Macrophages, with their long life span in vivo and their resistance to HIV-1-mediated cytopathic effect, might serve as viral reservoirs, contributing to virus persistence in an infected individual. Identification of host factors that increase the overall susceptibility of macrophages to HIV-1 might provide new therapeutic targets for the efficient control of viral replication in these cells and limit the formation of reservoirs in exposed individuals. In this study, we demonstrate the importance of p53 regulation by MDM2, which creates a cellular environment more favorable to the early steps of HIV-1 replication. Moreover, we show that p53 stabilization reduces virus infection in human macrophages, highlighting the important role of p53 in antiviral immunity.
KEYWORDS: human immunodeficiency virus, macrophages, mdm2, p53
ABSTRACT
The molecular basis for HIV-1 susceptibility in primary human monocyte-derived macrophages (MDMs) was previously evaluated by comparing the transcriptome of infected and bystander populations. Careful analysis of the data suggested that the ubiquitin ligase MDM2 acted as a positive regulator of HIV-1 replication in MDMs. In this study, MDM2 silencing through transcript-specific small interfering RNAs in MDMs induced a reduction in HIV-1 reverse transcription and integration along with an increase in the expression of p53-induced genes, including CDKN1A. Experiments with Nutlin-3, a pharmacological inhibitor of MDM2 p53-binding activity, showed a similar effect on HIV-1 infection, suggesting that the observed restriction in HIV-1 production results from the release/activation of p53 and not the absence of MDM2 per se. Knockdown and inhibition of MDM2 also both correlate with a decrease in the Thr592-phosphorylated inactive form of SAMHD1. The expression level of MDM2 and the p53 activation status are therefore important factors in the overall susceptibility of macrophages to HIV-1 infection, bringing a new understanding of signaling events controlling the process of virus replication in this cell type.
IMPORTANCE Macrophages, with their long life span in vivo and their resistance to HIV-1-mediated cytopathic effect, might serve as viral reservoirs, contributing to virus persistence in an infected individual. Identification of host factors that increase the overall susceptibility of macrophages to HIV-1 might provide new therapeutic targets for the efficient control of viral replication in these cells and limit the formation of reservoirs in exposed individuals. In this study, we demonstrate the importance of p53 regulation by MDM2, which creates a cellular environment more favorable to the early steps of HIV-1 replication. Moreover, we show that p53 stabilization reduces virus infection in human macrophages, highlighting the important role of p53 in antiviral immunity.
INTRODUCTION
Due to their inherent susceptibility to HIV-1 and ability to actively transfer the virus to other cell targets, macrophages play a significant role in the establishment and persistence of HIV-1 infection under in vivo situations (1, 2). Characteristics of tissue macrophages, such as their native resistance to the virus-mediated cytopathogenic effects, their capacity for self-renewal, and their relatively long life span, coupled with new evidence that they can harbor HIV-1 and can sustain long-term viral replication in the absence of lymphocytes in vivo (3, 4), suggest that these cells constitute a viral reservoir. Limiting their infection rates, self-renewal capabilities, or ability to replicate the virus could greatly increase the chances of a more successful virus eradication. Thus, it is important to understand the molecular basis of HIV-1 permissiveness and replication in myeloid cells.
Many cellular factors are known to impact the fate of the virus, both positively and negatively, upon infection. Restriction factors are cellular constituents that can interfere with the activity of viral proteins required for HIV-1 replication. As an example, the deoxynucleotide (dNTP) triphosphohydrolase SAM domain and HD domain-containing protein 1 (better known as SAMHD1), when unphosphorylated, reduces the pool of dNTPs available for viral replication, thereby inhibiting reverse transcription of the viral genome (5, 6). In contrast, expression of HIV-1 dependency factors, such as cyclin L2 or cyclin A2/CDK1, favors HIV-1 infection by controlling the abundance of SAMHD1 (7) or its antiviral activity via phosphorylation (8). To identify new potential regulators of HIV-1 infection, we previously compared the transcriptomic profiles of infected and bystander macrophages and revealed diverse pathways regulating HIV-1 infection in myeloid cells. Potential regulators were tested for their functional role in the establishment of HIV-1 infection in monocyte-derived macrophages (MDMs) with a short interfering RNA (siRNA) screening approach (9). A very careful analysis of these data identified Mouse Double Minute 2 (MDM2) as one of the few positive regulators of HIV-1 infection in MDMs.
MDM2 is a proto-oncogene with E3 ubiquitin ligase activity, found in both the nucleus and the cytoplasm (10), that regulates the turnover of various proteins by conjugating them with ubiquitin for proteasome-mediated degradation. Its main target is the transcription factor and tumor suppressor p53 (11). MDM2 can also directly inhibit the transcriptional activity of p53 by binding to its transactivation domain (12). However, upon cellular stress like DNA damage, MDM2 is phosphorylated by the ATM kinase, which leads to the release and stabilization of p53. In turn, phosphorylation of p53 itself at various serine and threonine residues induces or otherwise modulates its DNA-binding and transcriptional activities (13, 14). When stabilized and activated, p53 leads to the expression of a genetic program involved in cell cycle arrest, induction of apoptosis, DNA editing, and other stress-resolving factors. More specifically, p53 activation induces expression of the CDKN1A gene, coding for the cyclin inhibitor p21Waf1/Cip1 (CDKN1A; here called p21), which leads to cell cycle arrest in G1/S or G2/M and ultimately to cellular senescence if DNA damage cannot be repaired in a timely manner. Expression of apoptosis mediators such as FAS and BAX is also induced upon activation of p53 (15).
Several studies have reported an important role for p53 in the process of HIV-1 infection. For example, expression of this transcription factor is upregulated in activated CD4+ T cells from infected individuals (16), and an accumulation of the p53 protein can be detected specifically in the productively infected cell population (17). HIV-1-infected CD4+ T cells are also subject to apoptosis via p53 activation (18). On the other hand, macrophages, being noncycling terminally differentiated cells, are known to be more resistant both to HIV-1 infection and virus-mediated apoptosis (19, 20).
MDM2 was reported to play a conflicting role in the late events of the HIV-1 replicative cycle. For example, MDM2 can ubiquitinate the viral protein Vif, causing its degradation and, thus, removing its counteracting effect on the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G) restriction factor (21). MDM2 was also shown to ubiquitinate Tat, although this modification leads to an increase of its transcriptional activity instead of causing its degradation (22).
However, MDM2 expression was never reported to affect the early events of the HIV-1 replicative cycle, from viral binding and entry to integration of the proviral DNA into the genome. The cyclin inhibitor p21 was previously shown to inhibit HIV-1 infection in macrophages and dendritic cells not only by downregulating the expression of the RNR2 ribonucleotide reductase subunit, which blocks dNTP synthesis (23), but also by leading to the dephosphorylation and activation of SAMHD1 antiretroviral activity (24–27).
Following our siRNA screening that suggested that MDM2 acts as a positive regulator of HIV-1 infection in MDMs, we were interested in revealing which step(s) of the viral cycle is affected by MDM2 expression and how it controls HIV-1 replication in macrophages. Our results indicate that MDM2 performs as a master regulator of the early postentry events in the HIV-1 life cycle by controlling the stability and transcriptional activity of p53. By decreasing the level of MDM2 in MDMs, we induced resistance to HIV-1 infection which correlated with higher expression of transcripts usually associated with active p53, such as p21. As previously published, this increase in the p21 levels would lead to an accumulation of dephosphorylated SAMHD1, as observed here. Therefore, we propose that MDM2 is a dependency factor for HIV-1 that modulates reverse transcription and proviral DNA integration events in MDMs. A targeted control of the MDM2/p53 axis in macrophages could eventually limit viral spreading or favor eradication in this cellular reservoir.
RESULTS
Knockdown of MDM2 alters susceptibility of MDMs to HIV-1 infection but has no effect on virus gene expression at the single-cell level.
In order to further validate some of our previous siRNA screening results (9), we first transfected MDMs with a nonsense siRNA (siCtrl) or MDM2-specific siRNA (siMDM2) before infection with NL4.3-Bal-IRES-HSA, a previously described fully competent R5-tropic reporter virus expressing the glycosylphosphatidylinositol (GPI)-anchored murine protein HSA (here called NL/Bal-HSA) (17). We assessed the productive virus infection rate at 3 days postinfection (dpi) by flow cytometry analysis (i.e., HSA+ cells). This short time period of virus infection allowed us to estimate the effect of gene silencing on the susceptibility of MDMs to HIV-1 infection while limiting reinfection events.
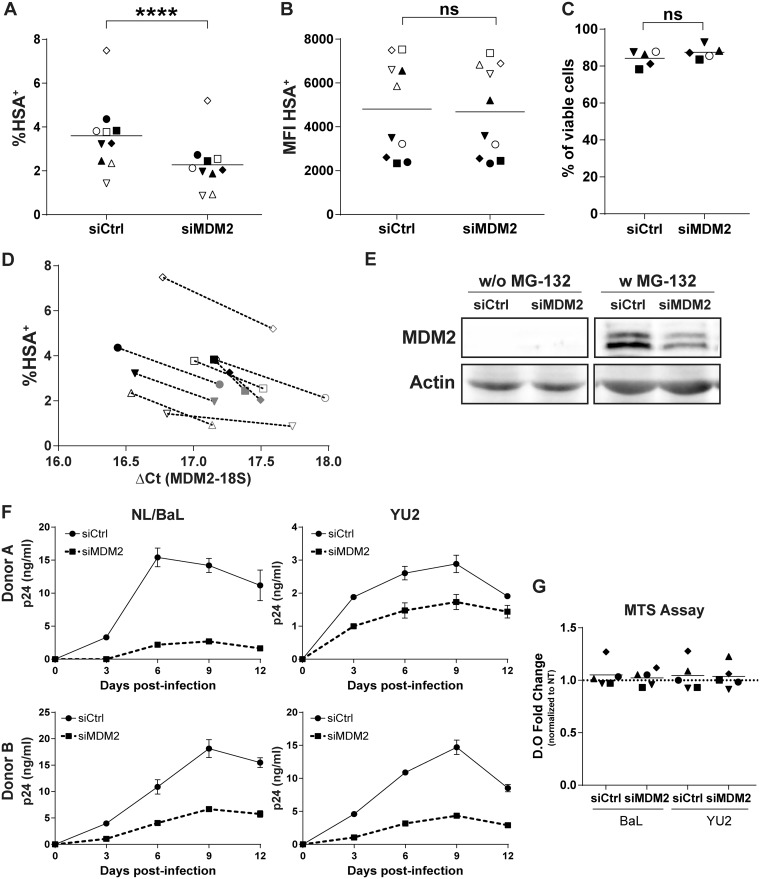
Silencing of MDM2 expression led to a statistically significant decrease in the percentage of MDMs expressing HSA for every donor tested (Fig. 1A), with a mean inhibition of 38%. However, the mean fluorescent intensity (MFI) of the HSA+ population was not modulated between control and gene-specific siRNAs (Fig. 1B). This could mean that viral gene expression at the single-cell level was not affected by MDM2 knockdown. This reduction in HSA+ cells correlates with a lower expression of MDM2 mRNA at the time of infection for every donor tested (Fig. 1D) and a decrease in the targeted protein level at the time of infection (Fig. 1E). Since MDM2 and p53 are ubiquitinated for proteasomal degradation (28), we analyzed each condition with or without exposure to the proteasome inhibitor MG-132 to stabilize the protein levels. This technical procedure allowed the detection of MDM2, which is undetectable otherwise in our protein extracts, and confirmed its silencing by the MDM2-specific siRNA. We also infected previously transfected MDMs with two distinct replication-competent HIV-1 strains (i.e., NL4.3-BalEnv [called NL/Bal] and YU2) and measured virus production over 12 days by a p24 enzyme-linked immunosorbent assay (ELISA). The production of de novo virions was decreased for both viral strains following the siRNA-mediated silencing of MDM2 (Fig. 1F), confirming our observations with the HSA-encoding virus. Toxicity of siRNAs was minimal, as monitored by flow cytometry with a fluorescent viability dye at 3 dpi and with a colorimetric 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay at 12 dpi (Fig. 1C and G).
FIG 1.
MDM2 knockdown reduces HIV-1 replication in MDMs. (A to D) Flow cytometry analysis of HSA expression on the surface of MDMs from 10 independent donors infected with NL/Bal-HSA following transfection with either a nonsense siRNA (siCtrl) or an MDM2-specific siRNA (siMDM2). The percentage of HSA+ cells (A) and the corresponding mean fluorescence intensity (MFI) (B) were monitored at 3 dpi. Statistical analyses were performed using the ratio-paired Student's t test. The asterisks denote statistically significant data (ns, not significant; ****, P ≤ 0.0001). (C) Cell viability was measured for 5 donors by flow cytometry at 3 dpi with the fixable viability dye eFluor780. No significant change in cell viability was detected (ratio-paired Student's t test, n = 5). (D) Infection rate of 9 donors with their respective mRNA level for MDM2 at the time of infection. Each symbol represents the siCtrl (black) and siMDM2 (gray) for an independent donor. (E) Western blot analyses for MDM2 and actin protein levels at the time of HIV-1 infection. MG-132 was added 6 h prior to protein extraction. Data shown are representative of 4 donors. (F) Virus production following infection with NL/Bal or YU2. The p24 content was measured by ELISA at the indicated time points. Data shown represent the means ± standard deviations (SD) from duplicates for 2 different donors, representative of a total of 5 donors. (G) Metabolic activity of MDMs was measured at 12 dpi with MTS assay. No significant change was seen compared to the untreated (NT) MDMs for each virus (one-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test, n = 5).
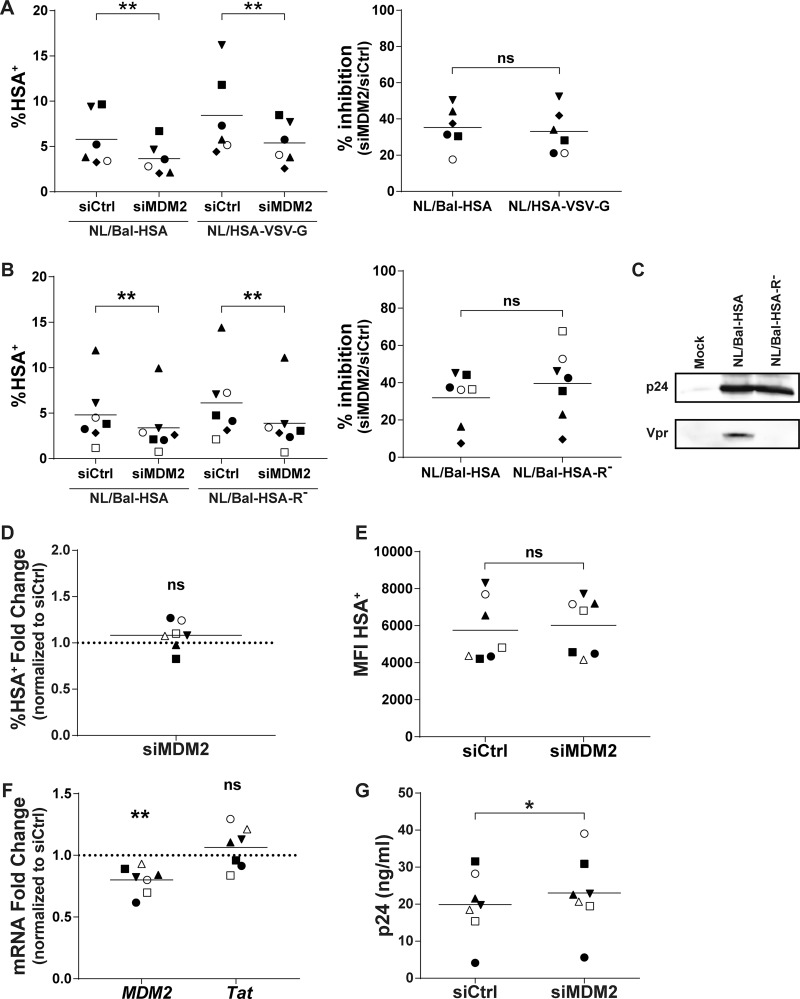
To identify the precise step(s) in the virus replicative cycle that is affected by MDM2, we first focused on some immediate-early events, especially those leading to proviral DNA integration, starting with binding and entry processes. We used two molecular clones of HIV-1 to infect, in a parallel manner, siRNA-transfected MDMs. The first one was the previously described NL/Bal-HSA virus construct, which expresses an R5-tropic gp120 that relies on CD4 and CCR5 to enter the cell and for which fusion is pH independent. The second molecular clone was the NL4.3-ΔENV-HSA vector, which produces envelope-deficient HSA-encoding viruses that were pseudotyped with vesicular stomatitis virus glycoprotein (NL/HSA-VSV-G). The VSV-G envelope glycoprotein relies on LDL receptor binding and allows virus entry in a pH-dependent entry mode. We measured the percentage of virus-infected cells by flow cytometry at 3 dpi and observed a decrease in the number of HSA+ MDMs for both viruses following MDM2 silencing. Since there is a high variation in the infection rate between donors, we compared the inhibition ratio upon MDM2 knockdown. The infection rate was inhibited by a mean of 35% and 33% for NL/Bal-HSA and NL/HSA-VSV-G, respectively, with maximal values reaching 50% and 52%, respectively (Fig. 2A).
FIG 2.
MDM2 gene silencing in MDMs does not affect HIV-1 entry or viral gene expression. (A and B) MDMs from distinct donors were transfected with siCtrl or siMDM2 and then infected for 3 days with NL/Bal-HSA, NL/HSA-VSV-G, or NL/Bal-HSA-R−. The line represents the mean number of biological replicates for each condition. Each symbol represents an independent donor. The percentage of HSA+ MDMs (left) and the calculated percentage of inhibition (right) are shown for cells infected either with NL/Bal-HSA and NL/HSA-VSV-G (n = 6) (A) or NL/Bal-HSA and NL/Bal-HSA-R− (n = 7) (B). (C) HEK293T cells were transfected with pcDNA3.1(-) (mock), NL4.3-Bal-IRES-HSA, or NL4.3-Bal-IRES-HSA-R− with the calcium-phosphate precipitation method for 2 days. Proteins were extracted and analyzed by Western blotting for p24 and Vpr expression. (D to G) MDMs from 7 different donors were infected for 3 days and then transfected for 2 days with siCtrl or siMDM2. Each symbol represents an independent donor and is conserved between panels. The line represents the mean number of biological replicates for each condition. When data are normalized, the dotted line represents the control. (D) Flow cytometry analysis of MDMs productively infected with HIV-1 on the basis of HSA surface expression. (E) Flow cytometry analysis of the MFI for HSA+ MDMs. (F) qRT-PCR analysis of MDM2 and Tat mRNA expression at 2 days posttransfection. (G) Quantification of the p24 content in cell-free supernatants from transfected and infected MDMs by p24 ELISA. Statistical analyses were performed using the ratio-paired Student's t test (A, B, D, E, and G) and paired Student's t test (F) (ns, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01).
HIV-1 Vpr can cause double-strand breaks (DSBs) (29) and structural alteration of the DNA (30), which would, in turn, activate the DNA damage response pathways. A significant decrease in the number of virus-infected MDMs was still detected at 3 dpi after MDM2 knockdown, and comparable inhibition ratios were obtained with both Vpr-encoding and Vpr-deleted viruses (Fig. 2B and C).
MDM2 was shown to increase the transactivation activity of Tat in some established cell lines via ubiquitination of lysine 71 (22). Such action by MDM2 in MDMs would result in a postintegration-mediated increase of viral replication. Therefore, we infected MDMs before siRNA transfection to investigate the effect of MDM2 knockdown on viral gene expression. We first exposed MDMs for 3 days with Bal-HSA and then transfected either siCtrl or siMDM2 in the presence of the reverse transcriptase inhibitor efavirenz (EFV) to prevent new HIV-1 infection events. After 48 h, we performed flow cytometry analysis to assess the percentage of HSA+ MDMs. We found that silencing of MDM2 in previously infected cells does not significantly alter the percentage of HIV-1-infected MDMs (Fig. 2D) or the MFI in HSA+ cells (Fig. 2E). In addition, we extracted total mRNA after 2 days of transfection (i.e., 5 dpi) to evaluate the expression of spliced Tat and MDM2 mRNA by quantitative reverse transcriptase PCR (qRT-PCR). While a knockdown of MDM2 expression in previously infected MDMs transfected with siMDM2 can indeed be detected, Tat mRNA was not significantly affected in the same samples (Fig. 2F). Finally, we measured the p24 viral content by ELISA in the cell-free supernatant of MDMs 2 days after transfection. A modest but statistically significant increase in p24 production was detected upon MDM2 silencing after cells were infected with HIV-1 (Fig. 2G).
Knockdown of MDM2 inhibits HIV-1 reverse transcription and integration.
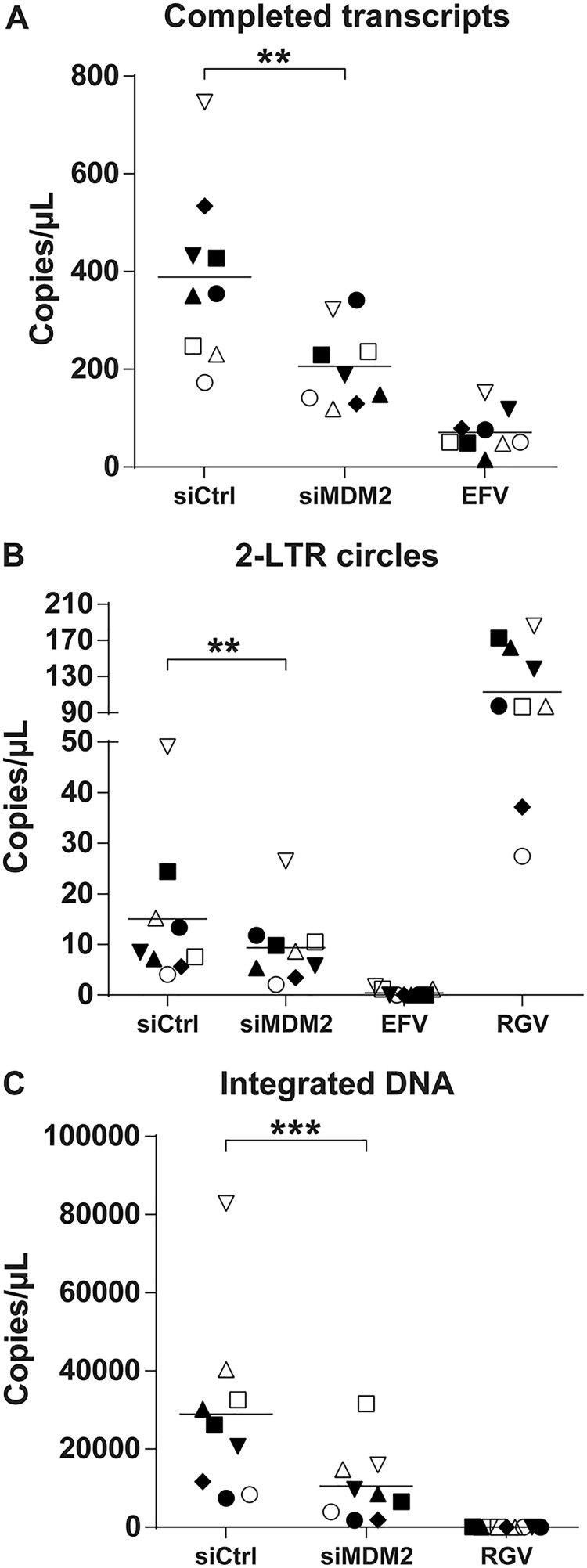
We next monitored the putative modulatory effect of MDM2 on some specific postentry steps of the virus life cycle, especially those that relate to reverse transcription and integration. Thus, we exposed siRNA-transfected MDMs to DNase-treated NL/Bal-HSA for 48 h before DNA extraction. EFV was added to the siRNA-transfected cells after 24 h of HIV-1 infection to ensure the measurement of a single round of infection. We finally evaluated the reverse transcription, nuclear import, and integration efficiency by quantitative real-time PCR (qPCR) using specific primers detecting the number of copies of completed reverse transcripts, 2-long terminal repeat (2-LTR) circles, and integrated proviral DNA, respectively. Samples treated with EFV or the integrase inhibitor raltegravir (RGV) were used as controls. As shown in Fig. 3, knockdown of MDM2 significantly affected all of these replication intermediates. The number of copies of completed reverse transcripts was reduced by a mean of 41% but with a significant variance across donors. The number of 2-LTR circles was also diminished, albeit to a smaller extent (average of 29%). A significant reduction was seen in the number of integrated DNA copies, with a mean decrease of 62% compared to the control.
FIG 3.
MDM2 knockdown decreases HIV-1 DNA integration events. MDMs were transfected with siCtrl or siMDM2 for 48 h or treated with EFV or RGV for 1 h before HIV-1 infection. Cells next were infected for 2 days with DNase-treated NL/Bal-HSA. DNA was extracted at 2 dpi and tested by qPCR to estimate the number of completed reverse transcripts (A), 2-LTR circles (B), and integrated DNA copies (C). EFV was added to the siRNA-transfected cells after 24 h of HIV-1 infection. Each symbol represents an independent donor and is conserved between the figures. Statistical analyses were achieved using the ratio-paired Student's t test (**, P ≤ 0.01; ***, P ≤ 0.001; n = 9).
Knockdown of MDM2 leads to induction of p53 activity in MDMs.
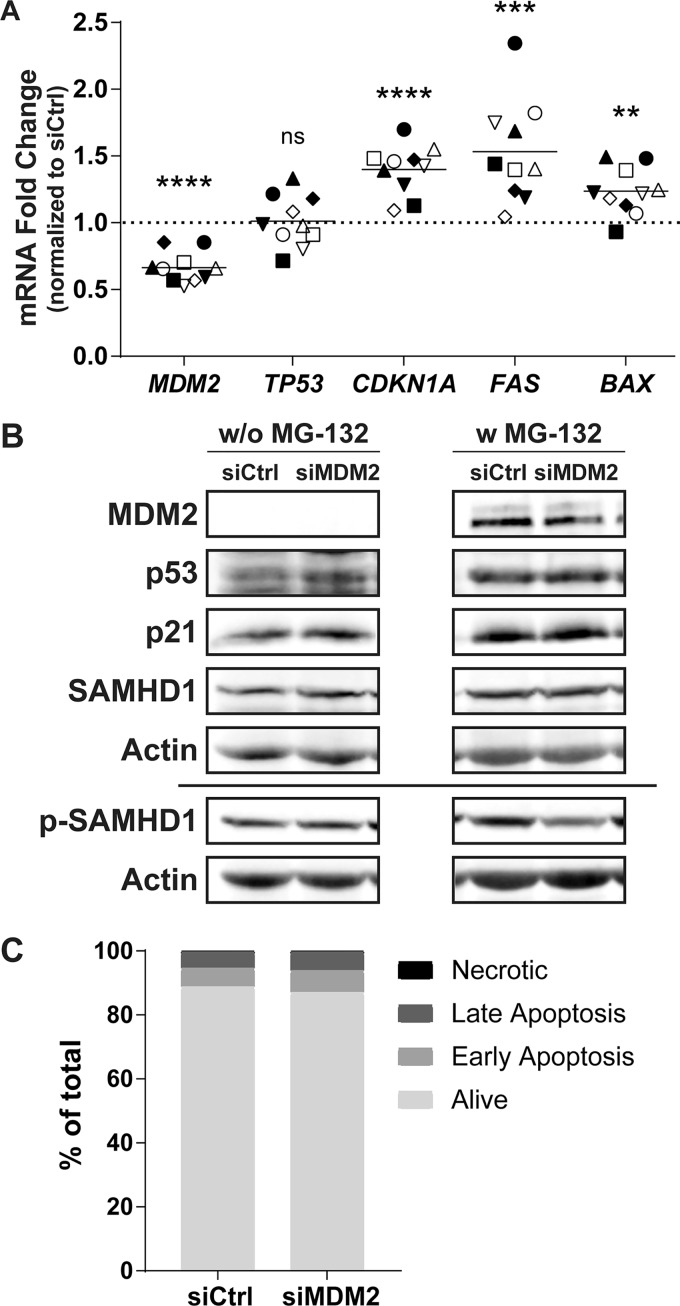
MDM2 is known to be the primary regulator of the transcription factor p53, tagging it for proteasome-mediated degradation via ubiquitination. Following an siRNA-mediated knockdown of MDM2, stabilization and accumulation of p53 should be observed. To define whether this scenario is occurring in MDMs, we extracted the mRNA after 48 h of transfection with siCtrl or siMDM2, which corresponds to the time of HIV-1 infection in the above-described experiments. We then assessed the relative expression of genes involved in the MDM2/p53 pathway by qRT-PCR. While the MDM2-specific siRNA efficiently reduces the expression level of MDM2 mRNA by an average of 34%, p53 (TP53) expression is not affected (Fig. 4A). However, expression levels of known p53-induced genes such as CDKN1A, FAS, and BAX are all increased following MDM2 knockdown, suggesting stabilization and activation of the p53 protein. At the time of virus infection, an increase of 40% for CDKN1A, 53% for FAS, and 24% for BAX mRNA can be observed without any major change in the number of apoptotic cells (Fig. 4C). Changes in expression of the studied genes were confirmed at the protein level at the time of HIV-1 infection (Fig. 4B). The reduction in MDM2 protein level correlates with a stabilization of p53 and an increase in p21, both seen only without exposure to MG-132. Finally, MDM2 knockdown does not affect SAMHD1 expression, but a reduction in SAMHD1 phosphorylated at threonine 592 is more readily observed when the proteasome is inhibited. This is probably caused by the increased stability of p53 in the presence of MG-132, enhancing the effect of the siRNA on SAMHD1 phosphorylation.
FIG 4.
MDM2 gene silencing induces some p53-regulated genes. (A to C) MDMs were transfected with siCtrl or siMDM2 for 48 h, which corresponds to the time of virus infection in previous figures. (A) qRT-PCR analysis was performed to monitor the relative mRNA expression for MDM2, TP53, CDKN1A, FAS, and BAX genes. Each symbol represents a different donor. The dotted line represents the gene expression of the siCtrl condition. Statistical analyses were performed using the paired Student's t test (ns, not significant; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n = 10). (B) Western blot analyses were carried out to quantify the amount of MDM2, p53, p21, total SAMHD1, p-T592 SAMHD1, and actin. Total SAMHD1 and p-T592 SAMHD1 were probed on a different membrane using the same samples and are shown with their respective actin loading controls. Some samples were treated with MG-132 for 6 h before cell lysis. Donor shown is representative of 4 distinct donors. (C) Apoptotic cells at the time of infection were measured by flow cytometry with annexin V and 7-AAD. siRNAs have only a mild effect on apoptosis compared to that of siCtrl. This donor is representative of a total of 2 donors.
Control of p53 by MDM2 favors HIV-1 integration in MDMs.
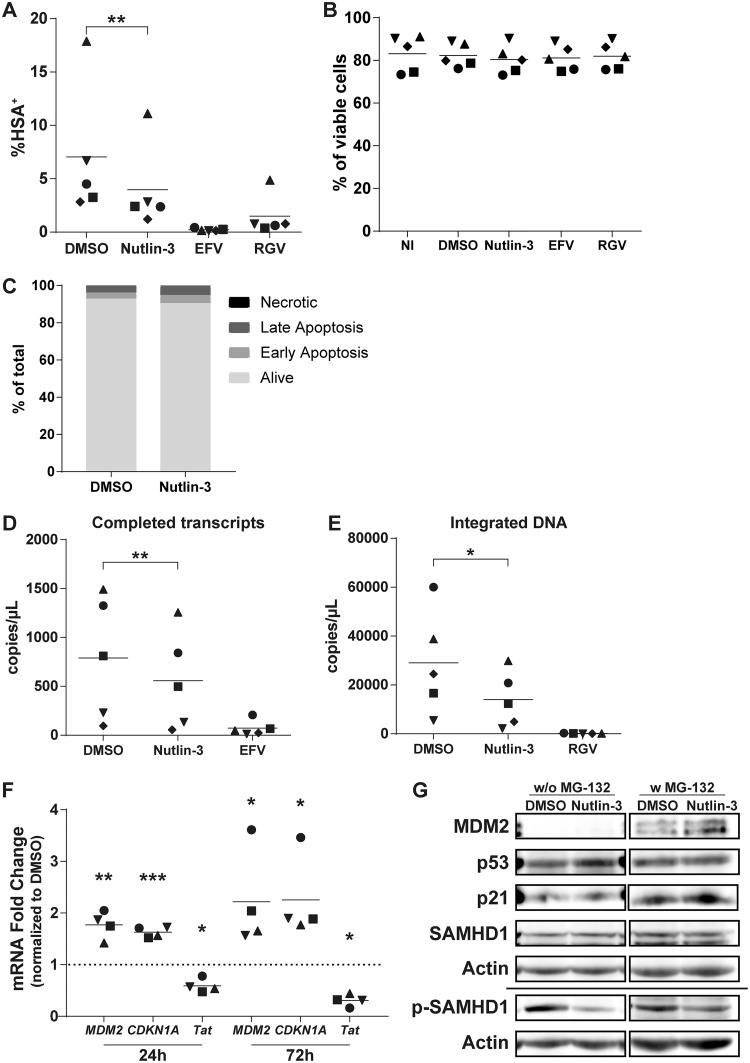
In order to confirm the involvement of MDM2 and p53 in the susceptibility of MDMs to HIV-1 infection, we used the chemical inhibitor Nutlin-3 to specifically abrogate the interaction between MDM2 and p53. This inhibitor leads to the dissociation of p53 proteins from MDM2 complexes and allows their stabilization without affecting the protein level of MDM2 (31). Thus, we treated MDMs with Nutlin-3 for 1 h before incubating them with DNase-treated Bal-HSA. We then measured infection by flow cytometry, detecting the HSA reporter protein at 3 dpi, or by qPCR, measuring the number of completed viral transcripts and integrated viral genomes at 2 dpi. Treatment with Nutlin-3 before HIV-1 infection induced a 45% decrease in the number of HSA+ MDMs (Fig. 5A), a 35% decrease in the number of completed reverse transcripts (Fig. 5D), and a 51% decrease in the number of integrated viral DNA copies (Fig. 5E). We did not detect any major induction of toxicity or apoptosis at the concentration used in our studies (Fig. 5B and C). We also analyzed the mRNA expression levels of some p53-induced genes and the viral Tat gene. We found that treatment with Nutlin-3 for 24 h resulted in an increase of MDM2 and CDKN1A mRNA levels of 77% and 63%, respectively (Fig. 5F). This augmentation was maintained during the entire course of the experiment (e.g., 2.2-fold for MDM2 and 2.3-fold for CDKN1A after 72 h of treatment), and the TP53 mRNA level was unaffected for both time points (data not shown). Nutlin-3 also decreased Tat expression by 41% and 69% compared to that of the control after 24 and 72 h of treatment, respectively, caused by the lower proportion of infected MDMs. At the protein level, Nutlin-3 induced accumulation of MDM2 after 24 h of treatment without exposure to HIV-1 in the presence of MG-132 (Fig. 5G). We also detected the accumulation of p53 without exposure to MG-132, but accumulation of p21 proteins could not be seen. However, a reduction in phosphorylated SAMHD1 was still detected with no effect on total SAMHD1, this time with or without treatment with MG-132.
FIG 5.
Release of p53 from MDM2 control is sufficient to reduce susceptibility of MDMs to HIV-1. (A, B, and D to F) MDMs were pretreated for 1 h with Nutlin-3, EFV, or RGV before infection with DNase-treated NL/Bal-HSA. The drug pressure was maintained for the entire period of virus infection. (A) The percentage of HSA+ MDMs was estimated by flow cytometry analysis at 3 dpi. (B) Cell viability was measured by flow cytometry at 3 dpi with the fixable viability dye eFluor780. No significant change in cell viability was detected (one-way ANOVA with Tukey’s multiple-comparison test, n = 5). (C) MDMs were treated with Nutlin-3 for 3 days without exposure to HIV-1. The percentage of apoptotic cells was measured by flow cytometry with annexin V and 7-AAD. Treatment with Nutlin-3 induces a small increase in apoptosis compared to the level for the DMSO control. This donor is representative of a total of 2 donors. (D and E) The number of completed reverse transcripts (D) and integrated proviral DNA copies (E) was defined by qPCR. (F) Relative mRNA levels of MDM2, CDKN1A, and Tat were measured at 24 and 72 h posttreatment by qRT-PCR. The dotted line represents the gene expression for the DMSO-treated MDMs. (G) The total protein levels of MDM2, p53, p21, SAMHD1, and phosphorylated SAMHD1 (Thr592) in MDMs after 24 h of treatment with Nutlin-3 without exposure to HIV-1 were determined by a Western blot assay. Indicated samples were treated with MG-132 for 6 h before protein extraction. Some proteins had to be exposed for a different time when treated with MG-132, as shown with the cropped membrane. The donor shown is representative of 5 distinct donors. Statistical analyses were done using the ratio-paired Student's t test (A, D, and E; n = 5) or paired Student's t test (F; n = 4) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
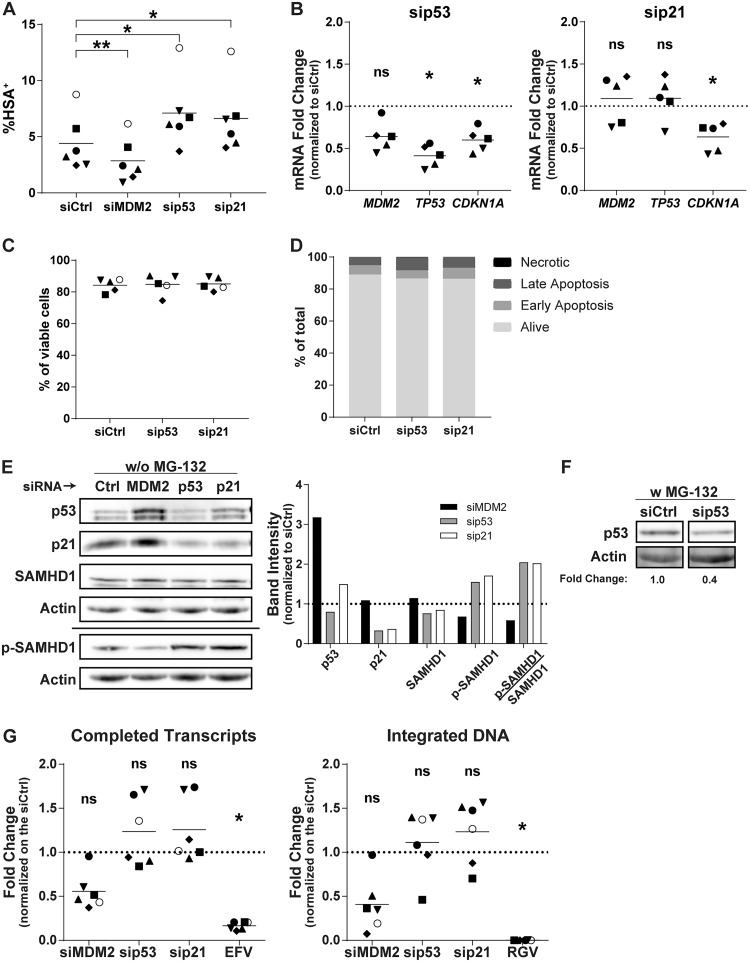
In an attempt to delineate the role of p53 and its downstream effectors in the susceptibility of MDMs to HIV-1 infection, we transfected siRNAs against p53 or p21. The rationale for targeting p21 is based on the idea that its expression is augmented following silencing of MDM2. The p53- and p21-specific siRNAs increased the number of HSA+ cells at 3 dpi to the same extent (Fig. 6A) without cell toxicity (Fig. 6C and D). We then assessed the mRNA level of p21 at the time of virus infection following each siRNA transfection. While the siRNA against MDM2 induced p21 expression (Fig. 4), the increase in the number of virus-infected cells seen in MDMs transfected with siRNAs targeting p53 and p21 correlates with a lower expression of p21 both at the mRNA (Fig. 6B) and protein levels (Fig. 6E). The efficacy of p53 knockdown was also analyzed at the protein level with exposure to MG-132 to increase its stability, facilitating its detection (Fig. 6F). Under these conditions, we saw a clear decrease in p53 protein expression with the corresponding siRNA. The knockdown of p53 and p21 enhanced the mean number of completed reverse transcripts but not for every donor (Fig. 6G), as was observed with the knockdown of MDM2. These siRNAs also increased the number of integrated proviral DNA copies but with significant variability.
FIG 6.
p53 and p21 knockdown lead to an increase in HIV-1 infection. (A) MDMs from 6 different donors were transfected with siCtrl or an siRNA specific for MDM2, p53, or p21 and next infected for 3 days with NL/Bal-HSA. The percentage of HSA+ cells then was assessed by flow cytometry. One-way ANOVA with Dunnett's multiple-comparison test was performed (*, P ≤ 0.05; **, P ≤ 0.01). (B) The relative mRNA expression levels for MDM2, TP53, and CDKN1A were monitored by qRT-PCR for 5 biological replicates at the time of HIV-1 infection. One-way ANOVA with Holm-Sidak's multiple-comparison test was performed (ns, nonsignificant; *, P ≤ 0.05). (C) MDMs were transfected for 2 days and then exposed to NL/Bal-HSA. Cell viability was measured by flow cytometry at 3 dpi with the fixable viability dye eFluor780. No significant change in cell viability was detected (one-way ANOVA with Tukey’s multiple-comparison test, n = 5). (D) MDMs were transfected for 2 days. Apoptotic cells at the time of infection were measured by flow cytometry with annexin V and 7-AAD. siRNAs have only a mild effect on apoptosis compared to that of the siCtrl. This donor is representative of a total of 2 donors. (E, left) The total protein levels of p53, p21, SAMHD1, and phosphorylated SAMHD1 (Thr592) at the time of virus infection were estimated by Western blot analysis. (Right) Band intensities were measured, normalized on the corresponding actin band, and compared to those of the siCtrl. Donor shown is representative of 4 independents donors. (F) The total protein levels of p53 and actin at the time of virus infection. Samples were treated with MG-132 for 6 h before cell lysis. Band intensities were measured, normalized on the corresponding actin band, and compared to those of the siCtrl. (G) The total number of completed reverse transcripts (left) and integrated proviral DNA copies (right) for each experimental condition was quantified by qPCR analysis in 6 biological replicates at 2 dpi. One-way ANOVA with Dunnett's multiple-comparison test was performed (ns, nonsignificant; *, P ≤ 0.05).
SAMHD1 degradation relieves the p53-dependent reduced susceptibility of MDMs to HIV-1 infection.
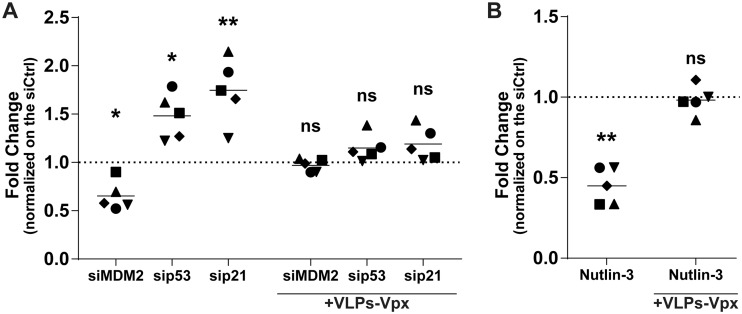
Considering that Nutlin-3 and the gene-specific siRNAs used in this study altered the level of phosphorylated SAMHD1 and susceptibility of MDMs to HIV-1 infection, we tested if reducing SAMHD1 levels could restore the virus infection rate. To this end, we used Vpx-containing virus-like particles derived from the simian immunodeficiency virus (VLPs-Vpx) to induce SAMHD1 degradation (6, 32, 33). We transfected MDMs with gene-specific siRNAs directed against MDM2, p53, and p21 for 2 days. Cells then were inoculated with VLPs-Vpx for 2 h and then infected with NL/Bal-HSA for 3 days. In some instances, MDMs were incubated with VLPs-Vpx, followed by a treatment with Nutlin-3 and, finally, infected with NL/Bal-HSA. As expected, HIV-1 infection was increased in the presence of Vpx for all conditions tested compared to levels for samples not treated with VLPs-Vpx (data not shown). However, when we compared the fold changes to the levels of their respective controls, the modulation in HIV-1 susceptibility caused by the gene-specific siRNAs was almost completely neutralized for cells inoculated with VLPs-Vpx (Fig. 7A). Similar results were obtained following a treatment with Nutlin-3, where its effect on HIV-1 infection was lost upon prior exposure to Vpx-containing VLPs (Fig. 7B).
FIG 7.
p53-mediated change in susceptibility of MDMs to HIV-1 is inhibited by Vpx-dependent degradation of SAMHD1. (A) Flow cytometry analysis showing fold changes of HSA expression on the surface of MDMs compared to that of the control from 5 biological replicates transfected for 48 h with control or gene-specific siRNA before either being left unexposed or being exposed to VLP-Vpx for 2 h, washed, and infected with NL/Bal-HSA for 3 days. One-way ANOVA with Dunnett's multiple-comparison test was performed (ns, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01). (B) Flow cytometry analysis showing fold changes of HSA expression on the surface of MDMs compared to that of the control from 5 biological replicates left unexposed or exposed to VLP-Vpx for 2 h and then to Nutlin-3 or DMSO for 1 h before being infected with NL/Bal-HSA for 3 days. Ratio-paired Student’s t tests were determined (ns, nonsignificant; **, P ≤ 0.01). (A and B) Data represent fold changes compared to the level of the respective control.
DISCUSSION
Macrophages consist of a heterogeneous group of cells, with some subtypes being more susceptible to HIV-1 infection than others. As these cells are natural cellular targets of HIV-1 and as they are suspected of playing an important role in the establishment and persistence of the infection, a better understanding of the cellular factors that dictate their susceptibility to HIV-1 could yield novel targets for treatment. An siRNA screen based on the most differentially expressed genes from our transcriptomic analysis in virus-infected MDMs further revealed that MDM2 acts as one of the rare positive regulators of HIV-1 infection in this cell type (9). Some previous studies performed with immortalized cell lines have shown that MDM2 can regulate HIV-1 infection by interacting with viral proteins such as Tat and Vif (21, 22, 34). However, our results suggest that MDM2 can shape the cellular environment by itself, thereby affecting the susceptibility of MDMs to productive HIV-1 infection.
We first demonstrated that downregulation of MDM2 expression led to a pronounced decrease in HIV-1 replication. The reduced HIV-1 infection could be explained by a less efficient transcription of viral proteins, as MDM2 was previously shown to enhance viral gene transcription through ubiquitination of Tat and potentiation of its transactivation activity (22). However, experiments performed specifically to test this hypothesis yielded negative results. Indeed, MDM2 knockdown in MDMs already infected with HIV-1 did not result in a decrease of virus gene expression. However, it should be noted that the knockdown of MDM2 was less efficient in cells previously infected with HIV-1, and a more significant gene silencing of MDM2 may lead to a greater effect on virus replication. We also eliminated the possibility that MDM2 exerts an effect at the level of virus entry by using VSV-G pseudotypes, which use a CD4/CCR5-independent entry pathway.
Knockdown of MDM2 consistently led to an increase in the expression of some known p53-dependent genes before HIV-1 infection without affecting TP53 mRNA. This strongly suggested a stabilization of p53 caused by MDM2 gene silencing. The accumulation of p21 mediated a decrease of the phosphorylated and inactive form of SAMHD1, a cellular factor known to exert an anti-HIV-1 effect during the reverse transcription step (5, 6, 33). In our work, MDM2 inhibition led to antiviral activity at the early postentry steps, ultimately resulting in a reduced amount of integrated proviral DNA. A similar study using a double-strand break inducer reported a block at the reverse transcription level caused by SAMHD1 activation (35), while another one, using topoisomerase inhibitors to induce DNA damage, also showed a post-reverse transcription effect despite activation of SAMHD1 and a concomitant reduction in intracellular dNTP levels (24). However, in contrast to these studies, we did not use chemicals to directly induce DNA damage. Importantly, as phosphorylation of SAMHD1 at tyrosine 592 lifts HIV-1 restriction but does not restore the level of dNTPs (27), it is still unclear whether the antiviral activity of SAMHD1 is due to its dNTP triphosphohyrolase activity, nuclease activity, or a combination of both (36–38). Another possible cause of resistance not investigated in this study is the repression of RNR2 by the accumulation of p21, inhibiting the dNTP biosynthesis pathway and resulting in restriction at the reverse transcription step (23).
Using the chemical inhibitor Nutlin-3, which disrupts the MDM2-p53 complex, we confirm that p53 stabilization causes resistance to HIV-1 infection through inhibition in postentry replicative steps leading to integration. Moreover, this decreased susceptibility to virus infection persists despite an increase in MDM2 expression, the association of which with p53 is hindered by the inhibitor, supporting the hypothesis that MDM2 affects HIV-1 infection via its binding and control of p53 activation. In contrast to siRNA-mediated silencing of MDM2, which yielded only partial inhibition of MDM2 expression, Nutlin-3 treatment, which targets a specific function of MDM2, showed a more consistent inhibition of HIV-1 reverse transcription and integration in MDMs.
There are several studies about the role of p53 in HIV-1 infection, but few have been performed in the macrophage’s cellular context. Knockdown of p53 before HIV-1 infection consistently increased the infection rate of MDMs. However, this was not always paralleled by an increase in the number of completed reverse transcripts and integrated proviral DNA. A possible explanation stems from the fact that knockdown of MDM2 affects p53 activity while silencing of p53 affects its transcriptional level. A very recent work showed similar results using siRNAs to study the biological function(s) of p53 and p21 in the context of HIV-1 infection (39). However, the role of p53 in virus replication intermediates was assessed in cell lines positive or negative for p53 expression instead of modulating its expression in primary human MDMs, as we did in the present work. Thus, there could be multiple roles of p53 in HIV-1 infection, which could depend on its expression level as well as the cellular environment. As an example, p53 was shown to favor HIV-1 expression in lymphoid cell lines (40), and cytoplasmic p53 was also shown to edit misincorporated nucleotides during reverse transcription of HIV-1 RNA (41). However, while these studies show a positive role of p53, our results demonstrate an inhibitory role of activated p53 in MDMs resulting from the induction of p21, an observation that was also reported in a recent study (24). Indeed, similar to p53, p21 knockdown consistently increased HIV-1 expression, but its effect on the preintegration stages was often conflicting. Similar observations were reported in another work on unstimulated macrophages, where the effect of a p21-specific siRNA on HIV-1 infection was not always consistent (23).
Lastly, since HIV-1 infection in macrophages was reported to induce CDKN1A expression through Vpr (42), we tested a Vpr-deficient HIV-1 isolate and observed a similar decrease in the susceptibility of MDMs to HIV-1 infection upon MDM2 knockdown, eliminating this mode of action.
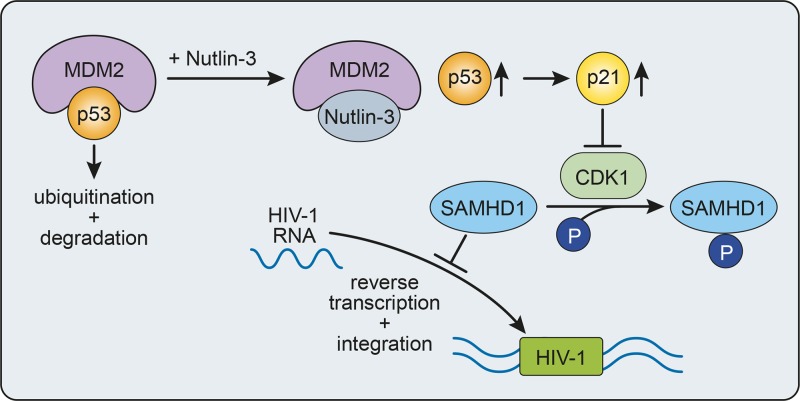
In summary, our results indicate that MDM2 is an important regulator of HIV-1 infection in macrophages, whose expression shapes the cellular environment to favor infection via control of p53 activity (Fig. 8). Important questions that remain unanswered include whether HIV-1, directly or indirectly, causes an increase of MDM2 to facilitate infection following entry into the cell or if the virus preferentially infects macrophages stochastically expressing a higher level of MDM2. Nonetheless, the present study shows that expression levels of MDM2, p53, and p21 are important factors for the susceptibility of MDMs to HIV-1 infection and that the sequestration of p53 by MDM2 favors HIV-1 reverse transcription and integration. Identification of cellular factors, such as MDM2, that act as master regulators for the susceptibility of macrophages to HIV-1 infection will provide new therapeutic targets for the efficient control of HIV-1 replication in these cells to limit the formation of reservoirs in exposed individuals and to efficiently purge infected cells, therefore eventually increasing the chances of a successful eradication strategy.
FIG 8.
Proposed model for MDM2-mediated regulation of HIV-1 infection in human macrophages. Inhibition of MDM2 p53-binding activity with Nutlin-3 before HIV-1 infection stabilizes and increases the level of p53, which in turn results in a higher level of p21 expression, interfering with the phosphorylation/inactivation of SAMHD1 levels by CDK1. This increase in active SAMHD1 leads to a lower number of HIV-1 replication intermediates. Thus, in HIV-1-susceptible MDMs, MDM2 maintains a low level of p53, leading to a high level of phosphorylated SAMHD1 and promoting viral reverse transcription and integration.
MATERIALS AND METHODS
Ethics statement and cell culture.
All experimental protocols involving human blood cells described in the present study were approved by the Bioethics Committee from the Centre Hospitalier Universitaire de Québec-Université Laval. Methods were performed in accordance with the institutional guidelines and regulations. All participants provided written free and informed consent. Peripheral blood samples were taken from healthy donors, and peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll-Hypaque gradient centrifugation. Monocytes were isolated by adhesion for 2 h at 37°C and then washed to remove unattached PBMCs. Monocytes were differentiated to macrophages for 3 days in complete RPMI 1640 culture medium (Corning Life Science, Tewksbury, MA) supplemented with 10% AB human serum (Valley Biomedical, Winchester, VA), antibiotics (Gibco, Thermofisher Scientific, Waltham, MA), and macrophage colony-stimulating factor (M-CSF) (25 ng/ml; GenScript, Piscataway, NJ). Cells were then kept in culture for 3 additional days in complete culture medium (without M-CSF) to obtain nonpolarized MDMs. After 6 days of differentiation, MDMs were treated with Accutase (Invitrogen, Thermofisher Scientific) for 1 h and detached with a soft cell scraper (Sarstedt, Nümbrecht, Germany). Cells were plated for 24 to 72 h in culture medium before treatment or virus infection either in hydrophobic-coating plates (Sarstedt) or ultralow attachment plates (Corning) for flow cytometry studies or with normal tissue culture-treated plates (Corning) when the detachment step was not required. HEK293T cells were kindly provided by Warner C. Greene (The J. Gladstone Institutes, San Francisco, CA) and cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) culture medium (with 10% fetal bovine serum [Corning] and antibiotics).
Virus stocks and infection.
Viruses were produced by calcium-phosphate transfection of HEK293T cells with the NL4.3-BalEnv plasmid coding for an R5-tropic HIV-1 molecular clone (called NL/Bal here) (43), the NL4.3-Bal-IRES-HSA clone expressing the HSA protein on the cell surface upon productive infection (called NL/Bal-HSA here), its Vpr-deficient variant, NL4.3-Bal-IRES-HSA-R− (NL/Bal-HSA-R−), or an R5-tropic molecular clone derived from the YU2 strain of HIV-1. A stock of HSA-expressing virus particles pseudotyped with the VSV-G envelope (NL/HSA-VSV-G) was also produced by cotransfecting an Env-deficient virus vector (NL4.3-IRES-HSAΔEnv) with the pHCMV-G vector expressing the VSV envelope glycoprotein. After 2 days, cell-free supernatant was filtered and ultracentrifuged at 100,000 × g for 1 h in an Optima L-90K ultracentrifuge (Beckman Coulter), and viral pellets were resuspended in endotoxin-free Dulbecco’s phosphate-buffered saline (DPBS). The p24 content of the viral preparation was measured with a homemade ELISA specific for the HIV-1 capsid antigen p24 (44). Adhered MDMs were infected by using a virus inoculum corresponding to 20 ng of p24 per 1 × 105 cells. Infection rate and viral production were monitored by flow cytometry and p24 ELISA, respectively. The pHCMV-G vector was kindly provided by T. Friedmann (University of California-San Diego, La Jolla, CA).
VLPs-Vpx were obtained by cotransfecting HEK293T cells with pHCMV-G and a Vpx-encoding plasmid (pSIV3+) (32, 33). To reduce SAMHD1 levels, MDMs were inoculated with an effective volume of VLPs-Vpx for 2 h in complete culture medium at 37°C, washed, and then infected with NL/Bal-HSA for 3 days. The pSIV3+ vector was a kind gift from F. Margottin-Goguet (Université Paris Descartes, Paris, France).
In order to generate the pNL4.3-Bal-IRES-HSA-R− molecular construct, the internal ribosomal entry site (IRES)-HSA insert was introduced by digestion of the pNL4.3ΔVpr and pNL4.3-Bal-IRES-HSA-WT with BamHI and XhoI restriction enzymes. pNL4.3ΔVpr encodes a nonfunctional truncated 27-amino-acid long Vpr (45). The fragment BamHI-XhoI from pNL4.3-Bal-IRES-HSA-WT was inserted into pNL4.3ΔVpr, giving the intermediate pNL4.3ΔVpr-IRES-HSA. Second, the envelope Bal was inserted by digesting the 2 vectors using SalI-BamHI. The SalI-BamHI fragment from pNL4.3-Bal-IRES-HSA-WT was inserted in pNL4.3ΔVpr-IRES-HSA, giving the final construct pNL4.3-Bal-IRES-HSA-R−.
Flow cytometry analyses were performed following infection of 1.5 × 105 target cells with HSA-expressing constructs for 3 days unless specified otherwise. MDMs were detached with a soft scraper after an incubation of 30 min at 37°C in phosphate-buffered saline (PBS) (pH 7.4)–2 mM ethylenediaminetetraacetic acid (EDTA)–0.5% (wt/vol) bovine serum albumin (BSA) and fraction V. Cell viability was evaluated with a fixable viability dye (FVD eFluor 450 or FVD eFluor 780; Invitrogen) by following the manufacturer’s instructions. After a blocking step of 30 min, cells were stained with phycoerythrin-conjugated rat anti-mouse HSA (30 ng per test; clone M1/69; Invitrogen) and fixed for 30 min at 4°C in 4% formaldehyde. Flow cytometry was performed on a BD FACSCelesta system and analyzed using FlowJo software, version 10 for Windows.
For p24 production kinetics, 1.5 × 105 cells were exposed to NL/Bal or YU2 for 2 h, extensively washed with DPBS, and further incubated in 1 ml of culture medium. Half of the supernatant was collected every 3 days (until 12 dpi) and replaced with fresh medium. The p24 content in cell-free supernatants was measured by ELISA. At 12 dpi, the metabolic activity of MDMs was assessed with a colorimetric MTS assay using a CellTiter 96 AQueous nonradioactive cell proliferation assay by following the manufacturer’s instructions (Promega, Madison, WI). Absorbance was measured at 490 nm with an ELX808 microplate reader (Biotek Instruments, Winooski, VT).
siRNA transfection.
siRNAs were transfected as described previously, with some minor modifications (9). Briefly, siRNAs were diluted in Lipofectamine RNAiMAX transfection reagent (Invitrogen) and Opti-MEM (Gibco) before being added, for a final concentration of 20 nM, to plated cells that had their culture medium replaced by RPMI 1640. MDMs were incubated for 2 h at 37°C in this transfection medium before an equal volume of RPMI containing 20% AB human serum was added to each well, for a final concentration of 10% AB human serum and 10 nM siRNAs. Cells were incubated for 48 h before performing further experiments.
Gene expression analysis.
After 48 h of transfection or 24 h of treatment with Nutlin-3 (500 nM; Tocris Bioscience, Bristol, UK), total mRNA was extracted from 1.5 × 105 MDMs using NucleoSpin RNA kits (Macherey-Nagel, Duren, Germany). The total RNA was reverse transcribed using a Moloney-murine leukemia virus RT polymerase kit (Promega), random primers (Roche, Basel, Switzerland), and dNTPs. The resulting cDNA was assessed by qRT-PCR with a Quantstudio 6 real-time PCR system (Applied Biosystems, Thermofisher Scientific) with PowerUp SYBR green master mix (Applied Biosystems) and specific primers (Integrated DNA Technologies, Skokie, IL) (Table 1 depicts the complete list of primer sequences) (46–49). mRNA expression was normalized with the 18S rRNA and analyzed using the 2−ΔΔCT method (50).
TABLE 1.
List of primers and probes
| Name | Sequence |
|---|---|
| 18S sense | TAGAGGGACAAGTGGCGTTC |
| 18S antisense | CGCTGAGCCAGTCAGTGT |
| MDM2 sense | GAGGGCTTTGATGTTCCTGA |
| MDM2 antisense | TGGCATTAAGGGGCAAACTA |
| TP53 sense | CTCACCATCATCACACTGGAA |
| TP53 antisense | TCATTCAGCTCTCGGAACATC |
| CDKN1A sense | AGGTGGACCTGGAGACTCTCAG |
| CDKN1A antisense | TCCTCTTGGAGAAGATCAGCCG |
| FAS sense | TGAAGGACATGGCTTAGAAGTG |
| FAS antisense | GGTGCAAGGGTCACAGTGTT |
| BAX sense | TGGCAGCTGACATGTTTTCTGAC |
| BAX antisense | TCACCCAACCACCCTGGTCTT |
| Spliced-Tat sense | GAAGCATCCAGGAAGTCAGC |
| Spliced-Tat antisense | GGAGGTGGGTTGCTTTGATA |
| Alu | TCCCAGCTACTCGGGAGGCTGAGG |
| M661 | CCTGCGTCGAGAGATCTCCTCTG |
| M667 | GGCTAACTAGGGAACCCACTGC |
| AA55 | CTGCTAGAGATTTTCCACACTGAC |
| HIV-1 probe | 5′-FAM-TAGTGTGTGCCCGTCTGTTGTGTGAC-BHQ-3′ |
| 2LTR-S | CCCTCAGACCCTTTTAGTCAGTG |
| 2LTR-AS | TGGTGTGTAGTTCTGCCAATCA |
| HIV-2LTR probe | 5′-FAM-TGTGGATCTACCACACACAAGGCTTCC-BHQ-3′ |
| Globin sense | TGGTCTATTTTCCCACCCT |
| Globin antisense | TGGCAAAGGTGCCCTTGA |
| Globin probe | 5′-VIC-TCTGTCCACTCCTGATGCTG-NFQ-MGB-3′ |
For protein expression, 8 × 105 MDMs were lysed with homemade T8 lysis buffer (7 M urea, 2 M thiourea, 3% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 20 mM dithiothreitol, 5 mM Tris(2-carboxyethyl)phosphine hydrochloride, and 50 mM Tris) supplemented with protease inhibitor cocktail (Sigma-Aldrich, Oakville, ON) and phosSTOP phosphatase inhibitor (Roche). For some conditions, MG-132 (Sigma-Aldrich) was added to the well 6 h before protein extraction (final concentration of 10 μM). Proteins were quantified with the Quick Start Bradford protein assay (Bio-Rad, Hercules, CA). Cell extracts were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. These membranes were probed overnight at 4°C with antibodies specific for MDM2 (SMP14; Novus Biologicals, Oakville, ON), p53 (DO-1; BioLegend, San Diego, CA), p21 (12D1; Cell Signaling Technology, Danvers, MA), SAMHD1 (I19-18; Millipore, Etobicoke, ON), phosphorylated SAMHD1 (Thr 592) (D702M; Cell Signaling Technology), and actin (C-2; Santa Cruz Biotechnology, Dallas, TX). Proteins were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) and revealed with Amersham ECL prime or ECL select (GE Healthcare Life Sciences, Mississauga, ON). Signal was captured with a Fusion FX7 Spectra chemical documentation system and analyzed with Fusion software (Vilber-Lourmat, Collégien, France).
For NL/Bal-HSA-R− plasmid validation, HEK293T cells were transfected with the plasmid for 2 days, and then proteins were extracted with T8 lysis buffer supplemented with a protease inhibitor cocktail. Cell extracts were separated as previously described, and the membrane was probed overnight with antibodies specific for HIV-1 p24 (183-H12-5C; NIH AIDS Reagent Program, Germantown, MD) and HIV-1 Vpr (Proteintech, Rosemont, IL). pcDNA3.1(-) and NL/Bal-HSA were used as negative and positive controls, respectively.
Apoptosis assay.
MDMs (1.5 × 105) were plated on ultralow attachment plates (24 wells) to avoid scraping and minimize cell death. Cells were transfected with the siRNAs for 48 h or treated with Nutlin-3 (500 nM) for 3 days. MDMs were detached after an incubation period of 15 min in PBS (pH 7.4)–2 mM EDTA–0.5% (wt/vol) BSA, fraction V, at 37°C. Cells were then stained with an annexin V-CF blue 7-aminoactinomycin D (7-AAD) apoptosis staining/detection kit (Abcam) by following the manufacturer’s instructions.
HIV-1 DNA quantification.
MDMs (4 × 105) were transfected for 2 days or treated for 1 h with chemical inhibitors before infection with DNase-treated Bal-HSA (80 ng of p24). The virus stocks were treated with DNase I (Roche) for 45 min at room temperature immediately before infection. EFV was added to the appropriate wells after 24 h of infection (100 nM) to ensure a single round of HIV-1 infection. After a total of 48 h, cells were washed thoroughly with PBS, and DNA was extracted using the NucleoSpin tissue extraction kit (Macherey-Nagel). Three forms of HIV-1 DNA were measured (i.e., completed reverse transcripts, 2-LTR circles, and proviral DNA copies) by qPCR using TaqMan fast advanced master mix (Applied Biosystems) and performed on a QuantStudio 6. All probes and primers are from Integrated DNA Technologies, except for the β-globin probe (Applied Biosystems). The number of completed reverse transcripts was quantified in 25 ng of cellular DNA using an HIV-1 TaqMan probe and M667/M661 primers (51). 2-LTR circles and integrated proviral DNA copies were measured as previously reported (52). 2-LTR circles, which are rapidly degraded in macrophages, were quantified using 250 ng of DNA with the 2-LTR TaqMan probe and 2LTR-S/2LTR-AS primers. Finally, the number of proviral DNA copies was assessed by Alu-HIV-1 PCR combined with nested qPCR. The first Alu-HIV-1 PCR was performed using Taq DNA polymerase (Invitrogen), 100 ng of genomic DNA, and Alu/M661 primers. The resulting samples were then diluted and assessed for the presence of proviral DNA with the AA55/M667 primers and the HIV-1 TaqMan probe. The number of DNA copies obtained for the 3 forms was normalized using the β-globin gene copy number for each diluted sample. Standard curves for HIV-1 replication intermediates were done with a serial dilution of the NL/Bal-HSA plasmid, starting at 6 × 106 copies. For the number of 2-LTR circles, a standard curve starting at 4 × 106 copies was made with the 2-LTR plasmid, kindly provided by Nicolas Chomont (Université de Montréal, Montréal, QC). EFV was kindly provided by the NIH AIDS Reagent Program. Raltegravir was purchased from Cayman Chemical (Ann Arbor, MI).
Statistical analysis.
All statistical tests were performed using GraphPad Prism, version 7.03, as specified in the figure legends. The number of independent donors (n) tested for each experiment is also indicated in the legends. Data were considered statistically significant for P values of ≤0.05.
ACKNOWLEDGMENTS
We thank Caroline Côté for technical support and Julie-Christine Lévesque at the Bioimaging Platform of the Infectious Disease Research Centre, which was funded by an equipment and infrastructure grant from the Canadian Foundation for Innovation. We acknowledge the clinical research team at the infectious disease unit for the blood samples and thank the participants, without whom we could not have performed this study.
This study was supported by funds allocated to M.J.T. from the Canadian HIV Cure Enterprise (CanCURE) via a Team Grant from the Canadian Institutes of Health Research (CIHR) in partnership with the Canadian Foundation for AIDS Research and the International AIDS Society (HIG-133050). M.J.T. is the recipient of the CIHR Canada Research Chair in Human Immuno-Retrovirology (Tier 1 level). We have no competing financial interests to declare.
Authors made the following contributions: conceptualization, Y.B., M.O., A.D., and M.J.T.; formal analysis, Y.B.; funding acquisition, M.J.T. and É.A.C.; investigation, Y.B., V.D., and C.T.; methodology, Y.B., A.D., and M.O.; resources, C.T. and É.A.C.; supervision, M.O.; original draft writing, Y.B. and M.O.; review and editing, Y.B., M.O., M.J.T., and É.A.C.
REFERENCES
- 1.Rodrigues V, Ruffin N, San-Roman M, Benaroch P. 2017. Myeloid cell interaction with HIV: a complex relationship. Front Immunol 8:1698. doi: 10.3389/fimmu.2017.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sattentau QJ, Stevenson M. 2016. Macrophages and HIV-1: an unhealthy constellation. Cell Host Microbe 19:304–310. doi: 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV. 2016. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Investig 126:1353–1366. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, Garcia JV. 2017. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 6.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyei GB, Cheng X, Ramani R, Ratner L. 2015. Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17:98–106. doi: 10.1016/j.chom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Deshiere A, Joly-Beauparlant C, Breton Y, Ouellet M, Raymond F, Lodge R, Barat C, Roy MA, Corbeil J, Tremblay MJ. 2017. Global mapping of the macrophage-HIV-1 transcriptome reveals that productive infection induces remodeling of host cell DNA and chromatin. Sci Rep 7:5238. doi: 10.1038/s41598-017-05566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J 17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 12.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 13.Shieh SY, Ikeda M, Taya Y, Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334. doi: 10.1016/S0092-8674(00)80416-X. [DOI] [PubMed] [Google Scholar]
- 14.Lazo PA. 2017. Reverting p53 activation after recovery of cellular stress to resume with cell cycle progression. Cell Signal 33:49–58. doi: 10.1016/j.cellsig.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. 2018. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ 25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedaghat AR, German J, Teslovich TM, Cofrancesco J Jr, Jie CC, Talbot CC Jr, Siliciano RF. 2008. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol 82:1870–1883. doi: 10.1128/JVI.02228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imbeault M, Lodge R, Ouellet M, Tremblay MJ. 2009. Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology 393:160–167. doi: 10.1016/j.virol.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Genini D, Sheeter D, Rought S, Zaunders JJ, Susin SA, Kroemer G, Richman DD, Carson DA, Corbeil J, Leoni LM. 2001. HIV induces lymphocyte apoptosis by a p53-initiated, mitochondrial-mediated mechanism. FASEB J 15:5–6. doi: 10.1096/fj.00-0336fje. [DOI] [PubMed] [Google Scholar]
- 19.Montaner LJ, Crowe SM, Aquaro S, Perno CF, Stevenson M, Collman RG. 2006. Advances in macrophage and dendritic cell biology in HIV-1 infection stress key understudied areas in infection, pathogenesis, and analysis of viral reservoirs. J Leukoc Biol 80:961–964. doi: 10.1189/jlb.0806488. [DOI] [PubMed] [Google Scholar]
- 20.Perno CF, Svicher V, Schols D, Pollicita M, Balzarini J, Aquaro S. 2006. Therapeutic strategies towards HIV-1 infection in macrophages. Antiviral Res 71:293–300. doi: 10.1016/j.antiviral.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Izumi T, Takaori-Kondo A, Shirakawa K, Higashitsuji H, Itoh K, Io K, Matsui M, Iwai K, Kondoh H, Sato T, Tomonaga M, Ikeda S, Akari H, Koyanagi Y, Fujita J, Uchiyama T. 2009. MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology 6:1. doi: 10.1186/1742-4690-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol 5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 23.Allouch A, David A, Amie SM, Lahouassa H, Chartier L, Margottin-Goguet F, Barré-Sinoussi F, Kim B, Sáez-Cirión A, Pancino G. 2013. p21-mediated RNR2 repression restricts HIV-1 replication in macrophages by inhibiting dNTP biosynthesis pathway. Proc Natl Acad Sci U S A 110:E3997–E4006. doi: 10.1073/pnas.1306719110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlcochova P, Caswell SJ, Taylor IA, Towers GJ, Gupta RK. 2018. DNA damage induced by topoisomerase inhibitors activates SAMHD1 and blocks HIV-1 infection of macrophages. EMBO J 37:50–62. doi: 10.15252/embj.201796880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauls E, Ruiz A, Riveira-Munoz E, Permanyer M, Badia R, Clotet B, Keppler OT, Ballana E, Este JA. 2014. p21 regulates the HIV-1 restriction factor SAMHD1. Proc Natl Acad Sci U S A 111:E1322–E1324. doi: 10.1073/pnas.1322059111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valle-Casuso JC, Allouch A, David A, Lenzi GM, Studdard L, Barre-Sinoussi F, Muller-Trutwin M, Kim B, Pancino G, Saez-Cirion A. 2017. p21 restricts HIV-1 in monocyte-derived dendritic cells through the reduction of deoxynucleoside triphosphate biosynthesis and regulation of SAMHD1 antiviral activity. J Virol 91:e01324–e01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparks A, Dayal S, Das J, Robertson P, Menendez S, Saville MK. 2014. The degradation of p53 and its major E3 ligase Mdm2 is differentially dependent on the proteasomal ubiquitin receptor S5a. Oncogene 33:4685–4696. doi: 10.1038/onc.2013.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tachiwana H, Shimura M, Nakai-Murakami C, Tokunaga K, Takizawa Y, Sata T, Kurumizaka H, Ishizaka Y. 2006. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res 66:627–631. doi: 10.1158/0008-5472.CAN-05-3144. [DOI] [PubMed] [Google Scholar]
- 30.Iijima K, Kobayashi J, Ishizaka Y. 2018. Structural alteration of DNA induced by viral protein R of HIV-1 triggers the DNA damage response. Retrovirology 15:8. doi: 10.1186/s12977-018-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 32.Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther 7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 33.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raja R, Ronsard L, Lata S, Trivedi S, Banerjea AC. 2017. HIV-1 Tat potently stabilises Mdm2 and enhances viral replication. Biochem J 474:2449–2464. doi: 10.1042/BCJ20160825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jauregui P, Landau NR. 2018. DNA damage induces a SAMHD1-mediated block to the infection of macrophages by HIV-1. Sci Rep 8:4153. doi: 10.1038/s41598-018-22432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. 2014. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med 20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonucci JM, St Gelais C, de Silva S, Yount JS, Tang C, Ji X, Shepard C, Xiong Y, Kim B, Wu L. 2016. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med 22:1072–1074. doi: 10.1038/nm.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi B, Sharifi HJ, DiGrigoli S, Kinnetz M, Mellon K, Hu W, de Noronha CMC. 2018. Inhibition of HIV early replication by the p53 and its downstream gene p21. Virol J 15:53. doi: 10.1186/s12985-018-0959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauls E, Senserrich J, Clotet B, Este JA. 2006. Inhibition of HIV-1 replication by RNA interference of p53 expression. J Leukoc Biol 80:659–667. doi: 10.1189/jlb.0306189. [DOI] [PubMed] [Google Scholar]
- 41.Akua T, Rahav G, Saragani Y, Hizi A, Bakhanashvili M. 2017. Removal of ribonucleotides by p53 protein incorporated during DNA synthesis by HIV-1 reverse transcriptase. AIDS 31:343–353. doi: 10.1097/QAD.0000000000001339. [DOI] [PubMed] [Google Scholar]
- 42.Vazquez N, Greenwell-Wild T, Marinos NJ, Swaim WD, Nares S, Ott DE, Schubert U, Henklein P, Orenstein JM, Sporn MB, Wahl SM. 2005. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J Virol 79:4479–4491. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dornadula G, Zhang H, Shetty S, Pomerantz RJ. 1999. HIV-1 virions produced from replicating peripheral blood lymphocytes are more infectious than those from nonproliferating macrophages due to higher levels of intravirion reverse transcripts: implications for pathogenesis and transmission. Virology 253:10–16. doi: 10.1006/viro.1998.9465. [DOI] [PubMed] [Google Scholar]
- 44.Bounou S, Leclerc JE, Tremblay MJ. 2002. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol 76:1004–1014. doi: 10.1128/JVI.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lodge R, Gilmore JC, Ferreira Barbosa JA, Lombard-Vadnais F, Cohen EA. 2017. Regulation of CD4 receptor and HIV-1 entry by microRNAs-221 and -222 during differentiation of THP-1 cells. Viruses 10:E13. doi: 10.3390/v10010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR, Ji H, Vojtesek B, Bourdon JC, Lane DP, Harris CC. 2009. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol 11:1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calderon MR, Verway M, An BS, DiFeo A, Bismar TA, Ann DK, Martignetti JA, Shalom-Barak T, White JH. 2012. Ligand-dependent corepressor (LCoR) recruitment by Kruppel-like factor 6 (KLF6) regulates expression of the cyclin-dependent kinase inhibitor CDKN1A gene. J Biol Chem 287:8662–8674. doi: 10.1074/jbc.M111.311605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korbakis D, Scorilas A. 2012. Quantitative expression analysis of the apoptosis-related genes BCL2, BAX and BCL2L12 in gastric adenocarcinoma cells following treatment with the anticancer drugs cisplatin, etoposide and taxol. Tumour Biol 33:865–875. doi: 10.1007/s13277-011-0313-z. [DOI] [PubMed] [Google Scholar]
- 49.Das H, Koizumi T, Sugimoto T, Chakraborty S, Ichimura T, Hasegawa K, Nishimura R. 2000. Quantitation of Fas and Fas ligand gene expression in human ovarian, cervical and endometrial carcinomas using real-time quantitative RT-PCR. Br J Cancer 82:1682–1688. doi: 10.1054/bjoc.2000.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. doi: 10.1016/0092-8674(90)90802-L. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki Y, Misawa N, Sato C, Ebina H, Masuda T, Yamamoto N, Koyanagi Y. 2003. Quantitative analysis of human immunodeficiency virus type 1 DNA dynamics by real-time PCR: integration efficiency in stimulated and unstimulated peripheral blood mononuclear cells. Virus Genes 27:177–188. doi: 10.1023/A:1025732728195. [DOI] [PubMed] [Google Scholar]