It is estimated that one-third of the world population have been exposed to hepatitis E virus (HEV). In developed countries and China, zoonotic HEV strains are responsible for almost all acute and chronic HEV infection cases. It is always of immediate interest to investigate the zoonotic potential of novel HEV strains. In 2016, we discovered a novel HEV genotype, HEV8, in Bactrian camels, but the epidemiology, zoonotic potential, and pathogenicity of the virus were unknown. In the present study, we demonstrated that HEV8 was circulating in multiple regions in China and was capable of infecting cynomolgus macaques, a surrogate for humans, posing high risk of zoonosis. Chronic hepatitis, systemic infection, and renal pathology were observed. Collectively, these data indicate that HEV8 exhibits a high potential for zoonotic transmission. Considering the importance of Bactrian camels as livestock animals, risk groups, such as camelid meat and milk consumers, should be screened for HEV8 infection.
KEYWORDS: Bactrian camel, chronic infection, cross-species transmission, hepatitis E virus, zoonotic infections
ABSTRACT
Hepatitis E virus (HEV) is zoonotic and a major cause of acute viral hepatitis worldwide. Recently, we identified a novel HEV genotype 8 (HEV8) in Bactrian camels in Xinjiang, China. However, the epidemiology, pathogenicity, and zoonotic potential of HEV8 are unclear. Here, we present the prevalence of HEV8 in China and investigate its pathogenicity and cross-species transmission in cynomolgus macaques. Fresh fecal and milk samples from Bactrian camels collected from four provinces/regions in China were screened for HEV RNA by reverse transcriptase PCR (RT-PCR). An HEV8-positive sample was used to inoculate two cynomolgus macaques to examine the potential for cross-species infection. The pathogenicity of HEV8 was analyzed by testing HEV markers and liver function during the study period and histopathology of liver biopsy specimens at 3, 13, and 25 weeks postinoculation. Extrahepatic replication was tested by using reverse transcriptase quantitative PCR (RT-qPCR) and immunofluorescence assays. The overall prevalence of HEV8 RNA in Chinese Bactrian camels was 1.4% (4/295), and positive samples were found in three different provinces/regions in China. Histopathology confirmed acute and chronic HEV8 infections in the two monkeys. Multiple tissues were positive for HEV RNA and ORF2 proteins. Renal pathology was observed in the monkey with chronic hepatitis. Whole-genome sequencing showed only 1 to 3 mutations in the HEV8 in the fecal samples from the two monkeys compared to that from the camel. HEV8 is circulating in multiple regions in China. Infection of two monkeys with HEV8 induced chronic and systemic infections, demonstrating the high potential zoonotic risk of HEV8.
IMPORTANCE It is estimated that one-third of the world population have been exposed to hepatitis E virus (HEV). In developed countries and China, zoonotic HEV strains are responsible for almost all acute and chronic HEV infection cases. It is always of immediate interest to investigate the zoonotic potential of novel HEV strains. In 2016, we discovered a novel HEV genotype, HEV8, in Bactrian camels, but the epidemiology, zoonotic potential, and pathogenicity of the virus were unknown. In the present study, we demonstrated that HEV8 was circulating in multiple regions in China and was capable of infecting cynomolgus macaques, a surrogate for humans, posing high risk of zoonosis. Chronic hepatitis, systemic infection, and renal pathology were observed. Collectively, these data indicate that HEV8 exhibits a high potential for zoonotic transmission. Considering the importance of Bactrian camels as livestock animals, risk groups, such as camelid meat and milk consumers, should be screened for HEV8 infection.
INTRODUCTION
Hepatitis E virus (HEV), the major causative agent of acute viral hepatitis worldwide, is a small, nonenveloped, single-stranded positive-sense RNA virus. It is mainly transmitted via the fecal-oral route and is well recognized as a zoonotic pathogen. In most patients, HEV infection induces an acute self-limiting hepatitis. However, in specific vulnerable groups, the outcome of HEV infection can be much more severe. In pregnant women, HEV infection has a 20% mortality rate (1), while immunocompromised individuals, such as transplant recipients (2) and HIV/AIDS patients (3), are prone to develop chronic hepatitis when infected with HEV. No specific antiviral agents are available for treating acute HEV infections. Although ribavirin therapy is currently recommended for treating chronic HEV infection (4), increasing case reports have identified viral mutations that can result in ribavirin treatment failure (5–7).
HEV is currently the sole member of the family Hepeviridae, genus Orthohepevirus, which comprises 4 species, Orthohepevirus A to D. To date, 7 genotypes of HEV (HEV1 to HEV7) in the Orthohepevirus A species have been recognized (8). HEV1 and HEV2 only infect humans and are transmitted via water in developing countries. HEV3 and HEV4 have been found both in humans in developed countries and in a variety of mammals worldwide, including domestic pigs (9), rabbits (10), wild boar (11), deer (12), cows (13), and goats (14). These four genotypes are the major genotypes that circulate in humans causing hepatitis E. HEV5 and HEV6 have been identified in wild boar in Japan, but the zoonotic risk of these two genotypes is still unknown (15). More recently, HEV7 has been isolated from dromedary camels in Dubai, United Arab Emirates (16), and several other countries (17). A case report of chronic infection of a human by HEV7 has appeared, indicating that zoonotic transmission of this HEV genotype can occur (18).
Camels are very important livestock animals. There are two surviving old-world camel species: Camelus dromedarius (dromedary or one-humped camel), which inhabits the Middle East and North and Northeast Africa, and Camelus bactrianus (Bactrian or two-humped camel), which is found mainly in northwestern China and southwestern Mongolia (18). In 2016, a novel HEV genotype was identified in Bactrian camels in Xinjiang, China (19). Sequence alignment revealed that the three Bactrian HEV strains detected represent a new genotype, and so they are currently classified as HEV8. These three HEV8 full genomes show >20% nucleotide sequence difference compared with the other seven HEV genotypes. In common with other HEV genotypes, the HEV8 strains have a genome size of 7.2 kb and have a typical Orthohepevirus A genome organization with three major open reading frames (ORFs) (19).
In contrast to the detailed information on the genome of HEV8, information remains limited on the epidemiology, capability of cross-species transmission, zoonotic risk, and pathogenicity of this genotype. The zoonotic potential of HEV8 requires urgent elucidation and is of immediate public health impact in regions where camels are important livestock. Therefore, this study focused on the prevalence of HEV8 and investigated the cross-species transmission and pathogenicity in cynomolgus macaques.
RESULTS
Prevalence of HEV8 in Chinese Bactrian camels.
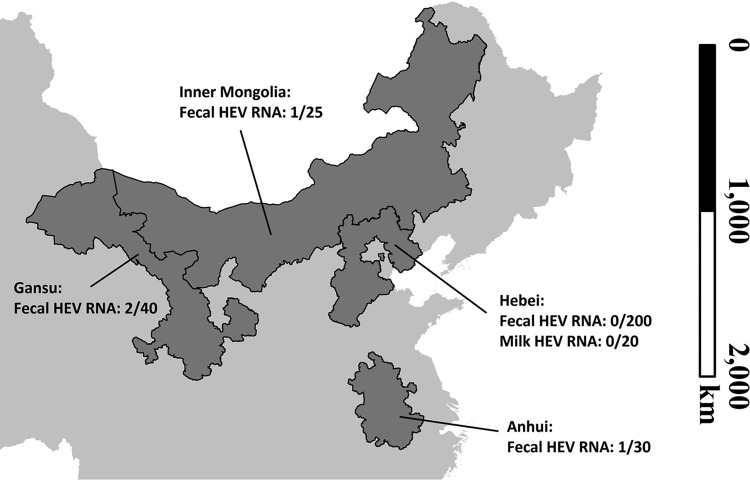
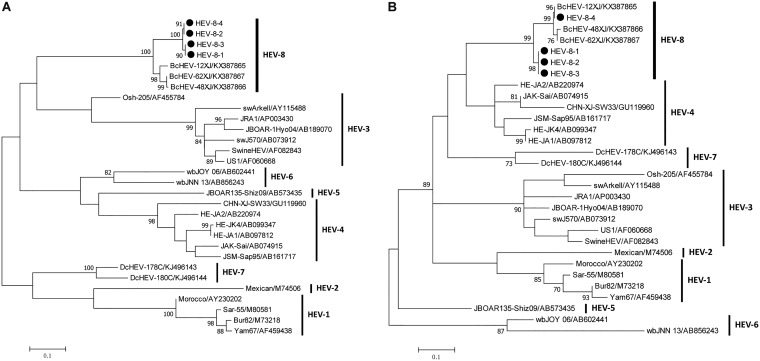
To investigate the prevalence of HEV in Chinese Bactrian camels, fresh fecal samples and milk samples were collected from camels from four provinces/regions in China (Fig. 1). A total of 295 fecal samples and 20 milk samples were obtained. All samples were screened for HEV RNA by heminested reverse transcriptase PCR (RT-PCR) using two sets of pan-HEV primers that amplified partial sequences of the ORF1 and ORF2 regions of the viral genomes of multiple HEV genotypes. Four of the 295 fecal samples (1.4%) were positive for HEV RNA, all of them were from farmed camels, including one from Anhui province (1/30 [3.3%]), two from Gansu province (2/40 [5.0%]), and one from the Inner Mongolia Autonomous Region (1/25 [4.0%]). The partial ORF1 and ORF2 sequences obtained from these four HEV strains shared 98.5% to 100% and 92.8% to 100% nucleotide identities, respectively. Phylogenetic analysis showed that they were clustered with the three HEV8 strains detected in Bactrian camels from Xinjiang in a previous study (Fig. 2) (19). No HEV RNA was detected in the milk samples. Repeat fecal samples were collected from the 40 Bactrian camels from Gansu province two months after initial sampling, but HEV RNA was not detected in any of these repeat samples. No other HEV genotype was detected in these samples.
FIG 1.
Sampling sites on a partial map of China. Four sampling sites and the positive/total number of hepatitis E virus (HEV) RNA in collected fecal/milk samples of Bactrian camels. Map was created only for this study by using an open source map data set, https://gadm.org/.
FIG 2.
Phylogenetic analysis of hepatitis E virus genotype 8 sequences. The 255 nt from the RNA-dependent RNA polymerase region in ORF1 (A) and 201 nt from the ORF2 (B), from this study. Reference sequences are of Orthohepevirus A genotypes 1 to 8. The phylogenetic tree was constructed by maximum likelihood method using the general time-reversible model with gamma distribution and invariant sites. Bootstrap values (%) of 1,000 repetitive analyses >70 are shown next to the nodes. The scale bar represents the genetic distance. New Bactrian camel HEV sequences (GenBank accession numbers MH423368 to MH423375) obtained in this study are indicated by black dots.
Genetic and phylogenetic analyses of HEV8 complete genome.
The consensus sequence of the genome of the HEV8 from a fecal sample collected from Gansu province (BcHEV-GP) was obtained by assembly of the sequences of RT-PCR products from the RNA extracted from the camel’s fecal suspension. We only sequenced one complete genome due to inadequate fecal materials left from the other three HEV8-positive samples.
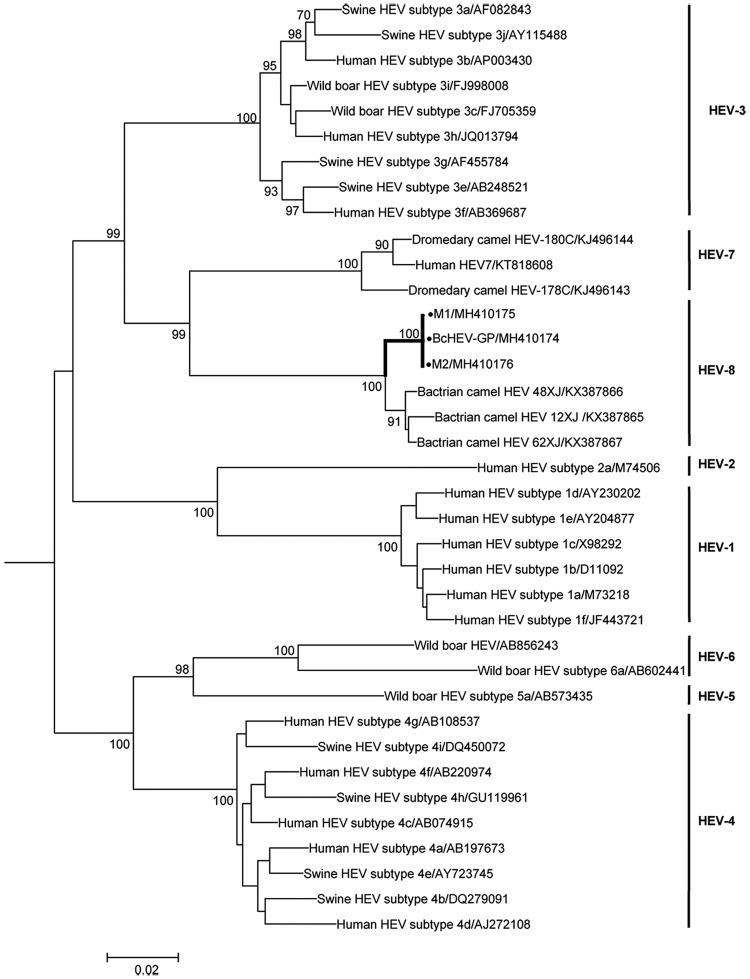
The genome size of BcHEV-GP was 7,223 bases, and the G+C content was 52%. Overall, the BcHEV-GP genome shared 85.5% to 85.6% nucleotide identities with the three previously reported Bactrian camel HEV8 strains (DcHEV-12XJ, -48XJ, and -62XJ). It contained three major ORFs, with typical genome organization and characteristics similar to other HEV genotypes of Orthohepevirus A. Both 5′ and 3′ ends contain short untranslated regions. Phylogenetic trees constructed from the concatenated ORF1/ORF2 excluding the hypervariable region (HVR) (Fig. 3) showed that BcHEV-GP was clustered with the three HEV8 strains previously reported from Bactrian camels (DcHEV-12XJ, -48XJ, and -62XJ). Amino acid distances based on the concatenated ORF1/ORF2 excluding the HVR region of BcHEV-GP and the previously reported HEV8 strains were ≤0.023, which was lower than the threshold (p-distance = 0.088) to demarcate intergenotype distance (19), confirming that BcHEV-GP was also a strain of HEV8.
FIG 3.
Phylogenetic analysis of ORF1/ORF2 proteins excluding the HVR of BcHEV-GP, M1, and M2. Reference sequences are of Orthohepevirus A genotypes 1 to 8. The tree was constructed by maximum likelihood method using JTT+G+I+F substitution model. Two thousand one hundred fifty-nine amino acid positions were included in the analysis. Bootstrap values (%) of 1,000 repetitive analyses >70 are shown next to the nodes. The scale bar represents the genetic distance. New HEV sequences obtained in this study are indicated by black dots.
Cross-species transmission of HEV8 to nonhuman primates.
The potential for transmission of HEV8 to humans is unknown. To make an initial assessment of the potential for such zoonotic transmission, cynomolgus monkeys were used as the nonhuman primate model and surrogate for humans. Two cynomolgus monkeys (M1 and M2) were successfully infected with the filtered fecal sample containing BcHEV-GP. Stable infection was established: viremia, urinary and fecal shedding of virus, elevation of liver enzymes, and seroconversion to anti-HEV IgM and IgG were all detected (Fig. 4). Jaundice was not observed during the study, but other less specific symptoms such as malaise and loss of appetite were found at 1 to 2 weeks postinoculation (wpi) for both monkeys.
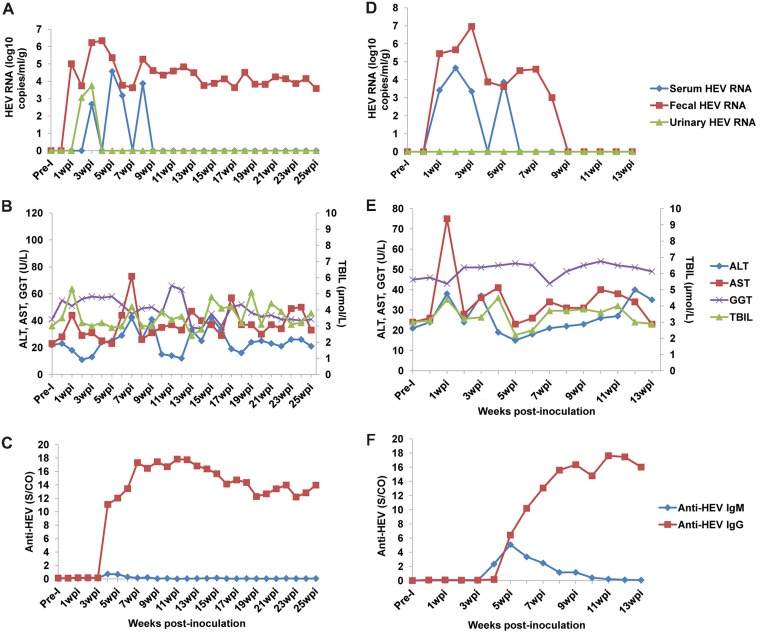
FIG 4.
Infectivity of HEV8 in monkeys. Detection of HEV RNA, liver function tests, and anti-HEV antibody detection in M1 (A to C) and M2 (D to F). Monkeys were intravenously inoculated with HEV8-positive inocula. Samples were collected weekly for immediate analysis.
Virus shedding was first detected in fecal samples from the two monkeys at 1 wpi. Intermittent viremia appeared at 1 and 3 wpi in M1 and M2, respectively. The HEV RNA titers decreased in serum and became undetectable after 6 and 9 wpi in M1 and M2, respectively (Fig. 4A and D). Urinary HEV was only seen in M1 at 3 and 4 wpi and at a relatively low titer (Fig. 4A). The fecal shedding of HEV was more stable and persisted longer than the viremia. The viral RNA titers in stool reached peaks at 4 wpi (2.21 × 106 copies/g) and 3 wpi (9.21 × 106 copies/g) in M1 and M2, respectively. In the case of M2, fecal shedding of virus was undetectable by 9 wpi (Fig. 4D). However, M1 persistently shed virus in feces at a relative low level (∼104 copies/g) for 25 weeks, and it unexpectedly died at 25 wpi (Fig. 4A).
Prior to inoculation, both monkeys were seronegative for anti-HEV and became seropositive for antibodies against HEV at 4 to 5 wpi (Fig. 4C and F). M2 tested positive for anti-HEV IgM at 5 to 10 wpi, but M1 tested negative (with a weak rise of the signal-to-cutoff [S/CO] value from 0.13 at 3 wpi to 0.73 at 4 wpi) during the whole study. Anti-HEV IgG began to rise a week later for M2 and remained positive thereafter. In the case of M1, detection of anti-HEV IgG started at 4 wpi and remained markedly elevated during the remaining 25 weeks until the animal’s unexpected death (Fig. 4C).
Dynamics of liver histopathology during acute and chronic infection of HEV8 in monkeys.
The two monkeys in this study presented two patterns of HEV infection. An acute-type HEV infection was observed in M2, and the infection spontaneously resolved within 9 weeks (Fig. 4D). Intriguingly, a chronic-type infection was observed in M1, and fecal shedding of virus persisted for 25 weeks (Fig. 4A). Biochemical analysis of samples from the two monkeys showed a >2-fold elevation of aspartate aminotransferase (AST) levels during the acute-phase of the infection (Fig. 4B and E). AST levels peaked at 7 wpi for M1 (73 U/liter) and at 1 wpi for M2 (75 U/liter).
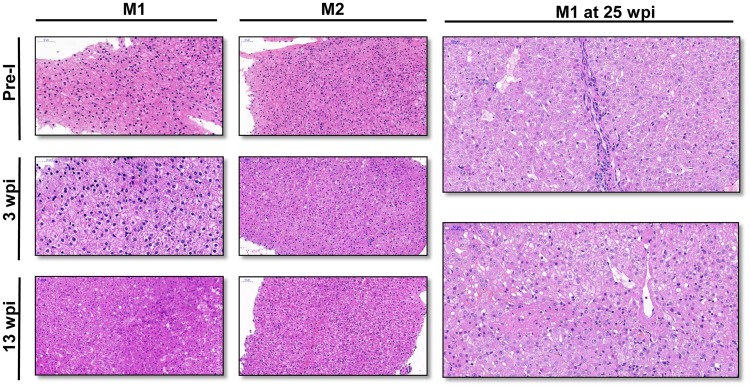
To better understand the histopathology changes in the liver during HEV infection, liver biopsy specimens were taken from the two monkeys at different time points. Biopsies were collected 1 week preinoculation and at 3 wpi and 13 wpi for the two monkeys. M1 died at 25 wpi and provided an opportunity to collect the liver tissue at this time point (Fig. 5).
FIG 5.
Histopathology of liver tissue in monkeys. (A) Liver biopsy specimens were collected from M1 and M2 1 week before inoculation (Pre-I), at 3 weeks postinoculation (wpi), and at 13 wpi. Hydropic degeneration and slight inflammatory cell infiltrates were found in biopsy specimens at 3 wpi for both monkeys. Focal necrosis of hepatic cells and dilation of the liver sinusoid were seen in the liver biopsy specimen of M1 at 13 wpi. (B) Liver tissues were collected at 25 wpi from M1, showing focal necrosis around the central vein and lobular inflammatory cell infiltrates. Ground-glass hepatocytes were also scattered in the liver. Bars, 50 μm.
Prior to inoculation, no obvious histopathological changes or lesions were observed in the livers of M1 and M2. At 3 wpi, hydropic degeneration was observed in both liver biopsy specimens and intrahepatic cholestasis was found in M1’s liver tissues. Both biopsy specimens showed slight infiltration of inflammatory cells. The histopathological changes at 13 wpi for M2 were similar to the mild lesions observed at 3 wpi. However, more severe histopathological changes, including hydropic degeneration, focal necrosis of hepatic cells, and dilation of liver sinusoid, were observed in liver biopsy specimens of M1 at 13 wpi (Fig. 5A). Liver samples from M1 were again collected at 25 wpi, and histopathology showed moderate lesions. Focal necrosis around the central vein and lobular inflammatory cell infiltrates were observed. Ground-glass hepatocytes were scattered in the liver (Fig. 5B). These histopathological changes seen for M1 are those associated with chronic hepatitis.
Extrahepatic replication and injury.
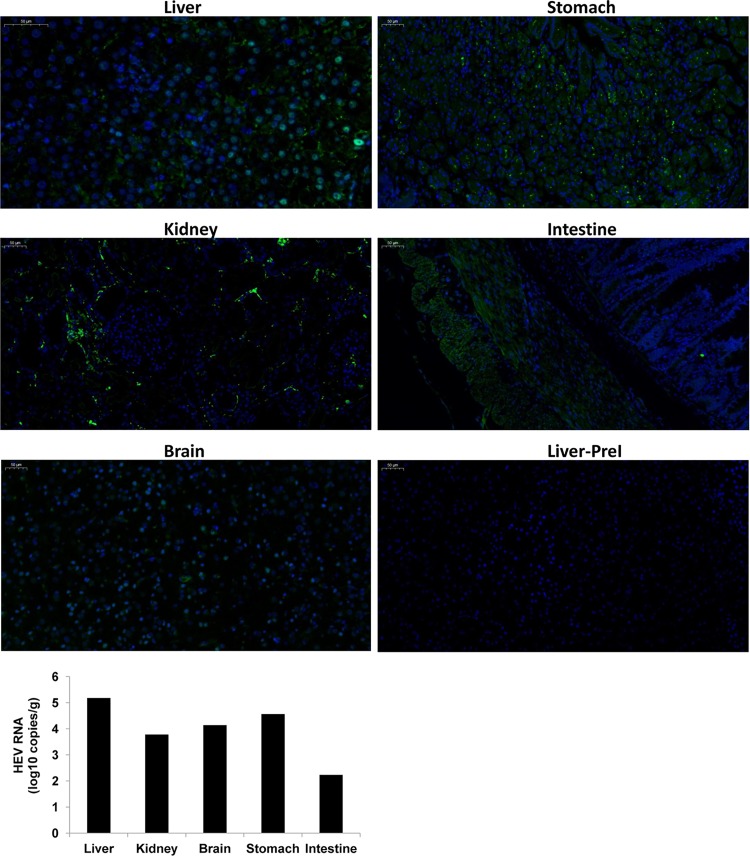
HEV ORF2 capsid protein was screened for in all tissues using an immunofluorescence assay. Positive staining was discovered in several extrahepatic samples from the kidney, brain, stomach, and intestine (Fig. 6). The liver biopsy specimen of M1 collected preinoculation served as a negative control. In addition, multiple tissues, including liver, kidney, brain, stomach, and intestine, were collected from M1 at 25 wpi. RNA was then extracted from supernatants of tissue homogenates. HEV8 RNA was detected in all tissues (Fig. 6).
FIG 6.
Extrahepatic replication of HEV in monkey M1. The viral RNA and ORF2 protein of HEV was detected using quantitative real-time PCR and an immunofluorescence assay, respectively, in liver, kidney, brain, stomach, and intestine tissues of monkey M1 at 25 wpi. Specific anti-HEV sera (bs-15457r; Bioss, Woburn, MA, USA) were used. The secondary antibody used for visualization was goat anti-rabbit IgG (green; Goodbio Technology, Wuhan, China). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue; Goodbio Technology, Wuhan, China). The liver biopsy specimen of M1 collected preinoculation (PreI) served as the negative control.
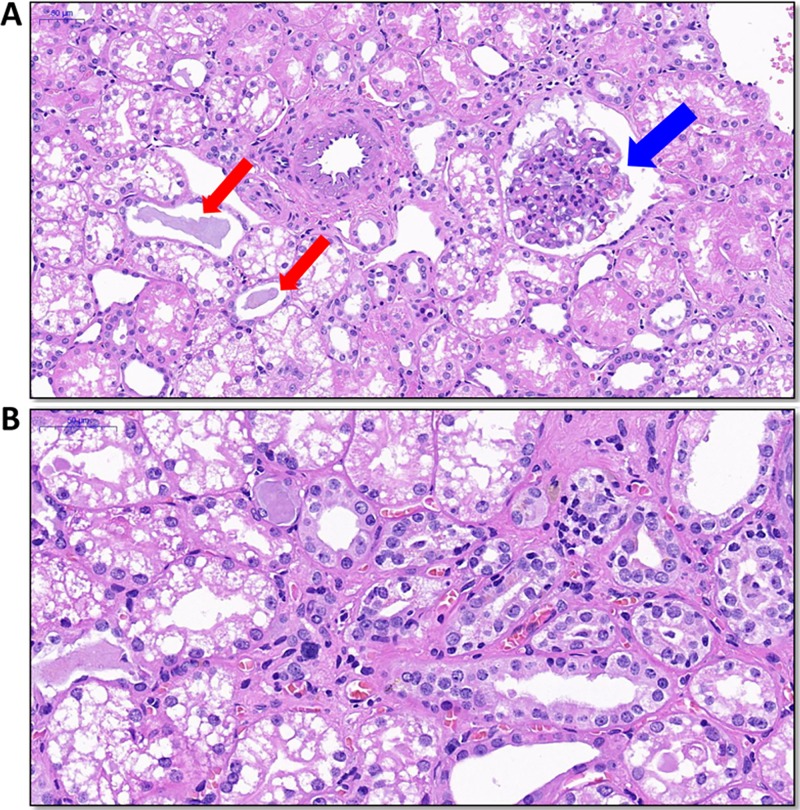
The histopathology of all tissues was also examined, and apparent lesions were observed in kidney tissues. Protein casts and proliferation of the glomerular mesangium were found (Fig. 7A). In the renal interstitium, thickening of tubular membranes and inflammatory cell infiltrates were seen (Fig. 7B).
FIG 7.
Histopathology of kidney tissue in monkey M1. Kidney tissue was collected at 25 weeks postinoculation. (A) Protein casts (red arrows) and proliferation of the glomerular mesangium were found (blue arrow). (B) In the renal interstitium, thickening of tubular membranes and inflammatory cell infiltrates were seen.
Genome comparisons of HEV8 during cross-species transmission.
The consensus sequences of M1 and M2 were obtained using fecal samples collected at 3 wpi. A sequence comparison revealed that the genomes of M1 and M2 shared an almost identical sequence (99.96% to 99.99% nucleotide identities) to that of BcHEV-GP. Using the genome of BcHEV-GP as the reference, three (nucleotide [nt] 221 C→T, nt 5,376 C→T, and nt 5,453 G→C) and one (nt 221 C→T, identical to the substitution in M1) substitutions were observed in the M1 and M2 genomes, respectively. For the three substitutions observed in M1, substitution nt 221 C→T was a synonymous substitution at position 65 (histidine [H]) in the ORF1, substitution nt 5,376 C→T caused no mutation at position 67 (threonine [T]) of ORF2 but led to a mutation at position 71 of ORF3, in which the proline (P) was changed to leucine (L) (P71L), and the substitution nt 5,453 G→C led to a mutation at position 93 of ORF2, in which the arginine (R) was changed to proline (P) (R93P), and a mutation at position 97 of ORF3, in which the alanine (A) was changed to proline (P) (A97P).
DISCUSSION
In the present study, HEV8 was found circulating in Bactrian camels in three regions of China, and most of them were domestic camels raised for tourism, meat, and dairy products. Both in the previous and inn the present study, it seems that HEV8 may be causing asymptomatic or relatively mild infections in its natural host, as the owners of the camel farms reported that the Bactrian camels were clinically healthy (19). Although there are no general criteria to define HEV subtypes for all HEV genotypes, according to previous studies on HEV1 to HEV4 (20), BcHEV-GP may constitute a potentially novel subtype of HEV8. Sequencing of additional HEV8 genomes would be necessary to evaluate this possibility. HEV RNA has previously been isolated from human (21) and cow (13) milk. However, HEV RNA was not detected in the 20 milk samples collected from Bactrian camels.
HEV7 and HEV8 are probably specific to their corresponding camel hosts. Bactrian camels and dromedary camels both belong to genus Camelus, but it seems that they are different as being hosts of many pathogens, such as Middle East respiratory syndrome (MERS) coronavirus (22) and West Nile virus (23). In all previous molecular surveillance studies on HEV in dromedary and Bactrian camels, as well as the present study, only HEV7 was found in dromedary camels and only HEV8 was detected in Bactrian camels (16, 17, 19), suggesting that dromedary and Bactrian camels are probably the sole natural hosts of HEV7 and HEV8, respectively. The separation of HEV genotypes may result from the species barrier and/or the geographical isolation of these two camel species.
The successful infection of two cynomolgus monkeys with HEV8 inocula from Bactrian camels suggests a strong zoonotic potential of HEV8. Nonhuman primates are a useful and desirable surrogate for humans and have been used in a number of HEV infection studies to assess the zoonotic risk posed by novel HEV genotypes (24). Several novel animal HEV strains, such as swine HEV (HEV3 and HEV4) (24), rabbit HEV (HEV3) (25, 26), and dromedary HEV (HEV7) (27, 28), successfully establishing infection in nonhuman primates were confirmed later to be able to infect humans. Although HEV mainly transmits via the fecal-oral route, in experimental assessments of cross-species transmission to nonhuman primates or pathogenicity studies, intravenous administration was used more often (25–27, 29, 30), as it is easy to control the total viral titers administered and easy to conduct. Many experimental studies have shown that oral administration is less efficient and usually induces no detectable fecal HEV shedding or viremia, even when the animals are the natural host of the HEV strain challenged (31). Besides, increasing studies highlight the blood-borne transmission of HEV in humans (1, 4). In this study, acute and chronic-type infections were observed in the two HEV8-infected monkeys. Protracted fecal shedding of HEV without viremia has been reported both in animals (25, 27, 32, 33) and in chronic hepatitis E patients (34, 35) and agrees well with the natural course of HEV infections (1). Although most chronic HEV infection cases occurred in immunocompromised patients, several studies have reported that occasionally, immunocompetent individuals can also develop chronic hepatitis E (36, 37). In animal studies, chronic HEV infection has been established in healthy rabbits (33), rhesus monkeys (29), and ferrets (38). The underlying mechanisms responsible for the different disease profiles remain unclear, but the findings of this study provide a potential alternative animal model for more detailed study of chronic HEV infection.
Extrahepatic replication was found in several tissues and organs of animal M1 at 25 wpi. Notably, obvious lesions were evident in the kidney. To our knowledge, there are 12 reported cases of HEV-associated renal manifestation; of them, nine cases were glomerulonephritis and the other three were either with no renal biopsy or showed no obvious injury (39). All but one patient were immunocompromised with chronic HEV infection. Renal biopsy specimens of the nine patients mainly showed glomerulitis with exudative proliferation and double contours in the glomerular basement membrane (39–41). Interstitial fibrosis and infiltrates of mononuclear cells were also observed in some cases (40, 42). The renal histology of M1 in our study is similar to that of human cases. The etiology of most forms of glomerulonephritis is likely infection (43). Infection-related immune responses and their direct effect on glomerular cells may cause the disease. As with the hepatitis C virus, HEV-associated kidney injury is presumed to be related to the glomerular deposition of immune complexes comprising the HEV antibodies and antigen (41, 43). The detection of HEV RNA and proteins in M1’s kidney suggests direct viral replication in renal cells may also play a role. The present macaque model may be used to elucidate the underlying mechanism in the future.
MATERIALS AND METHODS
Ethics statement.
The animal experimental protocol was approved by the Committee of Laboratory Animal Welfare and Ethics of Peking University Health Science Center (number LA2016332). All animal studies in this study were performed according to the Chinese Regulations of Laboratory Animals—The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People's Republic of China) and the Laboratory Animal Requirements of Environment and Housing Facilities (GB 14925–2010, National Laboratory Animal Standardization Technical Committee). Two cynomolgus monkeys were housed in adjoining individual cages allowing social interactions and with sufficient food and water (commercial monkey chow, treats, and fruits twice daily). All procedures were carried out under sodium pentobarbital anesthesia by trained personnel under the supervision of veterinary staff, and all efforts were made to ensure animal welfare and to minimize animal suffering in accordance with the recommendations of the Weatherall report, “The use of nonhuman primates in research (PDF).”
Sample collection and processing.
Between January and April 2017, fresh fecal samples were collected from adult Bactrian camels in four different provinces/regions in China (Fig. 1). Samples were taken from domestic Bactrian camels in Inner Mongolia Autonomous Region (25 samples), Gansu (40 samples), Hebei (200 samples from 2 farms), and Anhui (30 samples) provinces. In addition, 20 milk samples were collected from Bactrian camels in Hebei province. In June 2017, follow-up fecal samples were taken from the 40 animals in Gansu province. The fecal samples were all diluted with phosphate-buffered saline (PBS) to prepare 10% to 20% suspensions and centrifuged at 5,000 × g for 30 min. The supernatants obtained were immediately stored at −80°C until use.
Detection of HEV RNA.
All the samples collected were screened for the presence of HEV RNA. RNA was extracted from 100 μl of clarified supernatant, serum, urine, or milk using TRIzol reagent (Invitrogen, Burlington, ON, Canada). A heminested reverse transcription-PCR with two sets of primers that can detect all genotypes within Orthohepevirus species A, targeting ORF1 and ORF2 of the HEV genome, were used to screen for the presence of HEV RNA (19, 44). HEV-positive samples were commercially sequenced according to the manufacturer’s instructions (Beijing Genomics Institute, Beijing, China) on an automatic DNA sequencer (ABI model 3730 sequencer; Applied Biosystems, Foster City, CA, USA). Standard precautions were taken to avoid PCR contamination, and no false-positive result was observed for negative controls. All sequences were submitted to GenBank with accession numbers MH423368 to MH423375.
Quantification of HEV RNA was carried out using a commercial One-Step RT-qPCR kit (A6120; Promega, USA). The reaction conditions for the one-step RT-qPCR were 30 min at 50°C, 15-min incubation at 95°C, and 45 cycles of 10 s at 95°C, 20 s at 55°C, and 15 s at 72°C. A 10-fold serial dilution of capped HEV RNA (108 to 101 copies) was used as the standard for quantification of viral genome copy number (45).
Inoculation of monkeys and sample collection.
The animal experimental protocol was approved by the Committee of Laboratory Animal Welfare and Ethics of Peking University Health Science Center. Two 5-year-old female cynomolgus monkeys (Macaca fascicularis), weighing 2.0 to 2.5 kg (designated M1 and M2), were obtained from the Beijing Institute of Xieerxin Biology Resource (Beijing, China) for the cross-species infection and pathogenicity study. Both monkeys were negative for HEV RNA and anti-HEV IgM/IgG for two consecutive weeks prior to inoculation.
Inocula were prepared from the HEV8-positive fecal sample collected from Gansu province (BcHEV-GP). The fecal suspension was filtered through 0.45-μm and 0.22-μm filters prior to inoculation. Approximate viral titers of 4.50 × 104 genome copies/ml were determined by RT-qPCR. Each monkey was inoculated intravenously with 5 ml of inocula (approximately 2 × 105 copies of virus). The two monkeys were housed in individual cages, and serum, feces, and urine samples were collected weekly. All samples were subjected to RNA extraction and tested for HEV RNA and anti-HEV IgM/IgG.
Complete genome sequencing.
Three complete genomes of the HEV8 from the fecal sample collected from Gansu province (BcHEV-GP) and the two monkeys that were inoculated with the filtered fecal sample containing BcHEV-GP (M1 and M2) were amplified and sequenced using the RNA extracted directly from the original fecal specimens of the camel and monkeys, respectively, as the templates. The RNA was converted to cDNA by a combined random priming and oligo(dT) priming strategy. The cDNA was amplified by primers designed by multiple alignments of the genomes of other HEV8 with genome sequences available, using strategies described in our previous publication (19). Additional primers were designed from the results of the first and subsequent rounds of sequencing. The 5′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends using the SMARTer 5′/3′ RACE kit (Clontech, Mountain View, CA). Sequences were assembled and manually edited to produce final sequences of the viral genomes. All sequences were submitted to GenBank with accession numbers MH410174 to MH410176.
Genome and phylogenetic analyses of HEV sequences.
The nucleotide sequences of the genomes and the deduced amino acid sequences of the ORFs were compared to each other and to those of other HEVs using MatGAT 2.02 (19). Phylogenetic trees based on the nucleotide sequences of the PCR screening fragments and the amino acid sequences of concatenated ORF1/ORF2 excluding the HVR in ORF1 were constructed by maximum likelihood method using MEGA 6.0 (43), with bootstrap values calculated from 1,000 trees. The optimal substitution model for each ORF was selected by MEGA 6.0 (46). The mean amino acid genetic distances (p-distance) based on the concatenated ORF1/ORF2 excluding the HVR between BcHEV-GP, M1, and M2 and other HEV genotypes were calculated using MEGA 6.0 (43).
Biochemical analysis of blood samples from infected monkeys.
Several clinical chemistry parameters were monitored, including alanine aminotransferase, AST, total bilirubin, and γ-glutamyl transpeptidase, using a Hitachi Automatic Clinical Analyzer 7180 (Hitachi High-Technologies, Tokyo, Japan).
Detection of anti-HEV IgM and anti-HEV IgG.
Anti-HEV IgM and anti-HEV IgG were detected by enzyme immunoassay (EIA), using commercial kits (Wantai, Beijing, China) according to the manufacturer’s instructions. The method used was based on the E2 protein of genotype 1 HEV (amino acids 394 to 606 of HEV ORF2) (44). These commercial kits were widely used in studies of humans and animals infected with many different HEV genotypes, including genotype in Orthohepevirus C (4, 47, 48).
Histopathological and immunofluorescence assays.
To understand the pathogenicity of HEV8 in monkeys, liver biopsies were performed prior to inoculation, at 3 wpi and 13 wpi, using disposable core tissue biopsy needles (MN1610; Bard, USA). In addition, during the experiment, M1 suddenly died at 25 wpi, which presented the opportunity to collect extrahepatic tissue samples for further investigation. Tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Specimens were cut into 3- to 4-μm serial sections. Slides were stained with hematoxylin-eosin and subjected to histopathological microscopic examination. HEV proteins were visualized on paraformaldehyde-fixed tissues sections using HEV ORF2-specific antibodies (bs-15457r; Bioss, Woburn, MA, USA) and visualized using an inverted fluorescence microscope (Eclipse TI-SR; Nikon, Japan).
Approximately 100 mg of the fresh tissue or organs collected was homogenized in 1 ml of TRIzol reagent and clarified by centrifugation at 5,000 × g for 15 min at 4°C. The supernatants from this centrifugation were harvested and stored at −80°C for later RT-qPCR.
Accession number(s).
The sequences for HEV8 have been assigned GenBank accession numbers MH423368 to MH423375 and MH410174 to MH410176.
ACKNOWLEDGMENTS
This study was funded by the National Science Foundation of China (grant no. 81772175 and 81401746), the Beijing Natural Science Foundation (grant no. 7162103), the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Ministry of Education, China, the General Research Fund, University Grants Committee, Hong Kong, and the Peking University Health Science Center Innovative Fund for Doctoral Students.
We thank Malcolm A. McCrae from University of Warwick, UK, for proofreading the manuscript. We also thank Yuying Yuan from College of Urban and Environmental Sciences, Peking University, for editing the map figure.
REFERENCES
- 1.Nimgaonkar I, Ding Q, Schwartz RE, Ploss A. 2018. Hepatitis E virus: advances and challenges. Nat Rev Gastroenterol Hepatol 15:96–110. doi: 10.1038/nrgastro.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamar N, Selves J, Mansuy J, Ouezzani L, Péron J, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 3.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med 361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 4.Kamar N, Izopet J, Pavio N, Aggarwal R, Labrique A, Wedemeyer H, Dalton HR. 2017. Hepatitis E virus infection. Nat Rev Dis Primers 3:17086. doi: 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- 5.Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J. 2014. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147:1008–1011. doi: 10.1053/j.gastro.2014.08.040. [DOI] [PubMed] [Google Scholar]
- 6.Debing Y, Ramière C, Dallmeier K, Piorkowski G, Trabaud M, Lebossé F, Scholtès C, Roche M, Legras-Lachuer C, de Lamballerie X, André P, Neyts J. 2016. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol 65:499–508. doi: 10.1016/j.jhep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, Pischke S, Bremer B, Brown RJ, Manns MP, Cornberg M, Bock CT, Steinmann E, Wedemeyer H. 2016. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65:1733–1743. doi: 10.1136/gutjnl-2015-311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2016. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol 97:537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- 11.Tamada Y, Yano K, Yatsuhashi H, Inoue O, Mawatari F, Ishibashi H. 2004. Consumption of wild boar linked to cases of hepatitis E. J Hepatol 40:869–870. doi: 10.1016/j.jhep.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 12.Tei S, Kitajima N, Takahashi K, Mishiro S. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- 13.Huang F, Li Y, Yu W, Jing S, Wang J, Long F, He Z, Yang C, Bi Y, Cao W, Liu C, Hua X, Pan Q. 2016. Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis. Hepatology 64:350–359. doi: 10.1002/hep.28668. [DOI] [PubMed] [Google Scholar]
- 14.Di Martino B, Di Profio F, Melegari I, Sarchese V, Robetto S, Marsilio F, Martella V. 2016. Detection of hepatitis E virus (HEV) in goats. Virus Res 225:69–72. doi: 10.1016/j.virusres.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi M, Nishizawa T, Sato H, Sato Y, Jirintai, Nagashima S, Okamoto H. 2011. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J Gen Virol 92:902–908. doi: 10.1099/vir.0.029470-0. [DOI] [PubMed] [Google Scholar]
- 16.Woo PC, Lau SK, Teng JL, Tsang AK, Joseph M, Wong EY, Tang Y, Sivakumar S, Xie J, Bai R, Wernery R, Wernery U, Yuen KY. 2014. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis 20:1044–1048. doi: 10.3201/eid2006.140140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasche A, Saqib M, Liljander AM, Bornstein S, Zohaib A, Renneker S, Steinhagen K, Wernery R, Younan M, Gluecks I, Hilali M, Musa BE, Jores J, Wernery U, Drexler JF, Drosten C, Corman VM. 2016. Hepatitis E virus infection in dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015. Emerg Infect Dis 22:1249–1252. doi: 10.3201/eid2207.160168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapil S, Yeary T, Evermann JF. 2009. Viral diseases of new world camelids. Vet Clin North Am Food Anim Pract 25:323–337. doi: 10.1016/j.cvfa.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo PC, Lau SK, Teng JL, Cao KY, Wernery U, Schountz T, Chiu TH, Tsang AK, Wong PC, Wong EY, Yuen KY. 2016. New hepatitis E virus genotype in Bactrian camels, Xinjiang, China, 2013. Emerg Infect Dis 22:2219–2221. doi: 10.3201/eid2212.160979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol 16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 21.Rivero-Juarez A, Frias M, Rodriguez-Cano D, Cuenca-López F, Rivero A. 2016. Isolation of Hepatitis E virus from breast milk during acute infection. Clin Infect Dis 62:1464. doi: 10.1093/cid/ciw186. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, Wen Z, Wang J, Ge J, Chen H, Bu Z. 2015. Absence of Middle East respiratory syndrome coronavirus in Bactrian camels in the West Inner Mongolia Autonomous Region of China: surveillance study results from July 2015. Emerg Microbes Infect 4:e73. doi: 10.1038/emi.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joseph S, Wernery U, Teng JL, Wernery R, Huang Y, Patteril NA, Chan KH, Elizabeth SK, Fan RY, Lau SK, Kinne J, Woo PC. 2016. First isolation of West Nile virus from a dromedary camel. Emerg Microbes Infect 5:e53. doi: 10.1038/emi.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell RH, Engle RE, Govindarajan S, Herbert R, St CM, Elkins WR, Cook A, Shaver C, Beauregard M, Swerczek J, Emerson SU. 2013. Pathobiology of hepatitis E: lessons learned from primate models. Emerg Microbes Infect 2:e9. doi: 10.1038/emi.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Bu QN, Wang L, Han J, Du RJ, Lei YX, Ouyang YQ, Li J, Zhu YH, Lu FM, Zhuang H. 2013. Transmission of hepatitis E Virus from rabbits to cynomolgus macaques. Emerg Infect Dis 19:559–565. doi: 10.3201/eid1904.120827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abravanel F, Lhomme S, El CH, Schvartz B, Peron JM, Kamar N, Izopet J. 2017. Rabbit hepatitis E virus infections in humans, France. Emerg Infect Dis 23:1191–1193. doi: 10.3201/eid2307.170318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li TC, Zhou X, Yoshizaki S, Ami Y, Suzaki Y, Nakamura T, Takeda N, Wakita T. 2016. Production of infectious dromedary camel hepatitis E virus by a reverse genetic system: potential for zoonotic infection. J Hepatol 65:1104–1111. doi: 10.1016/j.jhep.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Lee G, Tan B, Chi-Yuan Teo E, Lim S, Dan Y, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, Purdy MA, Teo CG. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150:355–357. doi: 10.1053/j.gastro.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 29.Huang F, Yang C, Zhou X, Yu W, Pan Q. 2016. Rhesus macaques persistently infected with hepatitis E shed virus into urine. J Hepatol 64:1446–1447. doi: 10.1016/j.jhep.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 30.Geng Y, Zhao C, Huang W, Harrison TJ, Zhang H, Geng K, Wang Y. 2016. Detection and assessment of infectivity of hepatitis E virus in urine. J Hepatol 64:37–43. doi: 10.1016/j.jhep.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Yugo DM, Cossaboom CM, Meng XJ. 2014. Naturally occurring animal models of human hepatitis E virus infection. Ilar J 55:187–199. doi: 10.1093/ilar/ilu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao D, Cao QM, Subramaniam S, Yugo DM, Heffron CL, Rogers AJ, Kenney SP, Tian D, Matzinger SR, Overend C, Catanzaro N, LeRoith T, Wang H, Piñeyro P, Lindstrom N, Clark-Deener S, Yuan L, Meng XJ. 2017. Pig model mimicking chronic hepatitis E virus infection in immunocompromised patients to assess immune correlates during chronicity. Proc Natl Acad Sci U S A 114:6914–6923. doi: 10.1073/pnas.1705446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Xia J, Wang L, Wang Y. 2017. Experimental infection of rabbits with genotype 3 hepatitis E virus produced both chronicity and kidney injury. Gut 66:561–562. doi: 10.1136/gutjnl-2016-312023. [DOI] [PubMed] [Google Scholar]
- 34.Abravanel F, Lhomme S, Rostaing L, Kamar N, Izopet J. 2015. Protracted fecal shedding of HEV during ribavirin therapy predicts treatment relapse. Clin Infect Dis 60:96–99. doi: 10.1093/cid/ciu742. [DOI] [PubMed] [Google Scholar]
- 35.Ambrosioni J, Mamin A, Hadengue A, Bernimoulin M, Samii K, Landelle C, Negro F, Kaiser L. 2014. Long-term hepatitis E viral load kinetics in an immunocompromised patient treated with ribavirin. Clin Microbiol Infect 20:O718–O720. doi: 10.1111/1469-0691.12576. [DOI] [PubMed] [Google Scholar]
- 36.Grewal P, Kamili S, Motamed D. 2014. Chronic hepatitis E in an immunocompetent patient: a case report. Hepatology 59:347–348. doi: 10.1002/hep.26636. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez Tallon A, Moreira Vincente V, Mateos Lindemann ML, Achecar Justo LM. 2011. Chronic hepatitis E in an immunocompetent patient. Gastroenterol Hepatol 34:398–400. doi: 10.1016/j.gastrohep.2011.02.011 (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 38.Li TC, Yang T, Yoshizaki S, Ami Y, Suzaki Y, Ishii K, Kishida N, Shirakura M, Asanuma H, Takeda N, Wakita T. 2016. Ferret hepatitis E virus infection induces acute hepatitis and persistent infection in ferrets. Vet Microbiol 183:30–36. doi: 10.1016/j.vetmic.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Pischke S, Hartl J, Pas SD, Lohse AW, Jacobs BC, Van der Eijk AA. 2017. Hepatitis E virus: infection beyond the liver? J Hepatol 66:1082–1095. doi: 10.1016/j.jhep.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 40.Kamar N, Weclawiak H, Guilbeau-Frugier C, Legrand-Abravanel F, Cointault O, Ribes D, Esposito L, Cardeau-Desangles I, Guitard J, Sallusto F, Muscari F, Peron JM, Alric L, Izopet J, Rostaing L. 2012. Hepatitis E virus and the kidney in solid-organ transplant patients. Transplantation 93:617–623. doi: 10.1097/TP.0b013e318245f14c. [DOI] [PubMed] [Google Scholar]
- 41.Guinault D, Ribes D, Delas A, Milongo D, Abravanel F, Puissant-Lubrano B, Izopet J, Kamar N. 2016. Hepatitis E virus-induced cryoglobulinemic glomerulonephritis in a nonimmunocompromised person. Am J Kidney Dis 67:660–663. doi: 10.1053/j.ajkd.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 42.Taton B, Moreau K, Lepreux S, Bachelet T, Trimoulet P, De Ledinghen V, Pommereau A, Ronco P, Kamar N, Merville P, Couzi L. 2013. Hepatitis E virus infection as a new probable cause of de novo membranous nephropathy after kidney transplantation. Transpl Infect Dis 15:E211–E215. doi: 10.1111/tid.12143. [DOI] [PubMed] [Google Scholar]
- 43.Couser WG, Johnson RJ. 2014. The etiology of glomerulonephritis: roles of infection and autoimmunity. Kidney Int 86:905–914. doi: 10.1038/ki.2014.49. [DOI] [PubMed] [Google Scholar]
- 44.Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose SM, Adu-Sarkodie Y, Oppong SK, Kalko EKV, Osterman A, Rasche A, Adam A, Muller MA, Ulrich RG, Leroy EM, Lukashev AN, Drosten C. 2012. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86:9134–9147. doi: 10.1128/JVI.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Ge SX, Huang GY, Li SW, He ZQ, Wang YB, Zheng YJ, Gu Y, Ng MH, Xia NS. 2003. Evaluation of antibody-based and nucleic acid-based assays for diagnosis of hepatitis E virus infection in a rhesus monkey model. J Med Virol 71:518–526. doi: 10.1002/jmv.10523. [DOI] [PubMed] [Google Scholar]
- 48.Raj VS, Smits SL, Pas SD, Provacia LB, Moorman-Roest H, Osterhaus AD, Osterhaus AD, Haagmans BL. 2012. Novel hepatitis E virus in ferrets, the Netherlands. Emerg Infect Dis 18:1369–1370. doi: 10.3201/eid1808.111659. [DOI] [PMC free article] [PubMed] [Google Scholar]