Pathogen identification is vital for unknown infectious outbreaks, especially for dual or multiple infections. Sleeping disease (SD) in crabs causes great economic losses to aquaculture worldwide. Here we report the discovery and identification of a novel virus in mud crabs with multiple infections that was not previously detected by molecular, immune, or traditional electron microscopy (EM) methods. High-resolution structures of pathogenic viruses are essential for a molecular understanding and developing new disease prevention methods. The three-dimensional (3D) structure of the mud crab tombus-like virus (MCTV) and mud crab dicistrovirus (MCDV) determined in this study could assist the development of antiviral inhibitors. The identification of a novel virus in multiple infections previously missed using other methods demonstrates the usefulness of this strategy for investigating multiple infectious outbreaks, even in humans and other animals.
KEYWORDS: cryo-EM, virus, virion structure
ABSTRACT
Viruses associated with sleeping disease (SD) in crabs cause great economic losses to aquaculture, and no effective measures are available for their prevention. In this study, to help develop novel antiviral strategies, single-particle cryo-electron microscopy was applied to investigate viruses associated with SD. The results not only revealed the structure of mud crab dicistrovirus (MCDV) but also identified a novel mud crab tombus-like virus (MCTV) not previously detected using molecular biology methods. The structure of MCDV at a 3.5-Å resolution reveals three major capsid proteins (VP1 to VP3) organized into a pseudo-T=3 icosahedral capsid, and affirms the existence of VP4. Unusually, MCDV VP3 contains a long C-terminal region and forms a novel protrusion that has not been observed in other dicistrovirus. Our results also reveal that MCDV can release its genome via conformation changes of the protrusions when viral mixtures are heated. The structure of MCTV at a 3.3-Å resolution reveals a T= 3 icosahedral capsid with common features of both tombusviruses and nodaviruses. Furthermore, MCTV has a novel hydrophobic tunnel beneath the 5-fold vertex and 30 dimeric protrusions composed of the P-domains of the capsid protein at the 2-fold axes that are exposed on the virion surface. The structural features of MCTV are consistent with a novel type of virus.
IMPORTANCE Pathogen identification is vital for unknown infectious outbreaks, especially for dual or multiple infections. Sleeping disease (SD) in crabs causes great economic losses to aquaculture worldwide. Here we report the discovery and identification of a novel virus in mud crabs with multiple infections that was not previously detected by molecular, immune, or traditional electron microscopy (EM) methods. High-resolution structures of pathogenic viruses are essential for a molecular understanding and developing new disease prevention methods. The three-dimensional (3D) structure of the mud crab tombus-like virus (MCTV) and mud crab dicistrovirus (MCDV) determined in this study could assist the development of antiviral inhibitors. The identification of a novel virus in multiple infections previously missed using other methods demonstrates the usefulness of this strategy for investigating multiple infectious outbreaks, even in humans and other animals.
INTRODUCTION
Viruses cause serious illness and death in humans, animals, plants, and bacteria. To date, more than 30,000 viruses have been identified and classified into 125 different virus families (1), but many more are yet to be identified and investigated. The enormous number of different species brings great challenges for viral identification and antiviral precautions during epidemic outbreaks.
The mud crab Scylla paramamosain is an economically important aquaculture species in China, India, Australia, and many other countries. However, the prevalence of sleeping disease (SD) in this species results in significant economic losses. Previous molecular investigations revealed two major viral pathogens: mud crab reovirus (MCRV) and mud crab dicistrovirus (MCDV), with diameters of 72 and 30 nm, respectively (2–5).
MCDV belongs to the family Dicistroviridae, which contains three genera: Cripavirus, Triatovirus, and Aparavirus (6). The family derives its name from the two open reading frames (ORFs) or cistrons in the positive single-strand RNA (+ssRNA) genome. The 5′ ORF encodes the nonstructural proteins (viral polymerase, protease, and helicase), while the 3′ ORF codes for a polyprotein precursor of the mature capsid proteins VP1, VP2, VP3, and VP4 (6). Sixty copies of three of these major capsid proteins (VP1, VP2, and VP3) comprise the capsid shell and are arranged in a P=3 (pseudo-T=3) lattice.
Crabs are invertebrates that lack a well-established immune system capable of preventing virus infections. Thus, external agents and methods are essential for preventing viral diseases in crabs and other organisms lacking well-constituted immune systems. To assist in the design of an effective method to prevent future SD outbreaks, single-particle cryo-electron microscopy (cryo-EM) reconstruction was applied to study the structure of the two previously reported viruses. Due to its larger size, MCRV is easily recognized, and a low-resolution structure has been reported (2). During the process of resolving the structure of MCDV in the present study, a different virus with distinct features different from those of MCDV, but almost the same size as MCDV, was unexpectedly observed and identified. Interestingly, neither previous molecular assays nor traditional EM techniques were able to distinguish this novel virus in mud crabs simultaneously infected with three viruses.
RESULTS
Cryo-EM detection of two viruses of approximately equal size coinfecting mud crabs.
Previous reports revealed the existence of MCDV (3, 4) and MCRV (5) in mud crabs with SD, with diameters of ∼30 and ∼72 nm, respectively. Sequence analyses revealed that MCDV belongs to the family Dicistroviridae and harbors at least three different capsid proteins: VP1, VP2, and VP3. Based on the results of negative staining and cryo-EM, 30-nm particles were successfully isolated. The 30-nm particles were initially believed to be MCDV. Following reference-free 2D classification, initial model building, and reconstruction, we obtained a 3.8-Å density map (Fig. 1). At this resolution, the densities of protein main chains and some side chains are visible, and we were able to annotate the three proteins in the asymmetric unit. However, unexpectedly, atomic model building failed due to mismatches in side chain densities, and there was no density at all for some regions. When we tried to align the three proteins, they aligned surprisingly well except for the terminal regions. Most side chain densities were almost the same for the three proteins. These features suggested that the three proteins were chemically identical.
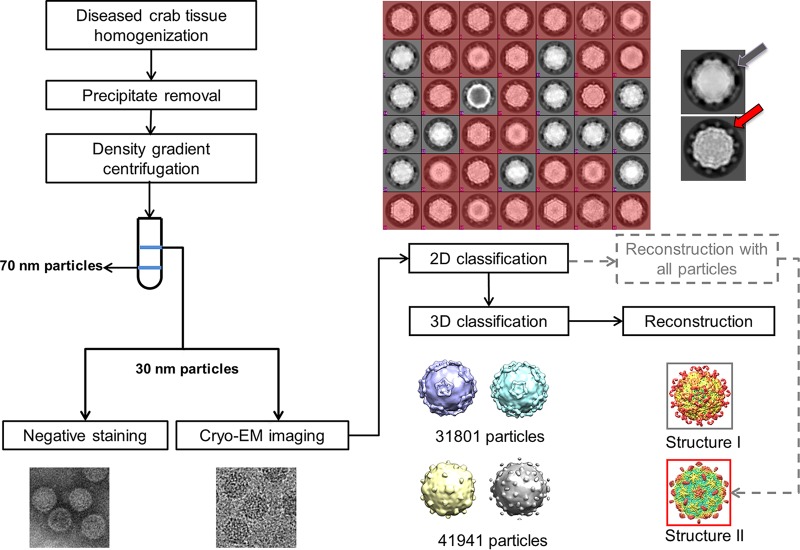
FIG 1.
Flow chart of sample isolation and data processing. Initial data processing steps indicated by gray dotted lines include 2D classification and reconstruction with all particles to obtain structure II. However, the result of 2D classification reveals two kinds of virus based on the shape of protrusions. One (colored red) has dot-like protrusions (red arrow), and the other (gray) has lamina-shaped protrusions (gray arrow). Therefore, the final data processing step included 2D and 3D classification, refinement, and reconstruction. The results confirm the presence of two kinds of virus with different protrusions.
These results implied that there might be another unknown virus in the purified sample similar in size to MCDV and composed of only one capsid protein, whereas MCDV is composed of at least three different capsid proteins.
To verify our hypothesis, the results of 25 iterations of reference-free two-dimensional (2D) classification using RELION 2.0 were carefully rechecked, and it became apparent that the sample probably contained two different viruses based on the shape of the protrusions, which formed “dot” shapes in some particles and a “lamina” shape in others (Fig. 1).
Further 3D classification and 50 cycles of refinement and reconstruction using RELION 2.0 also revealed two types of particles (Fig. 1). Two corresponding virus structures with different protrusions were eventually obtained, both ∼30 nm in diameter.
Finally, 31,801 particles were classified as one virus type (temporarily named type I virus) and 41,941 particles were sorted into the other virus type (temporarily named type II virus) (Fig. 1). Two different icosahedral 3D maps at 3.5- and 3.3-Å resolutions were obtained.
Identification of MCDV.
These structures allowed us to annotate each subunit of the capsids, and the results revealed four subunits (VP1 to 4) in each asymmetric unit for the type I virus (Fig. 2A to C), compared with three subunits (VPa to c) in each asymmetric unit for the type II virus. All subunits aligned well except for terminal regions in type II virus, as observed for the map initially obtained before 3D classification. These features suggested that VPa, VPb, and VPc were encoded by the same sequence. Therefore, the capsid of the type II virus is composed of one protein arranged in T=3 icosahedral symmetry.
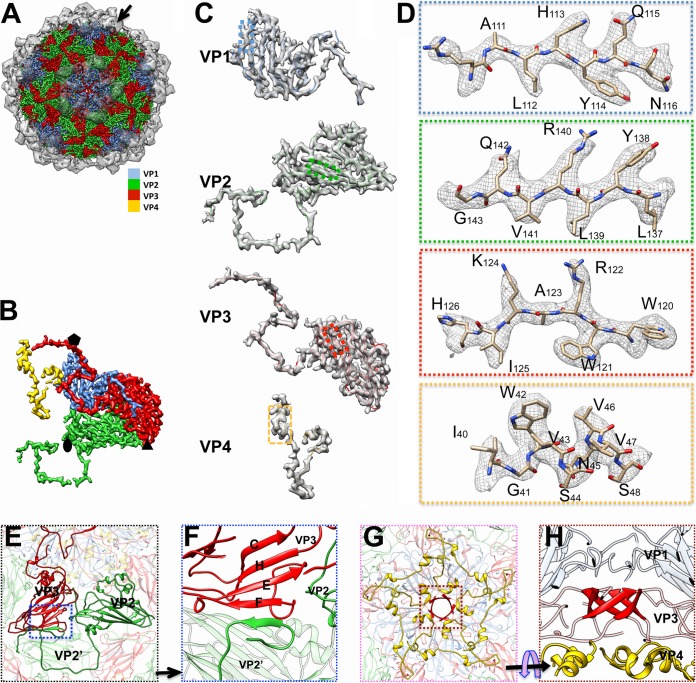
FIG 2.
Structural features of mud crab dicistrovirus (MCDV). (A) Overall structure of MCDV. Protrusions (black arrow), highlighted and rendered at an isodensity contour level of 1.5 σ, are shown as translucent. Capsid proteins VP1, VP2, VP3, and VP4 are colored cornflower blue, green, red, and gold, respectively, and rendered at an isodensity contour level of 3.5 σ. (B) Density map of the asymmetric unit of MCDV (inside view). The color scheme is the same as that in panel A. (C) Density maps and atomic models of VP1 to VP4. (D) Zoomed-in views of boxed regions in panel C illustrating residue features in the density map. (E) Interactions between VP2 and VP3. The N terminus of VP2 crosses the 2-fold axis and interacts with VP2 in another asymmetric unit (labeled VP2’). The N terminus of VP2 also interacts with VP3 (in the same asymmetric unit) via β-augmentation (blue boxed area) and approaches the 3-fold axis. (F) Zoomed-in view of β-augmentation boxed in panel E. (G) Structure of the disk feature (colored gold), which is composed of VP4, attached to the inner face of the 5-fold axis. (H) The β-cylinder structure (colored red) around the 5-fold axis, comprising the N termini of VP3 subunits.
Previous reports revealed the existence of MCDV in crabs with SD (4). Members of Dicistroviridae are usually composed of three or four capsid proteins (7, 8), but comparison of the structures of type I and type II viruses indicated that only the type I virus comprises four proteins.
Together, these results suggest that the type I virus may be MCDV. At this resolution, the densities of most large side chains can be easily traced (Fig. 2C and D). As expected, we succeeded in building an atomic model using the amino acid sequence of MCDV. Amino acids 3 to 190 of VP1, amino acids 4 to 253 of VP2, amino acids 3 to 57 of VP4, and the shell domain (S-domain) of VP3 could be traced (Fig. 2C). However, residues 253 to 447 of VP3, which is exposed on the capsid surface, could not be traced due to high flexibility.
Therefore, we assigned the type I virus as MCDV, while the type II virus appeared to be novel.
Structural features of MCDV and its constituent proteins.
The 3.5-Å map of MCDV demonstrates pseudo-T=3 icosahedral symmetry with 60 copies of each of the VP1, VP2, and VP3 structural proteins, all of which share a “jelly roll” fold with β-strands named B to I according to picornavirus convention (9). VP1 subunits form pentamers around 5-fold axes, whereas VP2 and VP3 subunits constitute alternating heterohexamers around the icosahedral 3-fold axes (Fig. 2A, B, and E).
Among the three major capsid proteins, the C terminus and CD loop of each VP1 subunit around the 5-fold axis stretch away from the major capsid surface (Fig. 3), while the N terminus of each VP1 subunit is inserted into the inner surface of the capsid. The N terminus of VP2 extends across the 2-fold axis and interdigitates with the neighboring VP2 subunit, then interacts with the adjacent VP3 via β-augmentation, and finally approaches the 3-fold axis (Fig. 2E to G, Fig. 3). Thus, the N terminus of VP2 binds the adjacent VP2 and VP3 subunits together. Five N-terminal regions of VP3 are twisted around the 5-fold axis and interact to form a cylindrical structure (Fig. 2G and H), which is a common feature of dicistroviruses and many picornaviruses (7, 8). One remarkable feature of MCDV is that VP3 has an additional 200 amino acids at the C terminus, and this longer feature forms a unique C-terminal protrusion that sticks out from the capsid surface around the 5-fold axes (Fig. 2A, Fig. 3).
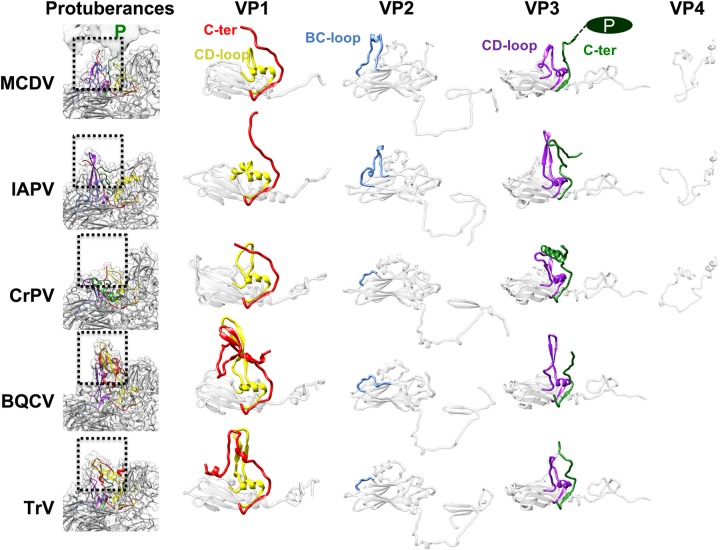
FIG 3.
Comparison of capsid proteins and finger-like protuberances among MCDV and other dicistroviruses. The densities of finger-like protuberances are shown in translucent gray, and models of subunits are shown in ribbon representation. The CD loop and C terminus of VP1 are colored yellow and red, respectively. The BC loop of VP2 is colored cornflower blue. The CD loop and C terminus of VP3 are purple and green, respectively. BQCV, IAPV, and TrV have obvious finger-like protuberances, which are inconspicuous in CrPV. In MCDV, the finger-like protuberances are not obvious and are covered by the C-terminal protrusions of VP3.
In previous studies, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) failed to confirm the existence of VP4 in MCDV (3, 4); however, we clearly identified obvious “disk-like” density features attached to the inner face of the 5-fold axis corresponding to VP4 in the virion structure (Fig. 2G and H).
As a representative dicistrovirus infecting crustaceans, MCDV shares 60.2% amino acid sequence identity with Taura syndrome virus (TSV) and significantly less identity (11% to 18%) with other dicistroviruses (4, 10). Besides MCDV, structures of four other viruses from the family Dicistroviridae have been determined to date (Fig. 3): Israeli acute bee paralysis virus (IAPV) (11), triatoma virus (TrV) (8), black queen cell virus (BQCV) (12), and cricket paralysis virus (CrPV) (7). Among these, a stable VP4 disk-like structure was seen only in IAPV and CrPV, while atomic models could not be built for TrV and BQCV.
Comparing MCDV with the aforementioned Dicistroviridae members, the most remarkable feature is clearly the unique C-terminal protrusion that covers the positions of finger-like protuberances (Fig. 2A, Fig. 3). In BQCV, the finger-like protuberances near the 5-fold axes are composed of the short C-terminal regions of VP1 and the CD loops of VP1 and VP3. These density features are most obvious in BQCV, less prominent in TrV and IAPV, and almost invisible in CrPV (12). The finger-like protuberances of TrV were speculated to play a role in virus-host interactions, in particular those involved in binding to the entry receptor (8). However, in MCDV, the positions of finger-like protuberances are covered by the density corresponding to the prominent C-terminal protrusions. Therefore, the C-terminal protrusions may play similar roles to the finger-like protuberances in other dicistroviruses. The presence of protrusions in MCDV may be related to its host; among reported dicistroviruses, only MCDV and TSV have an extralong C terminus, and both infect crustaceans, while most other dicistroviruses infect insects (10).
The genome of MCDV can be released by heating.
Previous studies revealed that the genome could be released by heating in some members of picornaviruses and dicistroviruses (11, 13, 14). We therefore heated a mixture of MCDV and the type II virus to 56°C for 0.5 h. After 2D classification, we first classified the mixture into MCDV and type II virus. MCDV particles could then be further classified into two (empty and full) groups. Finally, we obtained 6.8-Å heated full MCDV and 6.6-Å heated empty MCDV density maps. The results revealed very few empty type II virus particles after heating, while 44% of MCDV particles released their genome and were empty.
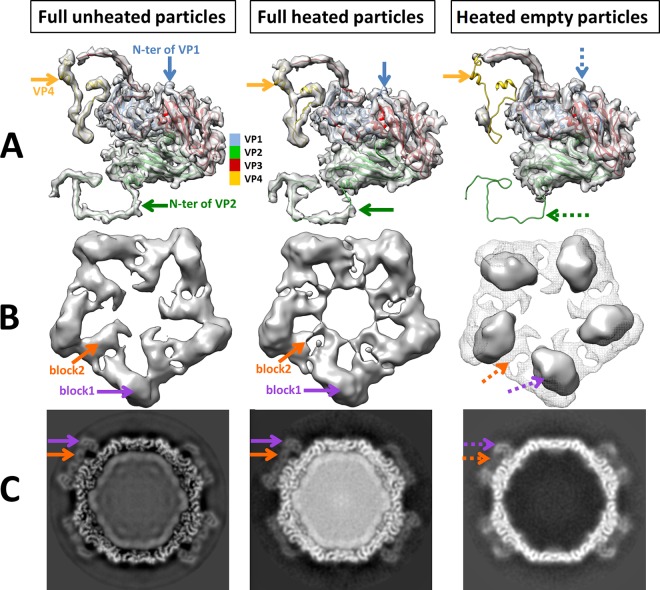
Comparing the maps of empty and full heated MCDV particles, densities corresponding to VP4 and the N termini of VP1 and VP2 were indiscernible in empty MCDV heated particles but presented in full particles, despite being heated at the same time (Fig. 4A). There was neither a credible diameter change between the empty and full heated particles nor an obvious pore or gap structure for RNA releases in both. One reason for this could be that genome release was too fast to be captured. Another possibility is that the holes or gaps for genome release are distributed asymmetrically and may therefore not be visible after imposition of icosahedral symmetry.
FIG 4.
Conformational changes between unheated full particles, heated full particles, and heated empty particles of MCDV. (A) Asymmetric units of particles. The densities of VP4 and the N-terminal regions of VP1 and VP2 (arrows) are missing in heated empty particles. (B) Top view of the protrusion. The density of full unheated particles (gray mesh) is superimposed on that of the empty heated particles to demonstrate the conformational changes. The map of the full unheated particles was low pass filtered to 6.8 Å, similar to that of heated particles. The density maps reveals two linked-block densities (orange and purple arrows) for each protrusion around the 5-fold axis in both full unheated and heated particles. However, in empty heated particles, there is only one block density (purple arrow) and more free space around the 5-fold axis. (C) Slice views of particles.
However, after heating, the MCDV protrusions were clearly different between empty and full heated particles (Fig. 4B and C). There are two “linked blocks” of density per protrusion in full heated particles, similar to the high-resolution map, while in empty particles, only one block can be seen, located between the two linked blocks in full particles (Fig. 4B and C). All these variations should be implicated in the genome release.
Identification of the type II virus.
To identify the type II virus, real-time quantitative PCR (RT-PCR) was reemployed using primers based on the protein mass spectrometry results for the capsid protein. As expected, a cDNA fragment was successfully obtained; the sequence shares 97% identity with that of the Wenzhou tombus-like virus 18 (GenBank accession number KX883013) (15). Further sequence determination and analyses were then performed. The results revealed that the type II virus harbors a single positive single-strand RNA (+ssRNA) genome. Unlike MCDV, which harbors a dicistron sequence and four capsid proteins, the genome of the type II virus harbors one sequence that contains three ORFs and encodes only one capsid protein. These results suggest that MCDV and the type II virus are entirely separate species. Using the determined sequence, we easily built an atomic model based on the density map of the type II virus, which we temporarily named the mud crab tombus-like virus (MCTV).
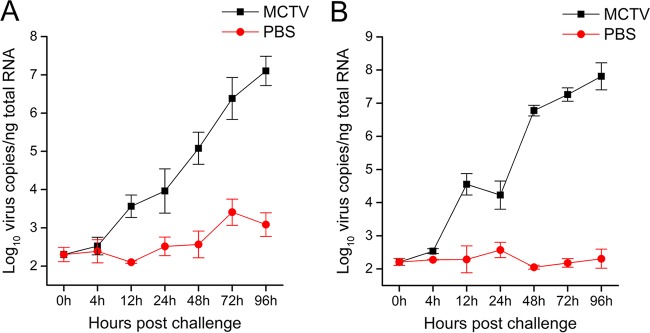
Though MCTV shares high identity with Wenzhou tombus-like virus 18, the two viruses were identified in crabs and bivalves, respectively. To confirm that MCTV is an infectious agent (rather than a contaminant) in crabs, we examined the proliferation of MCTV in vivo using an artificial-infestation experiment. Real-time PCR was used to measure MCTV loads in the hepatopancreas and hemocytes of MCTV-injected crabs at different times. As shown in Fig. 5, the copy number of MCTV in hepatopancreas and hemocytes increased during the course of infection. At 96 h post-MCTV infection, the mortality rate was about 60%; this was the point at which the MCTV copy number in surviving crabs reached its highest level. At the same time, the MCTV copy number in the control group from 0 to 96 h was faintly increased, mainly because the mud crabs used for the experiment carried MCTV themselves. The results suggest that the mud crab is a host for MCTV.
FIG 5.
Proliferation of MCTV in infected mud crabs. (A) Number of MCTV genome copies in the hepatopancreas of mud crabs at 4, 12, 24, 48, 72, and 96 h postinoculation. (B) Number of MCTV genome copies in hemocytes of mud crabs at 4, 12, 24, 48, 72, and 96 h postinoculation. Each bar represents the mean ± standard deviation of three samples.
Overall structure of the type II virus.
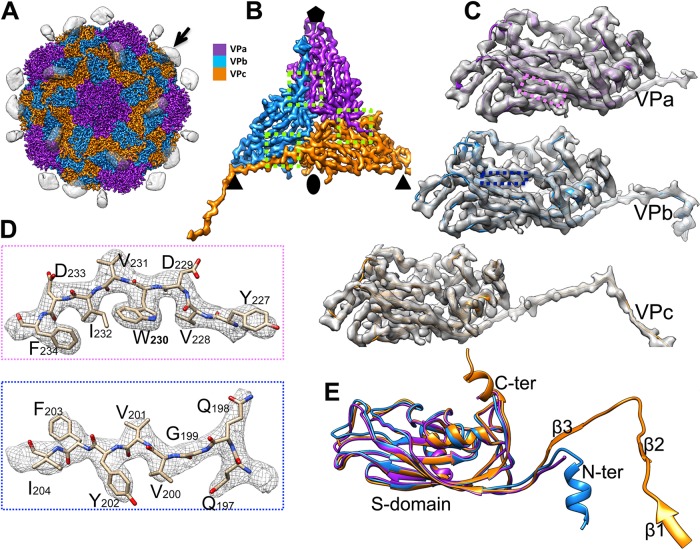
The 3.3-Å map of MCTV reveals a virion with T=3 icosahedral symmetry and 180 copies of the capsid protein. Five copies of VPa form pentamers around the 5-fold axes, while VPb and VPc constitute alternating hexamers around the icosahedral 3-fold axes (Fig. 6A and B).
FIG 6.
Overall structure of MCTV. (A) Overall structure of MCTV. Protrusions (black arrow) are highlighted, rendered at an isodensity contour level of 1.5 σ, and shown as translucent. Capsid proteins VPa, VPb, and VPc are colored purple, blue, and orange, respectively, and rendered at an isodensity contour level of 4 σ. (B) Density map of the asymmetric unit of MCTV (inside view). (C) Density maps and atomic models of VPa, VPb, and VPc. (D) Zoomed-in views of the boxed regions in panel C illustrating residue features in the density map. (E) Aligned models of VPa, VPb, and VPc. The β-barrels and most loops are nearly the same, except the N and C termini. There are three extra short β-strands in VPc, named β1, β2, and β3.
Most amino acids could be unambiguously identified in the 3.3-Å density map (Fig. 6C and D) except for residues 1 to 50 and 243 to 337 of VPa, 1 to 40 and 242 to 337 of VPb, and 1 to 29 and 250 to 337 of VPc, due to high flexibility. Alignment of the models of VPa, VPb, and VPc indicates that most copies are identical, except for the termini (Fig. 6E). The capsid protein of MCTV includes an N-terminal region, an S-domain, and a C-terminal P-domain (Fig. 6E). The S-domain has eight β-strands (βB to βI). The density of the N terminus could be only partly traced, but three short β-strands (β1 to β3) in the N-terminal region of VPc could be identified and form an N-arm (Fig. 6E). It was impossible to build accurate models of the P-domains due to high flexibility. Even so, the P-domain density of VPc could be clearly observed, and two such domains form a stable dimeric protrusion (Fig. 6A). However, the density of the P-domain for VPa and VPb was barely above the background noise, making tracing impossible.
Interactions among subunits within each asymmetric unit.
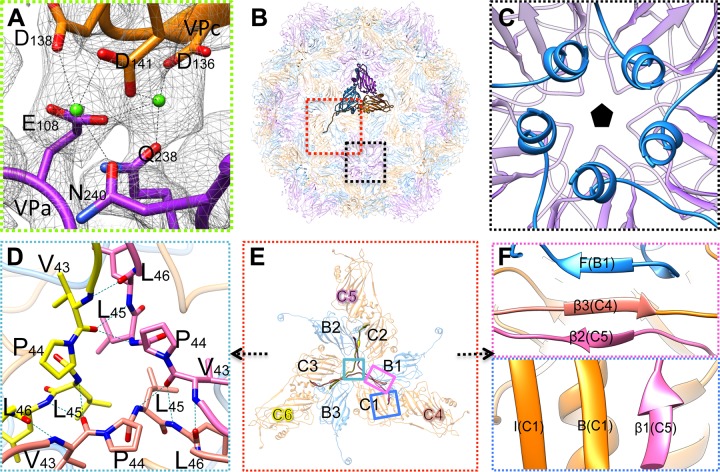
Extra densities for metal ions in positions between VPa and VPb, VPa and VPc, and VPb and VPc were evident when atomic models were fitted into the map (Fig. 6B, Fig. 7A). Two ions are chelated by the DxDxxD motif (Asp136, Asp138, and Asp141) of one subunit and Glu108, Gln238, and Asn240 of the neighboring subunit (Fig. 7A), all of which are conserved in a few members of Nodaviridae-like and Tombusviridae-like viruses Macrobrachium rosenbergii nodavirus (MrNV; GenBank accession number ACY26145), Penaeus vannamei nodavirus (PvNV; NCBI protein database accession number YP_004207809), Wenzhou crab virus 4 (NCBI protein database accession number YP_009337754), Wenzhou tombus-like virus 18 (NCBI protein database accession number YP_009342271), Wenling tombus-like virus 4 (NCBI protein database accession number YP_009337116), and Wenling tombus-like virus 5 (NCBI protein database accession number YP_009337263). This ion-binding motif is common among various tombusviruses, sobemoviruses, and nodaviruses. Most chelate one or two ions via a homologous motif at a similar position (16–18). Indeed, this is thought to be a key interaction for capsid assembly and stability.
FIG 7.
Interactions of the capsid proteins of MCTV. (A) Zoomed-in view of the interactions between neighboring subunits within one asymmetric unit (Fig. 6B). The density of the two metal ions between VPa and VPc is represented as green balls. These ions are chelated by D136, D138, and D141 of VP1 and E108, Q238, and N240 of VP3. (B) Inside view of the atomic models of MCTV. One asymmetric unit is highlighted, while others are translucent. Boxed regions in red and black are the areas around 3-fold and 5-fold axes, respectively. (C) Zoomed-in view of the area close to the 5-fold axis indicating the five helices of VP3 interacting with each other via hydrophobic interactions and the formation of the tunnel around the 5-fold axis. (D) Zoomed-in view of the area close to the 3-fold axis boxed in turquoise in panel E. Hydrogen bonds around Pro44 are shown as dotted lines. (E) Zoomed-in view of the interactions around the 3-fold axis. The hexamer is composed of three VPb subunits (B1, B2, and B3) and three VPc subunits (C1, C2, and C3). Three neighboring VPc subunits (C4, C5, and C6) interact with the hexamer via β-augmentation. Two types of β-augmentation can be formed, indicated in the two (magenta and blue) colored boxes. (F) Zoomed-in view of β-augmentation. One type (magenta box) is composed of βF of B2, β3 of C4, and β2 of C5. The other augmentation type (blue box) is composed of βI, βB of C1, and β1 of C5.
The N-terminal regions play different roles in MCTV.
VPa, VPb, and VPc of MCTV are chemically identical, but the structures of their N termini are different (the disordered N terminus of VPa could not be traced).
The N-terminal region of each VPb subunit forms a short helix attached to the inner surface, close to the 5-fold axis, and five helices together form an ∼8-Å-wide hydrophobic tunnel beneath the 5-fold vertex (Fig. 7C). The helices and the tunnel structure are unique among reported tombus-like viruses and nodaviruses.
The N-arm of each VPc subunit contains three extra short β-strands, and three N-arms of VPc subunits from different asymmetric units interdigitate to form a “trident” structure around the 3-fold axis (Fig. 7E). In the middle of the trident, a conserved proline close to the 3-fold axis plays an important role in stabilizing the structure, and few hydrogen bonds are formed around this proline (Fig. 7D). In addition to these hydrogen bonds close to the 3-fold axis, three short β-strands of the N-arms also interact with the hexamer via β-augmentation (Fig. 7E and F), which not only reinforces the stability of the hexamer but also establishes a steady network for binding neighboring hexamers together.
The interactions around the 3-fold axis are critical for the assembly and stability of the capsid. During heating of mixtures as described above, MCTV exhibited higher stability than MCDV; most MCTV particles did not lose their RNA genome, and capsids showed no observable changes following heating. This indicates the presence of powerful interactions between subunits.
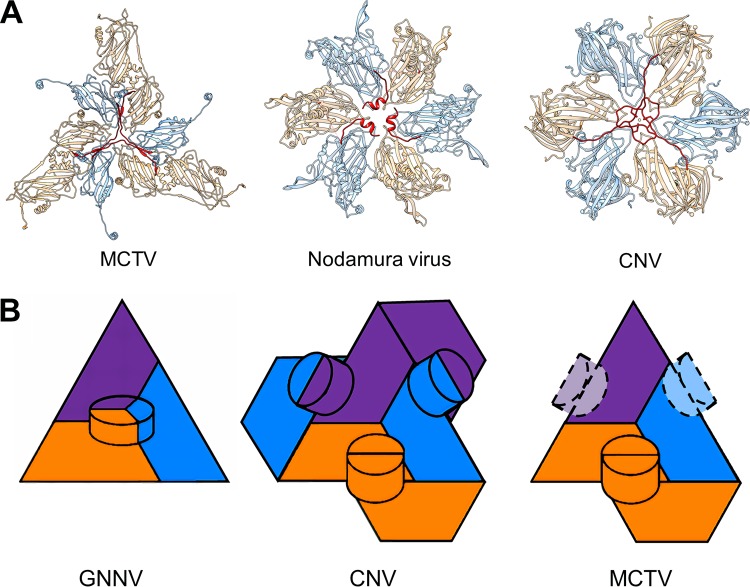
Structural features around the 3-fold axis are diverse in Nodaviridae-like and Tombusviridae-like viruses. In MCTV, grouper nervous necrosis virus (GNNV) (19), and Orsay virus (20), three N-arms form a trident structure as described above (18–20) (Fig. 7E, Fig. 8A). However, in some alphanodaviruses, such as Nodamura virus (PDB identifier [ID] 1NOV) (21), there are three α-helices around the 3-fold axes (Fig. 8A). Meanwhile, some tombusviruses possess no stable N-arms at all, and some, such as cucumber necrosis virus (CNV; PDB ID 4LLF), have a special structure that is distinct from the trident (22). The N-arms of VPc in CNV intertwine around a Zn2+ ion at the 3-fold axis (Fig. 8A). Thus, differences around the 3-fold axis may provide a basis for classification of these viruses.
FIG 8.
Structural comparison of MCTV with other viruses. (A) Comparison of the characteristic structural features close to the 3-fold axis. The VPbs and VPcs of the three viruses are colored blue and orange, respectively. The N-terminal regions of VPcs around 3-fold axes are colored red. MCTV forms a trident structure. Nodamura viruses have three helices. CNV has a distinctive structure at this location. (B) Organization of protrusions in GNNV, TBSV, and MCTV. The VPas, VPbs, and VPcs of the three viruses are colored purple, blue, and orange, respectively. Dashed lines indicate inconspicuous density.
Features of the dimeric protrusion of MCTV.
MCTV has 30 prominent dimeric protrusions extending from the surface at the 2-fold axis of the capsid (Fig. 6A). The resolution of the protrusions is somewhat lower than that of the major capsid structure, indicating higher flexibility. The density map, together with the atomic model, reveals that the dimeric protrusions are composed of the C termini (residues 250 to 337) of two VPc subunits at the 2-fold axis; hence, MCTV only has 30 obvious protrusions (Fig. 6A). By decreasing the threshold density to the mean of the map, extra inconspicuous extending density between VPa and VPb was revealed close to each 5-fold axis, and the C termini of VPa and VPb contribute to this inconspicuous density.
Protrusions of viruses usually play important roles in the organization of the capsid, attachment to host cells, and other important steps in the life cycle of viruses. In previously reported tombus-like and noda-like viruses, there are two different types of protrusions (Fig. 8B). In the first type, the C termini of VPa, VPb, and VPc protrude from the quasi-3-fold axis of the asymmetric unit, forming a “VPa-VPb-VPc” trimeric protrusion (Fig. 8B). The betanodavirus member GNNV VLP has 60 protrusions organized in this way (18, 19). In the second type, a pair of C termini from neighboring subunits from different asymmetric units protrudes and interacts with each other, forming a dimeric protrusion. Through these interactions, all 180 C termini of proteins in a single capsid form a coherent network with 90 protrusions (Fig. 8B). This protrusion type is common among members of Tombusviridae, such as tomato bushy stunt virus (TBSV) (23) and CNV (22). Among the 90 protrusions, there are two types of interactions: VPa-VPb and VPc-VPc (Fig. 8B). Protrusions composed of VPa and VPb are close to the 5-fold axis, while protrusions composed of the two termini of VPc are located at the 2-fold axis of the capsid (24). Strikingly, in MCTV, strong density was observed only for VPc-VPc protrusions at the 2-fold axis, although the resolution was relatively low due to high flexibility. Meanwhile, the density of the C termini of VPa and VPb was too weak to indicate the formation of protrusions. Therefore, MCTV has only 30 distinguishable dimeric protrusions (Fig. 6A, Fig. 8B).
DISCUSSION
The development of molecular and immunological techniques, such as enzyme-linked immunosorbent assay and PCR, has provided effective tools for high-throughput and high-sensitivity detection of various pathogens (25, 26). However, in the event of a disease caused by unknown pathogens, it can be difficult to select the appropriate reagent or probe for pathogen identification. Therefore, EM still plays an indispensable role in the detection of unknown infectious agents from unusual outbreaks and in the control of biosafety (27, 28), as demonstrated by the severe acute respiratory syndrome pandemic in 2003 (29, 30) and various monkeypox outbreaks in the United States (31). Furthermore, molecular probe- or antigen-based detection can often miss additional pathogens present in cases of dual or multiple infections. The present study investigated the application of single-particle cryo-EM to a multiple-infection case. The results suggest that this approach offers an effective complementary method for routine clinical or molecular diagnosis. Furthermore, our work suggests that the structures of intricate protein mixtures may also be determined using batch processing methods rather than complicated purification steps, particularly with the rapid development of cryo-EM.
MCDV shares several typical structural features with picornaviruses. However, on the capsid surface of human rhinovirus 14 and other similar picornaviruses, there is a hydrophobic pocket in VP1, which is a target of some inhibitors (32). Virus-receptor binding and genome release can be prevented when inhibitors bind to this pocket (33, 34). Considering the lack of the hydrophobic pocket in VP1, similar to other reported dicistroviruses (7, 8, 11, 12), MCDV shall have different receptor binding sites. A previous report has revealed that the finger-like protuberance of TrV plays roles in virus-host interactions (8), whereas the C terminus of MCDV VP3 occupies the location of finger-like protuberance. Furthermore, the conformation change of C-terminal protrusion observed in the heating experiments is involved in RNA release. Therefore, we believe that the C-terminal protrusion of VP3 plays important roles in both virus-receptor binding and genome release and can be a possible target for the design of inhibitors.
In this work, MCTV was detected by single-particle cryo-EM. Sequence analysis revealed that the genome of MCTV shares 97% identity with that of Wenzhou tombus-like virus 18, originally detected in bivalves (15). Analysis of genomic structures suggests that Wenzhou tombus-like virus 18 shares similarity with Tombusviridae, while the sequence of the capsid protein is much more similar to that of its counterparts in MrNV and PvNV, which infect crustaceans. Our results demonstrate some common structural features of tombusviruses and nodaviruses, while the N-arms of VPb are similar to those of GNNV, Orsay virus, and some sobemoviruses. However, MCTV also possesses some unique features, such as the 5-fold hydrophobic tunnel and dimeric protrusions. These structural features suggest that MCTV may belong to a novel virus class that is closely related to Tombusviridae and Nodaviridae in terms of taxonomy. However, virus taxonomy requires more extensive morphological and molecular biological evidence, and detailed taxonomy and evolutionary analysis of MCTV is needed.
MATERIALS AND METHODS
Virus isolation, purification, and identification.
The gills of SD mud crabs were homogenized in phosphate-buffered saline (PBS; 2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) at 4°C. To remove tissue and cell debris, homogenized samples were centrifuged for 1 h at 10,000 × g, and supernatants were then ultracentrifuged at 200,000 × g for 2 h. Pellets were resuspended in PBS and loaded onto a 15% to 45% (wt/wt) CsCl gradient and ultracentrifuged at 200,000 × g for 8 h at 4°C. The recovered fractions were diluted in PBS and ultracentrifuged for 2 h at 200,000 × g to obtain a workable concentration. Finally, pellets were resuspended in PBS and checked by negative staining with a transmission electron microscope (TEM; JEM 100CX II) to ensure that the concentration and purity were adequate for further cryo-EM imaging. For negative-stain EM, 3 μl of sample was applied to a glow-discharged grid covered with carbon film for 1 min. After removal of excess buffer, the grid was stained with 3% (wt/vol) phosphotungstic acid (PTA) for 1 min. Excess PTA was then removed with a filter paper, and the grid was air dried.
Quantification of MCTV by real-time PCR.
Two-month-old mud crabs (100 ± 10 g) were collected from a farm (Guangdong Province, China) and maintained in the aquarium facilities as described previously (35). Mud crabs (n = 200) were assigned randomly to experimental and control groups. The experimental group received an intramuscular (i.m.) injection of MCTV (104 copies/g of body weight), whereas the controls received PBS. Nine crabs were selected randomly at each time point (4, 12, 24, 48, 72, and 96 h postinoculation), and total RNA was extracted according to the manufacturer’s instructions. Reverse transcription was performed using the PrimeScript RT reagent kit (TaKaRa, Japan) according to the manufacturer’s instructions. Real-time quantitative PCR (RT-PCR) was conducted using SYBR Premix Ex Taq (TaKaRa, Japan) and a Roche LightCycler 480 (Roche). The primers used for RT-PCR were based on the MCTV sequence (MCTV-qPCR CGGAAGCGACTACCTTGGT and MCTV-qPCR CGATGGGCTCACGGATAAGA). Plasmid pMCTV (obtained by cloning a short sequence of MCTV into the pMD-19T vector) was serially diluted to obtain a standard curve, which was then used to quantify MCTV viral genomic copy numbers in the samples. Each assay was carried out in triplicate. The mean and standard deviation of the three replicate real-time PCR values were determined.
Virus heating.
The isolated virus mixture was incubated in a water bath at 56°C for 0.5 h for subsequent cryo-EM sample preparation.
Cryo-EM sample preparation and data acquisition.
R1.2/1.3 copper Quantifoil holey grids (Quantifoil Micro Tools, GmbH, Germany) were coated with a fresh layer of continuous thin carbon film immediately before cryo-EM sample freezing, and 2.5 μl of sample was applied, blotted, and flash-frozen in precooled liquid ethane using an Vitrobot Mark IV machine (FEI Company) at 100% humidity.
High-resolution cryo-EM data were collected on an 300 kV Titan Krios cryo-EM instrument (FEI Company) equipped with a Ultra-Scan4000 16-megapixel charge-coupled-device (CCD) camera (Gatan) at a nominal magnification of ×96,000, corresponding to a final pixel size of 0.933 Å. Data were collected from heated samples on an Tecnai F20 cryo-EM instrument (FEI Company) operated at 200 kV, equipped with an Eagle 16-megapixel CCD camera (FEI Company) at a nominal magnification of ×100,000, corresponding to a final pixel size of 1.09 Å. The total dose was ∼20 e−/Å2, and defocus values ranged from 1.0 to 3.0 μm (Table 1).
TABLE 1.
Parameters used for high-resolution image collection and model refinement
| Parameter | Value for: |
|
|---|---|---|
| MCDV | MCTV | |
| Pixel size, Å | 0.933 | 0.933 |
| Defocus range, μm | 1−3 | 1−3 |
| Acceleration voltage, kV | 300 | 300 |
| Dose, e/Å2 | 20 | 20 |
| Final resolution, Å | 3.5 | 3.3 |
| EMDB accession number | 9673 | 9754 |
| RMSDa bonds, Å | 0.0086 | 0.0082 |
| RMSD angles, Å | 1.14 | 1.12 |
| Avg B factor | 183.4 | 174.7 |
| Clash score | 10.74 | 9.5 |
| Good rotamers, % | 99.85 | 100 |
| Ramachandran favored, % | 91.20 | 90.79 |
| Ramachandran allowed, % | 6.50 | 7.57 |
| Ramachandran outliers, % | 2.30 | 1.64 |
| PDB ID | 6IIC | 6IZL |
RMSD, root mean square deviation.
Data processing.
A total of 74,880 particles (∼30 nm in diameter) were selected from 3,470 CCD cryo-EM frames captured by the Titan Krios instrument, and ∼14,000 heated particles from 552 CCD frames were collected using the Tecnai F20 instrument and selected using the e2boxer.py program in EMAN2 (36). Contrast transfer function parameters were determined using Gctf (37).
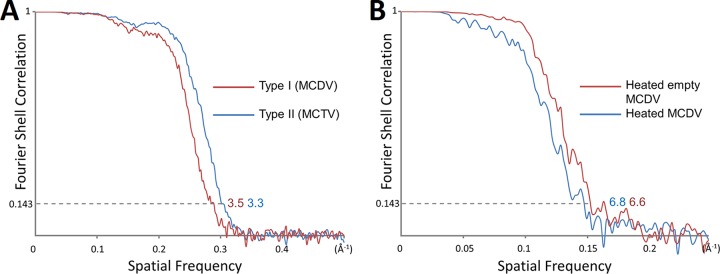
RELION 2.0 (38, 39) was used to perform reference-free 2D classification. An initial model was built by e2initialmodel.py in EMAN2, and 3D classification was then carried out by RELION 2.0. Two models were selected, and JSPR was used to refine the center, orientation, defocus, astigmatism, and magnification value of each particle and for density map reconstruction (40). The resolution of the maps was evaluated using the ‘‘gold standard’’ Fourier shell correlation (FSC) at 0.143 criterion (41, 42). The resolutions of the MCDV, MCTV, and heated full and empty MCDV are 3.5 Å, 3.3 Å, 6.8 Å, and 6.6 Å, respectively (Fig. 9).
FIG 9.
FSC curves of the reconstructions. (A) FSC curves MCDV (3.5 Å) and MCTV (3.3 Å). The curves for MCDV and MCTV are colored red and blue, respectively. (B) FSC curves of heated empty MCDV (6.6 Å) and heated full MCDV (6.8 Å). Heated empty MCDV and heated full MCDV are colored red and blue, respectively.
Model building and assessment.
The initial models of MCDV and MCTV were predicted using the Phyre2 server (43). After fitting into the density map of one virus, models were adjusted according to the densities of the amino acid side chains.
All manual adjustments were performed with Coot (44), followed by automatic refinement using Phenix (45). All parameters used for model refinement are listed in Table 1.
Data availability.
Density maps of MCTV and MCDV have been deposited in the EMDataBank (EMDB) under entries EMD-9754 and EMD-9673, respectively. Models for MCTV and MCDV have been deposited in the Protein Data Bank (PDB) under entries 6IZL and 6IIC, respectively. Density maps of heated full MCDV and heated empty MCDV have been deposited at EMDB under entries EMD-9755 and EMD-9756, respectively.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSFC; no. 31570736 and 31672677) and the China Agriculture Research System (no. CARS-48).
We thank Xiaojun Huang, Gang Ji, and colleagues at the Institute of Biophysics (IBP), Chinese Academy of Sciences, for access to their Titan Krios microscope. We also thank Junjie Zhang, Menqiu Jiang, and Alexander Rie from Texas A&M University and Xinghong Dai from the University of California Los Angeles (UCLA) for valuable suggestions regarding the manuscript.
Y. Gao, K. Li, and Q. Zhang collected cryo-EM data. Y. Gao and Q. Zhang processed the data. Y. Gao, Jian He, and Q. Zhang analyzed the structures. S. Liu, J. Huang, Q. Wang, Jianguo He, and S. Weng isolated and purified the virus and carried out sequence analysis and the biological experiments. Q. Zhang, Y. Gao, and S. Liu drafted the manuscript. Q. Zhang and S. Weng supervised the work and edited the final version of the manuscript.
REFERENCES
- 1.Lefkowitz EJ, Adams MJ, Davison AJ, Siddell SG, Simmonds P. 2017. ICTV virus taxonomy: classification and nomenclature of viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 2.Huang ZW, Deng XX, Li YY, Su HJ, Li KP, Guo ZX, Zheng PR, Xu HD, He JG, Zhang QF, Weng SP. 2012. Structural insights into the classification of mud crab reovirus. Virus Res 166:116–120. doi: 10.1016/j.virusres.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 3.Zhang R, He J, Su H, Dong C, Guo Z, Ou Y, Deng X, Weng S. 2011. Identification of the structural proteins of VP1 and VP2 of a novel mud crab dicistrovirus. J Virol Methods 171:323–328. doi: 10.1016/j.jviromet.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Guo ZX, He JG, Xu HD, Weng SP. 2013. Pathogenicity and complete genome sequence analysis of the mud crab dicistrovirus-1. Virus Res 171:8–14. doi: 10.1016/j.virusres.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Deng XX, Lu L, Ou YJ, Su HJ, Li G, Guo ZX, Zhang R, Zheng PR, Chen YG, He JG, Weng SP. 2012. Sequence analysis of 12 genome segments of mud crab reovirus (MCRV). Virology 422:185–194. doi: 10.1016/j.virol.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 6.Le Gall O, Christian P, Fauquet CM, King AMQ, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch Virol 153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 7.Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson JE. 1999. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol 6:765–774. doi: 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 8.Squires G, Pous J, Agirre J, Rozas-Dennis GS, Costabel MD, Marti GA, Navaza J, Bressanelli S, Guérin DMA, Rey FA. 2013. Structure of the Triatoma virus capsid. Acta Crystallogr D Biol Crystallogr 69:1026–1037. doi: 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Rueckert RR, Sherry B, Vriend G. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. [DOI] [PubMed] [Google Scholar]
- 10.Mari J, Poulos BT, Lightner DV, Bonami JR. 2002. Shrimp Taura syndrome virus: genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J Gen Virol 83:915–926. doi: 10.1099/0022-1317-83-4-915. [DOI] [PubMed] [Google Scholar]
- 11.Mullapudi E, Přidal A, Pálková L, de Miranda JR, Plevka P. 2016. Virion structure of Israeli acute bee paralysis virus. J Virol 90:8150–8159. doi: 10.1128/JVI.00854-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spurny R, Pridal A, Palkova L, Kiem HKT, de Miranda JR, Plevka P. 2017. Virion structure of black queen cell virus, a common honeybee pathogen. J Virol 91:e02100-16. doi: 10.1128/JVI.02100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butan C, Filman DJ, Hogle JM. 2014. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol 88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 a-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu JG, Holmes EC, Zhang YZ. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 16.Hopper P, Harrison SC, Sauer RT. 1984. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol 177:701–713. [DOI] [PubMed] [Google Scholar]
- 17.Tars K, Zeltins A, Liljas L. 2003. The three-dimensional structure of cocksfoot mottle virus at 2.7 A resolution. Virology 310:287–297. [DOI] [PubMed] [Google Scholar]
- 18.Chen NC, Yoshimura M, Guan HH, Wang TY, Misumi Y, Lin CC, Chuankhayan P, Nakagawa A, Chan SI, Tsukihara T, Chen TY, Chen CJ. 2015. Crystal structures of a piscine betanodavirus: mechanisms of capsid assembly and viral infection. PLoS Pathog 11:e1005203. doi: 10.1371/journal.ppat.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Li K, Gao Y, Huang R, Lai Y, Shi Y, Yang S, Zhu G, Zhang Q, He J. 2016. Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle. Vet Res 47:16. doi: 10.1186/s13567-015-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo YR, Hryc CF, Jakana J, Jiang H, Wang D, Chiu W, Zhong W, Tao YJ. 2014. Crystal structure of a nematode-infecting virus. Proc Natl Acad Sci U S A 111:12781–12786. doi: 10.1073/pnas.1407122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlotnick A, Natarajan P, Munshi S, Johnson JE. 1997. Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals. Acta Crystallogr D Biol Crystallogr 53:738–746. doi: 10.1107/S0907444997007427. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Kakani K, Katpally U, Johnson S, Rochon D, Smith TJ. 2013. Atomic structure of Cucumber necrosis virus and the role of the capsid in vector transmission. J Virol 87:12166–12175. doi: 10.1128/JVI.01965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson AJ, Bricogne G, Harrison SC. 1983. Structure of tomato busy stunt virus IV. The virus particle at 2.9 A resolution. J Mol Biol 171:61–93. [DOI] [PubMed] [Google Scholar]
- 24.Ho KL, Kueh CL, Beh PL, Tan WS, Bhella D. 2017. Cryo-electron microscopy structure of the Macrobrachium rosenbergii nodavirus capsid at 7 angstroms resolution. Sci Rep 7:2083. doi: 10.1038/s41598-017-02292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu FT, White PA, Takeda N. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol 78:1347–1353. doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, Wang Q, Saif YM. 2005. Development of a ssRNA internal control template reagent for a multiplex RT-PCR to detect turkey astroviruses. J Virol Methods 126:81–86. doi: 10.1016/j.jviromet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Roingeard P. 2008. Viral detection by electron microscopy: past, present and future. Biol Cell 100:491–501. doi: 10.1042/BC20070173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldsmith CS, Miller SE. 2009. Modern uses of electron microscopy for detection of viruses. Clin Microbiol Rev 22:552–563. doi: 10.1128/CMR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 30.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 31.Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, Kazmierczak JJ, Stratman EJ, Li Y, Fairley JA, Swain GR, Olson VA, Sargent EK, Kehl SC, Frace MA, Kline R, Foldy SL, Davis JP, Damon IK. 2004. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med 350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- 32.Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286–1293. [DOI] [PubMed] [Google Scholar]
- 33.Hadfield AT, Diana GD, Rossmann MG. 1999. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci U S A 96:14730–14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiremath CN, Grant RA, Filman DJ, Hogle JM. 1995. Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14. Acta Crystallogr D Biol Crystallogr 51:473–489. doi: 10.1107/S090744499401084X. [DOI] [PubMed] [Google Scholar]
- 35.Weng SP, Guo ZX, Sun JJ, Chan SM, He JG. 2007. A reovirus disease in cultured mud crab, Scylla serrata, in southern China. J Fish Dis 30:133–139. doi: 10.1111/j.1365-2761.2007.00794.x. [DOI] [PubMed] [Google Scholar]
- 36.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimanius D, Forsberg BO, Scheres SHW, Lindahl E. 2016. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo F, Jiang W. 2014. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol 1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL. 2012. Outcome of the first electron microscopy validation task force meeting. Structure 20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH, Chen S. 2012. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Density maps of MCTV and MCDV have been deposited in the EMDataBank (EMDB) under entries EMD-9754 and EMD-9673, respectively. Models for MCTV and MCDV have been deposited in the Protein Data Bank (PDB) under entries 6IZL and 6IIC, respectively. Density maps of heated full MCDV and heated empty MCDV have been deposited at EMDB under entries EMD-9755 and EMD-9756, respectively.