This paper reports the first biological characterization of a bona fide plant mitovirus in an important crop, Chenopodium quinoa, providing data supporting that mitoviruses have the typical features of cryptic (persistent) plant viruses. We, for the first time, demonstrate that plant mitoviruses are associated with mitochondria in plants. In contrast to fungal mitoviruses, plant mitoviruses are not substantially affected by the antiviral silencing pathway, and the most abundant mitovirus small RNA length is 16 nt.
KEYWORDS: plant virus, defence, mitochondria, mitovirus
ABSTRACT
Indirect evidence of mitochondrial viruses in plants comes from discovery of genomic fragments integrated into the nuclear and mitochondrial DNA of a number of plant species. Here, we report the existence of replicating mitochondrial virus in plants: from transcriptome sequencing (RNA-seq) data of infected Chenopodium quinoa, a plant species commonly used as a test plant in virus host range experiments, among other virus contigs, we could assemble a 2.7-kb contig that had highest similarity to mitoviruses found in plant genomes. Northern blot analyses confirmed the existence of plus- and minus-strand RNA corresponding to the mitovirus genome. No DNA corresponding to the genomic RNA was detected, excluding the endogenization of such virus. We have tested a number of C. quinoa accessions, and the virus was present in a number of commercial varieties but absent from a large collection of Bolivian and Peruvian accessions. The virus could not be transmitted mechanically or by grafting, but it is transmitted vertically through seeds at a 100% rate. Small RNA analysis of a C. quinoa line carrying the mitovirus and infected by alfalfa mosaic virus showed that the typical antiviral silencing response active against cytoplasmic viruses (21- to 22-nucleotide [nt] vsRNA peaks) is not active against CqMV1, since in this specific case the longest accumulating vsRNA length is 16 nt, which is the same as that corresponding to RNA from mitochondrial genes. This is evidence of a distinct viral RNA degradation mechanism active inside mitochondria that also may have an antiviral effect.
IMPORTANCE This paper reports the first biological characterization of a bona fide plant mitovirus in an important crop, Chenopodium quinoa, providing data supporting that mitoviruses have the typical features of cryptic (persistent) plant viruses. We, for the first time, demonstrate that plant mitoviruses are associated with mitochondria in plants. In contrast to fungal mitoviruses, plant mitoviruses are not substantially affected by the antiviral silencing pathway, and the most abundant mitovirus small RNA length is 16 nt.
INTRODUCTION
The family Narnaviridae comprises two genera of positive single-stranded RNA (ssRNA) viruses, Narnavirus and Mitovirus, both originally thought to be mycoviruses, because fungi are the main natural host in which they can replicate. Both are known to be naked viruses presenting only one open reading frame (ORF) that encodes one protein, the RNA-dependent RNA polymerase (RdRp) (1). The two genera are distinguished based on phylogenetic analysis (each of them belongs to a distinct, statistically well-supported clade) and on subcellular localization: the narnaviruses replicate and stay in the cytosol, whereas the mitoviruses typically replicate and persist in the mitochondrion. Evidence of their mitochondrial localization is provided by the fact that they fractionate with the mitochondrial fraction (2) and that for most of them the ORF is translated using the mitochondrial genetic code (1). The mitoviruses described so far are able to infect only mitochondria of filamentous fungi, and in some cases they are associated with hypovirulence (1). Sometimes it seems that the mitochondrial morphology is not affected by the virus, whereas in some other cases they can cause a morphological alteration (i.e., fibrous mitochondria) that possibly was associated with the induced hypovirulence (3).
Some fungal mitoviruses have the potential to use both the nuclear and mitochondrial genetic code for the translation of their genomes, and based on this, some authors have hypothesized promiscuous replication in both mitochondria and cytoplasm. However, bioinformatic analysis leads to a different explanation: in those host species where mitovirus RdRp can hypothetically be translated using both genetic codes, the mitochondrial genes have a strong bias for the tryptophan codon that is shared with the nuclear genetic code (UGG). This implies that the exclusive use of UGG codon for tryptophan in some mitoviruses is not because of a promiscuous lifestyle between mitochondria and cytoplasm but rather reflects the fact that UGA (the only differential codon between the nuclear and mitochondrial genetic codes in the fungal host species) is not present in general in genes carried by the mitochondria (4).
Intriguingly, species of genus Mitovirus are most closely related to the Leviviridae, the only taxonomically defined group of positive-stranded ssRNA bacterial phages (5, 6). Within the context of the theory of mitochondria derived from an alphaproteobacterial endosymbiont (7), this suggests that mitoviruses derived from an ancestral mitochondrial phage by losing the capsid protein (CP) (8), which is unnecessary due to the absence of an extracellular stage (9).
A further interesting observation related to mitoviruses comes from the availability of a great number of complete plant and fungal genomes that were mined to reveal the presence of nonretroviral endogenous RNA viral elements (NERVEs): complete genomes or partial/complete gene sequences of RNA viruses are present in almost all of the eukaryotic nuclear genomes (10–13). Specifically, Bruenn and coauthors demonstrated the widespread presence of mitoviral sequences in many plant nuclear and mitochondrial genomes (13). Two different hypotheses can explain this evidence. The first is diverse integration events of a fungal mitovirus, or of a native plant mitochondrial virus, into the mitochondrial genome, and from here to the nucleus as a result of mitochondrial DNA transfer (13, 14). The second proposes the integration of such sequences in plant genomes via fungus-mediated horizontal gene transfer (HGT) during the long-term coevolution of fungi and plants (15, 16). Very recently, indirect evidence of replicating plant mitoviruses was provided by mining the transcriptome of a number of plant species (17).
Here, we report the complete genome sequence and biological characterization of a replicating plant mitovirus detected in a number of Chenopodium quinoa accessions and designated Chenopodium quinoa mitovirus 1 (CqMV1). Furthermore, we provide evidence of differential small RNA (sRNA) processing of this virus compared to that of a cytoplasmic plant virus infecting the same plant, indicating the possible involvement of a still-uncharacterized new differential antiviral response inside the mitochondria.
RESULTS
In silico assembly of a mitovirus sequence from NGS analysis of total RNA from a C. quinoa leaf sample.
In order to identify a mechanically transmitted viral agent from a Hibiscus rosa-sinensis plant, we submitted for next-generation sequencing (NGS) analysis the total RNA (depleted of rRNA) of C. quinoa leaves showing chlorotic spots that were inoculated with sap from symptomatic H. rosa-sinensis plants (SRA accession no. SRR8169409). Our bioinformatics pipeline (18) identified two complete virus genomes: BLAST searches of viral databases identified a contig with high similarity to a tobamovirus, hibiscus latent ford pierce virus (19), and a second contig of 2,730 nucleotides (nt), which codes for a single putative protein, from nt 322 to nt 2644 (Fig. 1). A BLAST search identified the latter as a putative mitovirus RNA-dependent RNA polymerase (RdRp). Since such a contig also was present in uninoculated C. quinoa and not in the original hibiscus plant (data not shown), we decided to provisionally name the putative virus Chenopodium quinoa mitovirus 1 (CqMV1). Furthermore, to confirm absence of fungal contamination (from endophytes or pathogenic fungi), assembled contigs were analyzed in MEGAN 6 after DIAMOND processing for taxonomical placements of all the assembled contigs from the transcriptome sequencing (RNA-seq) experiment. No fungal contigs were detected (not shown).
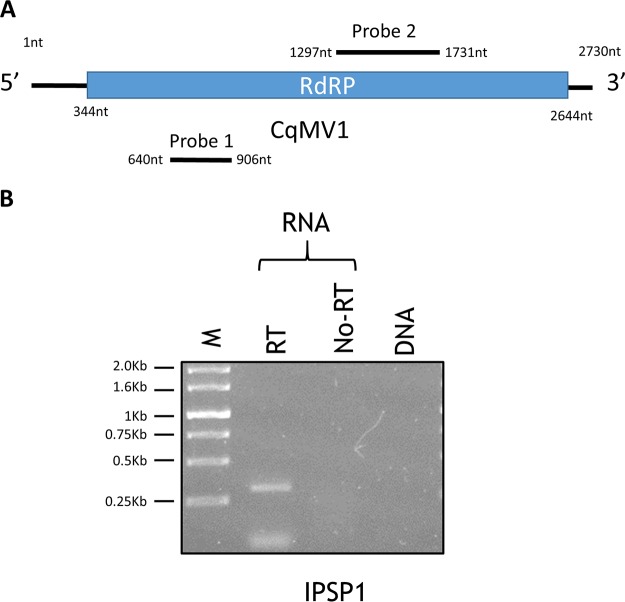
FIG 1.
CqMV1 genome organization and its replicative nature. (A) Genome organization of the single positive-strand genome with the RdRp ORF (blue box). The position of the two DNA segments amplified by reverse transcriptase PCR and used as probes in Northern hybridization experiments are shown as black lines (probe 1 and probe 2). nt, nucleotide position on the genome. (B) Ethidium bromide-stained Tris-acetate-EDTA gel (1%) was used to separate PCR products from RNA template and DNA template extracted from Chenopodium quinoa IPSP1. M, molecular weight marker; RT, reverse transcriptase; no-RT, RNA template without reverse transcriptase.
At the time when we identified the putative mitovirus (January 2017; deposited in the databases 22 June 2017), a tblastn search was performed by using the deduced C. quinoa mitovirus RdRp as the query, and when the total nr database was used, the first hits were those of a Solanum tuberosum mitochondrion gene (XP_006364252; E value of 0.0, 98% query coverage, 53% amino acid identity). A number of other nonretroviral endogenized RNA virus elements (NERVEs), described as mitovirus-like sequences, are also present in the list of hits obtained by this search. Repeating the tblastn search at the time of submission (October 2018), limited to annotated virus sequences, the highest score is to recently identified beta vulgaris mitovirus 1 (AVH76945.1; 56% identity at the amino acid level, 82% query cover) (17) that was deposited in the database 21 December 2017 and a still-uncharacterized Ocimum basilicum RNA virus 2 sequence (YP_009408146; 32% amino acid identity, 49% query cover) that was deposited in the database 3 June 2017. To exclude that such RNA mitoviral sequences resulted from transcription of a full-length viral genome endogenized in C. quinoa, we investigated the possible existence of a DNA fragment in the mitochondrial or nuclear DNA corresponding to the assembled viral sequence. For this purpose, we designed specific primers on the predicted ORF and performed a PCR protocol on both DNA and RNA to identify in which nucleic acid fraction the sequence is detectable. C. quinoa plants contained the viral sequence in the RNA fraction, but none of them showed any specific band in the DNA fraction (Fig. 1). These results were also confirmed in a more sensitive quantitative reverse transcription-PCR (qRT-PCR) protocol (not shown).
We then analyzed the CqMV1 amino acid sequence to identify conserved motifs. In addition to the conserved RdRp domain (GDD), we detected all six conserved motifs characteristic of mitoviruses (20). Due to the putative mitochondrial localization of the virus, we expected an AU content of >60% because of the A-U-rich nature of the mitochondrial genome (21, 22), but the observed A-U content is 58.39%. Whereas fungal mitoviruses typically use UGA to encode tryptophan rather than a stop codon, in our case all tryptophans are encoded by UGG.
Stem-loop secondary structures are characteristic of 5′ and 3′ untranslated region (UTR) sequences of the positive-strand mitovirus genomes (3). Here, we analyzed the presence of possible secondary structures with RNA-Fold software, which revealed the presence of these structures at the 5′ UTR (ΔG = 33 kcal/mol; not shown).
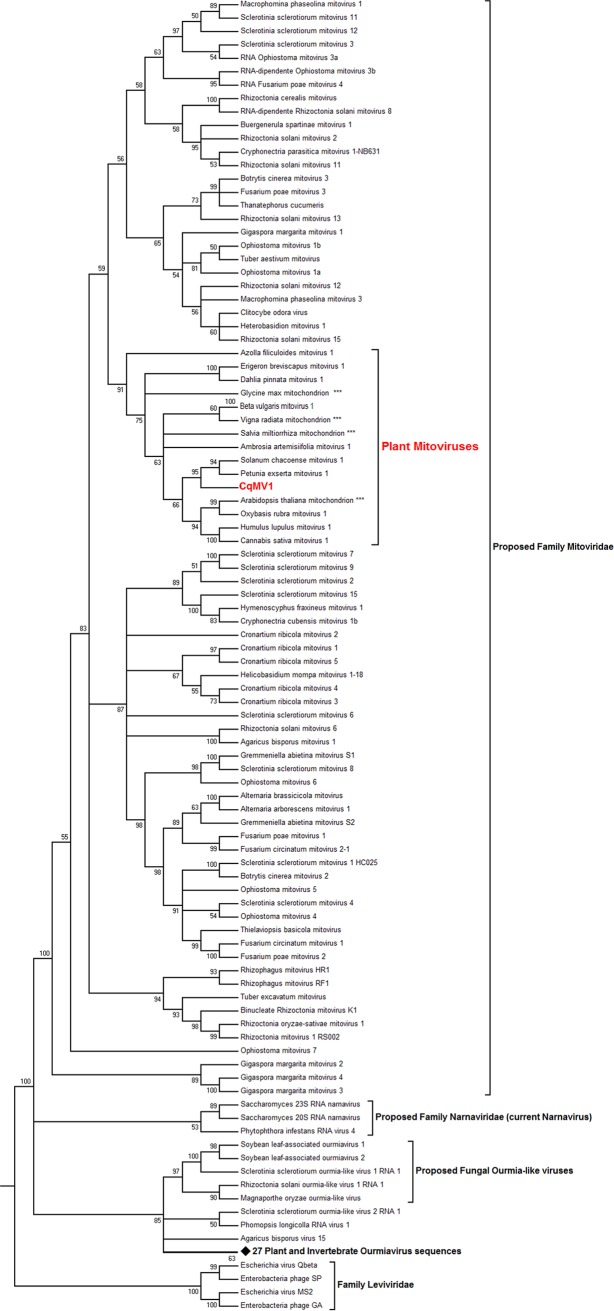
We then proceeded to carry out a phylogenetic analysis that included representatives of endogenized plant mitoviral sequences, characterized mitoviruses from fungi, and mitoviruses characterized from other plant transcriptomes; as outgroups, we included a representative of the Narnaviridae family and the recently proposed Ourmiaviridae-like family (Fig. 2). As observed by other authors, plant mitoviruses form a well-supported clade within the mitoviruses. Currently, known mitoviral sequences often group according to their specific host, consistent with coevolution and infrequent interspecific transmission.
FIG 2.
CqMV1 phylogenetic placement. Predicted ORFs encoding RdRp were used to build an alignment using MUSCLE implemented in MEGA 6 (83). The phylogenetic tree was built using the maximum likelihood method with 1,000 bootstrap replicates. Branches with bootstrap values of <50 have been collapsed. The analysis involved 125 amino acid sequences. All positions with less than 90% site coverage were eliminated. There were a total of 457 positions in the final data set. A list of the accession numbers of the viruses contained in the tree is shown in Table S2. The diamond symbol represents a node that was collapsed, which includes 27 RdRp sequences from a number of invertebrate and plant ourmiaviruses still awaiting taxonomical classification. Asterisks indicate endogenized plant mitochondrial sequences.
CqMV1 is differentially distributed in C. quinoa accessions and cultivars.
Chenopodium quinoa is known to plant virologists because it is historically a common host range test plant: it often gives local chlorotic or necrotic lesions upon mechanical inoculation with a number of plant viruses. This species is also a very important agricultural crop (23, 24) because of its resistance to a number of abiotic stresses and for its nutritional value (25, 26).
We therefore sought to determine how widespread CqMV1 is within C. quinoa germplasm by testing seed batches from different sources for the presence of the virus. For this purpose, initially we compared C. quinoa accessions from different plant virology laboratories (personal collection of Gancho Pasev, Maritsa Vegetable Crops Research Institute, Plovdiv, Bulgaria); later, we purchased a variety of accessions from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) and from the Collection of the U.S National Plant Germplasm System (NPGS). A sensitive and fast qRT-PCR protocol was established to screen newly germinated batches of 10 plantlets for each sample. We found that the C. quinoa strain from our laboratory, used as test plants in host range experiments (named IPSP1 in this work), some other accessions from the Pasev collection (see Table S1 in the supplemental material), and some common commercial cultivars (cv. Regalona and cv. Cherry vanilla) were positive for CqMV1. In contrast, CqMV1 was absent in 42 tested accessions from Peru and Bolivia (from the IPK collection) (Table S1). Accession PI614886 (NPGS collection) from Chile is the C. quinoa accession used for deriving the recently published genome sequence, and it carries CqMV1 (27).
CqMV1 genomic RNA accumulates preferentially in the mitochondrial fraction.
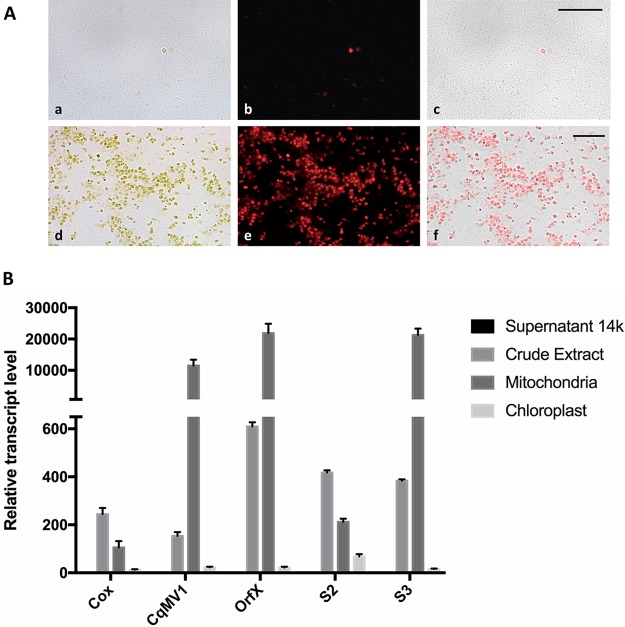
Translation of CqMV1 RdRp hypothetically could occur in both the cytoplasm and the mitochondria, since the same protein is encoded using both genetic codes. For this reason, we wanted to investigate if virus RNA accumulated preferentially in mitochondrion-enriched preparations compared to whole-cell extracts or to chloroplast-enriched fractions. We adapted a protocol for spinach mitochondrial enrichment and judged purity of the preparation based on chlorophyll fluorescence (Fig. 3A) and presence of specific genetic markers (ORF-X and S3 for mitochondria, S2 for a mitochondrial ribosomal protein encoded by a nuclear gene, and Cox encoded by a nuclear gene). RNA was extracted from the different fractions, and qRT-PCR was carried out to detect the virus and the mRNA corresponding to each of the marker genes. The virus copurifies with mitochondrial RNAs. It is >50-fold more abundant in the mitochondrial fraction than in either the chloroplastic or the soluble cytoplasmic RNA fractions (Fig. 3B).
FIG 3.
CqMV1 genomic RNA is enriched in mitochondrial fractions. (A) Fluorescent microscopy observation of mitochondrion (a to c) and chloroplast (d to f) fractions purified from Chenopodium quinoa leaves. (a and d) Bright-field images; (b and e) chlorophyll fluorescence; (c and f) overlay of the two images. Magnification bar, 50 μm. (B) Real-time quantification of RNA corresponding to CqMV1 and two nuclear (Cox and S2) and two mitochondrial (OrfX and S3) genes. All quantifications are relative to the amount of virus or mRNA present in the supernatant of the 14,000 × g centrifuge run; this amount was arbitrarily established as 1. Error bars represent standard errors of the means (n = 3).
Evidence of minus-strand CqMV1 RNA accumulation in leaf extracts.
The recent contention of “contemporary” mitoviruses infecting plants (17) relies on detection of RNA transcripts corresponding to the mitovirus genome (positive RT-PCR, after DNase treatment) in the absence of a corresponding DNA segment (negative PCR). This is robust indirect evidence. Nevertheless, more direct evidence of replicating mitoviruses could come from detection of a negative-strand full-length RNA corresponding to the viral genome by a PCR-independent method. Therefore, a Northern hybridization experiment relying on positive- and minus-strand runoff transcript probes to detect minus-strand genomic RNA and positive-strand genomic RNA, respectively, has been performed. We used the accession BO25 as a negative control and Regalona as a CqMV1-infected positive control. Our initial experiment with a first pair of plus- and minus-strand probes revealed a very abundant accumulation of a specific positive-strand CqMV1 genomic RNA band only in the virus-infected line. Attempts to detect a specific full-length negative-strand RNA band from leaf extracts failed; only shorter virus-specific RNA species could be detected by the positive-strand probe designed close to the 3′ end of the genome (Fig. 4A). The presence of specific full-length negative-strand viral genomic RNA is likely masked by the unspecific hybridization of the probe with a ribosomal band. We repeated the experiment with a second pair of probes designed in a different region of the genome (Fig. 1), and in this case, we were able to show a faint specific band hybridizing with the negative-sense genomic RNA after 15 days of membrane exposure to film (Fig. 4B), evidence of minimal CqMV1 replicative activity.
FIG 4.
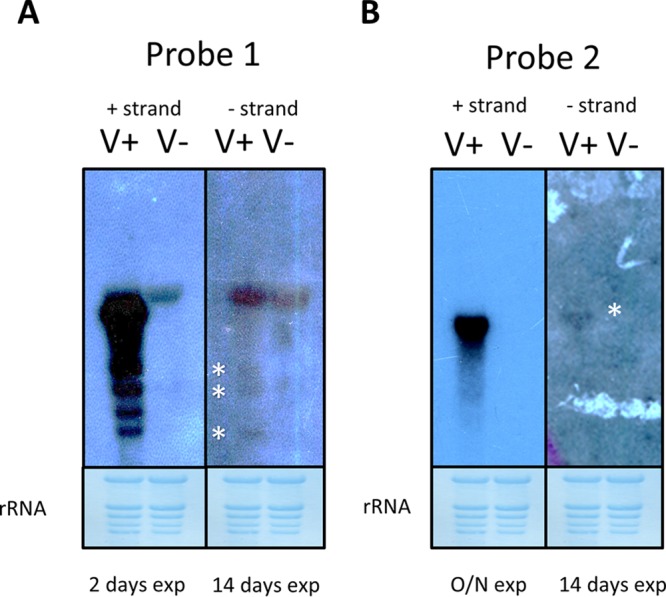
Evidence of minus-strand genomic RNA accumulation. Time of autoradiography exposure is indicated at the bottom of each panel (exp); RNA samples were extracted from leaves from uninfected Chenopodium quinoa accession BO25 (V−) and from CqMV1-infected Chenopodium quinoa cultivar Regalona (V+). In each panel, negative-sense probe reacting with the viral positive RNA strand is shown on the left, and positive-strand probe reacting with the viral negative RNA strand is shown on the right. Asterisks show weak specific signals given by positive probes targeting the viral negative RNA strand. (A) Signal from the two orientations of probe 1 (Fig. 1). (B) Signal given by probe 2 (Fig. 1). Lower panels are methylene blue-stained membranes showing rRNA loading.
Mechanical inoculation, seed transmission, and grafting experiments.
We then proceeded to investigate some basic biological properties of CqMV1. Most persistent viruses are not mechanically transmissible but are vertically transmissible through seeds with a 100% rate. We first sought to mechanically transmit CqMV1 from infected C. quinoa to accession BO25, which had tested negative for CqMV1. Out of 20 inoculated plants, none showed evidence of infection in either the inoculated leaf or systemically by following standard protocols that gave 100% infection with a mechanically transmissible control virus (alfalfa mosaic virus [AMV]; not shown).
We next investigated the vertical transmission rate of the virus in plantlets germinated from seeds obtained from infected plants (IPSP1 and cv. Regalona). We tested individually 100 plantlets for each accession by qRT-PCR, and all tested positive. Finally, we tested the possibility of transmitting CqMV1 through grafting, which often overcomes the mechanical transmission limitations of nonmechanically transmissible viruses. We grafted healthy BO25 on IPSP1 rootstocks (6 plants). We then tested the scion and rootstocks at 1 and 2 months after grafting. CqMV1 could not be detected in any grafted scion systemically, but its presence was confirmed in all the infected rootstocks.
Differential symptom severity of pathogenic virus infections on CqMV1-infected and CqMV1-free C. quinoa lines.
We focused our experiments on the accessions BO25 and BO78 as negative controls and IPSP1 and Regalona as CqMV1-infected positive controls, since they did not show any evident phenotypic differences under our environmental growth conditions. We wanted to test if the presence/absence of the mitovirus has any synergistic or antagonistic effect once the plants are inoculated with a disease-causing plant virus. In particular, the four accessions were infected with three different pathogenic viruses able to replicate, systemically infect, and induce symptoms on C. quinoa plants: (i) an isolate of AMV, (ii) an isolate of lettuce mosaic virus (LMV), and (iii) an isolate of hibiscus latent ringspot virus (HLRSV).
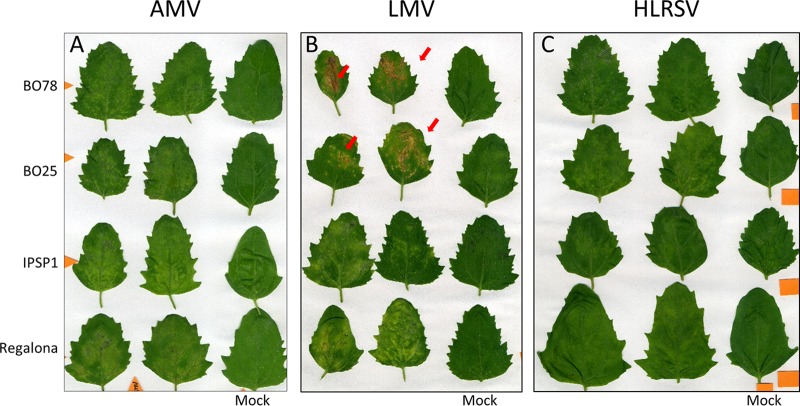
Leaves of virus-inoculated plants were compared to leaves of mock-inoculated plants 7 days postinfection (dpi). For both AMV and HLRV, we observed local chlorotic lesions indistinguishable between the CqMV1-infected lines and the CqMV1-free lines (Fig. 5A and C). In contrast, leaves of plants infected with LMV displayed local chlorotic lesions when the leaves came from CqMV1-infected lines but more severe local necrotic lesions in leaves from CqMV-free lines (Fig. 5B).
FIG 5.
Differential symptom severity of virus infections: local symptoms in leaves. Two CqMV1-infected accessions (cv. Regalona and IPSP1) and two CqMV1-free accessions (BO25 and BO75) were used to assess their responses when inoculated with pathogenic viruses. Observations were done at 7 days postinoculation (dpi). (A) AMV locally inoculated leaves did not show differences in terms of symptom severity between CqMV1-infected and CqMV1-free accessions. (B) Infection with LMV revealed differences between CqMV1-infected accessions, in which chlorotic lesions were observed, and CqMV1-free accessions, in which necrotic lesion (red arrows) were observed. (C) Infection with HLRSV did not reveal symptom differences among the four accessions.
We then repeated observation of symptoms on whole plants at 14 dpi. The CqMV1-infected lines IPSP1 and Regalona displayed milder systemic symptoms (Fig. 6D, E, F, J, and K to L) than the CqMV1-free lines BO25 and BO78 (Fig. 6A to C, G, H, and I). All four lines showed comparable mild growth impairment, leaf malformation, and mild mottling in upper uninoculated leaves. Nevertheless, the most evident specific differential phenotype associated with CqMV1 presence/absence is the red-violet stem pigmentation observed in CqMV1-free lines (Fig. 6M). Conversely, mock-inoculated or CqMV1-infected plants did not show stem pigmentation (Fig. 6N).
FIG 6.
Differential symptom severity of virus infections: systemic symptoms. Two CqMV1-infected accessions and two CqMV1-free accessions were used to assess responses when inoculated with three pathogenic viruses. Observations were done at 14 days postinoculation. In vertical rows are the virus species used in the experiments: AMV (A, D, G, and J), LMV (B, E, H, and K), and HLRSV (C, F, I, and L). In horizontal rows, the four Chenopodium quinoa accessions BO78, BO25, IPSP1, and Regalona (Regal.) are reported. A negative mock-inoculated plant of the same age is present next to two infected plants in each panel. All accessions show systemic symptoms of mild growth impairment, malformation, and mild mottling. CqMV1-free accessions BO78 (A, B, and C) and BO25 (D, E, and F) showed red violet pigmentation on stems (white arrows), whereas CqMV1-infected accessions IPSP1 (G, H, and I) and Regalona (J, K, and L) and mock-inoculated plants did not show any pigmentation. Inset of a pigmented stem from accession BO78 infected by LMV (B) is enlarged in panel M, whereas inset of a stem of cultivar Regalona also infected by LMV (K) is enlarged in panel N.
Differential sRNA accumulation and processing of AMV and CqMV1 in infected C. quinoa plants.
A number of fungal mitoviruses have been discovered through sRNA characterization, suggesting that, at least in fungi, they are subject to RNA interference (RNAi) processing (28, 29). In order to characterize the sRNA present in C. quinoa, we decided to compare three libraries of sRNA: (i) CqMV1-free BO25 infected by AMV (SRA accession no. SRR8169660); (ii) mock-inoculated cv. Regalona carrying CqMV1 (accession no. SRR8169658); and (iii) cv. Regalona mechanically infected with AMV (accession no. SRR8169659). Using a bioinformatic pipeline that was previously used to assemble de novo virus genomes (18, 30), we first confirmed that accession BO25 does not carry any virus other than AMV, whereas the same pipeline could assemble CqMV1 from cv. Regalona and AMV from AMV-infected cultivars. When we looked at the percentage of total sRNA that mapped to the two viral full-length sequences, we noticed that, for plant mitovirus, the percentage is very low compared to that for AMV (Table 1), even if the overall genomic RNA is actually more abundant based on qRT-PCR assessment and Northern blotting (not shown). We then proceeded to look at the size distribution of the sRNA. The availability of the C. quinoa genome (27, 31) allowed us to assess reads mapping to the host genome (with a characteristic peak at 24 nt common to most plants) and to each of the two viruses. In the case of AMV, a typical peak corresponding to 21 nt (47% of the reads) and a minor 22-nt peak (30% of the reads) are likely the hallmark of a Dicer- and RISC-mediated antiviral response (Fig. 7). Surprisingly, in the case of reads mapping to CqMV, a sharp peak for the 16-nt-long reads corresponding to 30% of the reads was present, whereas the peaks corresponding to 21- and 22-nt-long reads were both below 10% (Fig. 7). Availability of C. quinoa chloroplast (32) and nuclear (27) genomes, and our own selection of a number of mitochondrial genes from our transcriptome data, allowed us to check the read distribution lengths of host sRNA mapping to those genomes. sRNA mapping onto the chloroplast genome had a major peak at 27 nt (22%), whereas the second peak is at 22 nt; in the case of sRNA mapping to mitochondrial genes (in this case only the putative coding sequences were used), a sharp peak is present for 16 nt (67% of the sRNA). The N-terminal nucleotide distribution of reads mapping to CqMV and AMV were the following: for CqMV, A, 30.5%; C, 12.9%; G, 37.7%; and U, 18.9%; for AMV, A, 25.6%; C, 16.3%; G, 10.2%; and U, 47.9%.
TABLE 1.
sRNA reads mapping against each of the corresponding genomes as a percentage of total reads in Chenopodium quinoa cv. Regalona, cv. Regalona infected with AMV, and BO78 infected with AMV
| Strain | No. of sRNA reads in: |
Total no. of reads | ||||
|---|---|---|---|---|---|---|
| Chloroplast | CqMV1 | Nucleus | AMV | Mitochondria | ||
| Regalona | 14.017 | 0.006 | 84.071 | 0.000 | 1.906 | 4,119,721 |
| Regalona AMV | 14.783 | 0.111 | 78.580 | 4.583 | 1.945 | 7,240,581 |
| BO78 AMV | 15.262 | 0.000 | 80.333 | 2.489 | 1.916 | 3,758,683 |
FIG 7.
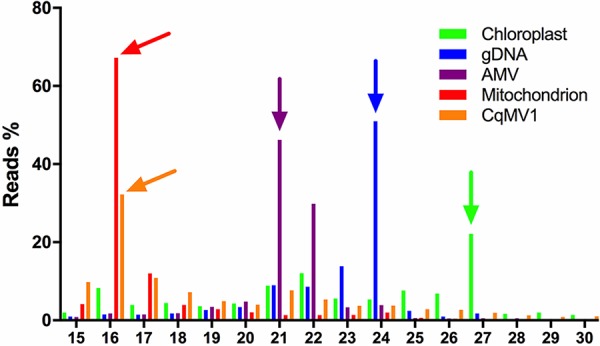
sRNA length distribution in virus-infected Chenopodium quinoa leaves. Reads from sRNA sequencing were mapped against genes encoded by chloroplast, nucleus, mitochondrion, AMV, and CqMV1 genomes. Abundance is expressed as percentage of reads of a particular length, and arrows above bars indicate the most abundant sRNA length inside the specific gene set. CqMV1 shared the same sRNA pattern distribution of genes encoded by the mitochondrial genomes, suggesting a mitochondrial localization and a specific but still uncharacterized RNA degradation pathway inside the mitochondria.
We then performed an sRNA miner analysis on reads mapping on the CqMV1 genome to reveal clustered organellar sRNA (cosRNA), which are putative footprints of RNA binding proteins (RBP). Using conservative settings, we were able to detect eight different footprints from 4 distinct peaks (Fig. S1). Therefore, sRNA analysis seems to confirm that most of the CqMV1 RNA is protected from Dicer/RISC-mediated viral antisilencing responses inside the mitochondria, where distinct processing occurs, resulting mostly in 16-nt sRNA.
DISCUSSION
Plant mitoviruses, a new class of cryptic (persistent) plant viruses.
Our work confirms the recent finding that plant mitochondrial viruses exist not only as widespread endogenized sequences in mitochondrial or nuclear genomes (13) but also as true virus-encoded RdRp-dependent replicating RNA elements (17). Here, we provide further molecular evidence through detection of minus-strand RNA. We can also exclude that such a replicating virus is carried by a fungal endophyte based on RNA-seq analysis, as our data set lacks any fungal reads. Our work is the first to provide evidence that bona fide plant mitoviruses are enriched in plant mitochondria.
This is not the first case of a virus associated with plant mitochondria. The plant virus carnation Italian ringspot virus replicates on mitochondrial external membranes and causes multivesicular body alterations of mitochondria (33), but particles and RNA readily accumulate in the cytoplasm and replicases (p35 and coterminal p95) targeted to the external membrane of mitochondria have N and C termini on the cytosolic side (33), so this virus is best viewed as cytoplasmic.
We also note that viruses have already been characterized from chloroplasts and mitochondria of the green alga Bryopsis species (34), specifically a mitochondrial virus related to fungal totiviruses (35) and a chloroplastic virus related to the partitiviruses (36); both viruses have a typical dsRNA genome and have no close phylogenetic relationship with any known bacterial virus. In contrast, plant mitoviruses, including CqMV1, are instead phylogenetically related to the phage family Leviviridae.
Our phylogenetic analysis suggests a comprehensive review of the taxonomy of mitoviruses. Inclusion of a number of well-characterized fungal and plant narna-like viruses and members of the family Leviviridae in the phylogenetic analysis makes it evident that mitoviruses are distributed in different clades. Their wide diversity warrants the establishment of a family taxon called Mitoviridae, separated from the family Narnaviridae. The newly established family would comprise a number of subfamilies and genera, including the very distantly related mitovirus species characterized from the arbuscular mycorrhyzal fungus Gigaspora margarita, which are included in a clade basal to existing characterized mitoviruses (37). As already observed by other authors (17), plant mitoviruses are nested in a specific fungal mitovirus clade, raising questions about the evolutionary trajectory of plant mitoviruses, which has been discussed at length elsewhere (17).
We provide here, for the first time, a basic biological characterization of a plant mitovirus, which has the typical features of cryptic viruses: they cannot be transmitted horizontally by mechanical inoculation or grafting, whereas they are transmitted vertically at a 100% rate through seeds. Lack of transmission through grafting is indeed an expected result for mitoviruses that replicate in plant mitochondria, since movement of mitochondria through the plant is likely minimal and will not replace existing populations of mitochondria. This is somewhat different from what was observed in fungi, where grafting (hyphal fusion through anastomoses) was shown to allow mitovirus transmission in some cases (38).
The high level of seed transmission observed in this study is also expected, as seeds formed on infected plants must inherit their parents’ infected mitochondria. Plants have mechanisms to ensure that mitochondria are only maternally inherited and to maintain low heteroplasmy levels. Nevertheless, there is evidence of biparental transmission of mitochondria in plants (39). C. quinoa is normally self-pollinating in nature, but studies of mitochondrial inheritance could be carried out through artificial mechanical emasculation and forced crosses (40). An important avenue of future work will therefore be the creation of reciprocal crosses with mitovirus-infected and mitovirus-free maternal and paternal lines to allow isolation of quasi-isogenic plants differing only in the mitochondrial/viral content.
The absence of a movement protein in mitovirus genomes and lack of a movement-complementing virus in CqMV1-infected C. quinoa also support the idea that they can infect all types of cells, including meristematic ones, as is the case for other cryptic viruses (41).
The existence of cryptic viruses in plants was known for at least four decades, since their discovery in a number of different plant species at the end of the 1970s (41). Currently known persistent/cryptic plant viruses include members of five families: Partitiviridae (42–44), Totiviridae (45, 46), Chrysoviridae (47, 48) Endornaviridae (49), and Amalgaviridae (50–53). Here, we provide convincing evidence that plant mitoviruses also should be defined as cryptic (persistent) viruses. The fact that CqMV1 is present in commercial varieties of C. quinoa (Regalona and Cherry Vanilla) but that a number of other accessions from Bolivia and Peru do not carry any mitovirus seems to support the idea that they cause no specific harm to their host. Moreover, the widespread occurrence of mitoviruses in domesticated material raises the possibility of their beneficial role in specific agro-ecological niches. Recent reviews and metagenomics studies unveiled the widespread occurrence of cryptic (persistent) viruses in plants, and other authors have discussed a possible beneficial role for their host (54–56). In particular, some studies have looked at the presence of endornavirus as it relates to the domestication of pepper (49). There is also growing evidence that other plant viruses, which are not defined as cryptic, since in some specific instances they can cause obvious symptoms, can indeed provide advantages to their host in resistance to abiotic stress. An example is grapevine rupestris stem pitting-associated virus (GRSPaV), where a molecular mechanism has also been proposed (57, 58).
A preliminary experiment to detect differential symptom reaction to a panel of viruses systemically infecting C. quinoa did not reveal any major synergistic or antagonistic effect caused by CqMV1 mitochondrial infection. Nevertheless, some interesting features in specific virus-plant interactions should be pointed out: the enhanced necrotic hypersensitive reaction and the stem pigmentation in the absence of CqMV1 implies that infection with the mitovirus can somehow ameliorate the symptoms of at least some other viruses. We can speculate that the presence of CqMV1 in the mitochondria alters the oxidative stress cellular signaling, resulting in necrosis and pigment accumulation. Further classes of biotic and abiotic stress should be tested, in view of the fact that mitochondria are central in a number of stress-related phenomena in plants, particularly in the roots, mediating tolerance of harsh environments (59).
Numerous attempts to isolate CqMV1 dsRNA from infected plants have failed using a protocol previously described (60) and using as a positive control tomato mosaic virus-infected C. quinoa and the fungal isolate MUT4330 previously described (60). Lack of detectable dsRNA in CqMV1-infected C. quinoa could be due to the extreme toxicity of dsRNA to plant mitochondria, as recently observed for dsRNA expressed in mitochondria in human cell lines: a specific degradation pathway prevents the export of dsRNA and the onset of a general antiviral defense driven by MDA5-dependent antiviral signaling (61).
Differential RNA processing inside mitochondria and evidence of an undefined antiviral response.
Our sRNA analysis indicates that the mitochondrial virus present in C. quinoa is not subject to the Dicer/argonaute-dependent antiviral silencing response that typically targets plant viruses (62). This is not due to a defective silencing response in C. quinoa, because we provided evidence that the typical Dicer/argonaute processing occurs in the same AMV-infected C. quinoa plant extract. This raises the question of what might limit mitochondrial virus replication in plants and fungi where mitoviruses are found. Silencing of fungal mitoviruses is not well characterized, but two recent studies show that mitoviral sRNA generated in fungi is not different from that generated from cytoplasmic viruses in either quality or quantity (28, 29). Our discovery that plants accumulate very small amounts of mitovirus sRNA and that these are most frequently 16 nt in length is a major difference with fungal systems and raises the question of what molecular pathway generates such a specific sRNA size distribution. Furthermore, the significant differences in the N-terminal nucleotide of reads mapping to CqMV and to AMV also point to distinct degradation machineries for the two viruses. Our data suggest that the 16-nt mitoviral sRNA result from a nonviral specific RNA degradation process, since size distribution of sRNA generated from mRNAs expressed from the mitochondrial genome shows the same peak at 16 nt. In this respect, the combined roles of exoribonucleases such as PNPase and RNR1 and RNA binding proteins in leaving footprints of various lengths could be at the basis of this differential sRNA accumulation, consistent with recent studies of size distribution footprints in chloroplast and mitochondrial RNA transcripts in Arabidopsis thaliana (63). Previous authors hypothesized a possible regulatory role of the sRNA resulting from processing by pentatricopeptide repeat proteins (PPR proteins, a subset of RBP proteins typical of plants) (63, 64), providing a testable model of antiviral defense based on intramitochondrial sRNA generation.
From an evolutionary perspective, it would be interesting to look at bacterial antiviral responses against RNA viruses (RNA phages). Recent work has shown that RNA bacteriophage diversity is much higher than previously thought (65): while current taxonomy has only two families of prokaryotic RNA viruses, the Leviviridae and the Cystoviridae, indirect evidence indicates that some Picobirnaviridae are bacterial viruses (66). In this respect, new antiviral defense systems are constantly unveiled (67). Although RNA-guided RNA cleavage by a specific clustered regularly interspaced short palindromic repeat (CRISPR) RNA-Cas system is known (68), its role in specific antiviral response in natural systems has yet to be shown (69, 70); nevertheless, in an in vitro heterologous system, a type III-A CRISPR-Cas system will restrict MS2 RNA phage infection (71).
Future work will pursue further biochemical characterization of the sRNA response to mitovirus infection in different biological systems (fungi and plants) and the analysis of possible differential physiological reactions linked to mitovirus infection in plants experiencing harsh environmental stress conditions.
MATERIALS AND METHODS
Plant material and RNA sequencing.
Plant seeds used in this study (accessions, seeds from personal collections, and from public repositories) are described in detail in Table S1 in the supplemental material.
Total RNA extraction from IPSP1 plants was performed using the Spectrum plant total RNA kit (Sigma-Aldrich, Saint Louis, MO) by following the manufacturer’s instructions. RNA quantification and quality were tested using a NanoDrop 2000 spectrophotometer (ThermoScientific, Waltham, MA). Macrogen Europe (Amsterdam, Netherlands) performed rRNA depletion using the Ribo-Zero plant kit (Epicentre, Madison, WI), library construction, and sequencing using an Illumina HiSeq4000.
For mechanical inoculation experiments, we used three different virus species, belonging to different families, that systemically infect C. quinoa: LMV, family Potyviridae, genus Potyvirus (Dim 60; PLAVIT collection); AMV, family Bromovididae, genus Alfamovirus (IFA 30; PLAVIT collection); and HLRSV, family Secoviridae, genus Nepovirus (VE 453; PLAVIT collection).
Bioinformatics analysis and molecular validation.
Raw reads obtained from total RNA sequencing were assembled into contigs using Trinity 2.3.2 (72), and viruses were identified as already described (18) using BLAST+ suite 2.6.0 (73), BWA 0.7.15-r1140 (74), and SAMtools 1.3 (75).
Once viral sequences were identified, specific primers were designed (Table S3) to reveal their molecular nature (if DNA, RNA, or both). To detect viral sequence possibly integrated into the host genome (both nuclear and mitochondrial), we performed total nucleic acid extraction using a phenol-chloroform protocol (76). We then performed, on half of the volume, a digestion with RNase A (4 h) to completely remove any trace of RNAs. The second half of the total nucleic acid extraction was subjected to a 4-h DNase treatment in order to completely remove all traces of DNA. The DNase-treated RNA was then used in a retrotranscription reaction in order to obtain cDNA suitable for PCR. We then performed PCR with specific primers for both an internal control (COX) and for the viral sequence using as the template the obtained DNA and cDNA samples from the four C. quinoa lines (IPSP1, Regalona, BO25, and BO78). The same templates were used in qRT-PCR with specific primers to evaluate the presence and quantities of the internal control and the viral sequence in the four different lines.
PCR products for viral sequence were cleaned with a DNA Clean & Concentrator-5 kit (Zymoresearch, CA), cloned into pGEM-T easy vector (Promega, Madison, WI), and sequenced at Bio-Fab Research (Rome, Italy).
The 5′- and 3′-terminal sequences were obtained through the rapid amplification of cDNA ends protocol. The presence of possible secondary structure was evaluated using RNAfold (77).
To identify possible contamination with fungal sequences in our RNA-seq experiments, we analyzed the taxonomic placement of all the assembled contigs using MEGAN6 software (78).
sRNA sequencing and analysis.
To detect CqMV1 sRNAs, we started from total RNA extraction of mock-inoculated accessions Regalona and Regalona infected by AMV. In parallel, the BO78 accession infected by AMV was used as a negative CqMV1 control. Total RNAs from the three samples were sent to the Italian Institute for Genomic Medicine (IIGM; Turin, Italy), where sRNAs were isolated and a library was constructed and then sequenced using a MiSeq System (Illumina Inc., San Diego, CA).
Raw reads were cleaned from the adaptor, quality filtered using the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html), and then assembled using Velvet (79) and Oases (80). Contigs were used in blastx and blastn searches against a custom database to identify viral sequences. To determine the sRNA size distribution on viral genomes (both CqMV1 and AMV), we used BWA (74) and SAMtools (75) to map raw reads against the two viral genomes and then filtered for read length through a custom Perl script. Due to the unexpected pattern of sRNA mapping on the CqMV1 genome, we decided also to map raw reads against C. quinoa genes. For nuclear and chloroplast gene representation, we used the genome assemblies GCF_001683475.1 and NC_034949.1. Due to the absence of an available mitochondrial genome assembly for C. quinoa, we created a custom database containing gene sequences of mitochondrial origin retrieved from the total RNA sequencing by comparison with the Arabidopsis thaliana mitochondrial genome (NC_037304.1). The relative frequency of each nucleotide at the 5′ N-terminal position of the small RNAs was calculated using Galaxy tools (81).
We performed an analysis for clustered organellar sRNA using sRNA miner, implemented in R/Bioconductor (82), using 40 as the parameter for the minimal reads for the end and 0.85 for the sharpness of the end.
Phylogenetic analysis.
Predicted ORFs encoding RdRp were used to build an alignment using the MUSCLE algorithm implemented in MEGA 6 (83). The phylogenetic tree was built using the maximum likelihood method, and aligned protein sequences were used to estimate the best substitution rate and parameter with MEGA 6. Substitution patterns and rates were estimated under the model designed by Dimmic et al. (+Gamma +Invar +Freq) (84). One thousand bootstrap replicates were performed, and branches with bootstrap values under 50 have been collapsed. A list of the accession numbers of the viruses contained in the tree is shown in Table S2.
Mitochondrial and chloroplast enrichment protocol and quantitative evaluation of CqMV1 and marker genes.
Chloroplast and mitochondrion-enriched fractions were obtained by a modified protocol already used for cucumber plants (85, 86). Ten grams of C. quinoa leaves was homogenized at a ratio of 1:10 in chilled extraction buffer (0.45 M sucrose, 15 mM morpholine propanesulfonic acid [MOPS], 1.5 mM EGTA, pH 7.4, with KOH) added with 0.6% polyvinylpyrrolidone (PVP), 0.2% bovine serum albumin (BSA), 10 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). After filtration on Miracloth (Calbiochem), the homogenate was centrifuged at 2,000 × g for 5 min to separate chloroplasts and cell debris. A pellet was used for the subsequent chloroplast purification; the supernatant was again centrifuged at 13,000 × g for 30 min to obtain the mitochondrial fraction in the pellet. From this point, the protocol followed two different methods.
Crude chloroplast pellets were suspended in 1 ml of sorbitol resuspension buffer (SRM; 1.65 M sorbitol, 250 mM HEPES, pH 8, with KOH), layered on a Percoll (Sigma-Aldrich) gradient (35% buffer and 80% in SRM), and centrifuged at 2,600 × g in a swing-out rotor (SW41; Beckman) for 10 min. After centrifugation, the fraction with chloroplast was collected with a Pasteur pipette, diluted in 30 ml of SRM buffer, and centrifuged at 2,000 × g for 5 min to remove all the Percoll. This step was repeated two times. The pellet was then suspended in resuspension buffer (RB; 0.4 M mannitol, 10 mM MOPS, 1 mM EGTA, pH 7.2) with 0.2 mM PMSF and checked on the microscope to evaluate the purity of the preparation. The crude mitochondrial pellet was resuspended in 1 ml of washing buffer (WB; 0.6 M sucrose, 20 mM MOPS, 2 mM EGTA, pH 7.2, with KOH) with 0.2 mM PMSF, layered on a Percoll gradient (18%, 23%, and 40% in WB), and centrifuged at 12,000 × g in an SW21 rotor (Beckmann) for 45 min. The mitochondrial fraction, between the 23% and 40% interface, was collected with a Pasteur pipette, and two washing steps were performed as already described for chloroplasts in WB. We resuspended pellet in about 0.1 ml and checked the quality of purification by observation with a fluorescence microscope. Mitochondrial and chloroplast-enriched fractions were stored at −80°C until use for RNA extraction and qRT-PCR analysis. In order to check which fraction contained virus genome enrichments, RNA was extracted using the Spectrum plant total RNA kit (Sigma-Aldrich, Saint Louis, MO) from preparations representing normalized amounts of each fraction. Complementary DNA (cDNA) was synthesized using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). We then tested the presence of virus genomic RNA and mRNA corresponding to a number of marker genes with quantitative real-time PCR using a CFX96 apparatus (Bio-Rad, Hercules, CA), iTaq Universal Probes, and iTaq Universal SYBR supermixes (Bio-Rad) by following protocols previously described (87). The marker genes corresponded to sequences of the cytochrome P450 oxidase (ANY30855.1); the S2 ribosomal protein, present in the mitochondria but encoded by the nuclear genome (88); the S3 ribosomal protein, encoded by plant mitochondrial genomes (89); the ORF-X protein, also encoded by mitochondrial genomes (90); and C. quinoa sequences related to the latter three genes were retrieved from our RNA-seq database. Oligonucleotides used for qRT-PCR are displayed in Table S3.
Northern blot analysis.
For Northern blot analyses, total RNA from leaves of different ages was prepared using total spectrum RNA reagent (Sigma-Aldrich) as suggested by the manufacturer. RNA samples were separated under denaturing conditions (glyoxal method) as detailed, using HEPES-EDTA buffer (91). Radiolabeled probes were prepared from linearized plasmid containing the cDNA clones (producing probes in both orientations) through T7 transcription using the Maxiscript T7 kit reagents (Thermo Fisher Scientific, Inc., Waltham, MA) as suggested by the manufacturer.
qRT-PCR fast screening method.
For a fast screening of viral infection, we applied modifications of a simple qRT-PCR protocol that uses crude extracts as the template (92). We placed 30 seeds to germinate in 90-mm-diameter petri dishes with wet filter paper and let it germinate for 3 days (or single plantlets in the case of the seed transmission assay). Plantlets of all the accessions tested were placed in extraction bags (Bioreba, Reinach, Switzerland) and diluted 1:20 (wt/vol) with carbonate buffer, pH 9.6 (93), supplemented with 2% PVP40, 0.2% BSA, 1% sodium metabisulfite, and 0.05% Tween 20. Raw extract was diluted 1:10 in sterile water and boiled for 10 min at 95°C. qRT-PCR screening was performed using a CFX96 real-time PCR detection system (Bio-Rad), and PCR mix was prepared with iTaq universal probes supermix (Bio-Rad), adding 3 U of reverse transcriptase from the high-capacity RNA-to-cDNA kit (Thermo Fisher Scientific) for each sample. Reactions were performed in a 10-μl total volume, adding 1 μl of boiled extract to 9 μl of PCR mix. The qRT-PCR protocol has a 30-min step at 37°C to perform the reverse transcription of the viral genome and then is followed by 1 min at 94°C and 40 steps of denaturation at 95°C for 10 s and annealing and extension at 60°C for 30 s.
Data availability.
The GenBank/eMBL/DDBJ accession number of the sequences reported in this paper is MF375475.
Supplementary Material
ACKNOWLEDGMENTS
We thank Caterina Perrone and Elena Zocca for excellent technical assistance in the greenhouse work, Riccardo Lenzi for mitochondrial enrichment preparation, Marco Chiapello for helping with the implementation of the sRNA miner analysis, and Doug Grubb for editing the manuscript.
This work was supported in part by the European Union’s Horizon H2020 Research and Innovation Action, grant agreement no. 773567.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01998-18.
REFERENCES
- 1.Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- 2.Polashock JJ, Hillman BI. 1994. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc Natl Acad Sci U S A 91:8680–8684. doi: 10.1073/pnas.91.18.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z, Wu S, Liu L, Cheng J, Fu Y, Jiang D, Xie J. 2015. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197:127–136. doi: 10.1016/j.virusres.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 4.Nibert ML. 2017. Mitovirus UGA(Trp) codon usage parallels that of host mitochondria. Virology 507:96–100. doi: 10.1016/j.virol.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turina M, Hillman BI, Izadpanah K, Rastgou M, Rosa C, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: ourmiavirus. J Gen Virol 98:129–130. doi: 10.1099/jgv.0.000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolja VV, Koonin EV. 2018. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res 244:36–52. doi: 10.1016/j.virusres.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raven PH. 1970. A multiple origin for plastids and mitochondria. Science 169:641–646. [DOI] [PubMed] [Google Scholar]
- 8.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin EV, Dolja VV. 2014. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo H, Chiba S, Suzuki N. 2015. Detection and analysis of non-retroviral RNA virus-like elements in plant, fungal, and insect genomes In Uyeda I, Masuta C (ed), Plant virology protocols. Methods in molecular biology (methods and protocols), vol 1236 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 11.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruenn JA, Warner BE, Yerramsetty P. 2015. Widespread mitovirus sequences in plant genomes. Peer J 3:e876. doi: 10.7717/peerj.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leister D, Kleine T. 2011. Role of intercompartmental DNA transfer in producing genetic diversity. Int Rev Cell Mol Biol 291:73–114. doi: 10.1016/B978-0-12-386035-4.00003-3. [DOI] [PubMed] [Google Scholar]
- 15.Marienfeld JR, Unseld M, Brandt P, Brennicke A. 1997. Viral nucleic acid sequence transfer between fungi and plants. Trends Genet 13:260–261. [DOI] [PubMed] [Google Scholar]
- 16.Goremykin VV, Salamini F, Velasco R, Viola R. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol 26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 17.Nibert ML, Vong M, Fugate KK, Debat HJ. 2018. Evidence for contemporary plant mitoviruses. Virology 518:14–24. doi: 10.1016/j.virol.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nerva L, Varese GC, Turina M. 2018. Different approaches to discover mycovirus associated to marine organisms In Pantaleo V, Chiumenti M (ed), Viral metagenomics. Methods in molecular biology, vol 1746 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 19.Nerva L, Vallino M, Turina M, Ciuffo M. 2018. Identification and characterization of Hibiscus latent Fort Pierce virus in Italy. J Plant Pathol 100:145–145. doi: 10.1007/s42161-018-0036-8. [DOI] [Google Scholar]
- 20.Hong Y, Dover SL, Cole TE, Brasier CM, Buck KW. 1999. Multiple mitochondrial viruses in an isolate of the Dutch elm disease fungus Ophiostoma novo-ulmi. Virology 258:118–127. doi: 10.1006/viro.1999.9691. [DOI] [PubMed] [Google Scholar]
- 21.Hong Y, Cole TE, Brasier CM, Buck KW. 1998. Evolutionary relationships among putative RNA-dependent RNA polymerases encoded by a mitochondrial virus-like RNA in the Dutch elm disease fungus, Ophiostoma novo-ulmi, by other viruses and virus-like RNAs and by the Arabidopsis mitochondrial genome. Virology 246:158–169. doi: 10.1006/viro.1998.9178. [DOI] [PubMed] [Google Scholar]
- 22.Paquin B, Laforest M-J, Forget L, Roewer I, Wang Z, Longcore J, Lang BF. 1997. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr Genet 31:380–395. [DOI] [PubMed] [Google Scholar]
- 23.Bazile D, Jacobsen S-E, Verniau A. 2016. The global expansion of quinoa: trends and limits. Front Plant Sci 7:622. doi: 10.3389/fpls.2016.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarbo K. 2015. From lost crop to lucrative commodity: conservation implications of the quinoa renaissance. Hum Organ 74:86–99. [Google Scholar]
- 25.Vega-Gálvez A, Miranda M, Vergara J, Uribe E, Puente L, Martínez EA. 2010. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: a review. J Sci Food Agric 90:2541–2547. doi: 10.1002/jsfa.4158. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen SE, Mujica A, Jensen CR. 2003. The resistance of quinoa (Chenopodium quinoa Willd.) to adverse abiotic factors. Food Rev International 19:99–109. doi: 10.1081/FRI-120018872. [DOI] [Google Scholar]
- 27.Jarvis DE, Ho YS, Lightfoot DJ, Schmockel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, Kharbatia NM, Rupper RR, Sharp AR, Dally N, Boughton BA, Woo YH, Gao G, Schijlen EGWM, Guo X, Momin AA, Negrao S, Al-Babili S, Gehring C, Roessner U, Jung C, Murphy K, Arold ST, Gojobori T, van der Linden CG, van Loo EN, Jellen EN, Maughan PJ, Tester M. 2017. The genome of Chenopodium quinoa. Nature 542:307–312. doi: 10.1038/nature21370. [DOI] [PubMed] [Google Scholar]
- 28.Donaire L, Ayllon MA. 2017. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol Plant Pathol 18:1127–1137. doi: 10.1111/mpp.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz-Adalia EJ, Diez JJ, Fernandez MM, Hantula J, Vainio EJ. 2018. Characterization of small RNAs originating from mitoviruses infecting the conifer pathogen Fusarium circinatum. Arch Virol 163:1009–1018. doi: 10.1007/s00705-018-3712-2. [DOI] [PubMed] [Google Scholar]
- 30.Margaria P, Miozzi L, Ciuffo M, Pappu HR, Turina M. 2015. The first complete genome sequences of two distinct European tomato spotted wilt virus isolates. Arch Virol 160:591–595. doi: 10.1007/s00705-014-2256-3. [DOI] [PubMed] [Google Scholar]
- 31.Zou C, Chen A, Xiao L, Muller HM, Ache P, Haberer G, Zhang M, Jia W, Deng P, Huang R, Lang D, Li F, Zhan D, Wu X, Zhang H, Bohm J, Liu R, Shabala S, Hedrich R, Zhu J-K, Zhang H. 2017. A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value. Cell Res 27:1327–1340. doi: 10.1038/cr.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong S-Y, Cheon K-S, Yoo K-O, Lee H-O, Cho K-S, Suh J-T, Kim S-J, Nam J-H, Sohn H-B, Kim Y-H. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front Plant Sci 8:1696. doi: 10.3389/fpls.2017.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber-Lotfi F, Dietrich A, Russo M, Rubino L. 2002. Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J Virol 76:10485–10496. doi: 10.1128/JVI.76.20.10485-10496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihara J, Pak JY, Fukuhara T, Nitta T. 1992. Association of particles that contain double-stranded RNAs with algal chloroplasts and mitochondria. Planta 187:475–482. doi: 10.1007/BF00199965. [DOI] [PubMed] [Google Scholar]
- 35.Koga R, Fukuhara T, Nitta T. 1998. Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol Biol 36:717–724. [DOI] [PubMed] [Google Scholar]
- 36.Koga R, Horiuchi H, Fukuhara T. 2003. Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol Biol 51:991–999. [DOI] [PubMed] [Google Scholar]
- 37.Turina M, Ghignone S, Astolfi N, Silvestri A, Bonfante P, Lanfranco L. 2018. The virome of the arbuscular mycorrhizal fungus Gigaspora margarita reveals the first report of DNA fragments corresponding to replicating non-retroviral RNA viruses in Fungi. Environ Microbiol 20:2012–2025. doi: 10.1111/1462-2920.14060. [DOI] [PubMed] [Google Scholar]
- 38.Polashock JJ, Bedker PJ, Hillman BI. 1997. Movement of a small mitochondrial double-stranded RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol Gen Genet 256:566–571. [DOI] [PubMed] [Google Scholar]
- 39.Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol 168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 40.Peterson A, Jacobsen S-E, Bonifacio A, Murphy K. 2015. A crossing method for quinoa. Sustainability 7:3230–3243. doi: 10.3390/su7033230. [DOI] [Google Scholar]
- 41.Boccardo G, Lisa V, Luisoni E, Milne RG. 1987. Cryptic plant viruses. Adv Virus Res 32:171–214. [DOI] [PubMed] [Google Scholar]
- 42.Natsuaki T, Yamashita S, Doi Y, Yora K. 1979. Radish Yellow Edge Virus, a seed-borne small spherical virus newly recognized in Japanese radish (Raphanus sativus L.). Jpn J Phytopathol 45:313–320. doi: 10.3186/jjphytopath.45.313. [DOI] [Google Scholar]
- 43.Kassanis B, White RF, Woods RD. 1977. Beet cryptic virus. J Phytopathol 90:350–360. doi: 10.1111/j.1439-0434.1977.tb03255.x. [DOI] [Google Scholar]
- 44.Boccardo G, Milne RG, Luisoni E, Lisa V, Accotto GP. 1985. Three seedborne cryptic viruses containing double-stranded RNA isolated from white clover. Virology 147:29–40. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Cao L, Huang Q, Qian Y, Zhou X. 2016. The complete genome sequence of a novel maize-associated totivirus. Arch Virol 161:487–490. doi: 10.1007/s00705-015-2657-y. [DOI] [PubMed] [Google Scholar]
- 46.Liu HQ, Fu YP, Xie JT, Cheng JS, Ghabrial SA, Li GQ, Peng YL, Yi XH, Jiang DH. 2012. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol Biol 12:91. doi: 10.1186/1471-2148-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Covelli L, Coutts RHA, Di Serio F, Citir A, Acikgoz S, Hernandez C, Ragozzino A, Flores R. 2004. Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I. Characterization of a new species in the genus Chrysovirus. J Gen Virol 85:3389–3397. doi: 10.1099/vir.0.80181-0. [DOI] [PubMed] [Google Scholar]
- 48.Li LQ, Liu JN, Xu AX, Wang T, Chen JS, Zhu XW. 2013. Molecular characterization of a trisegmented chrysovirus isolated from the radish Raphanus sativus. Virus Res 176:169–178. doi: 10.1016/j.virusres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Safari M, Roossinck MJ. 2018. Coevolution of a persistent plant virus and its pepper hosts. Mol Plant Microbe Interact 31:766–776. doi: 10.1094/MPMI-12-17-0312-R. [DOI] [PubMed] [Google Scholar]
- 50.Liu W, Chen J. 2009. A double-stranded RNA as the genome of a potential virus infecting Vicia faba. Virus Genes 39:126–131. doi: 10.1007/s11262-009-0362-1. [DOI] [PubMed] [Google Scholar]
- 51.Martin RR, Zhou J, Tzanetakis IE. 2011. Blueberry latent virus: an amalgam of the Partitiviridae and Totiviridae. Virus Res 155:175–180. doi: 10.1016/j.virusres.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 52.Sabanadzovic S, Valverde RA, Brown JK, Martin RR, Tzanetakis IE. 2009. Southern tomato virus: the link between the families Totiviridae and Partitiviridae. Virus Res 140:130–137. doi: 10.1016/j.virusres.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 53.Salem NM, Golino DA, Falk BW, Rowhani A. 2008. Complete nucleotide sequences and genome characterization of a novel double-stranded RNA virus infecting Rosa multiflora. Arch Virol 153:455–462. doi: 10.1007/s00705-007-0008-3. [DOI] [PubMed] [Google Scholar]
- 54.Roossinck MJ. 2014. Metagenomics of plant and fungal viruses reveals an abundance of persistent lifestyles. Front Microbiol 5:767. doi: 10.3389/fmicb.2014.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roossinck MJ. 2012. Persistent plant viruses: molecular hitchhikers or epigenetic elements?, p 177–186. In Viruses: essential agents of life. Springer, New York, NY. [Google Scholar]
- 56.Roossinck MJ. 2017. Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res 239:82–86. doi: 10.1016/j.virusres.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 57.Pantaleo V, Vitali M, Boccacci P, Miozzi L, Cuozzo D, Chitarra W, Mannini F, Lovisolo C, Gambino G. 2016. Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci Rep 6:20167. doi: 10.1038/srep20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perrone I, Chitarra W, Boccacci P, Gambino G. 2017. Grapevine-virus-environment interactions: an intriguing puzzle to solve. New Phytol 213:983–987. doi: 10.1111/nph.14271. [DOI] [PubMed] [Google Scholar]
- 59.Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, Taylor NL. 2012. Mitochondrial composition, function and stress response in plants. J Integr Plant Biol 54:887–906. [DOI] [PubMed] [Google Scholar]
- 60.Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M. 2016. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res 219:22–38. doi: 10.1016/j.virusres.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 61.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560:238–242. doi: 10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang M-B, Masuta C, Smith NA, Shimura H. 2012. RNA silencing and plant viral diseases. Mol Plant Microbe Interact 25:1275–1285. doi: 10.1094/MPMI-04-12-0093-CR. [DOI] [PubMed] [Google Scholar]
- 63.Ruwe H, Wang GW, Gusewski S, Schmitz-Linneweber C. 2016. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res 44:7406–7417. doi: 10.1093/nar/gkw466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 65.Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D. 2016. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol 14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnamurthy SR, Wang D. 2018. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology 516:108–114. doi: 10.1016/j.virol.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 71.Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. 2014. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell 56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 72.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turina M, Prodi A, Van Alfen NK. 2003. Role of the Mf1-1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet Biol 40:242–251. doi: 10.1016/S1087-1845(03)00084-7. [DOI] [PubMed] [Google Scholar]
- 77.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huson DH, Beier S, Flade I, Gorska A, El-Hadidi M, Mitra S, Ruscheweyh HJ, Tappu R. 2016. MEGAN community edition–interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol 55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 85.Vigani G, Bohic S, Faoro F, Vekemans B, Vincze L, Terzano R. 2018. Cellular fractionation and nanoscopic X-ray fluorescence imaging analyses reveal changes of zinc distribution in leaf cells of iron-deficient plants. Front Plant Sci 9:1112. doi: 10.3389/fpls.2018.01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vigani G, Faoro F, Ferretti AM, Cantele F, Maffi D, Marelli M, Maver M, Murgia I, Zocchi G. 2015. Three-dimensional reconstruction, by TEM tomography, of the ultrastructural modifications occurring in Cucumis sativus L. mitochondria under Fe deficiency. PLoS One 10:e0129141. doi: 10.1371/journal.pone.0129141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nerva L, Varese GC, Falk BW, Turina M. 2017. Mycoviruses of an endophytic fungus can replicate in plant cells: evolutionary implications. Sci Rep 7:1908. doi: 10.1038/s41598-017-02017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perrotta G, Grienenberger JM, Gualberto JM. 2002. Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol Biol 50:523–533. [DOI] [PubMed] [Google Scholar]
- 89.Kim Y, Kim HD, Kim J. 2013. Cytoplasmic ribosomal protein S3 (rpS3) plays a pivotal role in mitochondrial DNA damage surveillance. BBA Mol Cell Res 1833:2943–2952. doi: 10.1016/j.bbamcr.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 90.Suenkel S, Brennicke A, Knoop V. 1994. RNA editing of a conserved reading frame in plant mitochondria increases its similarity to two overlapping reading frames in Escherichia coli. Mol Gen Genet 242:65–72. [DOI] [PubMed] [Google Scholar]
- 91.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 92.Moretti M, Ciuffo M, Gotta P, Prodorutti D, Bragagna P, Turina M. 2011. Molecular characterization of two distinct strains of blueberry scorch virus (BlScV) in northern Italy. Arch Virol 156:1295–1297. doi: 10.1007/s00705-011-1018-8. [DOI] [PubMed] [Google Scholar]
- 93.Margaria P, Rosa C, Marzachi C, Turina M, Palmano S. 2007. Detection of Flavescence doree phytoplasma in grapevine by reverse-transcription PCR. Plant Dis 91:1496–1501. doi: 10.1094/PDIS-91-11-1496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GenBank/eMBL/DDBJ accession number of the sequences reported in this paper is MF375475.