain usually also acts as a protein–protein interaction module that can both self‐associate and interact with non‐BTB/POZ proteins (Geyer et al., 2003; Huynh and Bardwell, 1998). Further studies are needed to identify whether GmBTB/POZ can form dimers or polymers and how this complex takes part in the regulation of pathogen defence responses. Such studies should shed light on the functional mechanism of GmBTB/POZ in biotic stress responses in soybean.
Experimental Procedures
Plant materials and pathogen inoculation
Resistant cultivar ‘Suinong 10’, which is resistant to the dominant physiological race 1 of P. sojae in Heilongjiang, China (Zhang et al., 2010), was used for expression analysis and gene isolation in this study. ‘Suinong 10’ soybean plants were grown at 22 °C in a 16‐h/8‐h light/dark photoperiod with 70% relative humidity.
Susceptible cultivar ‘Dongnong 50’, a soybean cultivar with susceptibility to the dominant physiological race 1 of P. sojae, was obtained from the Key Laboratory of Soybean Biology in the Chinese Ministry of Education, Harbin, China, and used for P. sojae treatment and gene transformation experiments in this study.
Phytophthora sojae race 1 (PSR01) was isolated from infected soybean plants in Heilongjiang, China (Zhang et al., 2010) and cultivated at 25 °C for 7 days on V8 juice agar in a polystyrene dish.
GmBTB/POZ cloning, sequence analysis and vector construction
‘Suinong 10’ mRNA was used to clone the full‐length cDNA of GmBTB/POZ. The PCR products were ligated into the PMD18‐T vector (TaKaRa, Dalian, China). Phylogenetic analysis and amino acid sequence alignment of GmBTB/POZ and other BTB/POZs were performed using Molecular Evolutionary Genetics Analysis (MEGA) software 5.1 and DNAMAN, respectively. The ORF of GmBTB/POZ was cloned into the vector pCAMBIA3301 with the bar gene and 4 × Myc tag as the selectable marker under the control of the cauliflower mosaic virus 35S (CaMV35S) promoter to overexpress the GmBTB/POZ gene. The primers used for gene isolation are shown in Table S1 (see Supporting Information).
Soybean genetic transformation
Soybean ‘Dongnong 50’ was transformed via the Agrobacterium‐mediated transformation method described by Paz et al. (2004). The 35S:GmBTB/POZ recombinant plasmid was transferred into Agrobacterium tumefaciens strain LBA4404 via the freeze–thaw method as described by Holsters et al. (1978). Strain LBA4404 containing recombinant vector 35S:GmBTB/POZ with growth phases (OD600 = 0.6) and concentration (OD600 = 0.5) was used to infect the cotyledonary nodes of ‘Dongnong 50’ soybean as the explants. Then, the explants were cultured for 7 days on shoot induction medium. GmBTB/POZ‐OE plants were identified by PCR and immunoblotting using Myc antibody (Abmart, code number M20002M).
Quantitative RT‐PCR
The extraction of total RNA and reverse transcription were performed using TRIzol reagent (Invitrogen, Shanghai, China) and ReverTra Ace Kit (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. qRT‐PCR was employed to measure the gene expression levels using a real‐time RT‐PCR kit (Toyobo, Osaka, Japan) with a LightCycler® 96 System (Roche, California, USA). The gene expression levels were calculated by the 2−ΔΔCt method with GmEF1 (GenBank accession no. NM_001248778) as the internal control. The qPCR analyses were performed with three technical replicates. The primers used for expression analysis are shown in Table S1.
Subcellular localization
To determine the subcellular localization of GmBTB/POZ, the ORF of GmBTB/POZ was ligated into the vector pCAMBIA1302 under the control of the 35S promoter. The recombinant plasmid, 35S:GmBTB/POZ‐GFP, was introduced into Arabidopsis protoplasts by polyethylene glycol (PEG)‐mediated transfection (Yoo et al., 2007). The GFP signals were imaged using a TCS SP2 confocal spectral microscope imaging system (Leica, Solms, Germany). The primers used for subcellular localization analysis are shown in Table S1.
Transcriptional activation analysis in yeast cells
The ORF of GmBTB/POZ was amplified into pGBKT7, generating the fusion construct pGBKT7‐GmBTB/POZ. The yeast strain AH109 was transformed with pGBKT7‐53+pGADT7‐T, pGBKT7‐GmBTB/POZ, pGBKT7‐GmWRKY31 and pGBKT7. The transformed cells were grown on SD (‐Trp), SD (‐Trp/‐His/‐Ade) and SD (‐Trp/‐His/‐Ade/α‐gal). The transactivation activity of proteins was detected by the growth status and α‐gal activity. The primers used for transcriptional activation analysis are shown in Table S1.
Assessment of plant disease responses
To assay for plant resistance to pathogen infection, artificial inoculation procedures were performed as described by Dou et al. (2003) and Morrison and Thorne (1978) with minor modifications. Soybean roots and living cotyledons at the V1 stage were inoculated with P. sojae zoospores (approximately 1 × 105 spores/mL). Disease symptoms on infected roots and infected cotyledons were photographed with a Nikon D7000 camera. The lesions of the challenged cotyledons were measured by ImageJ software (https://imagej.nih.gov/ij/index.html).
Measurement of ROS generation and DAB staining
ROS levels were detected according to the instructions supplied with the Reactive Oxygen Species Assay Kit (Beyotime Institute of Biotechnology, Haimen, China), and fluorescence was assayed as described by Qian et al. (2009). H2O2 levels were visually detected with DAB as described by Zhu et al. (2014). The hypocotyls of soybean plants were treated with P. sojae zoospores to investigate the response of GmBTB/POZ‐OE soybean lines to oxidative stress. The artificial inoculation was performed according to the method described by Ward et al. (1979) and Morris et al. (1991).
Determination of antioxidant enzyme activity and plant hormone levels
SOD and POD enzymes were extracted from approximately 0.1 g of leaves using 0.8 mL of ice‐cold 25 mm HEPES buffer (pH 7.8) containing 0.2 mm ethylenediaminetetraacetic acid (EDTA), 2 mm ascorbate and 2% polyvinylpyrrolidone (PVP). The enzyme activities were measured as described by Wang et al. (2011). SA and JA levels were determined as described by Pan et al. (2010) and Zhu et al. (2014).
Statistical analysis
All experiments in this study were performed at least three times. Statistical significance between different measurements was examined by Student’s t‐test. A difference was considered to be statistically significant when *P < 0.05 or **P < 0.01. Bars indicate the standard error of the mean.
Author Contributions
P.X., S.Z. and C.Z. designed the research. C.Z., H.G., R.L. and D.H. performed the research. L.W. and J.W. analysed the data. P.X., S.Z. and C.Z. wrote the article.
Supporting information
Fig. S1 Bimolecular fluorescence complementation (BiFC) analyses of GmBTB/POZ interactions with GmLHP1. GmBTB/POZ‐YFPN+ and GmLHP1‐YFPC were co‐transfected into Arabidopsis protoplasts. Scale bars indicate 10 µm.
Fig. S2 Nucleotide and amino acid sequences of GmBTB/POZ. The BTB/POZ domain is shown in shadow.
Fig. S3 Phylogenetic analysis of GmBTB/POZ with orthologues from other plant species.
Fig. S4 Alignment of amino acid sequences of GmBTB/POZ with orthologues from other plant species.
Fig. S5 The expression analysis of GmPALs and GmNPR1 in GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) transgenic and wild‐type (WT) plants. The expression level of the control sample was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean.
Table S1 Primer sequences used in this study.
Acknowledgements
Thanks are due to Jeffrey D. Boehm Jr for revising the manuscript. This work was supported by National Key Research and Development Program of China, Ministry of Science and Technology of People's Republic of China (2017YFD0101300). National Natural Science Foundation of China (Grant Nos. 31171577 and 31671719). Outstanding Talents and Innovative Team of Agricultural Scientific Research, Ministry of Agriculture and Rural Affairs of the People's Republic of China. Young and Middle‐aged scientific and Technological innovation leader Ministry of Science and Technology of People's Republic of China. Academic backbone of Northeast Agricultural University (17XG21). Natural Science Foundation of Heilongjiang Province (Grant Nos. JC201308 and C2015010). Changjiang Scholar Candidates Program for Provincial Universities in Heilongjiang, Education Department of Heilongjiang Provincial (2013CJHB003). The authors have no conflicts of interest to declare.
References
- Ahmad, K.F. , Melnick, A. , Lax, S. , Bouchard, D. , Liu, J. , Kiang, C.L. , Mayer, S. , Takahashi, S. , Licht, J.D. and Prive, G.G. (2003) Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell, 12, 1551–1564. [DOI] [PubMed] [Google Scholar]
- Alazem, M. and Lin, N.S. (2015) Roles of plant hormones in the regulation of host‐virus interactions. Mol. Plant Pathol. 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher, R.G. , Erturk, N. and Heath, L.S. (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. (1999) Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Badawi, G.H. , Yamauchi, Y. , Shimada, E. , Sasaki, R. , Kawano, N. , Tanaka, K. and Tanaka, K. (2004) Enhanced tolerance to salt stress and water deficit by overexpressing superoxide dismutase in tobacco (Nicotiana tabacum) chloroplasts. Plant Sci. 166, 919–928. [Google Scholar]
- Bardwell, V.J. and Treisman, R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev. 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Baxter, A. , Mittler, R. and Suzuki, N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Bomont, P. , Cavalier, L. , Blondeau, F. , Hamida, C.B. , Belal, S. , Tazir, M. , Demir, E. , Topaloglu, H. , Korinthenberg, R. , Tüysüz, B. , Landrieu, P. , Hentati, F. and Koenig, M . (2000) The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 26, 370–374. [DOI] [PubMed] [Google Scholar]
- Brodersen, P. , Petersen, M. , Nielsen, H.B. , Zhu, S. , Newman, M.A. , Shokat, K.M. , Rietz, S. , Parker, J. and Mundy, J. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J. 47, 532–546. [DOI] [PubMed] [Google Scholar]
- Buscaill, P. and Rivas, S. (2014) Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. [DOI] [PubMed] [Google Scholar]
- Cao, H. , Glazebrook, J. , Clarke, J.D. , Volko, S. and Dong, X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Catinot, J. , Buchala, A. , Aboumansour, E. and Métraux, J.P. (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Lett. 582, 473–478. [DOI] [PubMed] [Google Scholar]
- Chandran, D. , Rickert, J. , Huang, Y. , Steinwand, M.A. , Marr, S.K. and Wildermuth, M.C. (2014) Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe. 15, 506–513. [DOI] [PubMed] [Google Scholar]
- Chen, Z. and Klessig, D.F. (1991) Identification of a soluble salicylic acid‐binding protein that may function in signal transduction in the plant disease‐resistance response. Proc. Natl. Acad. Sci. USA, 88, 8179–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Zheng, Z. , Huang, J. , Lai, Z. and Fan, B. (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav. 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians, M.J. , Gingerich, D.J. , Hua, Z. , Lauer, T.D. and Vierstra, R.D. (2012) The light‐response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 160, 118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet, J. and Hirt, H. (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226. [DOI] [PubMed] [Google Scholar]
- Collins, T. , Stone, J.R. and Williams, A.J. (2001) All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21, 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki, G. , Nagy, A. and Jaenisch, R. (2001) Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, G. , Alland, L. , Hong, S.H. , Wong, C.W. , Depinho, R.A. and Dejean, A. (1998) Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia‐associated PLZF protein. Oncogene, 16, 2549–2556. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Friedrich, L. and Ryals, J.A. (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA, 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Zhu, J. , Reid, A. , Strutt, P. , Guidez, F. , Zhong, H.J. , Wang, Z.Y. , Licht, J. , Waxman, S. , Chomienne, C. , Chen, Z. , Zelent, A. and Chen, S.J. (1996) Amino‐terminal protein‐protein interaction motif (POZ‐domain) is responsible for activities of the promyelocytic leukemia zinc finger‐retinoic acid receptor‐alpha fusion protein. Proc. Natl. Acad. Sci. USA, 93, 3624–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. (1998) SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Dou, D.L. , Wang, B.S. , Zhu, S.W. , Tang, Y.X. , Wang, Z.X. , Sun, J.S. , Ren‐Jing, L.I. and Zhang, Z.N. (2003) Transgenic tobacco with NDRl gene improved its resistance to two fungal diseases. Sci. Agric. Sin. 36, 1120–1124. [Google Scholar]
- Du, X.M. , Yin, W.X. , Zhao, Y.X. and Zhang, H. (2001) The production and scavenging of reactive oxygen species in plants. Sheng Wu Gong Cheng Xue Bao (Chin. J. Biotechnol.), 17, 121–125. [PubMed] [Google Scholar]
- Fan, S. , Dong, L. , Han, D. , Zhang, F. , Wu, J. , Jiang, L. , Cheng, Q. , Li, R. , Lu, W. and Meng, F. (2017) GmWRKY31 and GmHDL56 enhance resistance to Phytophthora sojae by regulating defense‐related gene expression in soybean. Front. Plant Sci. 8, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, M. , He, Y.J. , Borchers, C. and Xiong, Y. (2003) Targeting of protein ubiquitination by BTB‐Cullin 3‐Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Geyer, R. , Wee, S. , Anderson, S. , III, J.Y. and Wolf, D.A. (2003) BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell, 12, 783–790. [DOI] [PubMed] [Google Scholar]
- Gingerich, D.J. , Gagne, J.M. , Salter, D.W. , Hellmann, H. , Estelle, M. , Ma, L. and Vierstra, R.D. (2005) Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric‐a‐brac (BTB) protein family to form essential ubiquitin‐protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280, 18810–18821. [DOI] [PubMed] [Google Scholar]
- Gingerich, D.J. , Hanada, K. , Shiu, S.H. and Vierstra, R.D. (2007) Large‐scale, lineage‐specific expansion of a bric‐a‐brac/tramtrack/broad complex ubiquitin‐ligase gene family in rice. Plant Cell, 19, 2329–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- He, K. , Gou, X. , Yuan, T. , Lin, H. , Asami, T. , Yoshida, S. , Russell, S.D. and Li, J. (2007) BAK1 and BKK1 regulate brassinosteroid‐dependent growth and brassinosteroid‐independent cell‐death pathways. Curr. Biol. 17, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Hirt, H. (2006) Reactive oxygen species signaling in plants. Antioxid. Redox Signal. 8, 1757–1764. [DOI] [PubMed] [Google Scholar]
- Holsters, M. , De, W.D. , Depicker, A. , Messens, E. , Van, M.M. and Schell, J. (1978) Transfection and transformation of Agrobacterium tumefaciens . Mol. Gen. Genet. 163, 181–187. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Yuan, W. , Tang, M. , Wang, Y. , Fan, X. , Mo, X. , Li, Y. , Ying, Z. , Wan, Y. and Ocorr, K. (2010) KBTBD7, a novel human BTB‐kelch protein, activates transcriptional activities of SRE and AP‐1. BMB Rep. 43, 17–22. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Gu, M. , Lai, Z. , Fan, B. , Shi, K. , Zhou, Y.H. , Yu, J.Q. and Chen, Z. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven, R. and Kogel, K.H. (2003) Reactive oxygen intermediates in plant‐microbe interactions: who is who in powdery mildew resistance? Planta, 216, 891–902. [DOI] [PubMed] [Google Scholar]
- Huynh, K.D. and Bardwell, V.J. (1998) The BCL‐6 POZ domain and other POZ domains interact with the co‐repressors N‐CoR and SMRT. Oncogene, 17, 2473–2484. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kobayashi, A. , Kang, M.I. , Okawa, H. , Ohtsuji, M. , Zenke, Y. , Chiba, T. , Igarashi, K. and Yamamoto, M. (2004) Oxidative stress sensor keap1 functions as an adaptor for cul3‐based E3 ligase to proteasomal degradation of nrf2. Mol. Cell. Biol. 24, 7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, S. , Edwards, H. , Stacey, G. and Somerville, S. (2005) Loss‐of‐function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum . Plant Physiol. 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. , Senkevich, T.G. and Chernos, V.I. (1992) A family of DNA virus genes that consists of fused portions of unrelated cellular genes. Trends Biochem. Sci. 17, 213–214. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M.J. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lcvan, L. and Eavan, S. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Lin, W.C. , Lu, C.F. , Wu, J.W. , Cheng, M.L. , Lin, Y.M. , Yang, N.S. , Black, L. , Green, S.K. , Wang, J.F. and Cheng, C.P. (2004) Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13, 567–581. [DOI] [PubMed] [Google Scholar]
- Loon, L.C.V. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Makandar, R. , Essig, J.S. , Schapaugh, M.A. , Trick, H.N. and Shah, J. (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant‐Microbe Interact. 19, 123–129. [DOI] [PubMed] [Google Scholar]
- McKersie, B.D. and Jones, K.S. (1999) Winter survival of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol. 119, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchiorre, M. , Robert, G. , Trippi, V. , Racca, R. and Lascano, H.R. (2009) Superoxide dismutase and glutathione reductase overexpression in wheat protoplast: photooxidative stress tolerance and changes in cellular redox state. Plant Growth Regul. 57, 57–68. [Google Scholar]
- Melnick, A. , Ahmad, K.F. , Arai, S. , Polinger, A. , Ball, H. , Borden, K.L. , Carlile, G.W. , Prive, G.G. and Licht, J.D. (2000) In‐depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20, 6550–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meur, G. , Budatha, M. , Srinivasan, T. , Kumar, K.R.R. , Gupta, A.D. and Kirti, P.B. (2008) Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Physiol. Plant. 133, 765–775. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Morris, P.F. , Savard, M.E. and Ward, E.W.B. (1991) Identification and accumulation of isoflavonoids and isoflavone glucosides in soybean leaves and hypocotyls in resistance responses to Phytophthora megasperma f.sp. glycinea . Physiol. Mol. Plant Pathol. 39, 229–244. [Google Scholar]
- Morrison, R.H. and Thorne, J.C. (1978) Inoculation of detached cotyledons for screening soybeans against two races of Phytophthora megasperma Var. sojae 1. Crop Sci. 18, 1089–1091. [Google Scholar]
- Nandinip, S. , Hansjlyngs, J. , Jensdue, J. , Davidb, C. and Hshekar, S. (2008) Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280. [Google Scholar]
- Oyake, T. , Itoh, K. , Motohashi, H. , Hayashi, N. , Hoshino, H. , Nishizawa, M. , Yamamoto, M. and Igarashi, K. (1996) Bach proteins belong to a novel family of BTB‐basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF‐E2 site. Mol. Cell. Biol. 16, 6083–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Welti, R. and Wang, X. (2010) Quantitative analysis of major plant hormones in crude plant extracts by high‐performance liquid chromatography‐mass spectrometry. Nat. Protoc. 5, 986–992. [DOI] [PubMed] [Google Scholar]
- Paz, M.M. , Shou, H. , Guo, Z. , Zhang, Z. , Banerjee, A.K. and Wang, K. (2004) Assessment of conditions affecting Agrobacterium‐mediated soybean transformation using the cotyledonary node explant. Euphytica, 136, 167–179. [Google Scholar]
- Pintard, L. , Willems, A. and Peter, M. (2004) Cullin‐based ubiquitin ligases: Cul3–BTB complexes join the family. EMBO J. 23, 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, H. , Chen, W. , Sun, L. , Jin, Y. , Liu, W. and Fu, Z. (2009) Inhibitory effects of paraquat on photosynthesis and the response to oxidative stress in Chlorella vulgaris . Ecotoxicology, 18, 537–543. [DOI] [PubMed] [Google Scholar]
- Rasmussen, M.W. , Roux, M. , Petersen, M. and Mundy, J. (2012) MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci. 3, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal, H.C. , Singh, N.K. and Sharma, T.R. (2013) Conservation, divergence, and genome‐wide distribution of PAL and POX A gene families in plants. Int. J. Genomics, 2013, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rochon, A. , Boyle, P. , Wignes, T. , Fobert, P.R. and Despres, C. (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C‐terminal cysteines. Plant Cell, 18, 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas‐Vidal, E. , Meijer, A.H. , Cheng, X. and Spaink, H.P. (2005) Genomic annotation and expression analysis of the zebrafish Rho small GTPase family during development and bacterial infection. Genomics, 86, 25–37. [DOI] [PubMed] [Google Scholar]
- Schlueter, J. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Schneider, D.S. (2002) Plant immunity and film noir: what gumshoe detectives can teach us about plant‐pathogen interactions. Cell, 109, 537–540. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2003) The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. [DOI] [PubMed] [Google Scholar]
- Shetty, N.P. , Jørgensen, H.J.L. , Jensen, J.D. , Collinge, D.B. and Shetty, H.S. (2008) Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280. [Google Scholar]
- Shetty, N.P. , Kristensen, B.K. , Newman, M.A. Miller, K., Gregersen, P.L. and Jorgensen, H.J.L. (2003) Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol. Mol. Plant Pathol. 62, 333–346. [Google Scholar]
- Soosaar, J.L. , Burch‐Smith, T.M. and Dinesh‐Kumar, S.P. (2005) Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 3, 789–798. [DOI] [PubMed] [Google Scholar]
- Stogios, P.J. , Downs, G.S. , Jauhal, J.J. , Nandra, S.K. and Priv, G.G. (2005) Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari, R.K. , Kumar, P. and Sharma, P.N. (2006) Magnesium deficiency induced oxidative stress and antioxidant responses in mulberry plants. Sci. Hortic. 108, 7–14. [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. and Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. and Somssich, I.E. (2015) Transcriptional networks in plant immunity. New Phytol. 206, 932–947. [DOI] [PubMed] [Google Scholar]
- Vidhyasekaran, P. (2015) Plant hormone signaling systems in plant innate immunity. Plant Signal Commun. 2, 27–122. [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Wang, S.D. , Zhu, F. , Yuan, S. , Yang, H. , Xu, F. , Shang, J. , Xu, M.Y. , Jia, S.D. , Zhang, Z.W. and Wang, J.H. (2011) The roles of ascorbic acid and glutathione in symptom alleviation to SA‐deficient plants infected with RNA viruses. Planta, 234, 171–181. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Gao, P. , Huang, M. , Chen, H. , Yang, Z.R. and Sun, Q. (2015) Effects of high temperature on the activity and expression of antioxidative enzymes in rice flag leaves during the flowering stage. Plant Sci. J. 33, 355–361. [Google Scholar]
- Ward, E.R. , Uknes, S.J. , Williams, S.C. , Dincher, S.S. , Wiederhold, D.L. , Alexander, D.C. , Ahlgoy, P. , Métraux, J.P. and Ryals, J.A. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell, 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.W.B. , Lazarovits, G. , Unwin, C.H. and Buzzell, R.I. (1979) Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae . Phytopathology, 69, 951–955. [Google Scholar]
- Weber, H. , Bernhardt, A. , Dieterle, M. , Hano, P. , Mutlu, A. , Estelle, M. , Genschik, P. and Hellmann, H. (2005) Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ‐MATH protein family. Plant Physiol. 137, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Woodger, F.J. , Jacobsen, J.V. and Gubler, F. (2004) GMPOZ, a BTB/POZ domain nuclear protein, is a regulator of hormone responsive gene expression in barley aleurone. Plant Cell Physiol. 45, 945–950. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Wei, Y. , Reboul, J. , Vaglio, P. , Shin, T.H. , Vidal, M. , Elledge, S.J. and Harper, J.W. (2003) BTB proteins are substrate‐specific adaptors in an SCF‐like modular ubiquitin ligase containing CUL‐3. Nature, 425, 316–321. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu, B. and Liu, Y. (2003) Effects of salt stress on the metabolism of active oxygen in seedlings of annual halophyte Glycine soja . Acta Bot. Boreal‐Occidental. Sin. 23, 18–22. [Google Scholar]
- Zhang, C. , Wang, X. , Zhang, F. , Dong, L. , Wu, J. , Cheng, Q. , Qi, D. , Yan, X. , Jiang, L. and Fan, S. (2017) Phenylalanine ammonia‐lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 7, 7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Xu, P. , Wu, J. , Xue, A.G. , Zhang, J. , Li, W. , Chen, C. , Chen, W. and Lv, H. (2010) Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 94, 87–91. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Fan, W. , Kinkema, M. , Li, X. and Dong, X. (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR‐1 gene. Proc. Natl. Acad. Sci. USA, 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F. , Xi, D.H. , Deng, X.G. , Peng, X.J. , Tang, H. , Chen, Y.J. , Jian, W. , Feng, H. and Lin, H.H. (2014) The chilli veinal mottle virus regulates expression of the tobacco mosaic virus resistance gene N and jasmonic acid/ethylene signaling is essential for systemic resistance against chilli veinal mottle virus in tobacco. Plant Mol. Biol. Rep. 32, 382–394. [Google Scholar]
- Ziegelbauer, J. , Shan, B. , Yager, D. , Larabell, C. , Hoffmann, B. and Tjian, R. (2001) Transcription factor MIZ‐1 is regulated via microtubule association. Mol. Cell, 8, 339–349. [DOI] [PubMed] [Google Scholar]
- Zollman, S. , Godt, D. , Prive, G.G. , Couderc, J.L. and Laski, F.A. (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila . Proc. Natl. Acad. Sci. USA, 91, 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Bimolecular fluorescence complementation (BiFC) analyses of GmBTB/POZ interactions with GmLHP1. GmBTB/POZ‐YFPN+ and GmLHP1‐YFPC were co‐transfected into Arabidopsis protoplasts. Scale bars indicate 10 µm.
Fig. S2 Nucleotide and amino acid sequences of GmBTB/POZ. The BTB/POZ domain is shown in shadow.
Fig. S3 Phylogenetic analysis of GmBTB/POZ with orthologues from other plant species.
Fig. S4 Alignment of amino acid sequences of GmBTB/POZ with orthologues from other plant species.
Fig. S5 The expression analysis of GmPALs and GmNPR1 in GmBTB/POZ‐overexpressing (GmBTB/POZ‐OE) transgenic and wild‐type (WT) plants. The expression level of the control sample was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analysed using Student’s t‐test (*P < 0.05, **P < 0.01). Bars indicate the standard error of the mean.
Table S1 Primer sequences used in this study.


