Viruses have not previously been reported to act as chemotactic/chemoattractive agents. Rather, viruses as extracellular entities are generally viewed as non-metabolically active spore-like agents that await further infection events upon collision with appropriate host cells. That a virus might actively contribute to its fate via chemotaxis and change the behavior of an organism independent of infection is unprecedented.
KEYWORDS: chlorovirus, chemotaxis, giant virus, population dynamics, symbiosis
ABSTRACT
Chloroviruses exist in aquatic systems around the planet and they infect certain eukaryotic green algae that are mutualistic endosymbionts in a variety of protists and metazoans. Natural chlorovirus populations are seasonally dynamic, but the precise temporal changes in these populations and the mechanisms that underlie them have heretofore been unclear. We recently reported the novel concept that predator/prey-mediated virus activation regulates chlorovirus population dynamics, and in the current study, we demonstrate virus-packaged chemotactic modulation of prey behavior.
IMPORTANCE Viruses have not previously been reported to act as chemotactic/chemoattractive agents. Rather, viruses as extracellular entities are generally viewed as non-metabolically active spore-like agents that await further infection events upon collision with appropriate host cells. That a virus might actively contribute to its fate via chemotaxis and change the behavior of an organism independent of infection is unprecedented.
INTRODUCTION
Virus particles (virions) are generally considered to be inanimate, influencing cells only upon contact. Virions typically contact appropriate host cells through biological, mechanical, or other physical processes, but virions are not known to have their own mechanisms for attracting motile cells from a distance. Here, we report that chloroviruses can attract Paramecium bursaria from a distance by altering their movements. The action of a virus as a chemotactic agent has significant implications in biological systems from immune functions to predator-prey interactions.
Chloroviruses (family Phycodnaviridae) are large icosahedral (190 nm in diameter) double-stranded DNA (dsDNA) viruses (genomes of 290 to 370 kb) containing an internal single bilayered lipid membrane (1, 2). Chloroviruses infect certain eukaryotic green algae that are mutualistic endosymbionts (referred to as zoochlorellae) of organisms such as the protozoan Paramecium bursaria (Ciliophora) (3, 4). However, the zoochlorellae as endosymbionts are resistant to virus infection because the viruses have no way of reaching their hosts. For virus expansion to occur, the protective barrier provided by P. bursaria must be disrupted. We have determined that one mechanism for increasing the chlorovirus population is due to an ecological catalytic event driven by predators, including a cyclopoid copepod predator (Eucyclops agilis) that engulfs the entire P. bursaria (prey) during feeding (5) or the ciliate Didinium nasutum that disrupts the P. bursaria during feeding (referred to as messy feeding) (6). In the case of copepod consumption, when a fecal pellet is released into the water column, the virus replicates in the released zoochlorellae, and nascent virions subsequently diffuse from the fecal pellet, resulting in a localized high concentration of virus (5). Messy feeding by Didinium spp. releases algal cells into the water column, where they are infected (6). These catalytic processes can contribute to cycles of chlorovirus expansion in temperate lakes (see, e.g., reference 7). The efficiency of this process is enhanced because the chloroviruses reside on the outer surface of the paramecia, often at the base of the ciliary pits that can number in the thousands per cell (8–10). Previous estimates suggest that hundreds of infectious chloroviruses can be attached to the surface of a single cell (5). How so many virions accumulate on paramecium cells is unknown.
The accumulation of virions on the cell surface could occur through random contacts between the paramecia and virus particles as the paramecia move through the water. This process would be consistent with the view that virions cannot attract cells from a distance. If this is true, there should be no observable shifts in paramecium movement in response to gradients in virus density. In contrast, if chloroviruses are able to signal to paramecia from a distance through some chemical means, the paramecia should show detectable behavioral shifts as they orient toward the virus particles (chemotaxis). Here, we describe choice/no-choice experiments that reveal strong directional movement of P. bursaria toward concentrations of chloroviruses relative to alternative targets, demonstrating the chemotactic influence of a virus on cells from a distance.
RESULTS
We used a simple three-way system wherein P. bursaria placed in the center of a petri dish could move out into one of three arms toward different targets (Fig. 1). Targets were paper disks loaded with target agents. Each experiment was run with one of two possible sets of three choices (Fig. 1A and B) paired with a no-choice experiment as a negative control (Fig. 1C) and replicated 4 to 6 times. Each replicate experiment was analyzed with chi-square tests to assess differences in frequency of P. bursaria cells moving toward the three different targets, and outcomes were summarized across all replicates. We ran a series of trials that show that P. bursaria cells are not attracted to algal host cells but are attracted to infected cells, that P. bursaria respond more intentionally to higher densities of virions, and that P. bursaria cells respond to a variety of chlorovirus strains. P. bursaria cells showed no directional movements in control dishes.
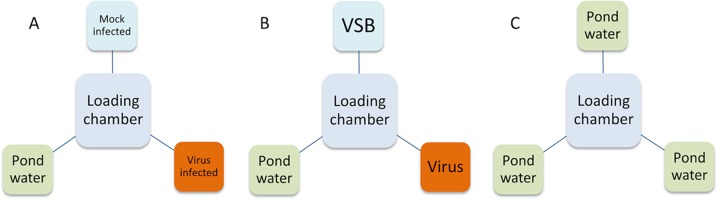
FIG 1.
Experimental scheme for evaluating potential of chemotaxis agents associated with chloroviruses. The microcosms are described in Materials and Methods. (A) In the first experiment, P. bursaria is provided with a choice of cell extracts of mock-infected cells, chlorovirus-infected cells, and pond water. (B) In later experiments, P. bursaria is provided a choice of purified virions suspended in virus stabilization buffer, virus stabilization buffer, or pond water. (C) All experiments were paired with a negative control where all targets were the same (pond water).
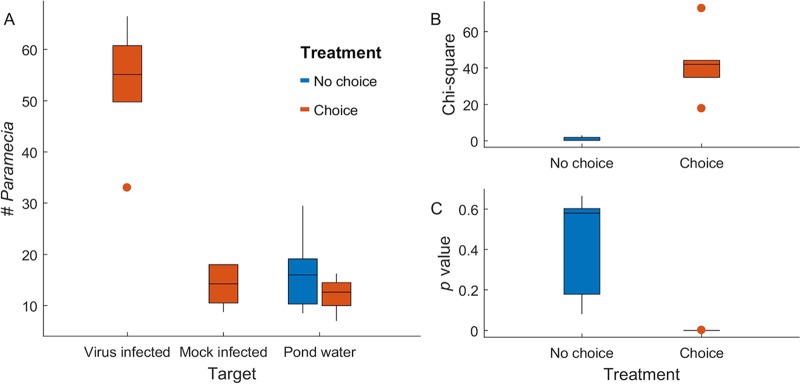
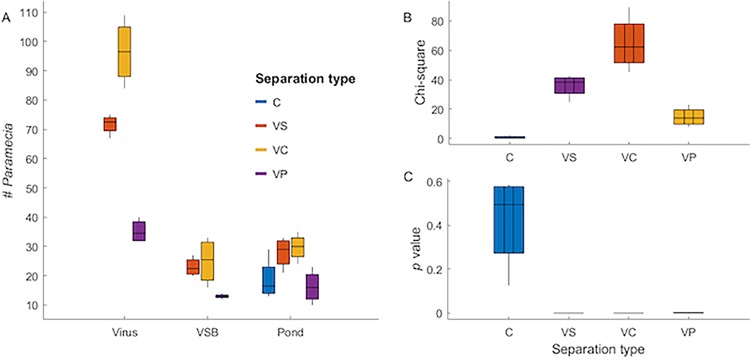
First, we determined that chlorovirus-infected cells could influence the movement behavior of P. bursaria. In these experiments, we used the paramecium-free zoochlorellae that are susceptible to the Chlorovirus Osy-NE-ZA1 (5, 6). We estimated that Osy-NE-ZA1 infection kinetics are similar to that of the type member of the genus Chlorovirus, P. bursaria chlorovirus 1 (PBCV-1) (1), indicating a burst size of ∼1,000 virus particles per cell, of which 20 to 30% are infectious (11). Thus, cell extracts were prepared from 4 h-infected zoochlorellae and used as the choice target in microcosm chambers, because intact infectious virus particles are inside host cells by 4 to 6 h postinfection (11, 12), as described in Materials and Methods. Cell extracts from mock-infected zoochlorellae and sterile pond water (here, pond water) were used as choice controls. The paramecium movement behavior was assessed by counting the population distribution after 12 h of free-ranging movement within the microcosm. We observed that paramecia were attracted to the virus-infected cell extract relative to the mock-infected cell extract or the pond water control (P = 10−17 to 10−5, n = 6; Table 1), whereas in microcosm chambers with no choices (all targets were pond water), the paramecia were equally distributed (P = 0.08 to 0.67, n = 5) (Fig. 2).
TABLE 1.
Paramecium distribution in the presence of cell extracts
| Treatment | Target or stat | Replicate expt avg valuea | |||||
|---|---|---|---|---|---|---|---|
| Cell extract isolate | Virus-infected | 33 | 57 | 49.75 | 60.75 | 53.25 | 66.5 |
| Mock-infected | 8.75 | 18 | 14.25 | 18 | 14.25 | 10.5 | |
| Pond water | 13.75 | 16.25 | 7 | 14.5 | 11.5 | 10 | |
| Chi-square | 17.72 | 34.90 | 44.23 | 42.67 | 41.41 | 72.74 | |
| P | 2.56E−05 | 3.47E−09 | 2.92E−11 | 6.48E−11 | 1.23E−10 | 1.48E−17 | |
| Mock-infected | Pond water 1 | 10.25 | 11 | 12.25 | 27.75 | 19.25 | |
| Pond water 2 | 10 | 16 | 9.75 | 18 | 22 | ||
| Pond water 3 | 8.5 | 17.5 | 10.5 | 29.5 | 18.75 | ||
| Chi-square | 0.19 | 1.56 | 0.30 | 3.06 | 0.31 | ||
| P | 0.67 | 0.21 | 0.58 | 0.08 | 0.58 | ||
Data are the measured values and associated statistical data from each of six microcosms for the virus-infected cell extracts and five microcosms for the mock-infected extracts; the values represent the average of the blind-coded readings by at least four individuals. The data are represented in Fig. 2.
FIG 2.
Paramecium bursaria showed significant movement toward cell extracts of virus-infected zoochlorellae relative to mock-infected cells and pond water. (A) Box plots showing numbers of P. bursaria organisms found at target sources in the counting zone. Boxes represent the central 50% of observations, and points are outliers, defined as greater than a box distance away from the box. (B) Box plots summarizing chi-square values across replicate choice and no-choice experiments. (C) Box plots summarizing P values across replicate experiments (all P < 0.00001), showing that all choice experiments revealed significant shifts in P. bursaria behavior and that none of the no-choice experiments showed significant orientation toward any target (all P > 0.08).
Pilot experiments with purified virions as the target showed that these movements of paramecia were directed toward the virus itself. We then varied the amount of virions in the target and showed that the movements were concentration dependent. Even at the lowest concentration tested (101 PFU), P. bursaria showed orientation toward the viruses (Fig. 3). As virion concentrations increased, the number of paramecia located at the virus target increased. This increase was confirmed by an increasing chi-square value, showing that an increase in virion numbers resulted in higher chi-square values and decreased P values; these results indicate that there is increasing confidence in the orientation toward the virus target as virion concentrations increase (Table 2). A dose-response curve revealed an apparent two-phase response, with a lower limit of efficacy of 101 to 103 PFU per disk and a secondary response above 105 PFU per disk.
FIG 3.
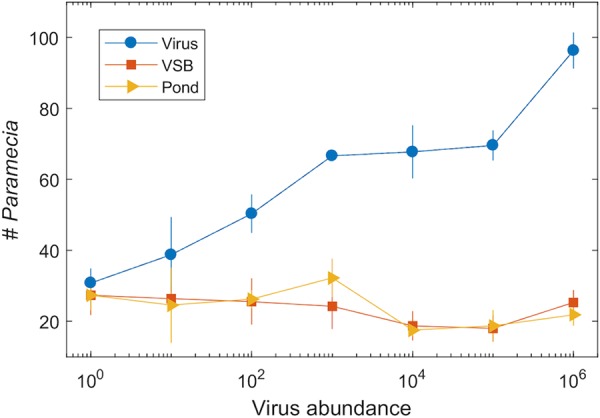
Dose-response curve showing an increasing response of P. bursaria cells to chlorovirus targets as the amount of virus loaded on the target increased. Blue circles represent the virus treatment, red boxes represent the virus stabilization buffer, and orange triangles represent pond water. Values are the mean of n = 4 for each concentration. Error bars are the standard deviation.
TABLE 2.
Paramecium distribution in the presence of various amounts of virions
| Treatment (PFU/disk) | Target or stat | Replicate expt avg valuea | |||
|---|---|---|---|---|---|
| 1 × 106 | Virus | 88.75 | 97.75 | 99.75 | 99 |
| VSB | 23.25 | 23.75 | 30.5 | 23.75 | |
| Pond water | 19.25 | 26 | 22.25 | 19.75 | |
| Chi-square | 69.61 | 72.06 | 71.28 | 83.92 | |
| P | 7.22E−17 | 2.09E−17 | 3.10E−17 | 5.14E−20 | |
| 1 × 105 | Virus | 64.25 | 68.25 | 74 | 71.75 |
| VSB | 13.75 | 18.25 | 22.75 | 17 | |
| Pond water | 19.75 | 24.5 | 16.25 | 14.25 | |
| Chi-square | 46.72 | 40.12 | 53.13 | 61.28 | |
| P | 8.21E−12 | 2.39E−10 | 3.12E−13 | 4.96E−15 | |
| 1 × 104 | Virus | 58.75 | 68.75 | 66.5 | 77 |
| VSB | 14.25 | 24.25 | 18.25 | 18 | |
| Pond water | 17.75 | 18 | 15 | 19 | |
| Chi-square | 40.48 | 41.40 | 50.03 | 60.05 | |
| P | 1.99E−10 | 1.24E−10 | 1.51E−12 | 9.24E−15 | |
| 1 × 103 | Virus | 67.25 | 65 | 67.75 | 66.5 |
| VSB | 21.25 | 33.75 | 22.5 | 19.5 | |
| Pond water | 31.75 | 35.25 | 37 | 24.75 | |
| Chi-square | 28.99 | 13.91 | 25.17 | 35.93 | |
| P | 7.26E−08 | 1.92E−04 | 5.24E−07 | 2.04E−09 | |
| 1 × 102 | Virus | 52 | 46.5 | 57.25 | 45.5 |
| VSB | 22.25 | 19.5 | 34.5 | 26 | |
| Pond water | 26.5 | 27.75 | 28 | 22.5 | |
| Chi-square | 15.42 | 12.25 | 11.82 | 9.80 | |
| P | 8.62E−05 | 4.65E−04 | 5.86E−04 | 1.74E−03 | |
| 1 × 101 | Virus | 45.25 | 23 | 45 | 41.75 |
| VSB | 30 | 23.5 | 28.5 | 23.5 | |
| Pond water | 28.75 | 8.75 | 28.5 | 32 | |
| Chi-square | 4.87 | 7.62 | 5.34 | 5.15 | |
| P | 0.027 | 0.006 | 0.021 | 0.023 | |
| Control (non-choice test) | Pond water 1 | 25.25 | 30 | 34.5 | 33.25 |
| Pond water 2 | 24.25 | 23 | 26.5 | 35.25 | |
| Pond water 3 | 22 | 28.75 | 26.25 | 32.25 | |
| Chi-square | 0.23 | 1.02 | 1.51 | 0.14 | |
| P | 0.63 | 0.31 | 0.22 | 0.71 | |
Data are the measured values and associated statistical data from each of four microcosms; the values represent the average of the blind-coded readings by at least four individuals. The data are represented in Fig. 3.
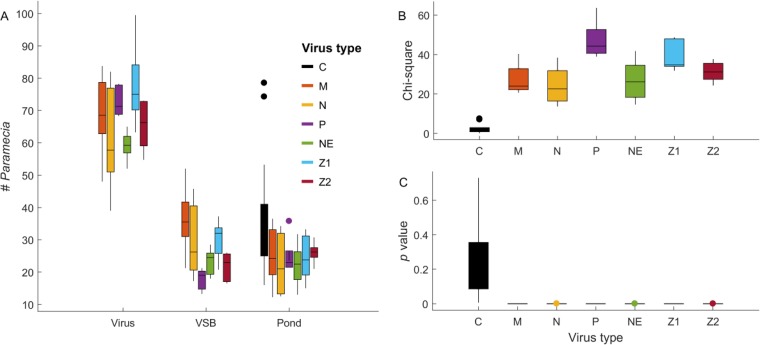
We used an Osy virus (Osy-NE-ZA1) as the target in the initial experiments because the P. bursaria cultures containing zoochlorellae were infected by Osy viruses (5, 6). Currently, our laboratory has four such algal/virus systems; they are Chlorella variabilis NC64A and its viruses (referred to as NC64A viruses), Chlorella variabilis Syngen 2-3 and its viruses (referred to as Osy viruses), Chlorella heliozoae SAG 3.83 and its viruses (referred to as SAG viruses), and Micractinium conductrix Pbi and its viruses (referred to as Pbi viruses) (1, 13–15). To determine if the behavior of P. bursaria was chlorovirus specific, we conducted identical experiments with other chlorovirus strains (PBCV-1, an NC64A virus; TN603, an SAG virus; and Chlorella virus 1 [CVM-1], a Pbi virus), and each chlorovirus attracted the paramecia (Fig. 4 and Table 3). Therefore, the chemotaxis was not chlorovirus type specific.
FIG 4.
P. bursaria showed chemotactic movements toward all tested chloroviruses. Panels are as in Fig. 3. Virus types are C (black, pond water, no-choice), M (red, Pbi-CVM-1), N (orange, SAG-TN603), P (purple, NC64A-PBCV-1), NE (green, Osy-NE5), Z1 (light blue, Osy-NE-ZA1), and Z2 (dark red, Osy-NE-ZA2). Each virus was evaluated at 104 PFU per target disk.
TABLE 3.
Paramecium distribution in the presence of various chloroviruses
| Virus (type) or other treatment | Target or stat | Replicate expt avg valuea | ||||
|---|---|---|---|---|---|---|
| CVM-1 (Pbi) | Virus | 48 | 68.5 | 67.75 | 77 | 83.75 |
| VSB | 35.5 | 52 | 38.25 | 21.25 | 34.25 | |
| Pond water | 12.25 | 21.5 | 24.25 | 32 | 36.5 | |
| Chi-square | 20.63 | 24.02 | 22.71 | 40.30 | 30.34 | |
| P | 5.59E−06 | 9.51E−07 | 1.88E−06 | 2.18E−10 | 3.62E−08 | |
| TN603 (SAG 3.83) | Virus | 75.25 | 57.75 | 82 | 55 | 39 |
| VSB | 45.75 | 26.25 | 38.75 | 17.25 | 21.75 | |
| Pond water | 34.25 | 21 | 31.25 | 12.5 | 13.5 | |
| Chi-square | 17.29 | 22.58 | 29.62 | 38.39 | 13.68 | |
| P | 3.22E−05 | 2.02E−06 | 5.25E−08 | 5.78E−10 | 2.17E−04 | |
| Osy-NE-ZA1 (Syngen 2-3) | Virus | 63.25 | 72.5 | 75 | 99.5 | 79 |
| VSB | 20.75 | 27.5 | 32 | 37.25 | 32.5 | |
| Pond water | 20.5 | 15 | 23.75 | 33.25 | 30.5 | |
| Chi-square | 34.77 | 47.72 | 34.75 | 48.71 | 31.82 | |
| P | 3.70E−09 | 4.92E−12 | 3.75E−09 | 2.97E−12 | 1.69E−08 | |
| Control (non-choice test) | Pond water 1 | 40.25 | 39 | 42.5 | 41 | 78.5 |
| Pond water 2 | 49.25 | 53.25 | 52.5 | 40.5 | 74.25 | |
| Pond water 3 | 33.75 | 29.5 | 39.75 | 33.75 | 49.25 | |
| Chi-square | 2.95 | 7.04 | 2.00 | 0.85 | 7.42 | |
| P | 0.086 | 0.008 | 0.157 | 0.356 | 0.006 | |
| Osy-NE5 (Syngen 2-3) | Virus | 58.5 | 52 | 61 | 59.25 | 65 |
| VSB | 18 | 25 | 24.5 | 19.75 | 28.5 | |
| Pond water | 13 | 24.5 | 22.5 | 19.25 | 31.75 | |
| Chi-square | 41.73 | 14.63 | 26.09 | 32.16 | 19.54 | |
| P | 1.04E−10 | 1.30E−04 | 3.25E−07 | 1.41E−08 | 9.81E−06 | |
| PBCV-1 (NC64A) | Virus | 68.25 | 78.25 | 71.25 | 69 | 77.75 |
| VSB | 20 | 21.25 | 19 | 15.25 | 13.25 | |
| Pond water | 21.5 | 35.75 | 23.5 | 21.5 | 23 | |
| Chi-square | 41.15 | 38.93 | 44.22 | 49.02 | 63.62 | |
| P | 1.41E−10 | 4.39E−10 | 2.93E−11 | 2.53E−12 | 1.51E−15 | |
| Control (non-choice test) | Pond water 1 | 25 | 16 | 33 | 25.75 | 31 |
| Pond water 2 | 28.75 | 17.25 | 31.25 | 16.75 | 19.25 | |
| Pond water 3 | 30 | 18 | 24.75 | 18.25 | 25.25 | |
| Chi-square | 0.49 | 0.12 | 1.27 | 2.30 | 2.74 | |
| P | 0.49 | 0.73 | 0.26 | 0.13 | 0.10 | |
Data are the measured values and associated statistical data from each of five microcosms; the values represent the average of the blind-coded readings by at least four individuals. All virus concentrations were 1 × 104 PFU/disk. The data are represented in Fig. 4.
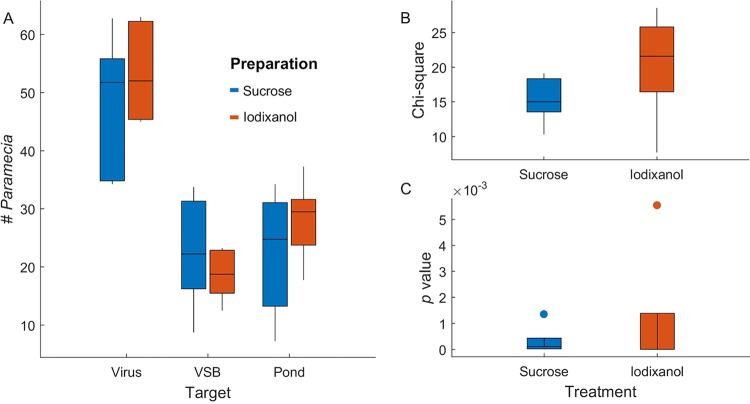
The observed chemotaxis was not due to reagents used in preparing the virus stocks. The chloroviruses were isolated using either sucrose or iodixanol density gradients. We evaluated the density gradient materials to determine if residuals of these chemicals influenced the ability of the virus to attract P. bursaria. No significant differences were detected using virions purified by either sucrose or iodixanol gradients to attract P. bursaria (Fig. 5 and Table 4). We also substituted disks soaked in 40% of either sucrose or iodixanol in place of virus for one of the three arms in the choice microcosms. P. bursaria cells did not show any preferential movement toward sucrose or iodixanol targets. Therefore, we conclude that the virus alone was responsible for attracting P. bursaria.
FIG 5.
Sucrose versus iodixanol. Paramecium bursaria showed no significant difference in movement toward virions prepared with sucrose density gradients compared to virions prepared with iodixanol density gradients. (A) Box plots showing numbers of P. bursaria found at target sources in the counting zone for preparations using either sucrose or iodixanol prepared virus, sucrose- or iodixanol-spiked VSB, or spiked pond water. Boxes represent the central 50% of observations, and points are outliers, defined as greater than a box distance away from the box. (B) Box plots summarizing chi-square values across replicate choice experiments. (C) Box plots summarizing P values across replicate experiments (all P < 0.001). The data indicate that all choice experiments revealed no significant shifts in P. bursaria behavior as a result of virus preparation.
TABLE 4.
Paramecium distribution in the presence of density gradient materials used in virion preparations
| Density gradient reatment | Target or stat | Replicate expt avg valuea | ||||
|---|---|---|---|---|---|---|
| Sucrose | Virus | 83.5 | 119 | 95.5 | 72.25 | 52.75 |
| VSB | 30.75 | 76 | 45.5 | 41.5 | 21.25 | |
| Pond water | 45.25 | 81.5 | 59 | 38.25 | 22.75 | |
| Chi-square | 19.12 | 10.29 | 15.02 | 18.09 | 14.64 | |
| P | 1.23E−05 | 1.34E−03 | 1.06E−04 | 2.11E−05 | 1.30E−04 | |
| Iodixanol | Virus | 81.25 | 61.75 | 86.75 | 82.75 | 86 |
| VSB | 33.5 | 26.5 | 18.25 | 33.25 | 14.25 | |
| Pond water | 44 | 37.25 | 48.75 | 52.75 | 26.75 | |
| Chi-square | 19.37 | 21.59 | 28.56 | 7.70 | 24.91 | |
| P | 1.08E−05 | 3.38E−06 | 9.08E−08 | 5.53E−03 | 6.02E−07 | |
Data are the measured values and associated statistical data from each of five microcosms; the values represent the average of the blind-coded readings by at least four individuals. The data are represented in Fig. 5.
To begin to evaluate the soluble nature of the chemoattractive agent, virions were separated from the aqueous phase by centrifugation so that we could recover and evaluate both the pellet and supernatant fractions (Fig. 6 and Table 5). The majority of the attractive “signal” was retained in the “wash” fraction that is essentially free of virus, indicating that there was a soluble agent(s) “leaking” from the particles. The virions (pellet fraction) remain intact as indicated by plaque assay, where essentially all of the initial PFU were recovered.
FIG 6.
Distribution of Paramecium bursaria in the presence of a soluble fraction of Chlorovirus Osy-NE-ZA1 virions to evaluate particle “leakage” of a chemotactic agent. (A) Distribution of the paramecia in the 3-chamber microcosms after an overnight incubation that allows the paramecia to roam throughout the microcosm space. The negative control (C) and the positive control (VC) were as observed previously (see, e.g., Fig. 4). Both the pellet fraction (VP) and the supernatant fraction (VS) attract paramecia relative to the VSB and pond water choices; however, the VS attracted more paramecia than did the VP. (B and C) Statistical support for the observations. C, no-choice negative control, where all targets are pond water (blue); VC, nontreated virus suspended in virus stabilization buffer (VSB) after 24 h incubation at room temperature (yellow); VP, virus after 24 h of incubation at room temperature and then centrifuged at 20,000 × g for 1 h and resuspended in VSB as the pellet fraction (which is intended to contain the vast majority of virions) (purple); VS, virus after 24 h of incubation at room temperature and then centrifuged at 20,000 × g for 1 h separated as the supernatant fraction (which is intended to be essentially free of virus) (orange). VC, VP, and VS were compared to VSB and pond water as choices. The data and statistical analyses for this experiment are found in Table 5.
TABLE 5.
Paramecium distribution in the presence of a soluble fraction from Osy-NE-ZA1 virions
| Treatment | Target or stat | Replicate expt avg valuea | |||
|---|---|---|---|---|---|
| Virus supernatant | Virus | 67 | 73 | 72 | 75 |
| VSB | 24 | 27 | 20 | 21 | |
| Pond water | 33 | 21 | 31 | 27 | |
| Chi-square | 24.89 | 40.13 | 36.63 | 42.73 | |
| P | 6.08E−07 | 2.37E−10 | 1.43E−09 | 6.28E−11 | |
| Virus control | Virus | 84 | 92 | 101 | 109 |
| VSB | 16 | 30 | 33 | 21 | |
| Pond water | 24 | 35 | 31 | 29 | |
| Chi-square | 66.84 | 45.34 | 57.75 | 89.36 | |
| P | 2.95E−16 | 1.66E−11 | 2.98E−14 | 3.29E−21 | |
| Virus pellet | Virus | 37 | 32 | 32 | 40 |
| VSB | 12 | 13 | 13 | 14 | |
| Pond water | 10 | 23 | 14 | 18 | |
| Chi-square | 23.02 | 7.97 | 11.63 | 16.33 | |
| P | 1.61E−06 | 4.75E−03 | 6.50E−04 | 5.31E−05 | |
| Control (non-choice test) | Pond water 1 | 17 | 24 | 16 | 14 |
| Pond water 2 | 13 | 24 | 13 | 17 | |
| Pond water 3 | 22 | 29 | 14 | 16 | |
| Chi-square | 2.35 | 0.65 | 0.33 | 0.30 | |
| P | 0.13 | 0.42 | 0.57 | 0.59 | |
Data are the measured values and associated statistical data from each of four treatments in four replicate microcosms that are evaluating untreated virions (virus control) and the soluble (virus supernatant) and pellet (virus pellet) fractions of the corresponding virus preparation, as described in Materials and Methods. The values represent the average of the blind-coded readings by four individuals. The data are represented in Fig. 6.
DISCUSSION
Collectively, our results indicate that P. bursaria could detect and move toward chloroviruses, and this intentional behavior was virus concentration dependent. These results stand in contrast to the paradigm that viruses do not signal to other cells from a distance and are dependent on biological, mechanical, or other physical processes to facilitate contacts with hosts and other intermediary cells. We infer that chloroviruses have associated chemical signals that are detected in low concentrations at a great distance by P. bursaria (roughly 400 cell lengths for cells ∼70 to 80 μm in length in the microcosms used in these studies). Although P. bursaria is not the host of the chloroviruses, attracting P. bursaria cells increases the possibility that chloroviruses can adhere to their outer membrane. These contacts would otherwise depend on substantial movement of P. bursaria cells through the water column to facilitate random encounters that lead to the effective accumulation of chloroviruses. This adherence in turn places the chloroviruses in the correct location to take advantage of ecological catalysts by predation that release zoochlorellae through either messy feeding or by passing fecal pellets. These processes may not be isolated to just this P. bursaria system, as zoochlorellae are also associated with other symbiotic hosts, such as the coelenterate Hydra viridis (see, e.g., reference 16) and the heliozoan Acanthocystis turfacea (see, e.g., reference 17).
Although paramecia moved toward the chloroviruses at potentially high ratios (up to ∼5:1 virus to alternative targets), not all individual paramecia chose to move toward the virus target. There was always a subset of the P. bursaria population that did not track to the virus target, independent of the virus amount. This suggests that individuals may differ in either their ability to detect the chemical cue or the motivation to respond to it. It is likely that individuals either vary in the number of viruses already on the cell surface at the time of the experiment, in their stage of the cell cycle, or in their physiology. Individual variation in propensity to seek out concentrations of chloroviruses could have significant implications for the evolution of chloroviruses and P. bursaria, as there may be fitness benefits to either carrying or shunning chlorovirus surface loads.
We do not know what the chemical nature of the chemotactic/chemoattractive signal is. Virus particle complexity tends to increase with the size of the genome, including virion size and shape, protein composition and diversity, membrane content and composition, small-molecule content, and charge-neutralizing agents, such as cations (18). All of these factors contribute to particle stability and infection potential as an extracellular agent that is animated upon contact with the host. Chloroviruses are large dsDNA-containing icosahedral particles composed of a glycoprotein outer capsid, which surrounds an inner lipid membrane. This shell encapsulates the >300-kb genome, along with many proteins that are mostly virus encoded (19). Additionally, the chlorovirus PBCV-1 contains several small molecules associated with polyamine biosynthesis, including putrescine, spermidine, homospermidine, and cadaverine (20, 21). These molecules were evaluated for their potential to neutralize the large negative charge associated with the virion DNA; however, the abundances measured indicate that the mole ratio was insufficient for this purpose. The best evidence for charge neutralization of the virion DNA is via divalent cations including calcium and magnesium, as well as certain cationic proteins (22). Thus, chloroviruses consist of both large and small molecules, some of which may contribute to the chemotactic properties of the virions reported here.
To begin to understand the chemical nature of chemotactic agent(s), virions were allowed to incubate in the VSB, the supernatant fraction was collected after centrifugation, and the pellet fraction was resuspended in an equivalent volume. These fractions were evaluated in the standard 3-chamber microcosm, as shown in Fig. 6. The paramecia migrated to both fractions but more so to the soluble fraction. The data support the hypothesis that there is a soluble agent that “leaks” from the virus particles. However, this is apparently not due to virus particle degradation, as indicated by a full recovery of PFU in the pellet fraction.
Virions have not previously been reported to act as chemotactic/chemoattractive agents. Rather, viruses as extracellular entities are generally viewed as non-metabolically active spore-like agents that await further infection events upon collisions with appropriate host cells. That a virus might actively contribute to its fate via chemotaxis and change the behavior of an organism independent of infection is unprecedented. However, a recent report indicates that at least some bacteriophage can communicate with one another by producing and sensing small peptides as chemical messages (23). It has not escaped our attention that other viruses may attract motile cells, e.g., macrophages, but this has not been demonstrated to date, to our knowledge. We must now rethink how virus particles can play an active role in directing their own fate. If one virus can do this, it is likely that others do as well.
MATERIALS AND METHODS
Preparing the paramecia.
Paramecia bursaria, a zoochlorella-bearing holobiont, was provided by John DeLong’s lab at the University of Nebraska—Lincoln. They were originally collected from Spring Creek Prairie Audubon Center pond water in Denton, NE, (global positioning system [GPS] coordinates: 40°41′37.6764″N, 96°51′12.2544″W). The paramecia were grown on protozoan medium (Carolina Biological Supply, Burlington, NC, USA) under constant light (light flux, 38 to 42 μmol/m2 s−1) at room temperature (∼25°C). Before each experiment, paramecia were transferred to pond water from the Spring Creek Prairie pond that had been filtered through Whatman no. 1 filter paper, aspirated through a 0.45-μm bottle-top filter (Sarstedt, Newton, NC, USA), autoclaved, and stored at room temperature. The paramecia were washed three times with pond water to remove unattached viruses and residue such as culture medium and algae (5).
Virus isolation.
The primary strain of virus tested in this project was Chlorella variabilis Syngen 2-3-infecting Chlorovirus OSY-NE-ZA1 (5). Other chlorovirus strains used were Chlorella variabilis NC64A-infecting Chlorovirus PBCV-1, Chlorella variabilis Syngen 2-3-infecting Chlorovirus OSY-NE-5, and C. heliozoae SAG 3.83-infecting Chlorovirus TN603 virus. All of these viruses were propagated using algal cells grown in modified Bold’s basal medium (MBBM) as described previously (11, 24), except for Micractinium conductrix Pbi-infecting chlorovirus CVM-1, which was propagated in FES medium (25).
Virus isolation: cell extracts.
Chlorella variabilis Syngen 2-3 cells (0.6 × 108 to 1.0 × 108 cells/ml) in basal Bold’s medium (BBM) were infected with Chlorovirus OSY-NE-ZA1 suspended in virus stabilization buffer (VSB; 50 mM Tris HCl, 10 mM MgCl2 [pH 7.8]) at a multiplicity of infection (MOI) of 3 PFU per cell, or mock-infected with VSB as a control. After 30 min of infection, unattached viruses were removed by low-speed centrifugation (1,900 × g for 3 min) of the virus- and mock-infected cells, and the pellet fractions containing the treated cells were resuspended in BBM. The infection was then allowed to proceed to 4 h under normal incubation conditions of light, temperature, and shaking. After this 4-h incubation, the cells were harvested by centrifugation at 3,000 × g for 5 min, washed one time (by resuspending and centrifugation) with pond water, and resuspended in pond water at a concentration of 0.9 × 109 to 1.3 × 109 cell/ml. Aliquots of 0.5 ml of infected cells were mixed with ∼0.5 g of glass beads (equal mixture of 0.25 to 0.30 mm and 0.45 to 0.5 mm [Glasperlen; B. Braun Melsungen AG, PA, USA]), extensively washed with deionized distilled water and then with sterilized pond water, and placed in a bead beater (Disruptor Genie; Scientific Industries, Bohemia, NY) for 15 min at 4°C. Disrupted cells were centrifuged for 1 min at 1,000 × g (to remove glass beads), and the supernatant fraction was collected and frozen in liquid nitrogen.
Chlorovirus purification.
All the algal cells were maintained at 25°C with continuous light and shaking (200 rpm) (26). The algal cell concentration was 1.5 × 107 cells/ml in 1,600 ml of culture with tetracycline (10 μg/ml). The cells were inoculated with 0.45-μm filter-sterilized virus (stock concentrations at ∼8 × 1010 PFU/ml) at an MOI of ∼0.005 PFU/cell and incubated for 2 days with continuous light at 25°C and shaking until complete cell lysis. The lysates were adjusted with Triton X-100 to a final concentration of 1% (stock concentration 20% [wt/vol]) to dissolve membranes, pigments, and lipids that trap virions. The suspensions were stirred at room temperature for approximately 1 h.
The detergent-treated lysates were filtered through Whatman no. 1 filter paper to remove cell debris, and the filtrates were collected. The filtered lysates were centrifuged in a Beckman type 19 ultracentrifuge rotor at 53,000 × g for 50 min at 4°C. The supernatant fractions were discarded, and the virion-enriched pellet fractions were resuspended in 1 ml of VSB. The resuspended virion fractions were further purified by density gradient fractionation. NC64A and Pbi viruses were separated on sucrose density gradients, whereas SAG and Osy-NE viruses were separated on iodixanol density gradients (OptiPrep; Axis-Shield, Oslo, Norway). The gradients were preestablished 10 to 40% linear sucrose or iodixanol equilibrated with VSB for the Beckman SW32 rotor. Three to 4 ml of virion suspensions was layered on the preformed gradients and then centrifuged at 72,000 × g for 20 min at 4°C. The virion bands were approximately 1/2 to 2/3 deep in the gradients, which were aspirated using a sterile bent cannula to remove the particles from the top. The suspended virions were transferred to a Beckman type 19 tube, diluted with VSB, and pelleted at 53,000 × g as described above. The supernatant fractions were discarded, and pellet fractions were resuspended overnight at 4°C with 2 ml of VSB. After fully resuspending the pelleted virions, the material was treated with proteinase K (final concentration, 0.02 mg/ml) and incubated at 45°C for 1 h to degrade non-virion-associated proteins (26). The proteinase K-treated virions were applied to another density gradient fractionation, as described above, and then collected, fully suspended in VSB, and pelleted with the Beckman type 19 rotor. This process was carried out two times, and then the viral pellets were suspended in 1 ml of VSB. The final gradient-purified virus particles were aspirated through a 0.45-μm bottle-top filter, and the virions were enumerated by a plaque assay to determine the virus concentrations in PFU per milliliter.
Soluble agent from virions.
Chlorovirus Osy-NE-ZA1 was freshly prepared as described above, evaluated for virus concentration by plaque assay, and stored at 4°C for 1 week. The 1-week-old virus prep (100 μl at a concentration of 2.5 × 1011 PFU/ml) was incubated on the benchtop at room temperature overnight and then was centrifuged for 1 h at 20,000 × g (S20 = 2,300 for the type virus PBCV-1 [27]). The supernatant fraction was removed, and the pellet fraction (containing the vast majority of virions) was resuspended in 100 μl of fresh VSB. The pellet and supernatant fractions were serially diluted to an equivalent of virus concentration of 1 × 108 PFU/ml, and then 10 μl of the diluted pellet and supernatant fractions was applied to the filter disks. Untreated virus at an equal concentration served as a positive control. The preparations were evaluated with our standard 3-chamber choice microcosm experimental design (described below); each condition was replicated four times, and each microcosm was read by four individuals. The infectivity of the virus in the resuspended pellet fraction was not affected by these manipulations, as measured by the plaque assay.
Microcosm setup.
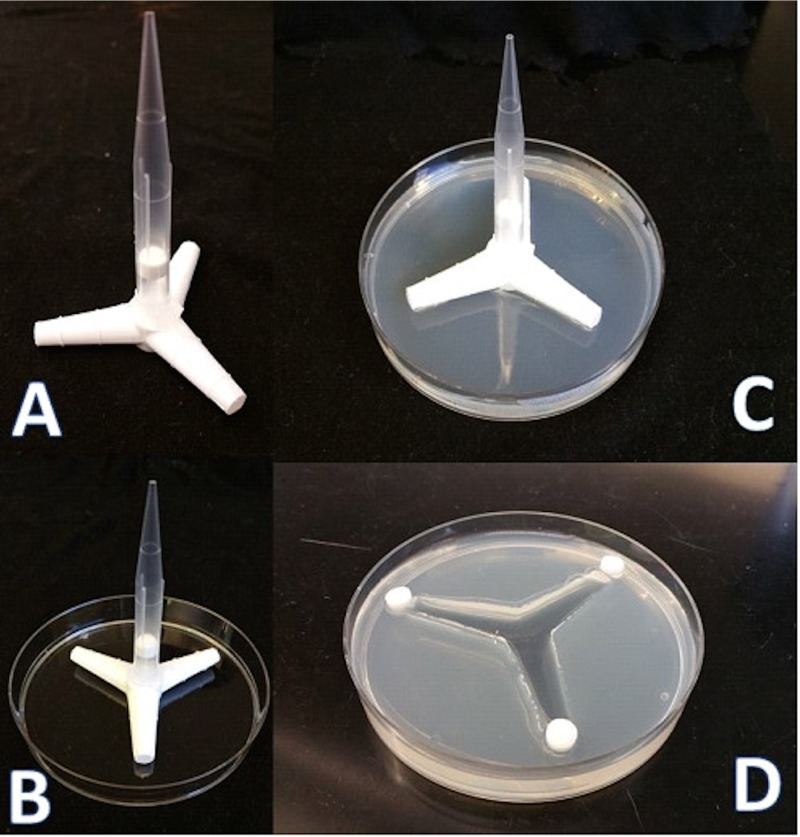
Paramecium movement behavior was evaluated by observing populations placed in a microcosm environment formed in petri dishes with molded sterile agar medium made in pond water (1.5% [wt/vol]; Thermo Fisher). We used a locally fabricated three-channel mold (Fig. 7A) that allowed us to provide a three-way choice for the paramecia while moving freely under aqueous conditions. To set up the microcosm chamber, a thin layer of agar was first added and allowed to cool in a level petri dish (92 by 16-mm petri dish; Sarstedt, Germany), and the mold was put in the middle of the dish. Then, hot liquid agar medium was added slowly (25 to 30 ml agar/dish) and left to solidify at room temperature. The mold was a three-channel triangle-like shape (plastic tubing connector-Y, tapered from 9.9 to 11.5 mm) (Fig. 7A and B); the end of each channel’s width was 7 mm, the entrance to the channel was 15 mm, and the length of each channel was 30 mm. Once solidified, the mold was carefully removed from the petri dish, and a small amount of hot liquid agar was added to smooth the channel and make them equal in all directions. The depth of each channel was 10 mm, and the agar thickness was 16 mm.
FIG 7.
Microcosm configuration. (A) Microcosms were molded into pond water agar using plastic tubing connectors fashioned with a pipette tip as a handle. (B and C) One-hundred-millimeter plastic petri dishes were used to contain the agar. When the three-channel mold (A) was removed from the agar, the microcosm consisted of three connected channels. (D) Chemotactic agents (experimental and control) were impregnated into Whatman paper disks, and these were placed at the terminal points of the open channels. At the time of incubation, paramecia were transferred to the open channels and allowed to swim freely.
Using three grade AA Whatman paper disks (6 mm from GE Healthcare, UK), 20 μl of each treatment condition was spotted onto the filter paper. Each disk was then placed at the end of each of the three channels (Fig. 7C). The paramecia (110 to 125 paramecia/ml in pond water) were counted, and 1 ml was added to the middle of the microcosm so that they could move freely in all directions. Each experimental treatment had five replicates. The choice-type microcosm included a chlorovirus, VSB, and pond water, whereas the non-choice-type microcosm contained three treatments of pond water as a control. The treatment conditions were blind-coded.
The microcosm chambers with ranging paramecia were left overnight, and then a microcoverglass (22 mm by 22 mm; VWR Scientific products, USA) was used to block each channel from the middle distance (15 mm between the filter paper disk to the petri dish center) (Fig. 7C) before we started counting. Counting of the paramecia was done using a dissection microscope (Leica Wild W3Z). At least four individuals counted the paramecia in each blind-coded microcosm chamber, and the mean values of these counts were used to represent the paramecium distributions within the microcosm chambers with respect to the treatment conditions.
Statistical analysis.
We used chi-square tests to assess differences in the movement of paramecia toward the three targets within each replicate experimental microcosm. We replicated each treatment and control 3 to 5 times. The frequencies were the counts of paramecia located at the three targets at the end of the experimental period. We then compiled results across replicates to show the consistency of chemotactic movement toward each target type.
ACKNOWLEDGMENTS
This material is based upon work supported by the National Science Foundation under grant 1736030 (to J.L.V.E., D.D.D., and J.P.D.) and the University of Nebraska—Lincoln Agricultural Research Division and the Office of Research and Economic Development (to D.D.D.), and was partially supported by the Ministry of Higher Education & Scientific Research, Republic of Iraq, and the Iraqi Cultural Office in Washington, DC (to Z.A.-A.).
D.D.D. was responsible for the project concept and contributed to the experimental design, analyses, and manuscript writing; M.A.-S. contributed to the experimental design and data collection; Z.A.-A. contributed to the experimental data collection and critical materials development; I.V.A. contributed to the initial pilot experiments, experiment materials, and data analyses; J.P.D. contributed to the experimental design, data reduction, statistical analysis, and manuscript writing; and J.L.V.E. contributed to the experimental design and manuscript development.
We declare no competing interests.
REFERENCES
- 1.Van Etten JL, Dunigan DD. 2012. Chloroviruses: not your everyday plant virus. Trends Plant Sci 17:1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Etten JL, Dunigan DD. 2016. Giant chloroviruses–five easy questions. PLoS Pathog 12:e1005751. doi: 10.1371/journal.ppat.1005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karakashian SJ, Karakashian MW. 1965. Evolution and symbiosis in the genus Chlorella and related algae. Evolution 19:368–377. doi: 10.1111/j.1558-5646.1965.tb01728.x. [DOI] [Google Scholar]
- 4.Kodama Y, Suzuki H, Dohra H, Sugii M, Kitazume T, Yamaguchi K, Shigenobu S, Fujishima M. 2014. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics 15:183. doi: 10.1186/1471-2164-15-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLong JP, Al-Ameeli Z, Duncan GA, Van Etten JL, Dunigan DD. 2016. Predators catalyze an increase in chloroviruses by foraging on the symbiotic hosts of zoochlorellae. Proc Natl Acad Sci U S A 113:13780–13784. doi: 10.1073/pnas.1613843113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong JP, Al-Ameeli Z, Lyon S, Van Etten JL, Dunigan DD. 2018. Size-dependent catalysis of chlorovirus population growth by a messy feeding predator. Microb Ecol 75:847–853. doi: 10.1007/s00248-017-1106-8. [DOI] [PubMed] [Google Scholar]
- 7.Quispe CF, Sonderman O, Seng A, Rasmussen B, Weber G, Mueller C, Dunigan DD, Van Etten JL. 2016. Three-year survey of abundance, prevalence, and genetic diversity of Chlorovirus populations in a small urban lake. Arch Virol 161:1839–1847. doi: 10.1007/s00705-016-2853-4. [DOI] [PubMed] [Google Scholar]
- 8.Kleene SJ, Van Houten JL. 2014. Electrical signaling in motile and primary cilia. Bioscience 64:1092–1102. doi: 10.1093/biosci/biu181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakami H, Kawakami N. 1978. Behavior of a virus in a symbiotic system Paramecium bursaria–zoochlorella. J Protozool 25:217–225. doi: 10.1111/j.1550-7408.1978.tb04399.x. [DOI] [Google Scholar]
- 10.Yashchenko VV, Gavrilova OV, Rautian MS, Jakobsen KS. 2012. Association of Paramecium bursaria chlorella viruses with Paramecium bursaria cells: ultrastructural studies. Eur J Protistol 48:149–159. doi: 10.1016/j.ejop.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Meints RH, Lee K, Van Etten JL. 1986. Assembly site of the virus PBCV-1 in a chlorella-like green alga: ultrastructural studies. Virology 154:240–245. doi: 10.1016/0042-6822(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 12.Van Etten JL, Burbank DE, Xia Y, Meints RH. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126:117–125. doi: 10.1016/0042-6822(83)90466-X. [DOI] [PubMed] [Google Scholar]
- 13.Yamada T, Onimatsu H, Van Etten JL. 2006. Chlorella viruses. Adv Virus Res 66:293–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunigan DD, Fitzgerald LA, Van Etten JL. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res 117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Quispe CF, Esmael A, Sonderman O, McQuinn M, Agarkova I, Battah M, Duncan GA, Dunigan DD, Smith TPL, De Castro C, Speciale I, Ma F, Van Etten JL. 2017. Characterization of a new Chlorovirus type with permissive and nonpermissive features on phylogenetically related strains. Virology 500:103–113. doi: 10.1016/j.virol.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch TC. 2012. What hydra has to say about the role and origin of symbiotic interactions. Biol Bull 223:78–84. doi: 10.1086/BBLv223n1p78. [DOI] [PubMed] [Google Scholar]
- 17.Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L. 2011. The systematics of zoochlorella revisited employing an integrative approach. Environ Microbiol 13:350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 18.Glasgow J, Tullman-Ercek D. 2014. Production and applications of engineered viral capsids. Appl Microbiol Biotechnol 98:5847–5858. doi: 10.1007/s00253-014-5787-3. [DOI] [PubMed] [Google Scholar]
- 19.Dunigan DD, Cerny RL, Bauman AT, Roach JC, Lane LC, Agarkova IV, Wulser K, Yanai-Balser GM, Gurnon JR, Vitek JC, Kronschnabel BJ, Jeanniard A, Blanc G, Upton C, Duncan GA, McClung OW, Ma F, Van Etten JL. 2012. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J Virol 86:8821–8834. doi: 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser A, Vollmert M, Tholl D, Graves MV, Gurnon JR, Xing W, Lisec AD, Nickerson KW, Van Etten JL. 1999. Chlorella virus PBCV-1 encodes a functional homospermidine synthase. Virology 263:254–262. doi: 10.1006/viro.1999.9972. [DOI] [PubMed] [Google Scholar]
- 21.Baumann S, Sander A, Gurnon JR, Yanai-Balser GM, Van Etten JL, Piotrowski M. 2007. Chlorella viruses contain genes encoding a complete polyamine biosynthetic pathway. Virology 360:209–217. doi: 10.1016/j.virol.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wulfmeyer T, Polzer C, Hiepler G, Hamacher K, Shoeman R, Dunigan DD, Van Etten JL, Lolicato M, Moroni A, Thiel G, Meckel T. 2012. Structural organization of DNA in Chlorella viruses. PLoS One 7:e30133. doi: 10.1371/journal.pone.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. 2017. Communication between viruses guides lysis-lysogeny decisions. Nature 541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicols HW, Bold HC. 1965. Trichosarchina polymorpha gen.et. sp. nov. J Phycol 1:34–38. doi: 10.1111/j.1529-8817.1965.tb04552.x. [DOI] [Google Scholar]
- 25.Reisser W, Becker B, Klein T. 1986. Studies on ultrastructure and host range of a Chlorella attacking virus. Protoplasma 135:162–165. doi: 10.1007/BF01277009. [DOI] [Google Scholar]
- 26.Agarkova IV, Dunigan DD, Van Etten JL. 2006. Virion-associated restriction endonucleases of chloroviruses. J Virol 80:8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Etten JL, Meints RH, Kuczmarski D, Burbank DE, Lee K. 1982. Viruses of symbiotic Chlorella-like algae isolated from Paramecium bursaria and Hydra viridis. Proc Natl Acad Sci U S A 79:3867–3871. doi: 10.1073/pnas.79.12.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]