Two billion people have been infected with HBV worldwide; about 240 million infected patients developed chronic hepatitis B (CHB), and 650,000 die each year from liver cirrhosis (LC) or hepatocellular carcinoma (HCC). This work elucidates a mechanism underlying the control of HBV replication. HBV infection activates HoxA10, a regulator of cell differentiation and cancer progression, in human cells and patients with CHB and HCC. HoxA10 subsequently inhibits HBV replication in human tissue culture cells and mice. Additionally, HoxA10 interacts with p38 MAPK to repress the activation of p38 MAPK and STAT3 and recruits and facilitates SHP-1 to catalyze the dephosphorylation of p38 MAPK and STAT3. Moreover, HoxA10 competes with STAT3 for binding of the HBV X promoter to repress HBV transcription. Thus, this work reveals a negative regulatory mechanism underlying the control of HBV replication and provides new insights into the development of potential agents to control HBV infection.
KEYWORDS: hepatitis B virus infection and pathogenesis, homeobox protein A10, HoxA10, SH2-containing protein tyrosine phosphatase 1, SHP-1, signal transducer and activator of transcription 3, p38 MAPK-STAT3 signaling pathway, mitogen-activated protein kinase
ABSTRACT
Hepatitis B virus (HBV) infection is the leading cause of chronic hepatitis B (CHB), liver cirrhosis (LC), and hepatocellular carcinoma (HCC). This study reveals a distinct mechanism underlying the regulation of HBV replication. HBV activates homeobox A10 (HoxA10) in human hepatocytes, leukocytes, peripheral blood mononuclear cells (PBMCs), HepG2-NTCP cells, leukocytes isolated from CHB patients, and HBV-associated HCC tissues. HoxA10 in turn represses HBV replication in human hepatocytes, HepG2-NTCP cells, and BALB/c mice. Interestingly, we show that during early HBV infection, p38 mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription 3 (STAT3) were activated to facilitate HBV replication; however, during late HBV infection, HoxA10 was induced to attenuate HBV replication. Detailed studies reveal that HoxA10 binds to p38 MAPK, recruits SH2-containing protein tyrosine phosphatase 1 (SHP-1) to facilitate SHP-1 in catalyzing dephosphorylation of p38 MAPK/STAT3, and thereby attenuates p38 MAPK/STAT3 activation and HBV replication. Furthermore, HoxA10 binds to the HBV enhancer element I (EnhI)/X promoter, competes with STAT3 for binding of the promoter, and thereby represses HBV transcription. Taken together, these results show that HoxA10 attenuates HBV replication through repressing the p38 MAPK/STAT3 pathway by two approaches: HoxA10 interacts with p38 MAPK and recruits SHP-1 to repress HBV replication, and HoxA10 binds to the EnhI/X promoter and competes with STAT3 to attenuate HBV transcription. Thus, the function of HoxA10 is similar to the action of interferon (IFN) in terms of inhibition of HBV infection; however, the mechanism of HoxA10-mediated repression of HBV replication is different from the mechanism underlying IFN-induced inhibition of HBV infection.
IMPORTANCE Two billion people have been infected with HBV worldwide; about 240 million infected patients developed chronic hepatitis B (CHB), and 650,000 die each year from liver cirrhosis (LC) or hepatocellular carcinoma (HCC). This work elucidates a mechanism underlying the control of HBV replication. HBV infection activates HoxA10, a regulator of cell differentiation and cancer progression, in human cells and patients with CHB and HCC. HoxA10 subsequently inhibits HBV replication in human tissue culture cells and mice. Additionally, HoxA10 interacts with p38 MAPK to repress the activation of p38 MAPK and STAT3 and recruits and facilitates SHP-1 to catalyze the dephosphorylation of p38 MAPK and STAT3. Moreover, HoxA10 competes with STAT3 for binding of the HBV X promoter to repress HBV transcription. Thus, this work reveals a negative regulatory mechanism underlying the control of HBV replication and provides new insights into the development of potential agents to control HBV infection.
INTRODUCTION
Hepatitis B virus (HBV) is the leading cause of chronic hepatitis B (CHB), liver cirrhosis (LC), and hepatocellular carcinoma (HCC) and is the second leading cause of cancer mortality worldwide (1). Approximately 240 million people are estimated to be chronically infected by HBV, and more than 650,000 people die annually due to HBV-associated liver failure (2). HBV is a prototype member of the hepadnavirus family with a compact DNA genome replicating by the viral reverse transcriptase from an RNA intermediate (3). The virus genome is a circular, partially double-stranded DNA containing four open reading frames (ORFs) encoding seven proteins: three surface proteins (LHBs, MHBs, and SHBs), hepatitis B core antigen (HBcAg), hepatitis B e antigen (HBeAg), the viral polymerase (HBp), and the X protein (HBx) (4). Transcription of the HBV genome is initiated by four promoters, core, pre-S1, pre-S2, and X, which direct the transcriptions of 3.5-kb mRNAs for HBcAg, HBeAg, and HBp; 2.4-kb mRNA for LHBs; 2.1-kb mRNAs for MHBs and SHBs; and 0.7-kb mRNA for HBx (5, 6). These promoters are regulated by two liver-specific enhancers (enhancer element I [EnhI] and EnhII) (7, 8). EnhI spans a sequence of about 300 bp overlapping the X promoter, which consists of multiple transcription factor-binding sites. EnhII is 148 bp long and is located upstream of the core promoter (9, 10). Transcriptional activities of HBV promoters are mostly regulated by cellular factors or cytokines, including hepatocyte nuclear factor 4 (HNF-4), peroxisome proliferator-activated receptor (PPAR), retinoid X receptor (RXR), and signal transducer and activator of transcription 3 (STAT3) (11).
Homeobox genes encode a set of highly conserved transcription factors that are arranged in four clusters (clusters A to D), and among them, homeobox A10 (HoxA10) belongs to the A cluster (12). The homeobox domain of HoxA10 is structurally related to the helix-turn-helix motif of prokaryotic DNA-binding proteins, which regulates gene expression during differentiation and cancer progression (13). Disordered expression of HoxA10 has been found in leukemia and breast, cervical, ovarian, and pancreatic cancers (14–16). However, the role of HoxA10 in pathogen infection and HCC development has not been reported.
HBV infection activates mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinase (JNK), and p38 MAPK (17–19). p38 MAPK is activated following phosphorylation at Thr180/Tyr182, which is mediated primarily by upstream mitogen-activated protein kinase kinase 3 (MKK3) and MKK6. Phosphorylated p38 MAPK (phosphor-p38 MAPK) translocates to the nucleus and mediates the specific binding of STAT3, hepatocyte nuclear factor 3 (HNF3), HNF4, PPAR, and RXR to HBV EnhI/X (20), leading to an overall stimulation of HBV gene expression.
Tyrosyl phosphorylation is a reversible process whereby dephosphorylation is catalyzed by a set of protein-tyrosine phosphatases (PTPs), including SH2-containing protein tyrosine phosphatase 1 (SHP-1), SHP-2, and PTP1B (21–23). Among them, SHP-1 is a dephosphorylase that was implicated in the regulation of cell functions, including nuclear localization (24), lipid raft targeting (25), and cell signaling regulation (26), and is recognized as a negative regulator of signal transduction pathways (27, 28). SHP-1 has the capacity for dephosphorylation and acts as an inhibitory modulator of MAPKs, as implicated in the JNK and p38 pathways (29, 30). Dephosphorylation of phosphor-p38 MAPK results in the loss of MAPK activity and thus suppresses the activation of signal transduction.
In this study, we demonstrate that HBV infection initially activates p38 MAPK phosphorylation and subsequently induces HoxA10 production, which in turn attenuates HBV replication through a unique negative regulatory mechanism. Interestingly, HoxA10 interacts with p38 MAPK and then recruits SHP-1 to facilitate this tyrosine phosphatase in catalyzing the dephosphorylation of p38 MAPK and STAT3, leading to the repression of HBV replication. Moreover, HoxA10 binds to the X/EnhI promoter and competes with STAT3 for binding of the promoter, resulting in the repression of HBV transcription. This study provides new insights into the mechanism underlying the regulation of HBV replication and chronic HBV infection.
RESULTS
HBV infection induces HoxA10 expression both in vitro and in vivo.
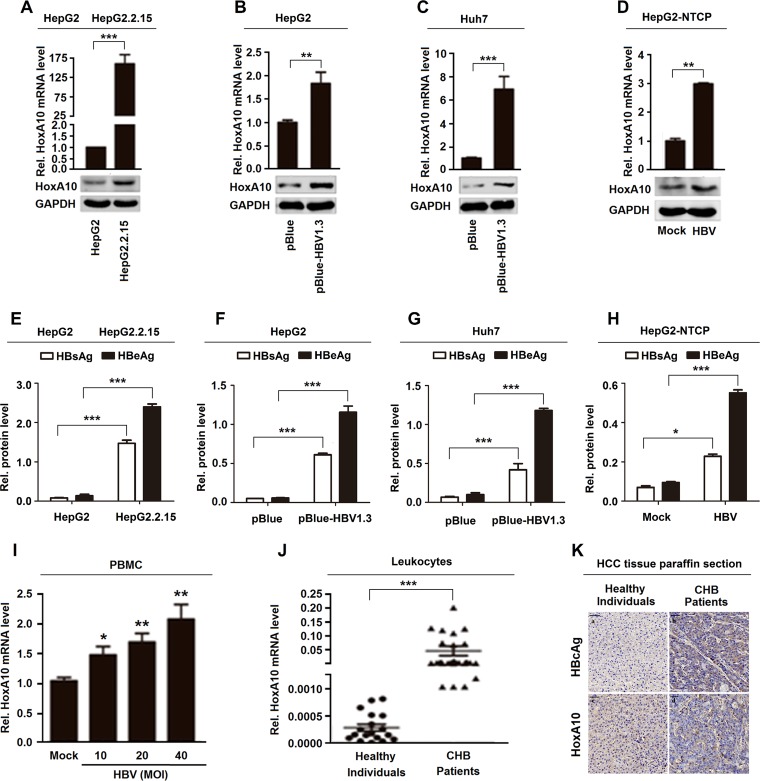
We initially investigated the effect of HBV infection on HoxA10 expression in vitro and in vivo. HoxA10 mRNA and HoxA10 protein levels were significantly higher in HepG2.2.15 cells (carrying an integrated HBV genome) than in HepG2 cells (Fig. 1A), promoted in pBlue-HBV1.3-transfected HepG2 cells (Fig. 1B), significantly induced in pBlue-HBV1.3-transfected Huh7 cells (Fig. 1C), and promoted in HBV-infected HepG2-NTCP cells (a well-established HBV infection system) (Fig. 1D). HBsAg and HBeAg were detected in HepG2.2.15 cells but not in HepG2 cells (Fig. 1E), expressed in pBlue-HBV1.3-transfected HepG2 cells but not in pBlue-transfected HepG2 cells (Fig. 1F), produced in pBlue-HBV1.3-transfected Huh7 cells but not in pBlue-transfected cells (Fig. 1G), and expressed in HBV-infected HepG2-NTCP cells but not in mock-infected HepG2-NTCP cells (Fig. 1H), confirming robust HBV replication or infection in the cells. Collectively, these results suggest that HBV facilitates HoxA10 expression in hepatocytes. In addition, HoxA10 mRNA was induced by HBV in human peripheral blood mononuclear cells (PBMCs) in a dose-dependent manner (Fig. 1I), revealing that HBV infection induces HoxA10 production in human PBMCs. Additionally, HoxA10 is significantly activated in leukocytes isolated from CHB patients (n = 32) compared to healthy individuals (n = 18) (Table 1 and Fig. 1J). Furthermore, HBcAg and HoxA10 levels were significantly higher in HBV-associated HCC tissue paraffin sections (n = 2) than in liver tissue paraffin sections from healthy individuals (n = 2) (Table 2 and Fig. 1K). These results demonstrate that HBV infection activates HoxA10 production both in vitro and in vivo.
FIG 1.
HBV induces HoxA10 expression in cultured cells and patients. (A to D) HoxA10 mRNA and HoxA10 protein expressed in HepG2 and HepG2.2.15 cells (A), HepG2 cells transfected with pBlue-HBV1.3 for 48 h (B), Huh7 cells transfected with pBlue-HBV1.3 for 48 h (C), and HepG2-NTCP cells infected with HBV by inoculation with the supernatants of HepaAD38 cells for 96 h (D) were measured by qPCR (top) and Western blotting (bottom), respectively. (E to H) HBsAg and HBeAg proteins produced in HepG2 and HepG2.2.15 cells (E), HepG2 cells transfected with pBlue-HBV1.3 for 48 h (F), Huh7 cells transfected with pBlue-HBV1.3 for 48 h (G), and HepG2-NTCP cells infected with HBV by inoculation with the supernatants of HepaAD38 cells for 96 h (H) were measured by an ELISA to confirm HBV infection and replication. (I) PBMCs (4 × 106) isolated from healthy individuals were incubated with supernatants of HepG2 cells or supernatants of HepaAD38 cells for 48 h. HoxA10 mRNAs were measured by qPCR. MOI, multiplicity of infection. (J) HoxA10 mRNAs in the leukocytes of CHB patients (n = 32) and healthy individuals (n = 18) were measured by qPCR. (K) Immunohistochemical staining of HoxA10 in paraffin sections of HBV-associated HCC tissues (n = 2) and liver tissues from healthy individuals. (n = 2). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 1.
Characteristics of HBV-infected patients and healthy individualsa
| Characteristic | Value for group |
|
|---|---|---|
| Patients (CHB) (n = 32) | Healthy individuals (n = 18) | |
| No. (%) of individuals positive for HBeAg | 32 (100) | 0 (0) |
| No. (%) of individuals positive for anti-HBeAg | 0 (0) | —b |
| No. (%) of individuals positive for HBsAg | 32 (100) | —b |
| Mean ALT level (U/ml) | >30 | <30 |
| No. (%) of male individuals | 24 (75) | 9 (50) |
ALT, alanine aminotransferase; CHB, chronic hepatitis B.
Not tested.
TABLE 2.
Characteristics of CHB patients and healthy individuals
| Sample | Gender of individual | Country | Presence of HBcAg | Organ |
|---|---|---|---|---|
| N0008D1900-B30-9 | Male | China | No | Liver |
| N0004D1900-B30-4 | Male | China | No | Liver |
| D19A1261-B30-C2 | Male | China | + | Liver |
| D19A3812-B30-C01 | Male | China | + | Liver |
HoxA10 represses HBV replication in hepatocytes.
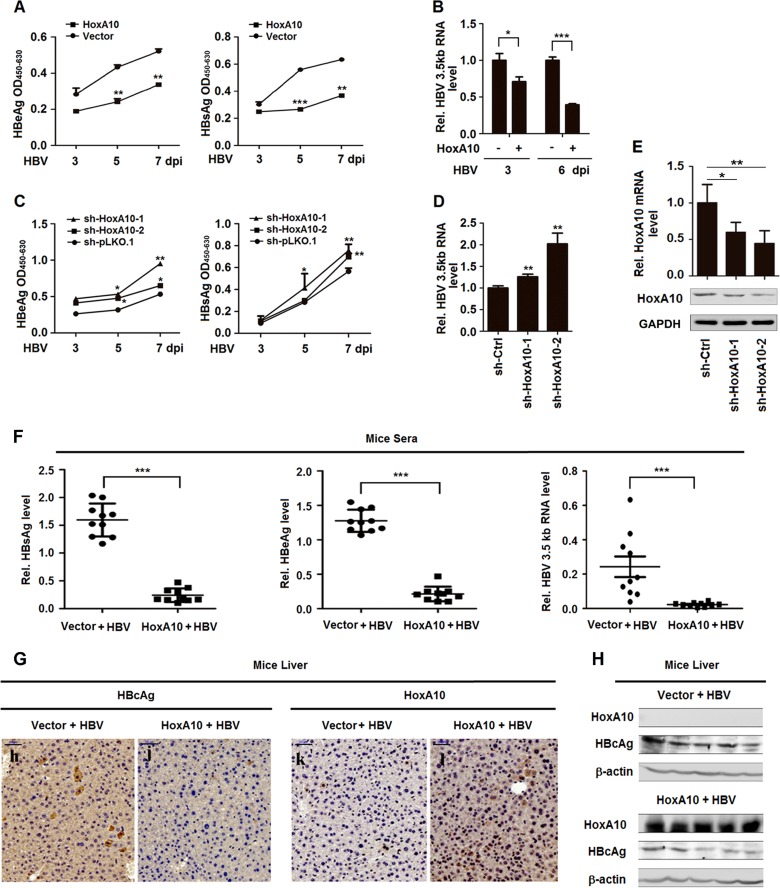
Although HoxA10 plays important roles in regulating multiple biological activities (31, 32), its function in viral replication has not been reported. Since HoxA10 is regulated by HBV, we speculated that HoxA10 may play a role in HBV replication. Initially, the effect of HoxA10 overexpression on HBV replication was determined in HepG2 cells and Huh7 cells cotransfected with pBlue-HBV1.3 and pFlag-HoxA10 or pCMV-3×Flag. The results showed that HBV RNAs (Fig. 2A), secreted HBeAg and HBsAg proteins (Fig. 2B), HBV core-associated DNA (Fig. 2C), and HBV covalently closed circular DNA (cccDNA) (Fig. 2D) were downregulated by HoxA10 in HepG2 and Huh7 cells, suggesting that HoxA10 attenuates HBV replication. HoxA10 protein was detected in HoxA10-transfected HepG2 cells and Huh7 cells but not in untreated cells (Fig. 2E and F), confirming that HoxA10 transfection is effective.
FIG 2.
HoxA10 represses HBV replication in hepatocytes. (A to F) HepG2 and Huh7 cells were cotransfected with different concentrations of Flag-HoxA10 and pBlue-HBV1.3 for 48 h. (G, H, K, and L) HepG2 and Huh7 cells were cotransfected with different concentrations of si-HoxA10-1 and pBlue-HBV1.3. (I, J, and M) HepG2 and Huh7 cells were cotransfected with sh-HoxA10-1 or sh-HoxA10-2 and pBlue-HBV1.3 for 48 h. (A and G) Total RNAs were extracted from the cells, and HBV RNAs were measured by Northern blot analysis. (B and H) Secreted HBsAg and HBeAg in cell culture supernatants were detected by using an ELISA kit. (C and I) HBV core-associated DNAs were extracted and measured by qPCR. (D and J) HBV cccDNAs were extracted from the cells and measured by qPCR. (E to L) HoxA10 and GAPDH proteins were detected by Western blotting using the corresponding antibodies. (M) The levels of HoxA10 mRNA were measured by RT-PCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Additionally, the effect of HoxA10 knockdown on HBV replication was evaluated by generating and analyzing a small interfering RNA (siRNA) specifically targeting HoxA10 (siR-HoxA10) and a short hairpin RNA (shRNA) specifically targeting HoxA10 (sh-HoxA10). HepG2 cells and Huh7 cells were cotransfected with pBlue-HBV1.3 and siR-HoxA10 or control siRNA (siR-Ctrl) or treated with sh-HoxA10 or sh-Ctrl. HBV RNAs (Fig. 2G), secreted HBeAg and HBsAg proteins (Fig. 2H), core-associated DNA (Fig. 2I), and HBV cccDNA (Fig. 2J) were facilitated by sh-HoxA10 in HepG2 and Huh7 cells, indicating that knockdown of HoxA10 facilitates HBV replication. The levels of HoxA10 protein were downregulated by siR-HoxA10 in HepG2 and Huh7 cells (Fig. 2K and L), revealing that siR-HoxA10 is effective. Similarly, the levels of HoxA10 mRNAs were attenuated by sh-HoxA10-1 and sh-HoxA10-2 in HepG2 and Huh7 cells (Fig. 2M), demonstrating that sh-HoxA10 is effective. Taken together, these results reveal that HoxA10 represses HBV replication in hepatocytes.
HoxA10 inhibits HBV infection in the HepG2-NTCP infection system and mice.
The effect of HoxA10 on HBV infection was further determined in the HepG2-NTCP infection system and in mice. Initially, HepG2-NTCP cells were transfected with pFlag-HoxA10 and then infected with HBV by inoculation with the supernatants of HepaAD38 cells, which were concentrated, purified, and identified as being free of other contaminants. In HBV-infected HepG2-NTCP cells, HBV HBeAg and HBsAg proteins (Fig. 3A) and HBV 3.5-kb RNA (Fig. 3B) were downregulated by HoxA10, indicating that the overexpression of HoxA10 leads to the attenuation of HBV infection. Additionally, HepG2-NTCP cells were transfected with sh-HoxA10-1 or sh-HoxA10-2 and then infected with HBV. In treated cells, HBeAg and HBsAg proteins (Fig. 3C) and HBV 3.5-kb RNA (Fig. 3D) were upregulated by sh-HoxA10-1 and sh-HoxA10-2, suggesting that the knockdown of HoxA10 results in the promotion of HBV infection. The levels of HoxA10 mRNA (Fig. 3E, top) and HoxA10 protein (Fig. 3E, bottom) were attenuated by sh-HoxA10-1 and sh-HoxA10-2 in HepG2-NTCP cells, revealing that both sh-HoxA10-1 and sh-HoxA10-2 are effective. Therefore, we reveal that HoxA10 represses HBV infection in the HepG2-NTCP infection system.
FIG 3.
HoxA10 attenuates HBV replication in HepG2-NTCP cells and mice. (A to E) HepG2-NTCP cells were transfected with Flag-HoxA10 for 24 h (A and B) or with sh-Ctrl, sh-HoxA10-1, or sh-HoxA10-2 for 24 h (C to E) and then inoculated with HBV for different times, as indicated, in the presence of 4% PEG 8000. (A and C) The cells were collected at the indicated times, washed five times with PBS, and cultured in PMM, and the levels of HBsAg and secreted HBeAg in the supernatants were determined by using an ELISA kit. OD450-630, optical density at 450 to 630 nm. (B and D) Total cellular RNAs were extracted from the cells, and HBV 3.5-kb RNA was measured by qPCR. (E) Total cellular RNAs were extracted, HoxA10 mRNA was quantitated by qPCR (top), and HoxA10 protein was detected by Western blotting (bottom). (F to H) BALB/c mice (10 in each group) were coinjected with pAAV-HBV and pCMV-3×Flag-HoxA10 or pCMV-3×Flag by hydrodynamic injection. Six days after injection, the mice were sacrificed, and livers and blood were processed for analyses. (F) Levels of secreted HBsAg (left) and HBeAg (middle) in mouse sera were determined by using an ELISA kit, and HBV RNA levels (right) in mouse liver tissues were determined by qPCR. (G) Immunohistochemical staining of HBcAg and HoxA10 in mouse livers. (H) Levels of HoxA10 and HBcAg proteins expressed in mouse liver were determined by Western blotting. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Moreover, the effect of HoxA10 on HBV infection was investigated in mice. BALB/c mice (10 in each group) were treated with pAAV-HBV and pFlag-HoxA10 through hydrodynamic injection. At 6 days postinjection, the mice were sacrificed, and blood and livers were collected. In the sera of treated mice, HBsAg, HBeAg, and HBV 3.5-kb RNA were significantly repressed by HoxA10 (Fig. 3F). Additionally, in the liver of treated mice, the HBcAg protein level was significantly reduced in the presence of HoxA10 (Fig. 3G and H). Taken together, our results demonstrate that HoxA10 inhibits HBV infection in HepG2-NTCP cells and mice.
HoxA10 attenuates HBV replication by repressing the p38 MAPK/STAT3 pathway.
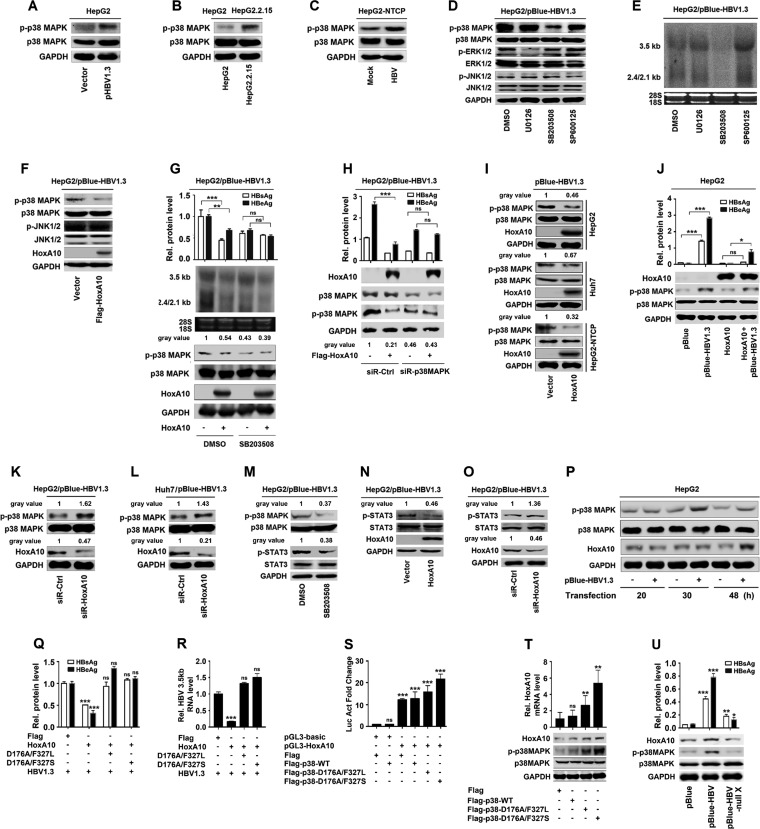
The molecular mechanism by which HoxA10 represses HBV replication was investigated. Previous studies reported that HBV infection activates the MAPK pathways (33–35). Here, we show that the phosphorylation of p38 MAPK was upregulated by HBV in HepG2 cells (Fig. 4A), significantly stimulated in HepG2.2.15 cells compared to HepG2 cells (Fig. 4B), and enhanced by HBV infection in HepG2-NTCP cells (Fig. 4C), suggesting that HBV induces p38 MAPK phosphorylation. HBV-induced phosphorylation of ERK, JNK, and p38 MAPK was attenuated by U0126 (ERK1/2 inhibitor), SB203508 (p38 MAPK inhibitor), or SP600125 (JNK1/2 inhibitor) (Fig. 4D). HBV RNAs (3.5-kb and 2.4/2.1-kb RNAs) were upregulated by U0126 and SP600125 but downregulated by SB203508 (Fig. 4E), revealing that p38 MAPK is involved in the activation of HBV replication.
FIG 4.
HoxA10 inhibits HBV replication by repressing p38 MAPK activity. (A) HepG2 cells were transfected with pBlue-HBV1.3 for 48 h. (B) HepG2 cells and HepG2.2.15 cells were cultured for 48 h. (C) HepG2-NTCP cells were mock infected or infected with HBV at 1,000 GEq for 96 h. Levels of phosphorylated p38 MAPK protein, total p38 MAPK, and GAPDH proteins were determined by Western blot analyses using the corresponding antibodies. (D and E) HepG2 cells were transfected with pBlue-HBV1.3 and treated with 1% DMSO or 20 μM inhibitors of ERK (U0126), p38 MAPK (SB203508), or JNK (SP600125) for 2 h. (D) Phosphorylated and total ERK1/2, JNK1/2, p38 MAPK, and GAPDH were detected by Western blotting. (E) HBV RNA levels (3.5 kb and 2.4 kb/2.1 kb) were measured by Northern blotting, and 18S and 28S rRNAs were used as loading controls. (F) HepG2 cells were cotransfected with pBlue-HBV1.3 and Flag-HoxA10. Levels of phosphorylated p38 MAPK (p-p38) and JNK1/2 (p-JNK1/2) and total p38 MAPK, JNK1/2, HoxA10, and GAPDH proteins were determined by Western blot analyses using the corresponding antibodies. (G) HepG2 cells were transfected with pBlue-HBV1.3 and Flag-HoxA10 or pCMV-3×Flag and treated with SB203508 or 1% DMSO. (Top) Levels of secreted HBsAg and HBeAg in supernatants were determined by using an ELISA kit. (Middle) HBV RNA levels in treated cells were measured by Northern blotting. (Bottom) HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were detected by Western blotting. (H) HepG2 cells were cotransfected with pBlue-HBV1.3 and Flag-HoxA10 or pCMV-3×Flag and treated with siR-p38 MAPK or siR-Ctrl. (Top) Secreted HBsAg and HBeAg levels in supernatants were determined by using an ELISA kit. (Bottom) HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were detected by Western blotting. (I, top and middle) HepG2 and Huh7 cells were cotransfected with pBlue-HBV1.3 and Flag-HoxA10 or the vector for 48 h. (Bottom) HepG2-NTCP cells were treated with Flag-HoxA10 or the vector and infected with HBV for 96 h. Phosphorylated p38 MAPK, total p38 MAPK, HoxA10, and GAPDH were detected with specific antibodies by Western blotting. (J) HepG2 cells were transfected with pBlue and pBlue-HBV1.3 and cotransfected with HoxA10 and pBlue or pBule-HBV1.3. (Top) Secreted HBsAg and HBeAg in the cell culture supernatants were detected by using an ELISA kit. (Bottom) HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were detected by Western blotting. (K and L) HepG2 cells (K) and Huh7 cells (L) were transfected with pBlue-HBV1.3 and siR-HoxA10 or siR-Ctrl (50 nM) for 48 h. Protein levels of HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were determined by Western blotting. (M to O) HepG2 cells were transfected with pBlue-HBV1.3 and treated with DMSO or SB203508 for 2 h (M), cotransfected with pBlue-HBV1.3 and Flag-HoxA10 for 48 h (N), or cotransfected with pBlue-HBV1.3 and siR-HoxA10 for 48 h (O). Phosphorylated p38 MAPK and STAT3, total p38 MAPK and STAT3, HoxA10, and GAPDH were detected with specific antibodies by Western blotting. (P) Effects of different times after HBV infection on the p38 MAPK pathway and expression of HoxA10. HepG2 cells were transfected with 1 μg pBlue or pBlue-HBV1.3 for different times, as indicated. The cells were collected, and the cell lysates were prepared. The levels of phosphorylated p38 MAPK, total p38 MAPK, HoxA10, and GAPDH were determined by Western blotting using the corresponding antibodies. (Q and R) HepG2 cells were cotransfected with pBule-HBV1.3 and HoxA10, double mutants of p38 (D176A/327L and D176A/327S), or Flag for 48 h. (Q) Secreted HBsAg and HBeAg in the cell culture supernatants were detected by using an ELISA kit. (R) Total RNAs were extracted from the cells, and HBV RNA levels were measured by qPCR. (S) HepG2 cells were cotransfected with pGL3-HoxA10 reporter plasmids, double mutants of p38 (D176A/327L and D176A/327S), and wild-type p38 (p38-wt) for 48 h. Luciferase activity was measured and normalized to the control. (T) HepG2 cells were transfected with Flag, p38-wt, and double mutants of p38 (D176A/327L and D176A/327S) for 48 h. (Top) Total RNAs were extracted from the cells, and HoxA10 mRNA levels were measured by qPCR. (Bottom) HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were detected by Western blotting. (U) HepG2 cells were transfected with pBlue, pBlue-HBV1.3, and pBlue-HBV-null-X for 72 h. HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were detected by Western blotting. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, the role of HoxA10 in the regulation of p38 MAPK was evaluated. HoxA10 significantly attenuated HBV-induced phosphorylation of p38 MAPK but not HBV-induced phosphorylation of JNK1/2 (Fig. 4F), suggesting that HoxA10 specifically represses HBV-induced activation of p38 MAPK. HepG2 cells were cotransfected with pBlue-HBV1.3 and Flag-HoxA10 or pFlag and treated with SB203508 or dimethyl sulfoxide (DMSO). HBsAg and HBeAg (Fig. 4G, top, lane 2 versus lane 1), HBV RNAs (Fig. 4G, middle, lane 2 versus lane 1), and p38 MAPK phosphorylation (Fig. 4G, bottom, lane 2 versus lane 1) were downregulated by HoxA10; however, in the presence of SB203508, HBsAg and HBeAg proteins (Fig. 4G, top, lane 4 versus lane 3), HBV RNAs (Fig. 4G, middle, lane 4 versus lane 3), and p38 MAPK phosphorylation (Fig. 4G, bottom, lane 4 versus lane 3) were relatively unaffected by HoxA10. Similarly, HBsAg and HBeAg (Fig. 4H, top) and p38 MAPK phosphorylation (Fig. 4H, bottom) were downregulated by HoxA10; however, in the presence of siR-p38 MAPK, HBsAg and HBeAg (Fig. 4H, top) and p38 MAPK phosphorylation (Fig. 4H, bottom) were relatively unaffected by HoxA10. Collectively, these results indicate that HoxA10 attenuates HBV replication through repressing p38 MAPK activation. HBV-induced phosphorylation of p38 MAPK was downregulated by HoxA10 in HepG2 cells (Fig. 4I, top), Huh7 cells (Fig. 4I, middle), and HepG2-NTCP cells (Fig. 4I, bottom). In addition, HBsAg and HBeAg (Fig. 4J, top) and p38 MAPK phosphorylation (Fig. 4J, bottom) were activated by HBV but downregulated by HoxA10, and HBV-activated p38 MAPK phosphorylation was attenuated by HoxA10 in HepG2 cells (Fig. 4J). Moreover, HBV-induced phosphorylation of p38 MAPK was upregulated by siR-HoxA10 in HepG2 cells (Fig. 4K) and Huh7 cells (Fig. 4L). Collectively, these results indicate that HoxA10 attenuates HBV replication through repressing p38 MAPK activation.
The p38 MAPK mediates the phosphorylation of STAT3, which then binds to the X/EnhI promoter of HBV to promote virus replication (11, 20). Here, we show that HBV activated the phosphorylation of p38 MAPK and STAT3, and HBV-mediated phosphorylation of STAT3 was attenuated by SB203508 (p38 MAPK inhibitor) (Fig. 4M), confirming that p38 MAPK is involved in HBV-mediated phosphorylation of STAT3. In addition, HBV-mediated phosphorylation of STAT3 was downregulated by HoxA10 (Fig. 4N) but upregulated by siR-HoxA10 (Fig. 4O). Thus, we demonstrate that HoxA10 attenuates HBV replication through repressing the p38 MAPK/STAT3 pathway. Furthermore, the effects of HBV on the temporal activation of endogenous p38 MAPK and HoxA10 were evaluated in HepG2 cells transfected with pBlue-HBV1.3. Interestingly, phosphorylation of endogenous p38 MAPK was not induced by HBV at 20 h posttransfection, significantly activated by HBV at 30 h posttransfection, and slightly induced by HBV at 48 h posttransfection, whereas production of endogenous HoxA10 was not induced by HBV at 20 and 30 h posttransfection but significantly induced by HBV at 48 h posttransfection (Fig. 4P). These results suggest that HBV initially activates p38 MAPK phosphorylation at 30 h posttransfection and subsequently induces HoxA10 production at 48 h posttransfection, which in turn attenuates p38 MAPK phosphorylation.
To explore whether and how p38 MAPK phosphorylation regulates HBV replication and HoxA10 expression, we constructed and analyzed two phosphomimetic mutants of p38 MAPK, D176A/F327L and D176A/F327S (36). The results showed that HBsAg and HBeAg (Fig. 4Q) and HBV 3.5-kb RNA (Fig. 4R) were repressed by HoxA10 but relatively unaffected by HoxA10 in the presence of p38 D176A/F327L and p38 D176A/F327S (Fig. 4Q and R), indicating that p38 MAPK phosphorylation is involved in the regulation of HBV replication. In addition, the HoxA10 promoter was subcloned into pGL3-Basic to generate the luciferase (Luc) reporter pGL3-HoxA10. The activity of the Luc-HoxA10 promoter was upregulated by p38 D176A/F327L and p38 D176A/F327S (Fig. 4S), and HoxA10 mRNA was significantly activated by p38 D176A/F327L and p38 D176A/F327S (Fig. 4T), suggesting that p38 MAPK phosphorylation participates in the regulation of the HoxA10 promoter and regulates HoxA10 at the transcriptional level. Furthermore, we speculated that the X protein of HBV is involved in the regulation of HoxA10 and p38 MAPK. The results revealed that HoxA10 protein production and p38 MAPK phosphorylation were induced by wild-type (WT) HBV but not by HBV-null X (Fig. 4U), revealing that the X protein is required for the regulation of HoxA10 production and p38 MAPK phosphorylation.
HoxA10 interacts with p38 MAPK to form a complex in nuclei.
The mechanism by which HoxA10 represses the p38 MAPK pathway was elucidated. An interaction between HoxA10 and p38 MAPK may exist, as predicted previously based on affinity purification mass spectrometry (AP-MS) (37). Here, we show that HoxA10 and p38 MAPK interacted with each other (Fig. 5A), endogenous HoxA10 interacted with endogenous p38 MAPK (Fig. 5B), and HoxA10 was pulled down by purified glutathione S-transferase (GST)-p38 MAPK (Fig. 5C). All the results confirm that HoxA10 directly interacts with and binds to p38 MAPK. To determine whether the phosphorylation sites of p38 MAPK play roles, if any, in the interaction with HoxA10, three phosphorylation site mutants (Thr180Ala, Tyr182Ala, and Thr/Tyr180/182Ala/Ala) of p38 MAPK were generated. Like wild-type p38 MAPK, the mutant p38 MAPK proteins interacted with HoxA10 (Fig. 5D), indicating that the phosphorylation sites of p38 MAPK are not involved in the interaction with HoxA10. Laser scanning confocal microscopy showed that in the presence of a single protein, HoxA10 mainly localized in nuclei (Fig. 5Ea to d), and p38 MAPK was distributed in the cytoplasm and nuclei (Fig. 5Ee to h), whereas in the presence of two proteins, HoxA10 and a large proportion of p38 MAPK colocalized in the nuclei (Fig. 5Ei to l), suggesting that HoxA10 interacts with p38 MAPK to form a complex in the nuclei. Moreover, HoxA10 could not interact with STAT3 (Fig. 5F), and similarly, SHP-1 failed to interact with STAT3 (Fig. 5G). Therefore, our results suggest that HoxA10 recruits SHP-1 to dephosphorylate p38 MAPK, leading to suppression of HBV transcription.
FIG 5.
HoxA10 interacts with p38 MAPK in nuclei. (A to D) HEK293T cells were cotransfected with hemagglutinin (HA)-p38 MAPK and Flag-HoxA10. (A) Immunoprecipitation (IP) of cell extracts was performed by using anti-Flag/HA antibody. (B) Huh7 cells were collected and lysed. Lysates were immunoprecipitated using HoxA10 antibody. (C) GST pulldown assay. Expression of GST-p38 MAPK or GST alone was induced by incubation with GST-protein A/G beads. After incubation, the beads were incubated with Flag-HoxA10 cell extracts. (D) HEK293T cells were cotransfected with HA-HoxA10 and Flag-p38 MAPK (wt), Flag-p38-Thr180Ala, Flag-p38-Tyr182Ala, or Flag-p38-Thr/Tyr180/182Ala/Ala. Cells lysates were precipitated by using anti-HA/Flag antibody. (E) Coimmunoprecipitation of HoxA10/p38 MAPK was detected by Western blotting. HepG2 cells were fixed and immunostained with HoxA10 and p38 MAPK antibody. After the nucleus was stained by DAPI (4′,6-diamidino-2-phenylindole), the locations of the HoxA10 and p38 MAPK proteins were analyzed by immunofluorescence staining. (F and G) HEK293T cells were cotransfected with HA-STAT3 and Flag-HoxA10 (F) and with HA-STAT3 and Flag–SHP-1 (G). Immunoprecipitation of cell extracts was performed by using anti-HA/IgG antibody. HoxA10, STAT3, SHP-1, and HoxA10 were detected by Western blotting. IB, immunoblot.
HoxA10 recruits and facilitates SHP-1 to catalyze the dephosphorylation of p38 MAPK.
SHP-1 acts as an inhibitory modulator of MAPK signal pathways (26, 28). We show that HBV-induced phosphorylation of p38 MAPK was downregulated by SHP-1 (Fig. 6A), demonstrating that SHP-1 attenuates p38 MAPK phosphorylation. HBV-induced phosphorylation of p38 MAPK was significantly reduced by HoxA10 in the absence of siR-SHP-1 but was not affected by HoxA10 in the presence of siR-SHP-1 (Fig. 6B and C), revealing that SHP-1 is required for HoxA10-mediated dephosphorylation of p38 MAPK. p38 MAPK interacted with SHP-1 in the presence of HoxA10 but failed to interact with SHP-1 in the absence of HoxA10 (Fig. 6D), suggesting that HoxA10 is required for the interaction of SHP-1 with p38 MAPK. HoxA10 was pulled down by purified GST–SHP-1 (Fig. 6E), confirming that HoxA10 binds SHP-1. Taken together, our results demonstrate that HoxA10 recruits SHP-1 to facilitate SHP-1 in catalyzing the dephosphorylation of p38 MAPK.
FIG 6.
HoxA10 recruits SHP-1 to facilitate p38 MAPK dephosphorylation. (A and B) HepG2 cells were cotransfected with pBlue-HBV1.3 and different concentrations of HA–SHP-1 (A) or with pBlue-HBV1.3 and siR-SHP-1 and/or Flag-HoxA10 (B). Levels of SHP-1, HoxA10, phosphorylated p38 MAPK, total p38 MAPK, and GAPDH were measured by Western blotting with specific antibodies. (C) HepG2 cells were transfected with siR-Ctrl or si-SHP-1 at different concentrations, as indicated. The levels of SHP-1 mRNA and protein were quantified by qPCR (top) and Western blotting (bottom). (D) HEK293T cells were cotransfected with Flag-p38 MAPK/HA–SHP-1 and HA-HoxA10 or pCAGG-HA. Lysates were immunoprecipitated using anti-Flag or anti-IgG. Coimmunoprecipitation of HoxA10/p38 MAPK/SHP-1 was detected by Western blotting with anti-HA, anti-Flag, and anti-SHP-1. (E) GST pulldown assay. Purified GST–SHP-1 protein or GST protein was incubated with GST-protein A/G beads. The beads were then incubated with Flag-HoxA10-transfected cell extracts. Coimmunoprecipitation of HoxA10/SHP-1 was detected by Western blotting. *, P < 0.05; **, P < 0.01.
HoxA10 represses HBV replication by binding to the X/EnhI promoter.
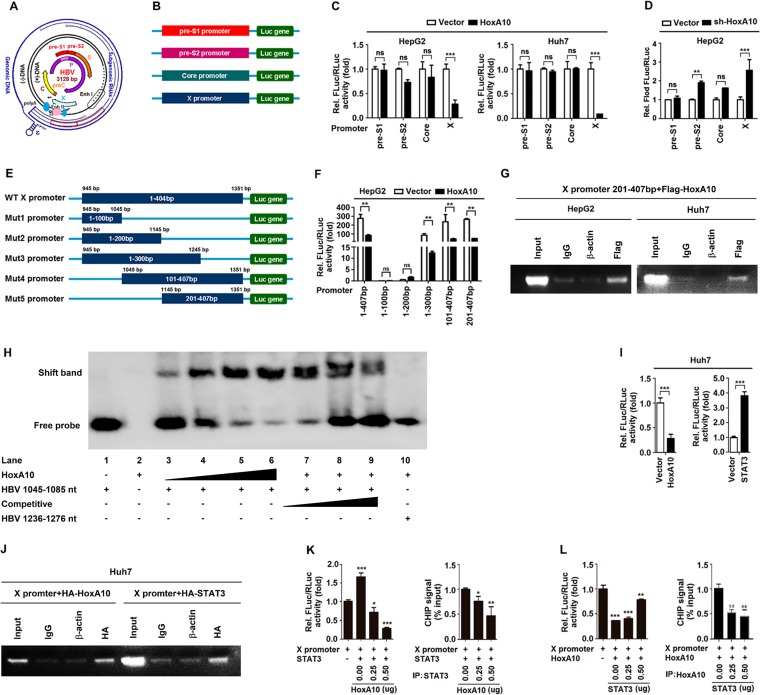
HBV genome transcription was controlled by four promoters: pre-S1, pre-S2, core, and X (Fig. 7A). The pre-S1, pre-S2, core, and X promoters were subcloned into pGL3-Basic to generate four reporters (Fig. 7B). We show that HoxA10 significantly downregulated the X promoter (contains enhancer I) activity but not the pre-S1, pre-S2, or core promoter in HepG2 cells (Fig. 7C, left) and Huh7 cells (Fig. 7C, right), whereas sh-HoxA10 significantly upregulated X promoter activity but not the pre-S1, pre-S2, or core promoter in HepG2 cells (Fig. 7D), revealing that HoxA10 represses X promoter activation. The X promoter regulates HBx gene transcription, which potentiates HBV replication, regulates HBV oncogenicity, and affects multiple cellular functions (38–41). We constructed the X/EnhI promoter and its five promoter-truncated mutants and then subcloned these constructs into the pGL3-Basic vector to generate six reporters (Fig. 7E). The activities of the WT X/EnhI promoter (bp 1 to 407), the mutant 3 (mut3) X/EnhI promoter (bp 1 to 300), the mut4 X/EnhI promoter (bp 101 to 407), and the mut5 X/EnhI promoter (bp 201 to 407) were reduced by HoxA10 (Fig. 7F), indicating that bp 201 to 407 of the X/EnhI promoter are required for the negative regulation of HBV replication mediated by HoxA10. Moreover, chromatin immunoprecipitation (CHIP) analyses demonstrated that HoxA10 could bind to the segment from bp 201 to 407 of the X/EnhI promoter (Fig. 7G). Thus, we reveal that HoxA10 downregulates HBV replication by binding to the X promoter to repress viral RNA transcription. Furthermore, an electrophoretic mobility shift assay (EMSA) was performed to detect the binding of HoxA10 to HBV X/EnhI. The results indicated that HoxA10 could directly bind to the DNA fragment from bp 101 to 141 of the X/EnhI promoter (nucleotides [nt] 1045 to 1085) (Fig. 7H). Therefore, we reveal that HoxA10 downregulates HBV replication by direct and indirect binding to the X promoter to repress viral RNA transcription.
FIG 7.
HoxA10 represses HBV replication by binding to the X promoter. (A) Schematic representations of HBV genomic DNA and RNAs, enhancer I and II, and four promoters, S1, S2, core, and X. DR represents the direct repeat. HBV promoters and enhancers are indicated. Each viral transcript is labeled with a poly(A) tail at the 3′ end. Multiple transcription initiation sites are indicated at the 5′ ends of the 3.5-, 2.1-, 2.4-, and 0.7-kb RNAs. (B) The four promoters were generated and subcloned into the pGL3-Basic vector to construct luciferase reporters. Luciferase activity and the activity of the inserted promoter were positively correlated. (C) HepG2 cells and Huh7 cells were cotransfected with Flag-HoxA10 and pre-S1, pre-S2, core, and X reporter plasmids for 48 h. (D) HepG2 cells were cotransfected with sh-HoxA10 and pre-S1, pre-S2, core, and X reporter plasmids for 48 h. Luciferase activity was measured and normalized to the control. (E) Schematic diagram of the WT X promoter and its truncated mutants (mut1 to mut5). The promoters were subcloned into the pGL3-Basic vector to generate the corresponding reporters. (F) HepG2 cells were cotransfected with the X promoter or its truncated mutants and HoxA10, as indicated. Luciferase activity was measured and normalized to the control. (G) HepG2 cells (left) and Huh7 cells (right) were cotransfected with the mut5 truncated mutant promoter (bp 201 to 407) and Flag-HoxA10. Immunoprecipitation of cell extracts was performed by using anti-Flag/IgG antibody. Protein-bound DNA fragments were extracted and detected by CHIP assays. (H) Electrophoretic mobility shift assays were performed to detect the binding of HoxA10 to HBV X/EnhI. Lanes 1 to 6, 5′ biotin-labeled HBV (nt 1045 to 1085) probe incubated with different concentrations of HoxA10 overexpression nuclear lysates from HEK293T cells; lanes 7 and 9, competition with different concentrations of an unlabeled cold probe for HBV (nt 1045 to 1085); lane 10, 5′ biotin-labeled HBV (nt 1236 to 1276) probe incubated with HoxA10 overexpression nuclear lysates. (I and J) Huh7 cells were cotransfected with the X promoter reporter and HA-HoxA10 or HA-STAT3. (I) Luciferase activity were measured and compared with that of vector cells. (J) CHIP assays were performed to analyze the ability of HoxA10 or STAT3 to bind the X promoter. (K) Huh7 cells were cotransfected with the X promoter reporter and HA-STAT3 and then with different concentrations of Flag-HoxA10 for 48 h. Immunoprecipitation of cell extracts was performed by using anti-STAT3 antibody. (L) Huh7 cells were cotransfected with the X promoter reporter and Flag-HoxA10 and then with different concentrations of HA-STAT3 for 48 h. Immunoprecipitation of cell extracts was performed by using anti-HoxA10 antibody. X promoter activities were measured by a luciferase assay (left), and CHIP signals were detected by qPCR (right). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
A previous report showed that STAT3 binds the X/EnhI promoter to promote HBV replication (11). Interestingly, we revealed that the X/EnhI promoter was significantly repressed by HoxA10 (Fig. 7I, left) but activated by STAT3, as expected (Fig. 7I, right), and both HoxA10 and STAT3 could bind the X/EnhI promoter (Fig. 7J). Moreover, luciferase assays demonstrated that X/EnhI promoter activity was induced by STAT3, but such activation was attenuated by HoxA10 in a dose-dependent manner. Luciferase assays (Fig. 7K, left) and CHIP analyses revealed that STAT3 could bind to the X/EnhI promoter, but such binding was attenuated by HoxA10 in a dose-dependent fashion (Fig. 7K, right). Furthermore, luciferase assays showed that HoxA10 attenuated X/EnhI promoter activity but that such repression was suppressed by STAT3 in a dose-dependent fashion (Fig. 7L, left), and CHIP analyses revealed that HoxA10 could bind to the X/EnhI promoter but that such repression was suppressed by STAT3 in a dose-dependent fashion (Fig. 7L, right). These results demonstrate that HoxA10 and STAT3 compete with each other for binding to the X/EnhI promoter and thereby regulate HBV replication.
Taken together, our results reveal that HoxA10 acts as a host negative regulator to attenuate HBV replication through repressing the p38 MAPK/STAT3 pathway by two approaches (Fig. 8). HoxA10 interacts with p38 MAPK and recruits SHP-1 to facilitate the dephosphorylation of p38 MAPK/STAT3 and thereby attenuates HBV replication. HoxA10 also directly binds to the HBV EnhI/X promoter and competes with STAT3 for binding to the promoter and thereby represses HBV transcription.
FIG 8.
Proposed mechanism of HoxA10-mediated inhibition of HBV replication. (Left) HBV life cycle. HBV virions enter the cell through the sodium taurocholate cotransporting polypeptide (NTCP) receptor and release rcDNA-containing nucleocapsids into cytoplasm. rcDNA is transported to the nucleus and forms cccDNA. Transcription of cccDNA by RNA polymerase (Pol) produces 4 pregenomic RNAs (pgRNAs) (3.5 kb, 2.4 kb, 2.1 kb, and 0.7 kb) encoding surface antigens, core or HBeAg, X, and DNA polymerase. The pgRNA is encapsidated, together with polymerase, and reverse transcribed into negative-strand DNA [(−)DNA] inside the nucleocapsid. Positive-strand DNA [(+)DNA] synthesis from (−)DNA generates rcDNA and forms HBV virions, which are released from the cells. (Right) HBV replication activation. In response to HBV infection or other stimuli, p38 MAPK is activated by phosphorylation, enters the nucleus, and phosphorylates STAT3. Activated STAT3 binds the EnhI/X promoter to facilitate HBV gene expression. (Middle) HBV replication repression. HBV activates HoxA10, which subsequently inhibits HBV replication through several strategies. First, HoxA10 directly interacts with p38 MAPK to repress the signaling pathway. Second, HoxA10 recruits SHP-1 to facilitate p38 MAPK dephosphorylation, which leads to repression of STAT3. Third, HoxA10 directly binds the HBV X promoter to compete with STAT3 and repress HBV genome transcription. TGF-β, transforming growth factor β; GPCR, G-protein-coupled receptor.
DISCUSSION
Although HBV infection is an important public health problem worldwide (39), the mechanisms of HBV pathogenesis are largely unknown. The aim of this study is to reveal the mechanism underlying the regulation of HBV replication. This work identifies a previously unrevealed mechanism by which HoxA10 represses HBV replication through attenuating the p38 MAPK/STAT3 pathway. We initially show that HBV infection induces HoxA10 production in cultured cells and patients. HoxA10 has multiple functions through binding to DNA and protein to regulate gene expression (42, 43), cell signaling (12, 44), development (45), reproduction (46), and the development of many cancers (14, 16, 47, 48). However, the role of HoxA10 in the regulation of pathogen infection and HCC development has not been reported. Here, for the first time, we provide evidence that HoxA10 represses HBV replication in cultured cells and in mice.
Previous studies reported that HBV activates MAPK signaling pathways (33, 34); p38 MAPK is activated by HBsAg, and inhibition of the pathway attenuates HBsAg production (33); HBx activates the p38 MAPK pathway, which in turn facilitates HBx-mediated STAT3 phosphorylation (35); and STAT3 directly binds to HBV X/EnhI to promote HBV replication (11). Here, we reveal that phosphorylation of p38 MAPK is induced by HBV in HepG2, HepG2.2.15, and HepG2-NTCP cells, confirming that HBV activates the p38 MAPK pathway. Interestingly, we show that HBV-mediated activation of p38 MAPK is attenuated by HoxA10 and thus suggest that HoxA10 plays an important role in the repression of p38 MAPK signaling upon HBV infection. Since HoxA10 regulates the p38 MAPK pathway to promote cell invasion in pancreatic cancer cells (49), we speculate that HoxA10 may also regulate p38 MAPK to promote cell invasion in hepatocytes.
In evaluating the mechanism by which HoxA10 regulates p38 MAPK, we initially demonstrate that HoxA10 and p38 MAPK interact with each other, and HBV-mediated phosphorylation of p38 MAPK is attenuated by HoxA10, suggesting that HoxA10 interacts with p38 MAPK to repress p38 MAPK activation. Further studies reveal that HBV-induced phosphorylation of p38 MAPK is repressed by the tyrosine phosphatase SHP-1, and p38 MAPK phosphorylation is attenuated by HoxA10 in the presence of SHP-1 but not in the absence of SHP-1, suggesting that HoxA10 attenuates p38 MAPK phosphorylation through regulating SHP-1. Interestingly, the interaction between p38 MAPK and SHP-1 occurred in the presence of HoxA10, indicating that HoxA10 binds to p38 MAPK to recruit SHP-1 and facilitate tyrosine phosphatase in catalyzing the dephosphorylation of p38 MAPK. It has been reported that sorafenib inhibits HBx-induced androgen receptor (AR) activity through activating SHP-1 (15). Thus, we suggest that SHP-1 catalyzes the dephosphorylation of p38 MAPK and plays a role in HoxA10-mediated repression of HBV replication. Moreover, evaluation of the effects of HBV on the temporal regulation of p38 MAPK and HoxA10 revealed that p38 MAPK phosphorylation was activated by HBV at 30 h posttransfection, whereas HoxA10 production was induced by HBV at 48 h posttransfection, suggesting that HBV initially activates p38 MAPK and subsequently induces HoxA10, which in turn attenuates p38 MAPK activation.
The transcription of the HBV genome is initiated by four promoters and controlled by two enhancers (4, 7, 17, 50). EnhI spans a sequence of about 300 nt adjacent to the X promoter and upregulates transcription in an orientation-independent manner. Several cellular factors, including HNF-1, HNF-4, PPAR, RXR, and STAT3, bind the X/EnhI promote to regulate HBV replication. SHP-1 regulates HoxA10 DNA-binding activity by direct interaction with HoxA10 (51). This study demonstrates that X promoter activity is inhibited by HoxA10. Moreover, HoxA10 and STAT3 compete with each other for binding of X/EnhI, thereby regulating X/EnhI activity. Thus, we propose that HoxA10 binds to X/EnhI to repress HBV RNA transcription.
In conclusion, we reveal a negative-feedback regulatory mechanism underlying the regulations of HBV replication and HoxA10 production (Fig. 8). HBV initially activates p38 MAPK phosphorylation, which in turn facilitates HBV replication during early infection; however, HBV subsequently induces HoxA10 production, which in turn represses HBV replication during late infection. HoxA10 attenuates p38 MAPK/STAT3 activation and HBV replication through two approaches. It interacts with p38 MAPK, recruits SHP-1 to facilitate SHP-1 in catalyzing the dephosphorylation of p38 MAPK/STAT3, and thereby attenuates HBV replication. HoxA10 also binds to the HBV EnhI/X promoter, competes with STAT3 for binding of the promoter, and thereby represses HBV RNA transcription. Therefore, the function of HoxA10 in the repression of HBV replication is similar to the action of interferon (IFN) signaling in the inhibition of HBV infection. However, the mechanism underlying HoxA10-mediated repression of HBV replication is different from the mechanism underlying IFN-induced inhibition of HBV replication. This work reveals a negative regulatory mechanism underlying the control of HBV replication, demonstrates that HoxA10 exhibits a feedback-regulatory role relevant for the maintenance of persistent viral infection, and provides new insights into the development of potential agents to control HBV infection.
MATERIALS AND METHODS
Clinical analyses.
Sera of HBV-infected patients (n = 32) and healthy individuals (n = 18) were provided by Hubei Provincial Hospital (Wuhan, China) (Table 1). Patients were seropositive for hepatitis B surface antigen (HBsAg) and positive for HBV DNA. All healthy individuals were seronegative for HBsAg and negative for HBV DNA. Human HBV-associated HCC tissue paraffin sections (n = 2) and liver tissue paraffin sections from healthy individuals (n = 2) were provided by the National Human Genetic Resources Sharing Service Platform (Shanghai, China) (Table 2). Written informed consent was obtained from each individual according to the National Human Genetic Resources Sharing Service Platform.
Animal study.
BALB/c mice at 6 to 8 weeks of age were hydrodynamically injected with the HoxA10 plasmid and pAAV/HBV1.3 (adeno-associated virus carrying 1.3 copies of the HBV genome, genotype D, serotype ayw) according procedures described previously (54, 55). The founders were screened by analyzing serum HBsAg and HBeAg, and the one that produced high levels of HBsAg and HBeAg with active intrahepatic HBV replication was chosen for the present study. The mice were sacrificed at 6 days postinjection. Livers and blood were processed for analyses. All the BALB/c mice were kept in a specific-pathogen-free room.
Ethics statement.
The study was conducted according to the principles of the Declaration of Helsinki and approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, in accordance with its guidelines for the protection of human subjects. The Institutional Review Board of the College of Life Sciences, Wuhan University, approved the collection of blood samples for this study, and it was conducted in accordance with guidelines for the protection of human subjects. Written informed consent was obtained from each participant. The animal study was approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, and conducted in accordance with guidelines for the protection of animal subjects. All procedures involving mice and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the College of Life Sciences, Wuhan University.
Human PBMCs.
To isolate PBMCs, blood cells isolated from healthy individuals were separated from blood samples and diluted in RPMI 1640 purchased from Gibco (Grand Island, NY, USA). Diluted blood cells (5 ml) were added gently to a 15-ml centrifuge tube containing 5 ml lymphocyte separation medium (catalog no. 50494) purchased from MP Biomedicals (Santa Ana, CA, USA) and centrifuged at 2,000 × g for 10 min at room temperature (RT). The middle layer was transferred to a new centrifuge tube and diluted with RPMI 1640. The remaining red blood cells were removed using red blood cell lysis buffer (Sigma-Aldrich, St. Louis, MO, USA). The purified PBMCs were centrifuged at 1,500 × g for 10 min at RT and cultured in RPMI 1640.
Viruses and infection.
The ∼100-fold-concentrated supernatant of HepAD38 cells was used for HBV inoculation. For infection, HepG2-NTCP cells were seeded in collagen I-coated 24-well plates and inoculated overnight, and medium was changed to protoplast maintenance medium (PMM) with 2% fetal bovine serum for 12 h. PMM is Williams’ E medium (Gibco, USA) supplemented with ITS (insulin, transferrin, selenium; catalog no. I3146; Sigma, Corning, NY, USA), 2 mM l-glutamine, 10 ng/ml of human epidermal growth factor (EGF) (PeproTech, NJ, USA), 18 g/ml of hydrocortisone (Selleckchem, Houston, TX, USA), 40 ng/ml of dexamethasone (Sigma, USA), 2% dimethyl sulfoxide (DMSO; Sigma, USA), 100 U/ml of penicillin, and 100 g/ml of streptomycin. HepG2-NTCP cells were then infected with approximately 1,000 genome equivalents (GEq)/cell of HBV-containing 4% polyethylene glycol 8000 (PEG 8000) in medium for 24 h. The virus-containing medium was removed, and cells were washed five times and further incubated in PMM (56).
GST pulldown assays.
Bacterial cultures expressing GST fusion proteins were harvested and resuspended in phosphate-buffered saline (PBS)–Triton X-100 lysis buffer (2 mM KH2PO4, 10 mM Na2HPO4, 2.7 mM KCl, 137 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol [DTT], 100 μg/ml lysozyme, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The GST-tagged recombinant protein and GST protein were purified by using glutathione-agarose beads. After two washes with 1 ml lysis buffer, the beads were incubated with extracts of transfected HoxA10 plasmid cells overnight at 4°C. Beads were then washed five times with PBS–Triton X-100 buffer, proteins were eluted in SDS loading buffer, and protein levels were determined by Western blot analysis.
Northern blotting.
Total RNA was isolated using an Ultrapure RNA kit (Cwbio, Beijing, China) according to the manufacturer’s instructions. Fifteen micrograms of total RNA was separated in a 1.5% formaldehyde-agarose gel containing MOPS (morpholinepropanesulfonic acid) buffer, transferred onto a positively charged nylon membrane (GE Healthcare, PA, USA), and immobilized using UV cross-linking. The membranes were hybridized with a digoxigenin (DIG)-labeled RNA probe and detected with the DIG Northern starter kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions. The amounts of 28S and 18S rRNAs were used as loading controls.
HBV DNA analysis.
At 96 h posttransfection, cells were lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, and 1% NP-40) at 4°C for 30 min and centrifuged. The supernatants were collected and digested by adding DNase I plus 10 mM MgCl to remove plasmids and DNA not protected by HBV core. Protein was digested with proteinase K containing 0.5% SDS at 55°C overnight, and core-associated DNA was isolated by phenol-chloroform extraction and ethanol precipitation. HBV DNA was further detected by real-time PCR with a LightCycler instrument (Roche, Basel, Switzerland) by using the following primers: HBV forward primer 5′-AGAAACAACACATAGCGCCTCAT-3′ and HBV reverse primer 5′-TGCCCCATGCTGTAGATCTTG-3′.
Chromatin immunoprecipitation assay.
Briefly, cross-linked chromatin was sonicated into 200- to 1,000-bp fragments. Chromatin was immunoprecipitated using anti-HoxA10. Normal goat immunoglobulin G (IgG) was used as a negative control. PCR was performed, and DNA was separated by using a DNA agarose gel. HBV-specific primer sequences are forward primer 5′-AGAGTCTAGCCAGGAGGACTGCTCGCGGGC-3′ and reverse primer 5′-TGGGCCGGAGGTTCCAGCCCCGAGCC-3′.
Analysis of secreted HBV antigens.
Cell culture supernatants were collected to measure the HBsAg and HBeAg concentrations by using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Kehua Bio-Engineering, Shanghai, China) according to the manufacturer’s protocol.
Nuclear and cytoplasm extraction reagents.
Cells in 10-cm dishes were collected after transfection for 48 h and washed twice with cold PBS, 600 μl cytoplasmic extraction reagent I was added, the mixture was incubated for 10 min on ice, 33 μl cytoplasmic extraction reagent II was added, and the mixture was incubated for 1 min on ice. The cells were centrifuged at 16,000 × g for 10 min, and supernatants (cytoplasmic extract) were collected. Three hundred microliters of nuclear extraction reagent was added, and the mixture was incubated for 40 min on ice, with inversion of the tube to mix the contents. Nuclei were clarified by centrifugation at 16,000 × g for 10 min, and supernatants (nuclear extract) were transferred to a clean prechilled tube. Protease inhibitor cocktails were added to each type of buffer.
Electrophoretic mobility shift assay.
An EMSA was performed with 10-μl reaction mixtures by using a chemiluminescent EMSA kit as recommended by the manufacturer. Nuclear lysates were obtained by using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions, on HEK293T cells, which were transfected with a HoxA10 plasmid. The HoxA10 nuclear extract mixture, probe, and competitive probe were incubated at 25°C for 20 min. The sample was loaded onto a 6% polyacrylamide gel and run at 100 V for 1 h in 0.5× Tris-borate-EDTA (TBE) buffer. The polyacrylamide gel was transferred onto a nylon membrane at 180 mA for 2 h in 0.5× TBE buffer.
HBV cccDNA analysis.
DNA was extracted from cells transfected with the pBlue-HBV1.3 plasmid using an Ezup column animal genomic DNA purification kit (Sangon Biotech, Shanghai, China). To enhance the specificity of cccDNA detection, Plasmid-Safe ATP-dependent DNase (PSAD; Epicentre Biotechnologies, Madison, WI, USA) was used to degrade relaxed circular DNA (rcDNA) and single-stranded DNA (ssDNA) prior to quantitative PCR (qPCR) analysis, according to the manufacturer’s instructions. The levels of cccDNA in cells were measured using qPCR analysis as described previously (52). Primers for cccDNA amplification were forward primers 5′-GCGGTCTCCCCGTCTGTGCC-3′ (NCCC1 nt 1553 to 1562) and DRF1 (5′-GTCTGTGCCTTCTCATCTGC-3′) and reverse primer 5′-GTCCATGCCCCAAAGCCACCCAAGCCACC-3′ (CCCAS2 nt 1909 to 1891). Real-time PCR was performed in a LightCycler instrument (Roche, Grenoble, France) using a 20-μl reaction mixture volume containing 20 ng of DNA. Serial dilutions of a plasmid containing an HBV monomer served as a quantification standard. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA (housekeeping gene) was detected in order to count the number of cells (53).
Statistical analysis.
Statistical analyses were performed with GraphPad Prism 5. Individual experiments were performed in triplicate, and repetitive assays with similar results were performed to confirm the reproducibility of the results. Data are presented as the means ± standard deviations (SD). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81730061, 31230005, 81471942, 31200134, and 31270206), the National Health and Family Planning Commission of China (National Mega Project on Major Infectious Disease Prevention) (2017ZX10103005 and 2017ZX10202201), the Guangdong Province Pearl River Talent Plan Innovation and Entrepreneurship Team Project (2017ZT07Y580), and the Postdoctoral Science Foundation of China (grant no. 2018T110923).
We thank Ying Zhu of Wuhan University, China, for kindly providing the HepG2-NTCP cells and HepaAD38 cells.
We declare no competing financial interests.
REFERENCES
- 1.El-Serag HB. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2015. Guidelines for the prevention, care and treatment of persons with chronic hepatitis B infection. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 3.Beck J, Nassal M. 2007. Hepatitis B virus replication. World J Gastroenterol 13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganem D, Varmus HE. 1987. The molecular biology of the hepatitis B viruses. Annu Rev Biochem 56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 5.Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengel R, Cattaneo R, Schaller H. 1987. Replication strategy of human hepatitis B virus. J Virol 61:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quasdorff M, Protzer U. 2010. Control of hepatitis B virus at the level of transcription. J Viral Hepat 17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. [DOI] [PubMed] [Google Scholar]
- 7.Bock CT, Kubicka S, Manns MP, Trautwein C. 1999. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology 29:1236–1247. doi: 10.1002/hep.510290426. [DOI] [PubMed] [Google Scholar]
- 8.Yee JK. 1989. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science 246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]
- 9.Honigwachs J, Faktor O, Dikstein R, Shaul Y, Laub O. 1989. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J Virol 63:919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trujillo MA, Letovsky J, Maguire HF, Lopez-Cabrera M, Siddiqui A. 1991. Functional analysis of a liver-specific enhancer of the hepatitis B virus. Proc Natl Acad Sci U S A 88:3797–3801. doi: 10.1073/pnas.88.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waris G, Siddiqui A. 2002. Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J Virol 76:2721–2729. doi: 10.1128/JVI.76.6.2721-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Bei L, Shah CA, Horvath E, Eklund EA. 2011. HoxA10 influences protein ubiquitination by activating transcription of ARIH2, the gene encoding Triad1. J Biol Chem 286:16832–16845. doi: 10.1074/jbc.M110.213975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bei L, Shah C, Wang H, Huang W, Roy R, Eklund EA. 2012. Beta-catenin activates the HOXA10 and CDX4 genes in myeloid progenitor cells. J Biol Chem 287:39589–39601. doi: 10.1074/jbc.M112.402172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, Ozer HG, Carramusa L, Avigdor A, Volinia S, Buchberg A, Mazo A, Kollet O, Largman C, Croce CM, Nakamura T, Lapidot T, Canaani E. 2011. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci U S A 108:7956–7961. doi: 10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang SH, Yeh SH, Shiau CW, Chen KF, Lin WH, Tsai TF, Teng YC, Chen DS, Chen PJ. 2015. Sorafenib action in hepatitis B virus X-activated oncogenic androgen pathway in liver through SHP-1. J Natl Cancer Inst 107:djv190. doi: 10.1093/jnci/djv190. [DOI] [PubMed] [Google Scholar]
- 16.Han Y, Lu S, Wen YG, Yu FD, Zhu XW, Qiu GQ, Tang HM, Peng ZH, Zhou CZ. 2015. Overexpression of HOXA10 promotes gastric cancer cells proliferation and HOXA10(+)/CD44(+) is potential prognostic biomarker for gastric cancer. Eur J Cell Biol 94:642–652. doi: 10.1016/j.ejcb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Chin R, Earnest-Silveira L, Koeberlein B, Franz S, Zentgraf H, Dong X, Gowans E, Bock CT, Torresi J. 2007. Modulation of MAPK pathways and cell cycle by replicating hepatitis B virus: factors contributing to hepatocarcinogenesis. J Hepatol 47:325–337. doi: 10.1016/j.jhep.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Chang WW, Su IJ, Lai MD, Chang WT, Huang W, Lei HY. 2008. Suppression of p38 mitogen-activated protein kinase inhibits hepatitis B virus replication in human hepatoma cell: the antiviral role of nitric oxide. J Viral Hepat 15:490–497. doi: 10.1111/j.1365-2893.2008.00968.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Li YX, Fu HJ, Ren YL, Zou L, Shen SZ, Chen P, Sun T, Huang CH. 2017. Hepatitis B virus core antigen stimulates IL-6 expression via p38, ERK and NF-kappaB pathways in hepatocytes. Cell Physiol Biochem 41:91–100. doi: 10.1159/000455954. [DOI] [PubMed] [Google Scholar]
- 20.Xie J, Zhang Y, Zhang Q, Han Y, Yin J, Pu R, Shen Q, Lu W, Du Y, Zhao J, Han X, Zhang H, Cao G. 2013. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology 57:2369–2377. doi: 10.1002/hep.26303. [DOI] [PubMed] [Google Scholar]
- 21.Frank C, Keilhack H, Opitz F, Zschornig O, Bohmer FD. 1999. Binding of phosphatidic acid to the protein-tyrosine phosphatase SHP-1 as a basis for activity modulation. Biochemistry 38:11993–12002. doi: 10.1021/bi982586w. [DOI] [PubMed] [Google Scholar]
- 22.Pao LI, Badour K, Siminovitch KA, Neel BG. 2007. Nonreceptor protein-tyrosine phosphatases in immune cell signaling. Annu Rev Immunol 25:473–523. doi: 10.1146/annurev.immunol.23.021704.115647. [DOI] [PubMed] [Google Scholar]
- 23.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. 2004. Protein tyrosine phosphatases in the human genome. Cell 117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Craggs G, Kellie S. 2001. A functional nuclear localization sequence in the C-terminal domain of SHP-1. J Biol Chem 276:23719–23725. doi: 10.1074/jbc.M102846200. [DOI] [PubMed] [Google Scholar]
- 25.Fawcett VC, Lorenz U. 2005. Localization of Src homology 2 domain-containing phosphatase 1 (SHP-1) to lipid rafts in T lymphocytes: functional implications and a role for the SHP-1 carboxyl terminus. J Immunol 174:2849–2859. doi: 10.4049/jimmunol.174.5.2849. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Somani AK, Siminovitch KA. 2000. Roles of the SHP-1 tyrosine phosphatase in the negative regulation of cell signalling. Semin Immunol 12:361–378. doi: 10.1006/smim.2000.0223. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, Hong H, Kawakami Y, Kato Y, Wu D, Yasudo H, Kimura A, Kubagawa H, Bertoli LF, Davis RS, Chau LA, Madrenas J, Hsia CC, Xenocostas A, Kipps TJ, Hennighausen L, Iwama A, Nakauchi H, Kawakami T. 2009. Tumor suppression by phospholipase C-beta3 via SHP-1-mediated dephosphorylation of Stat5. Cancer Cell 16:161–171. doi: 10.1016/j.ccr.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenwa C, Kumar A, Rego D, Konarski Y, Nilchi L, Wright K, Kozlowski M. 2013. SHP-1-Pyk2-Src protein complex and p38 MAPK pathways independently regulate IL-10 production in lipopolysaccharide-stimulated macrophages. J Immunol 191:2589–2603. doi: 10.4049/jimmunol.1300466. [DOI] [PubMed] [Google Scholar]
- 29.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 30.Lee K, Esselman WJ. 2002. Inhibition of PTPs by H(2)O(2) regulates the activation of distinct MAPK pathways. Free Radic Biol Med 33:1121–1132. doi: 10.1016/S0891-5849(02)01000-6. [DOI] [PubMed] [Google Scholar]
- 31.Bei L, Huang W, Wang H, Shah C, Horvath E, Eklund E. 2011. HoxA10 activates CDX4 transcription and Cdx4 activates HOXA10 transcription in myeloid cells. J Biol Chem 286:19047–19064. doi: 10.1074/jbc.M110.213983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JJ, Taylor HS, Akbas GE, Foucher I, Trembleau A, Jaffe RC, Fazleabas AT, Unterman TG. 2003. Regulation of insulin-like growth factor binding protein-1 promoter activity by FKHR and HOXA10 in primate endometrial cells. Biol Reprod 68:24–30. doi: 10.1095/biolreprod.102.009316. [DOI] [PubMed] [Google Scholar]
- 33.Jan RH, Lin YL, Chen CJ, Lin TY, Hsu YC, Chen LK, Chiang BL. 2012. Hepatitis B virus surface antigen can activate human monocyte-derived dendritic cells by nuclear factor kappa B and p38 mitogen-activated protein kinase mediated signaling. Microbiol Immunol 56:719–727. doi: 10.1111/j.1348-0421.2012.00496.x. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Sun XH, Zhu XJ, Jin SG, Zeng ZJ, Zhou ZH, Yu Z, Gao YQ. 2012. HBcAg induces PD-1 upregulation on CD4+ T cells through activation of JNK, ERK and PI3K/AKT pathways in chronic hepatitis-B-infected patients. Lab Invest 92:295–304. doi: 10.1038/labinvest.2011.157. [DOI] [PubMed] [Google Scholar]
- 35.Tarn C, Zou L, Hullinger RL, Andrisani OM. 2002. Hepatitis B virus X protein activates the p38 mitogen-activated protein kinase pathway in dedifferentiated hepatocytes. J Virol 76:9763–9772. doi: 10.1128/JVI.76.19.9763-9772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diskin R, Askari N, Capone R, Engelberg D, Livnah O. 2004. Active mutants of the human p38alpha mitogen-activated protein kinase. J Biol Chem 279:47040–47049. doi: 10.1074/jbc.M404595200. [DOI] [PubMed] [Google Scholar]
- 37.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. 2015. The BioPlex network: a systematic exploration of the human interactome. Cell 162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. 2009. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Feng J, Yang G, Zhang S, Liu Y, Bu Y, Sun M, Zhao M, Chen F, Zhang W, Ye L, Zhang X. 2017. HBx-elevated MSL2 modulates HBV cccDNA through inducing degradation of AP88OBEC3B to enhance hepatocarcinogenesis. Hepatology 66:1413–1429. doi: 10.1002/hep.29316. [DOI] [PubMed] [Google Scholar]
- 40.Ruan P, Zhou B, Dai X, Sun Z, Guo X, Huang J, Gong Z. 2014. Predictive value of intrahepatic hepatitis B virus covalently closed circular DNA and total DNA in patients with acute hepatitis B and patients with chronic hepatitis B receiving anti-viral treatment. Mol Med Rep 9:1135–1141. doi: 10.3892/mmr.2014.1972. [DOI] [PubMed] [Google Scholar]
- 41.Bouchard MJ, Wang LH, Schneider RJ. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 42.Shah CA, Bei L, Wang H, Platanias LC, Eklund EA. 2012. HoxA10 protein regulates transcription of gene encoding fibroblast growth factor 2 (FGF2) in myeloid cells. J Biol Chem 287:18230–18248. doi: 10.1074/jbc.M111.328401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada T, Shimizu T, Sakurai T, Nanashima N, Kihara-Negishi F, Suzuki M, Fan Y, Akita M, Oikawa T, Tsuchida S. 2009. Physical and functional interactions between hematopoietic cell-specific ETS transcription factors and homeodomain proteins. Leuk Res 33:483–489. doi: 10.1016/j.leukres.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Sugimoto Y, Nakamura S, Okinaka K, Hirano I, Ono T, Shigeno K, Shinjo K, Ohnishi K. 2008. HOXA10 expression induced by Abl kinase inhibitors enhanced apoptosis through PI3K pathway in CML cells. Leuk Res 32:962–971. doi: 10.1016/j.leukres.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 45.Pearson JC, Lemons D, McGinnis W. 2005. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 46.Eklund EA, Jalava A, Kakar R. 2000. Tyrosine phosphorylation of HoxA10 decreases DNA binding and transcriptional repression during interferon gamma-induced differentiation of myeloid leukemia cell lines. J Biol Chem 275:20117–20126. doi: 10.1074/jbc.M907915199. [DOI] [PubMed] [Google Scholar]
- 47.Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu Y-F, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, Wong I, Sousa P, Zhu TN, Ditadi A, Keller G, Engelman AN, Snapper SB, Doulatov S, Daley GQ. 2017. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545:432–438. doi: 10.1038/nature22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanwar PS, Kaneko-Tarui T, Lee HJ, Zhang L, Teixeira JM. 2013. PTEN loss and HOXA10 expression are associated with ovarian endometrioid adenocarcinoma differentiation and progression. Carcinogenesis 34:893–901. doi: 10.1093/carcin/bgs405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X, Wang SK, Tian XS. 2014. HOXA10 promotes cell invasion and MMP-3 expression via TGFbeta2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci 59:1442–1451. doi: 10.1007/s10620-014-3033-6. [DOI] [PubMed] [Google Scholar]
- 50.Doitsh G, Shaul Y. 2004. Enhancer I predominance in hepatitis B virus gene expression. Mol Cell Biol 24:1799–1808. doi: 10.1128/MCB.24.4.1799-1808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eklund EA, Goldenberg I, Lu Y, Andrejic J, Kakar R. 2002. SHP1 protein-tyrosine phosphatase regulates HoxA10 DNA binding and transcriptional repression activity in undifferentiated myeloid cells. J Biol Chem 277:36878–36888. doi: 10.1074/jbc.M203917200. [DOI] [PubMed] [Google Scholar]
- 52.Bowden S, Jackson K, Littlejohn M, Locarnini S. 2004. Quantification of HBV covalently closed circular DNA from liver tissue by real-time PCR. Methods Mol Med 95:41–50. doi: 10.1385/1-59259-669-X:41. [DOI] [PubMed] [Google Scholar]
- 53.Saeed U, Kim J, Piracha ZZ, Kwon H, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. 19 December 2018. Parvulin 14 and parvulin 17 bind to HBx and cccDNA and upregulate HBV replication from cccDNA to virion in a HBx-dependent manner. J Virol doi: 10.1128/JVI.01840-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Bian Y, Zhang Z, Sun Z, Zhao J, Zhu D, Wang Y, Fu S, Guo J, Liu L, Su L, Wang FS, Fu YX, Peng H. 2017. Vaccines targeting preS1 domain overcome immune tolerance in HBV carrier mice. Hepatology 66:1067–1082. doi: 10.1002/hep.29239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang PL, Althage A, Chung J, Chisari FV. 2002. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci U S A 99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]