Influenza A viruses (IAV) constitute a major public health issue, causing illness and death in high-risk populations during seasonal epidemics or pandemics. IAV are known to modulate cellular pathways to promote their replication and avoid immune restriction via the targeting of several cellular proteins. One of these proteins, p53, is a master regulator involved in a large panel of biological processes, including cell cycle arrest, apoptosis, or senescence. This “cellular gatekeeper” is also involved in the control of viral infections, and viruses have developed a wide diversity of mechanisms to modulate/hijack p53 functions to achieve an optimal replication in their hosts. Our group and others have previously shown that p53 activity is finely modulated by different multilevel mechanisms during IAV infection. Here, we characterized IAV nonstructural protein NS1 and the cellular factor CPSF4 as major partners involved in the IAV-induced modulation of the TP53 alternative splicing that was associated with a strong modulation of p53 activity and notably the p53-mediated antiviral response.
KEYWORDS: CPSF30, antiviral response, influenza viruses, p53, splicing, virus-host interactions
ABSTRACT
Influenza A viruses (IAV) are known to modulate and “hijack” several cellular host mechanisms, including gene splicing and RNA maturation machineries. These modulations alter host cellular responses and enable an optimal expression of viral products throughout infection. The interplay between the host protein p53 and IAV, in particular through the viral nonstructural protein NS1, has been shown to be supportive for IAV replication. However, it remains unknown whether alternatively spliced isoforms of p53, known to modulate p53 transcriptional activity, are affected by IAV infection and contribute to IAV replication. Using a TP53 minigene, which mimics intron 9 alternative splicing, we have shown here that the NS1 protein of IAV changes the expression pattern of p53 isoforms. Our results demonstrate that CPSF4 (cellular protein cleavage and polyadenylation specificity factor 4) independently and the interaction between NS1 and CPSF4 modulate the alternative splicing of TP53 transcripts, which may result in the differential activation of p53-responsive genes. Finally, we report that CPSF4 and most likely beta and gamma spliced p53 isoforms affect both viral replication and IAV-associated type I interferon secretion. All together, our data show that cellular p53 and CPSF4 factors, both interacting with viral NS1, have a crucial role during IAV replication that allows IAV to interact with and alter the expression of alternatively spliced p53 isoforms in order to regulate the cellular innate response, especially via type I interferon secretion, and perform efficient viral replication.
IMPORTANCE Influenza A viruses (IAV) constitute a major public health issue, causing illness and death in high-risk populations during seasonal epidemics or pandemics. IAV are known to modulate cellular pathways to promote their replication and avoid immune restriction via the targeting of several cellular proteins. One of these proteins, p53, is a master regulator involved in a large panel of biological processes, including cell cycle arrest, apoptosis, or senescence. This “cellular gatekeeper” is also involved in the control of viral infections, and viruses have developed a wide diversity of mechanisms to modulate/hijack p53 functions to achieve an optimal replication in their hosts. Our group and others have previously shown that p53 activity is finely modulated by different multilevel mechanisms during IAV infection. Here, we characterized IAV nonstructural protein NS1 and the cellular factor CPSF4 as major partners involved in the IAV-induced modulation of the TP53 alternative splicing that was associated with a strong modulation of p53 activity and notably the p53-mediated antiviral response.
INTRODUCTION
Influenza A viruses (IAV) constitute a major public health issue, causing illness and death in high-risk populations during seasonal epidemics or pandemics (1, 2). IAV belong to the family Orthomyxoviridae of enveloped viruses and contain a segmented genome of single-stranded negative-sense RNA (3). In contrast to most RNA viruses, IAV have a nuclear infectious cycle, therefore requiring direct access to numerous nuclear host factors and machineries to successfully achieve their replication (4). These viruses have notably developed mechanisms to reassign the host spliceosome to simultaneously alter host cellular expression and enable an optimal expression of specific spliced viral products throughout infection (reviewed in reference 5). Although the regulation of viral splicing has been extensively studied, the intricate interactions between IAV and the host spliceosome, as well as the host splicing regulators, remain to be further deciphered.
Among IAV proteins, the nonstructural protein NS1 is known to be a key “multitool” protein with which the virus can extensively alter the host cell responses (6–9). Indeed, NS1 can counteract the antiviral interferon (IFN) response using several mechanisms (6–8). In particular, NS1 acts as a modulator of numerous cellular host processes, including mRNA processing (5, 10), nuclear trafficking/export (11, 12), and translation (13, 14). These multiple accessory functions are well illustrated by the considerable number of NS1 host interactors (more than 50) and the numerous binding sites within the NS1 protein, which include, for example, binding sites for double-stranded RNA (15, 16), p85β for activation of phosphatidylinositol 3-kinase (PI3K) signaling (17), RIG-I, which inhibits the induction of the type I IFN response (IFN-I) (18–20), and cleavage and polyadenylation specificity factor 4 (CPSF4; also known as CPSF30) (21). Among the NS1-interacting host proteins, CPSF4 is a component of the CPSF complex, which is involved in the last step of mRNA maturation and polyadenylation (22, 23). It has been demonstrated that IAV NS1 interacts with CPSF4, thus preventing CPSF4 binding to RNA and, consequently, inhibiting the 3′-end cleavage and polyadenylation of host pre-mRNA (21). It is assumed that this interaction contributes to the NS1-dependent regulation of several host genes and particularly to the inhibition of the IFN response (24–27). Furthermore, it has been recently suggested that the CPSF complex might promote alternative splicing events. The CPSF complex can be recruited as a cofactor by known splicing regulators at sites distal to the polyadenylation site simultaneously to its usual mRNA binding sites (28–30). These data suggest that the interaction between NS1 and CPSF4 could also affect host gene expression at another level of regulation, such as host mRNA splicing.
The host tumor suppressor p53 is a master regulatory transcription factor that is activated in response to various cellular stresses and that regulates a broad range of biological processes, such as cell cycle arrest, apoptosis, and senescence (31, 32). An increasing number of reports suggest that p53 is also a key player in the regulation of immune, inflammatory, and antiviral responses, including that of influenza viruses (33–42). Our current understanding is that the IAV-induced modulation of p53 endogenous expression, and subsequently of p53 transcriptional activity, contributes to maintain a cellular state that favors viral replication during the time of infection (41, 43). We and others have indeed demonstrated that NS1 plays a major role in the functional interplay between IAV and the host p53 pathway (37, 42). We have shown that NS1 interacts with the endogenous p53 protein and alters the binding of p53 to its responsive genes in a promoter-dependent manner (37). However, the complete mechanisms of NS1-induced modulation of p53 activity are still not fully understood. In addition to the canonical full-length p53 protein (also called TAp53α or p53α), the TP53 gene expresses about twelve p53 protein isoforms that result from combinational use of alternative promoters, splicing sites, and/or translational initiation sites (44, 45). In particular, three C-terminal variants (α, β, and γ) that result from alternative splicing of TP53 intron 9 (TP53-i9) have been described. Exclusion of the entire intron 9 generates the canonical full-length p53 protein (or p53α), whereas inclusion of alternative exons 9b and 9g contained in intron 9 gives rise to p53β and p53γ protein isoforms that present short residue sequences in place of the usual oligomerization domain present in p53α, resulting in shorter forms (47 kDa instead of 53 kDa) (44, 45).
The p53 isoforms have been shown to contribute to the cell fate decision in response to stress but also to the maintenance of cell homeostasis, mainly by regulating transcriptional activity of the full-length p53α protein (44, 46, 47). We have notably demonstrated that increased expression of the C-terminal p53β and p53γ isoforms, resulting from fine control of the alternative splicing of TP53-i9, promotes G1 cell cycle arrest and apoptosis in a p53α-dependent manner (47). Interestingly, we reported that IAV differentially affect the endogenous expression of p53 isoforms at both transcriptional and posttranscriptional levels. In response to influenza infection, we observed an increased expression of the p53β isoform, known to be generated by partial retention of TP53-i9 (46). Reciprocally, modulations of the endogenous expression of p53 isoforms affect viral replication in a p53α-dependent manner (38).
Here, we further characterized the functional connections between IAV and the host p53 pathway at the level of C-terminal spliced p53 isoforms. Using a minigene construct, we demonstrated that TP53 splicing is strongly modulated during the time course of IAV infection. In addition, we characterized the viral protein NS1 and the host factor CPSF4 as major partners involved in the IAV-induced modulation of the TP53 alternative splicing that was associated with a strong modulation of p53 transcriptional activity. Our study highlights for the first time a role for a cell factor, CPSF4, in TP53 alternative splicing in response to viral infection, and we have identified a novel regulator of TP53 alternative splicing under basal conditions. In the context of IAV infection, our results suggest that not only spliced p53β and p53γ isoforms but also CPSF4 may contribute to the p53-mediated antiviral type I IFN response.
RESULTS
Influenza virus infection and NS1 protein modulate the alternative splicing of TP53-i9.
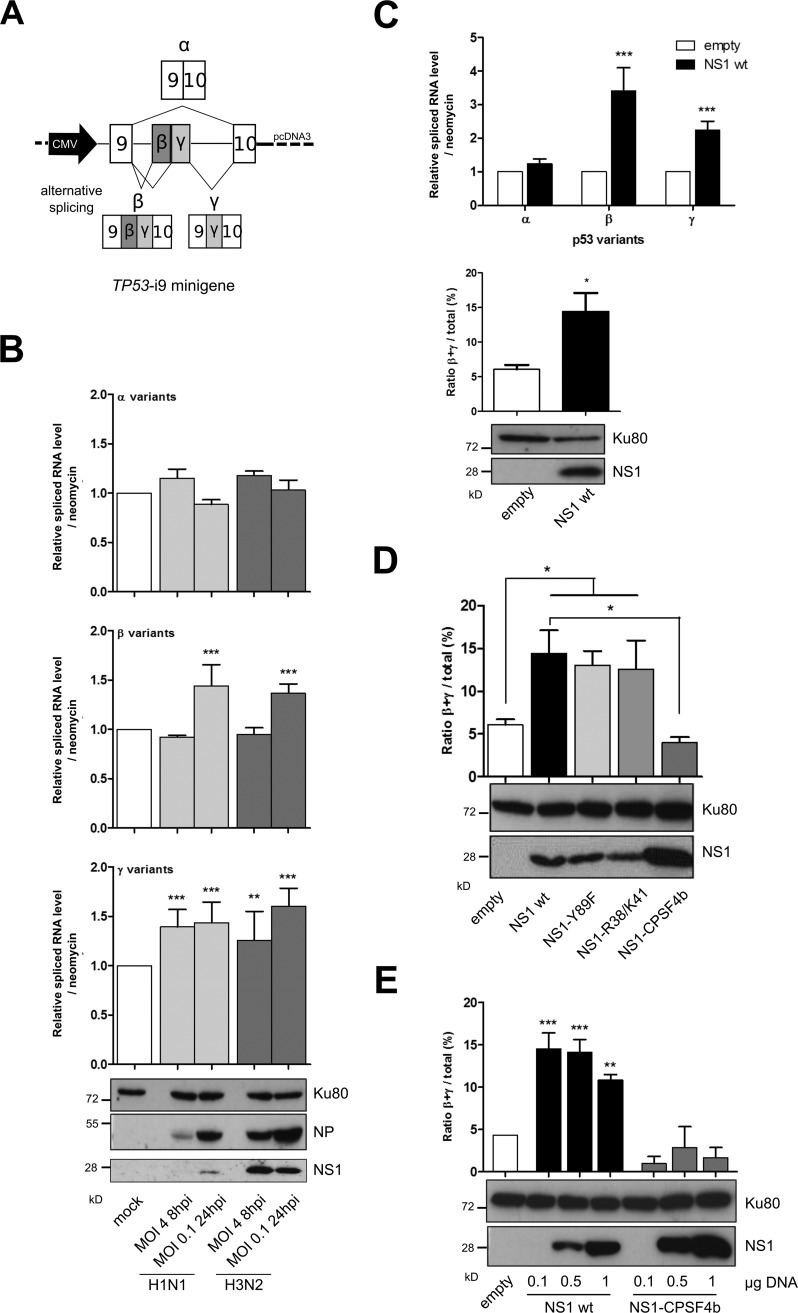
To investigate the impact of infection on the alternative splicing of TP53-i9, we used a previously described minigene approach (38). This TP53-i9 minigene mimics alternative splicing of TP53-i9 and allows analysis of the exclusion and/or partial retention of intron 9, which leads to the production of C-terminal α, β, or γ variants (Fig. 1A). H1299 p53-null cells were transfected with TP53-i9 minigene plasmids for 36 h and then infected with two IAV strains (A/Puerto Rico/8/34 [H1N1] or A/Moscow/10/99 [H3N2]) at a multiplicity of infection (MOI) of 4 or 0.1 for 8 or 24 h postinfection (hpi), respectively. IAV infection was validated by monitoring the expression of NP and NS1 proteins by Western blot analysis, reflecting only slight differences of viral kinetics (Fig. 1B). Relative levels of spliced RNA of the exogenous α, β, and γ isoforms of p53, reflecting the three C-terminal spliced variants, were then quantified by reverse transcription-quantitative PCR (RT-qPCR). Both H1N1 and H3N2 infections affected the levels of α, β, and γ p53 isoforms compared with mock-infected cells (Fig. 1B). At 8 hpi (MOI of 4), we observed significant increases in the p53γ isoform levels, 1.39- and 1.27-fold, in cells infected by H1N1 and H3N2, respectively, compared with that in the mock infection. However, no significant change was observed for both the α and β isoforms. In the context of multiple viral cycles (that is, MOIs of 0.1 and 24 hpi), we observed a broader effect of infection on the retention of TP53-i9. In addition to significant 1.44- and 1.36-fold increases in the level of the p53γ isoform for H1N1 and H3N2, respectively, the level of the β isoform was also significantly increased in IAV-infected cells compared with mock-infected cells (1.43- and 1.60-fold increases for H1N1 and H3N2, respectively) (Fig. 1B). In contrast, the level of the major α isoform remained unaffected by H3N2 infection or was slightly decreased by H1N1 infection. These results suggest that IAV infection promotes the retention of TP53-i9, leading to the generation of β and γ variants of p53 and, thus, could explain the IAV-induced expression of the p53β isoform previously described (38).
FIG 1.
Influenza viruses and the viral NS1 protein alone modulate TP53-i9 minigene alternative splicing. (A) Schematic of the TP53-i9 minigene. Intron 9 of p53 was inserted into pCDNA3 plasmids with part of exon 9 and 10 on both sides, and transcription of mRNA corresponding to each spliced variant was under the control of the CMV promoter (47). (B) Levels of mRNA corresponding to p53 spliced variants α, β, and γ were measured in H1299 cells (p53 null) in which the TP53-i9 minigene plasmid was transfected 36 h before mock infection or infections by H1N1 or H3N2 influenza viruses. Cells lysates were harvested at 8 hpi for infection at an MOI of 4 or at 24 hpi for infection at an MOI of 0.1, and relative levels of mRNA corresponding to α, β, and γ p53 spliced variants were measured by specific RT-qPCR. The mRNA level of each variant was normalized against the neomycin resistance gene, which is also present in the TP53-i9 minigene. Viral protein expression (NP and NS1) was monitored by Western blotting. (C) The relative spliced mRNA level of p53α, p53β, and p53γ isoforms was measured in H1299 cells 48 h after cotransfection of the TP53-i9 minigene and 1 μg of an empty pCI plasmid or a plasmid containing wt NS1 (pCI NS1 wt, from influenza strain A/Moscow/10/99 [H3N2]) and was used to calculate the proportion of β+γ isoforms out of the total α, β, and γ variant p53 mRNA expression. Mean values ± standard deviations of results for at least three independent experiments are shown, and statistical tests compared each condition with its control condition using two-way analysis of variance (ANOVA) (**, P < 0.01; ***, P < 0.001). (D and E) In H1299 cells, the TP53-i9 minigene was cotransfected with 1 μg of empty pCI, pCI NS1wt (results extracted from Fig. 1C), or pCI NS1-Y89F, pCI NS1-R38A/K41A, and pCI NS1-CPSF4b mutants (D) or with increasing amounts (0.1, 0.5, or 1 μg) of pCI NS1 wt or pCI NS1-CPSF4b plasmid (E). Forty-eight hours after cotransfection, levels of α, β, and γ variant mRNA were measured (Fig. S1) and used to estimate the β+γ proportion. The efficacy of NS1 transient expression was validated by Western blotting. Mean values ± standard deviations of results from more than three independent experiments are shown, and statistical tests compared each condition with its control empty condition or NS1 wt condition using Student's t test (*, P < 0,05; **, P < 0.01; ***, P < 0.001).
As the multifunctional NS1 protein is known to affect the host splicing machinery, we then investigated whether NS1 could modulate, by itself, the retention of TP53-i9. The TP53-i9 minigene plasmid was cotransfected with the wild-type (wt) NS1-expressing pCI plasmid (H3N2), which induces NS1 expression, as verified by Western blotting (Fig. 1C, bottom panel). RT-qPCR analyses revealed that NS1 affected the retention of TP53-i9. Indeed, NS1 expression was associated with a significant increase of both p53β and p53γ isoform levels compared with the empty-plasmid transfection condition, the mean fold increases reaching 3.40 and 2.24, respectively (Fig. 1C, top panel). Similar to that observed in the above-mentioned context of IAV infection, the level of p53α isoforms remained unaffected by transient NS1 expression. As the β and γ variants are known to be expressed at lower levels than the major α variant under basal conditions (36), we also determined the percentage of the combined β and γ (β+γ) isoforms out of the three C-terminal p53 isoforms to determine the relative proportion of intron 9 retention. NS1 expression induced a significant >2-fold increase of the β+γ isoform proportion compared with the empty-plasmid condition (14.42% versus 6.06%, respectively) (Fig. 1C, bottom panel). Overall, these results indicate that IAV infection, and also NS1 alone, can modulate the alternative splicing of TP53-i9 and favors the generation of β and γ isoforms of p53.
The CPSF4-binding domain of NS1 is critical to NS1-mediated modulation of TP53-i9 retention.
The impact of different previously described NS1 mutants (16, 17, 21) on TP53-i9 retention was assessed using a TP53-i9 minigene in H1299 p53-null cells to determine the mechanisms underlying NS1-induced retention of TP53-i9. Three pCI plasmids were used to express H3N2 NS1 mutants that are deficient in their binding with either the PI3K p85β subunit (NS1-Y89F), RNA (NS1-R38A/K41A), or CPSF4 (NS1-CPSF4b), all of which had detectable amounts of the various NS1 proteins (Fig. 1D). Similar to wt NS1, there was a strong and significant 2-fold increase in the proportion of β+γ isoforms in NS1-Y89F or NS1-R38A/K41A mutant-expressing cells, compared with that of the empty-plasmid condition (13.03% and 12.58% versus 5%, respectively) (Fig. 1D; see Fig. S1A in the supplemental material). These results suggest that both RNA-binding and p85β-binding domains of NS1 are not involved in the NS1-induced retention of TP53-i9. In contrast, the increased β+γ proportion observed in response to wt NS1 expression is abrogated when the NS1-CPSF4b mutant is expressed (Fig. 1D), which suggests that NS1 mutants that could not bind to CPSF4 lost the capacity to induce TP53-i9 retention. Of note, the relative RNA levels of α, β, and γ forms generated by the TP53-i9 minigene were significantly lower in NS1-CPSF4b-expressing cells than under the empty-plasmid condition, indicating an additional layer of regulation (Fig. S1A).
Increasing numbers of wt NS1 and NS1-CPSF4b-expressing plasmids were then cotransfected with the TP53-i9 minigene in H1299 cells. Under the wt NS1 condition, no dose-dependent effect was observed, and even minimal quantities of wt NS1 plasmid, resulting in barely detectable protein levels, were sufficient to significantly increase the exogenous β+γ isoform proportion (Fig. 1E and Fig. S1B). In contrast, the generation of α, β, and γ forms from the TP53-i9 minigene was progressively reduced with the NS1-CPSF4b mutant as the introduced plasmid quantity increased (Fig. S1B). The augmentation of the β+γ isoform proportion was abrogated at each NS1-CPSF4b dose used, which suggests that the CPSF4-binding domain of NS1 has a role in regulating TP53-i9 retention (Fig. 1E). At endogenous levels in A549 cells expressing the wt TP53 gene, we observed concordant patterns of p53β protein augmentation with wt NS1 versus its absence with NS1-CPSF4b, at least when small amounts of plasmid were transfected (Fig. S1D), in accordance with previous results (38). All together, these results suggest that the CPSF4-binding domain of NS1 is critical for the NS1-induced retention of TP53-i9.
IAV-mediated regulation of p53 expression is affected in NS1 CPSF4 mutant influenza virus.
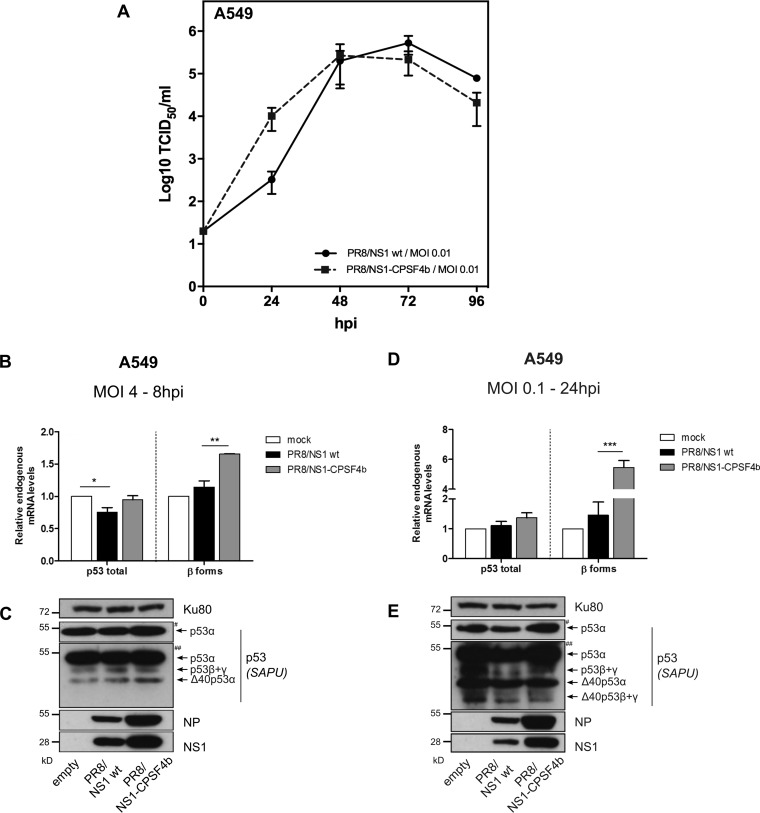
We then investigated the impact of NS1-CPSF4b on the expression of spliced p53 isoforms in the context of IAV infection. The NS1-CPSF4b mutant virus was previously described as having strongly attenuated replication (48). To minimize this limitation, we used the reverse genetics system of the highly replicative A/PuertoRico/8/33 virus (H1N1, PR8) (49) to produce viruses expressing the wt NS1 or mutant NS1-CPSF4b proteins. Virus recovery from pHW2000 plasmid transfection was achieved, and recombinant PR8/NS1-wt and PR8/NS1-CPSF4b viruses were generated and titrated. These two viruses presented comparable growth kinetics and infectivity (Fig. 2A). A549 cells (p53 wt) were infected with either virus, and relative levels of endogenous mRNA of p53 isoforms were measured by RT-qPCR and Western blotting. At 8 hpi, the PR8/NS1 wt virus induced a significant 25% reduction in endogenous p53 total mRNA levels compared with that of mock infection and a slight but not significant increase in both p53β isoform mRNA and protein levels (Fig. 2B and C). Thus, infection with PR8/NS1 wt virus reproduced what was observed with the endogenous p53 isoform at mRNA and protein levels (38) and in H1299 NS1-transfected cells (Fig. 1C). In contrast to PR8/NS1 wt virus, no variation in total p53 mRNA levels was observed with PR8/NS1-CPSF4b virus at 8 hpi compared with mock infection, while a significant 1.65-fold upregulation of β mRNA variants and a slight increase of p53β protein levels were detected, suggesting that the CPSF4-binding domain of NS1 has a role in modulating p53 isoform expression (Fig. 2B and C). At 24 hpi, no changes were observed in the mRNA levels in PR8/NS1 wt virus-infected cells, whereas a drastic and significant increase in β mRNA variants was induced by PR8/NS1-CPSF4b infection compared with mock infection (Fig. 2D). However, this increase in mRNA levels was not matched by a similar increase in p53β protein levels, as observed by Western blotting (Fig. 2E). This discrepancy can result from other regulations occurring during the several viral cycles taking place over the time course of 24 hpi. Nevertheless, these data showed that endogenous p53 isoform expression is differentially affected by influenza virus expressing either the wt NS1 or NS1-CPSF4b mutant protein, underlying the critical role of the CPSF4 binding domain in NS1-induced modulation of p53 isoform expression.
FIG 2.
IAV-regulated p53 expression is affected by NS1 CPSF4-binding mutant. (A) Comparative viral kinetics between recombinant IAV PR8/NS1 wt or PR8/NS1-CPSF4b viruses. A549 cells were infected with the two viruses at an MOI of 0.01, and supernatants were harvested at 24, 48, 72, and 96 hpi for the determination of viral titers. (B to E) A549 cells were mock infected or infected with either recombinant IAV PR8/NS1 wt or PR8/NS1-CPSF4b viruses. Cells lysates were harvested at 8 hpi (MOI of 4 [B and C]) or 24 hpi (MOI of 0.1 [D and E]). Detection of total p53 and p53β isoforms was performed by RT-qPCR and Western blotting (SAPU antibody). Viral proteins NP and NS1 were also detected. # and ## indicate short and long exposures, respectively. Mean values ± standard deviations of results of experimental duplicates are shown, and statistical tests compared each condition with its control condition using two-way ANOVA and Dunnett’s posttest (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Interestingly, we observed that CPSF4 itself affects spliced p53 isoform expression under basal conditions. In fact, we observed a slight increase of α variant mRNA in conjunction with a significant 40% reduction of β variant mRNA levels in p53-null H1299 cells, previously transfected with the TP53-i9 minigene plasmid, when treated with a pool of small interfering RNA (siRNA) that targeted CPSF4 (si-CPSF4), in comparison with a nonspecific siRNA (si-ctrl) (Fig. S2A). In accordance, a similar observation was made at endogenous levels in si-CPSF4-treated A549 cells with an increase in total p53 mRNA levels but no change in the proportion of β variant mRNA levels in comparison to those of si-ctrl-treated cells (Fig. S2B). Thus, these results suggest that the silencing of the cellular CPSF4 factor contributes to the exclusion of TP53-i9, which modulates the spliced p53 isoform ratio and favors the increase of p53α variant mRNA levels, the major form expressed by the TP53 gene. All together, these results demonstrate that the CPSF4-binding domain of NS1 is critical for the IAV-mediated regulation of p53 isoform expression. Moreover, it suggests a role of the NS1 interactant, CPSF4, in p53 isoform expression under both mock and infected conditions.
CPSF4, NS1, and the CPSF4-binding domain of NS1 protein play a critical role in p53 transcriptional activity.
Since we observed that silencing of CPSF4 modulates p53 isoform expression patterns under basal conditions, we investigated the effect of CPSF4 silencing on p53 transcriptional activity in noninfected cells. We used a panel of luciferase reporter vectors that reflect either the intrinsic p53 transcriptional activity (pG13-Luc, a de novo-generated promoter containing 13 consecutive p53-responsive elements) or promoter activity of well-characterized p53 target genes (MDM2-Luc, p21-Luc, or Bax-Luc). Compared with si-ctrl treatment, si-CPSF4 treatment increased pGL13-Luc luciferase activity, indicating that CPSF4 modulates p53 transcriptional activity (Fig. S2C). These results support that CPSF4 contributes to the modulation of the transcriptional activity of p53 in a promoter-dependent manner.
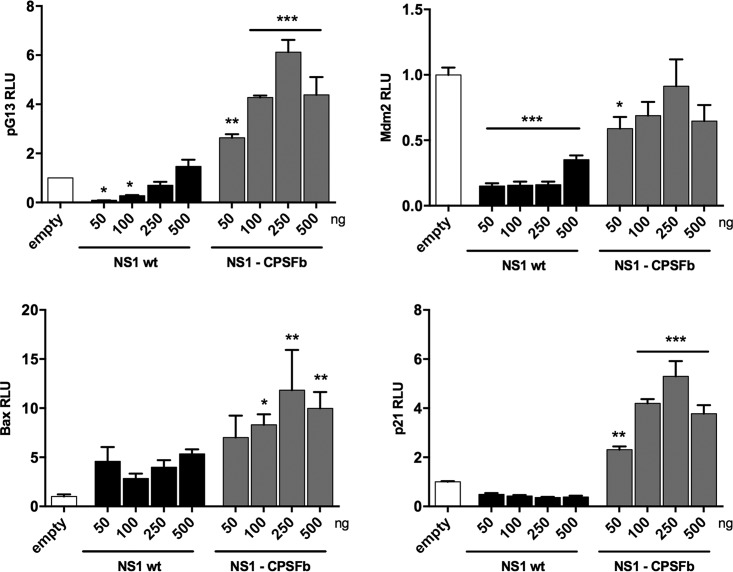
We then investigated whether the NS1-mediated modulation of the p53 isoform expression pattern affected p53 transcriptional activity and whether such modulation was dependent on the CPSF4-binding domain of NS1 (Fig. 3). Increasing amounts of wt NS1- or NS1-CPSF4-expressing vectors were cotransfected with either a pG13-Luc, MDM2-Luc, p21-Luc, or Bax-Luc luciferase reporter vector in A549 cells (NS1 expression was verified by Western blotting [Fig. S1C]). In wt H3N2 NS1-transfected cells, we observed a slight decrease in the pG13-Luc luciferase activity together with a significant increase in Bax and a reduction in MDM2 and p21 promoter activities, in comparison with the empty-plasmid condition. This suggests that NS1 modulates p53 transcriptional activity (Fig. 3), in accordance with previously published studies (including ours), demonstrating that NS1 modulates p53 transcriptional activity in a promoter-specific manner (37, 42). In cells expressing the NS1-CPSF4b mutant, pG13-Luc luciferase activity was significantly increased compared with that of the control and wt NS1 conditions, resulting in a dose-dependent increase in relative luciferase activity that reached a maximum of a 6-fold induction. This was supported by the observations that the NS1-CPSF4b mutant differentially affected MDM2-Luc, p21-Luc, and Bax-Luc promoter activities compared with wt NS1 (Fig. 3). These results indicate that the CPFS4-binding domain of NS1 plays a critical role in the regulation of p53 transcriptional activity.
FIG 3.
The CPSF4-binding domain of NS1 protein plays a crucial role in the alteration of p53 isoform expression and p53 transcriptional activity. A549 cells were transfected with pG13-Luc, Mdm2-Luc, Bax-Luc or p21-Luc reporter plasmids together with increasing concentrations of an NS1-expressing plasmid (either H3N2 NS1 wt or H3N2 NS1-CPSF4b mutant). p53 transactivation activity was measured after 48 h, in triplicate in two independent experiments, and expressed in relative luciferase units (RLU) compared with the empty-plasmid condition. Mean values ± standard deviations are shown, and statistical tests compared each condition with the empty-plasmid condition using one-way ANOVA and Dunnett’s posttest (*, P < 0.05; **, P < 0.01 ***, P < 0.001).
All together, these data indicate that the CPSF4-binding domain of NS1, and CPSF4 itself, affects p53 transcriptional activity and p53-responsive promoters. In addition, the modulation of the p53 isoform expression pattern, resulting from alternative splicing of TP53-i9 and induced by NS1 through its CPSF4-binding domain or by CPSF4 itself, occurred in parallel with the modulation of p53 transcriptional activity, suggesting a functional impact on viral replication.
The silencing of spliced p53 isoforms impairs IAV replication.
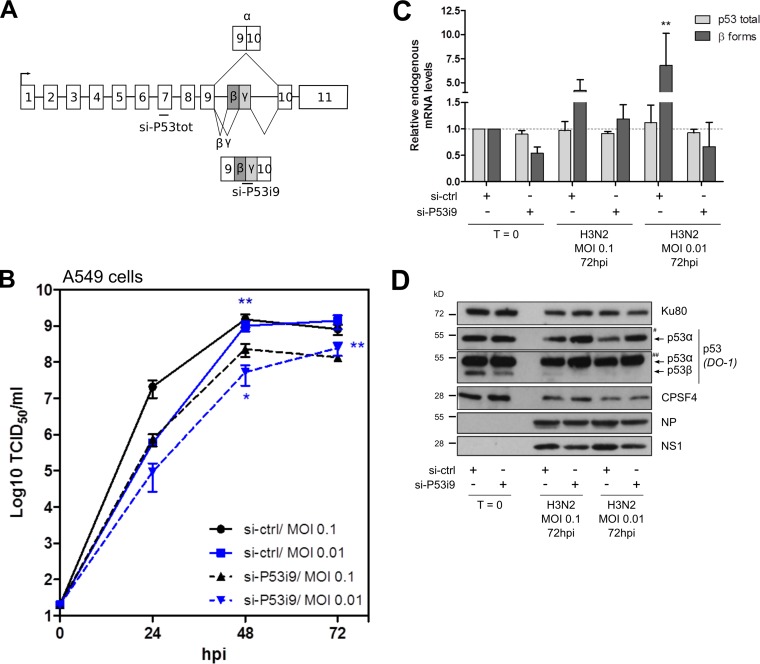
To support the hypothesis of an interplay between NS1, CPSF4, and p53 activity and its functional impact on viral replication, we first determined whether a combined endogenous silencing of the TP53-i9 alternatively spliced p53β and p53γ isoforms could affect IAV replication. We treated A549 cells with either si-ctrl or an siRNA that targets alternatively spliced TP53-i9 (si-P53i9) prior to infection with H3N2 influenza virus (Fig. 4A). First, we verified the silencing of spliced p53 isoforms in response to si-P53i9 treatment (Fig. 4C and D). In noninfected cells, si-P53i9 treatment decreased both β mRNA variant and p53β protein levels by about 40%. As previously described (28), infection alone (si-ctrl condition) did not affect endogenous p53 total mRNA levels but induced a significant and strong increase in endogenous β mRNA variant levels at 72 hpi (4.23- and 6.80-fold increases compared with mock infections for MOIs of 0.1 and 0.01, respectively) (Fig. 4C). Such induction was abrogated in si-P53i9-treated cells, demonstrating the si-RNA treatment efficacy along the infection time course. Second, we analyzed IAV cell titers under these conditions (Fig. 4B). Silencing of TP53-i9 spliced p53 isoforms was associated with a significant decrease in viral titers, particularly at 48 hpi, with 6.5- and 19-fold decreased production levels of influenza virus (for MOIs of 0.1 and 0.01, respectively) compared with that of si-ctrl treatment. Accordingly, viral expression of NP and NS1 proteins decreased in si-P53i9-treated cells compared with si-ctrl-treated cells at 72 hpi, suggesting that spliced p53 isoforms have proviral activity (Fig. 4D). These data are in accordance with our previously published study, which highlighted p53β as a proviral factor (38).
FIG 4.
Silencing of the TP53-i9 alternatively spliced β and γ isoforms impairs IAV replication. (A) A549 cells were treated twice with either the control nonspecific si-RNA (si-ctrl) or a specific si-RNA targeting the p53β and p53γ spliced isoforms. (B) Twenty-four hours after the last si-RNA treatment, cells were infected with H3N2 virus at an MOI of 0.1 or 0.01. Supernatants were harvested at 24-h intervals over 3 days, and the viral replication was determined by endpoint TCID50 titration in MDCK cells (measured in quadruplicate in two independent experiments). (C) Cell lysates were harvested before infection (T = 0) or at 72 hpi to quantify p53 total and p53β mRNA expression levels by RT-qPCR, normalized to actin expression. (D) Cellular and viral proteins were detected by Western blotting. # and ## indicate short and long exposures, respectively. Data represent independent experimental duplicates. Mean values ± standard deviations are shown, and statistical tests compared each condition with the si-ctrl T = 0 condition using two-way ANOVA (*, P < 0.05; **, P < 0.01).
Interestingly, levels of p53α protein were reduced in IAV-infected cells compared with those in mock-infected cells (si-ctrl condition) at both 48 and 72 hpi (Fig. 4D). Surprisingly, while si-P53i9 treatment did not change the level of p53α protein in mock-infected cells, we clearly observed more p53α in infected cells and in si-P53i9-treated A549 cells than in si-ctrl-treated cells. As these variations at the protein level were not observed at the mRNA level, these observations suggest that C-terminal spliced p53 isoforms contribute to the IAV reduction of p53α regulation through a posttranscriptional mechanism (Fig. 4D).
As CPSF4 affects the spliced p53 isoform expression pattern and p53 transcriptional activity (Fig. S2) and as the CPSF4 promoter contains p53 response elements (data not shown), we analyzed CPSF4 expression under si-P53i9-treated conditions. Interestingly, silencing of the spliced β and γ mRNA variants was associated with an increase in CPSF4 protein, under both noninfected and infected conditions, while it decreased CPSF4 mRNA levels, suggesting that there is regulation of CPSF4 at the transcriptional level and that CPSF4 protein stabilization in IAV-infected cells is dependent upon spliced p53 isoforms (Fig. 4D and Fig. S3). These data suggest a regulatory loop between CPSF4 and p53 isoforms that could modulate IAV infection.
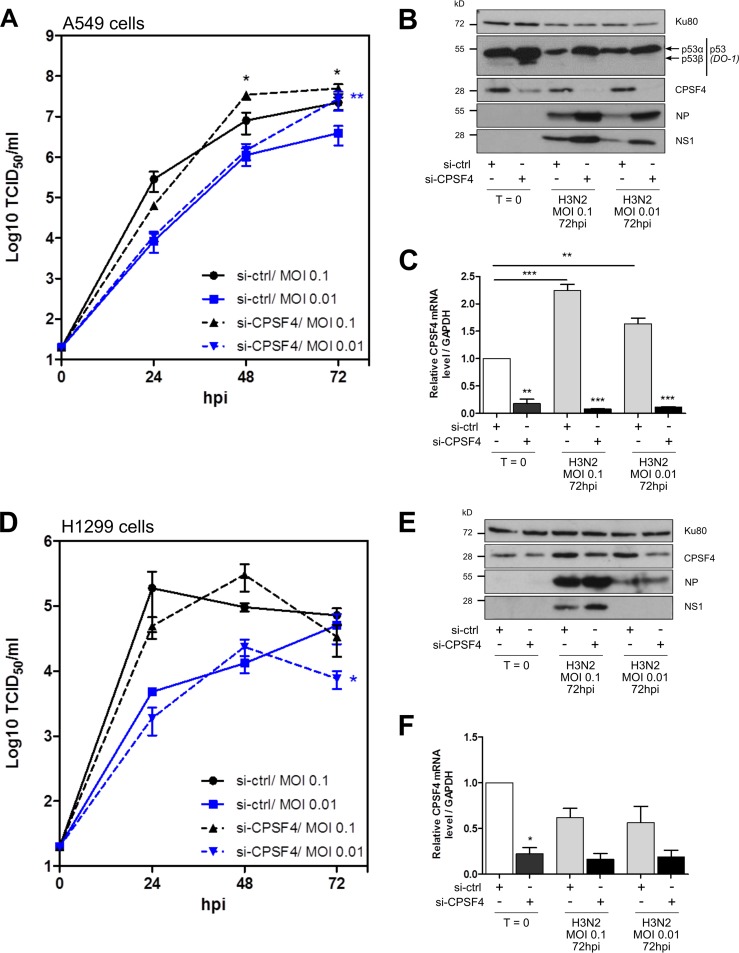
Silencing of CPSF4 also impacts IAV replication.
Owing to the multilayer interplay between p53 isoforms and CPSF4, as well as between NS1 and CPSF4, we investigated the impact of cellular CPSF4 silencing on IAV replication. First, we compared IAV growth kinetics in A549 cells treated with si-CPSF4 or si-ctrl (Fig. 5A). When CPSF4 expression was drastically reduced by si-CPSF4 treatment, at mRNA and protein levels (Fig. 5B and C), a significant increase in viral replication was observed in comparison with that in si-ctrl cells, with viral titers increased by up to 7 times at 72 hpi (MOI of 0.01) (Fig. 5A). Accordingly, viral expression of NP and NS1 proteins was increased in si-CPSF4-treated cells compared with that in si-ctrl-treated cells (Fig. 5B). These data suggest that CPSF4 exhibits an antiviral effect in an independent manner. Inversely, influenza virus (si-ctrl condition) decreased CPSF4 protein expression in A549 cells while it increased CPSF4 mRNA levels, suggesting that there is a regulation of CPSF4 at posttranscriptional levels in response to infection (Fig. 4D and Fig. S3). Moreover, as expected regarding our previous results, si-CPSF4 treatment altered endogenous p53 expression by enhancing levels of both p53α and p53β proteins at 72 hpi (Fig. 5B).
FIG 5.
The silencing of CPSF4 impacts IAV replication in a partially p53-dependent manner. A549 (A to C) or H1299 (D to F) cells were treated twice with either a control nonspecific siRNA (si-ctrl) or a pool of siRNAs targeting cellular CPSF4. Twenty-four hours after the last treatment, cells were infected with H3N2 at an MOI of 0.1 or 0.01. Supernatants were harvested at 24-h intervals over 3 days, and the viral kinetics in A549 cells (A) or H1299 cells (D) were determined by endpoint TCID50 titration in MDCK cells (measured in quadruplicate in two independent experiments). Cell lysates were harvested before infection (T = 0) or at 72 hpi, and cellular p53 and CPSF4, together with viral NP and NS1 proteins, were detected by Western blotting (B and E). CPSF4 mRNA expression was also measured by RT-qPCR (normalized against GAPDH expression) at T = 0 and 72 hpi (C and F). All data represent independent experimental duplicates. Mean values ± standard deviations are shown, and statistical tests compared each condition with the si-ctrl at T = 0 control using two-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Second, we measured IAV growth kinetics in H1299 p53-null cells treated with si-CPSF4 or si-ctrl (Fig. 5D). In clear contrast with results obtained in A549 cells, the si-CPSF4 treatment had no significant impact on viral growth and very limited impact on viral expression (Fig. 5E). Interestingly, the increased CPSF4 mRNA levels seen in response to infection in A549 cells (Fig. 5C) was inhibited in the absence of endogenous p53 expression (si-ctrl conditions) (Fig. 5F). However, at the protein level, CPSF4 protein expression remained elevated in p53-null cells in both noninfected and infected cells, suggesting that CPSF4 protein stability is dependent upon p53 (Fig. 5E). These results support a regulatory loop between CPSF4 and p53 during IAV infection and suggest that the CPSF4 antiviral effect on IAV replication is p53 dependent.
TP53 spliced p53β and p53γ isoforms together with CPSF4 contribute to a p53-mediated type I IFN response to IAV infection and extracellular stresses.
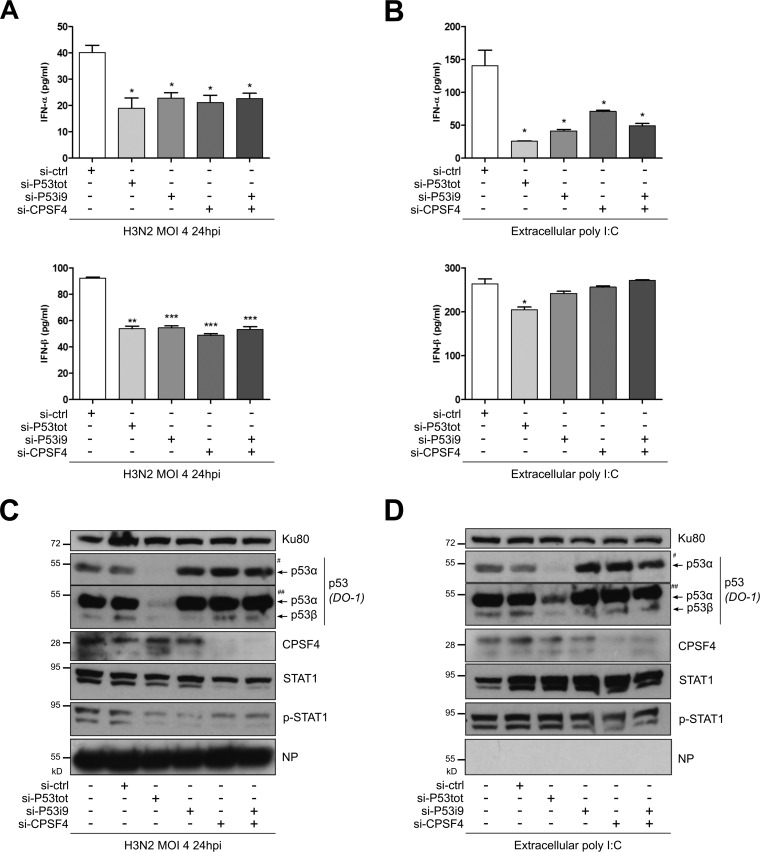
Given that p53 is a key player in the regulation of innate immune responses (reviewed in reference 33), and that the CPSF4-binding domain of NS1 has been recently associated with the type I IFN regulation mechanism during infection (50), we wanted to further investigate the role of the TP53 gene pathway, as well as that of CPSF4, on type I IFN responses in the context of infection, using an siRNA-based approach (Fig. 6). A549 cells were transfected with either a pool of siRNA targeting all p53 isoforms (si-P53tot) (Fig. 4A), siRNA targeting spliced p53β and p53γ isoforms (si-P53i9), siRNA targeting CPSF4 (si-CPSF4), or a nonspecific siRNA (si-ctrl). Treated cells were then infected with influenza virus H3N2 at an MOI of 4. Supernatants were harvested at 24 hpi to monitor IFN-α and IFN-β levels (Fig. 6A), while the siRNA efficiency was controlled by Western blotting using cell lysates (Fig. 6C).
FIG 6.
p53β and p53γ isoforms together with CPSF4 contribute to the p53-mediated IFN-I response to IAV infection and extracellular stresses. A549 cells were treated twice with nonspecific si-RNA (si-ctrl), an siRNA targeting all p53 forms (si-P53tot) (Fig. 4A), a specific siRNA targeting alternatively spliced p53β and p53γ isoforms (si-P53i9) (Fig. 4A), a pool of si-RNAs targeting CPSF4 (si-CPSF4), or a combination of si-P53i9 and si-CPSF4. Twenty-four hours later, cells were infected with H3N2 at an MOI of 4 (A and C) or extracellular poly(I·C) was added to induce a stress through TLR3 activation (B and D). (A and B) Supernatants were harvested 24 h after treatment, and IFN-α and IFN-β levels were quantified. (C) Cell lysates were also collected to monitor infection, si-RNA efficiency, and type I IFN response via STAT1 phosphorylation by Western blotting. # and ## indicate short and long exposures, respectively, with DO-1 antibody. Mean values ± standard deviations of results of experimental duplicates are shown, and the two-way ANOVA test compared each condition with the si-ctrl condition (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
First, our results indicated that the knockdown of all p53 isoforms (including full-length p53α) strongly reduced the production of IFN-I compared with that of cells treated with si-ctrl, with more than 50% and 40% reductions of IFN-α and IFN-β, respectively (Fig. 6A). This result concurred with previous reports and underlined the contribution of TP53 to the regulation of the IFN-mediated antiviral response (51). Interestingly, we observed a very similar impact on IFN production under the si-P53i9-treated condition, with around 50% reduction of IFN-α and IFN-β (Fig. 6A). More importantly, since the reductions of IFN were similar using either si-P53tot or si-P53i9, it suggested that the spliced β and γ mRNA variants are the major, if not the only, component of the global p53-mediated regulation of the IFN response. Regarding CPFS4, a significant 50% reduction of IFN-α and IFN-β was observed in si-CPSF4-treated cells compared with si-ctrl-treated cells, suggesting that CPSF4 is also an important player, by itself, in the regulation of IFN-I production. Furthermore, the combination of si-P53i9 and si-CPSF4 treatments also decreased the level of IFN production; however, the reduction of IFN was in a range similar to that seen with single siRNA treatments, without any cumulative effect. These observations were also supported by Western blotting, using the phosphorylation of signal transducer and activator of transcription 1 (STAT1) as an indicator of the IFN response (Fig. 6C). These data suggest that both the spliced p53β and p53γ isoforms and CPSF4 are involved in the promotion of IFN-I production in response to IAV infection, certainly through a common pathway.
To determine whether the CPSF4- and spliced p53 isoform-induced IFN production also occurs in an extracellular stress context, distinct from IAV infection, we performed similar experiments whereby we measured the response of siRNA-treated cells exposed to poly(I·C) treatment, which mimics nonspecific viral signaling through Toll-like receptor 3 (TLR3) (Fig. 6B and D). Interestingly, we obtained results similar to those seen with IAV infection using the range of siRNA treatment, but only in the context of IFN-α. Indeed, compared with si-ctrl-treated cells, only a limited but statistically significant reduction in IFN-β was achieved in si-P53tot-treated cells (Fig. 6B). These data suggest that both CPSF4-induced IFN production and spliced p53 isoform-induced IFN production are specifically dependent upon IAV infection.
All together, these results clearly indicate that spliced p53β and p53γ isoforms are major contributors to the global p53-mediated regulation of type I IFN. Interestingly, our data suggest that CPSF4/spliced p53 isoforms are part of the same pathway, stimulating type I IFN production during IAV infection.
DISCUSSION
During their replication cycle, IAV are known to “hijack” the host splicing machinery to process their smallest gene segments (M and NS). To do this, they have developed accurate regulation mechanisms to appropriate the host spliceosome to enable the expression of specific spliced IAV products throughout infection (5). Indeed, several genome-wide screening studies, dedicated to pinpointing important host factors for IAV replication, have highlighted a large number of cellular proteins involved in RNA maturation and splicing processes, including several splicing factors (5, 52–54). However, the impact of IAV infection on the regulation of splicing of host mRNA has not yet been extensively investigated.
The transcription factor p53, also named the “guardian of the genome” or the “cellular gatekeeper,” is most of the time, if not always, targeted by viruses during the time course of infection (34, 55). This feature is a powerful way for viruses to modulate or hijack p53-mediated cellular functions, including those involved in the regulation of the immune and inflammatory responses (33, 39). In the context of IAV, we and others have demonstrated that IAV target p53 at several transcriptional and posttranscriptional regulatory levels (36–38, 40–42). Among these, we demonstrated a functional interplay between IAV viral production and p53 isoforms, notably the p53β isoform, known to be generated by partial retention of TP53-i9 (38, 56). Here, we describe another layer of interplay, showing that IAV infection modulates the alternative splicing of TP53 at the level in intron 9 that favors underrepresented spliced forms such as p53β, which concurs with that previously observed (38). Interestingly, the nonstructural protein NS1 appears to be a major player in this mechanism, as experiments involving transient NS1 expression recapitulated our initial observation on TP53 splicing in the context of infection (Fig. 1C). Among the multiple functions of NS1, we currently know that NS1 interferes with several cellular signaling pathways, including p53, but also has an important role in the modulation of mRNA maturation and splicing (6, 7), and our results illustrate well how these different functions overlap during infection.
One striking result in our study was that the CPSF4-binding domain of NS1 is involved in the NS1-mediated modulation of TP53-i9 retention, as demonstrated by transient expression experiments (Fig. 1 and 4) and the use of NS1 recombinant viruses (Fig. 2). In addition, our results suggest a regulatory loop between IAV NS1 and host factors CPSF4/p53 during IAV infection (Fig. 5 and 7). Interestingly, a large part of the literature dedicated to CPSF4 originates from studies performed in the context of IAV infection (21). The first described role of CPSF4 is participation in the maturation and polyadenylation of cellular pre-mRNA. The binding of NS1 to CPSF4 inhibited these activities, contributing to the NS1-mediated regulation of host gene expression and inhibition of the type I IFN response (24, 25, 27, 57). In line with these features, our results indicate that CPSF4 contributes to the p53-mediated type I IFN response to IAV infection (Fig. 6), supporting the hypothesis of a link between CPSF4 and the innate immune response. More recently, several reports have investigated the functional contribution of CPSF4 to the spliceosome machinery, acting as a cofactor for splicing regulators, such as RBFOX2 or HNRNPA1, suggesting that CPSF4 is more directly involved in the regulation of alternative splicing (28–30). Once again, our results are in good agreement with this emerging role for CPSF4. Indeed, we demonstrated that CPSF4 is involved in the regulation of TP53 splicing, showing that the use of a specific si-RNA targeting CPSF4 contributes to the exclusion of TP53-i9 in favor of the expression of the p53α variant, even under basal conditions, outside an IAV infection context (Fig. S2). Thus, we identified a novel splicing regulator of the TP53 alternative splicing at the level of its intron 9. Future investigations will be necessary to understand whether this CPSF4-mediated modulation of splicing is specific to TP53 (or to a subset of host genes) or rather results from a more global impact on host splicing.
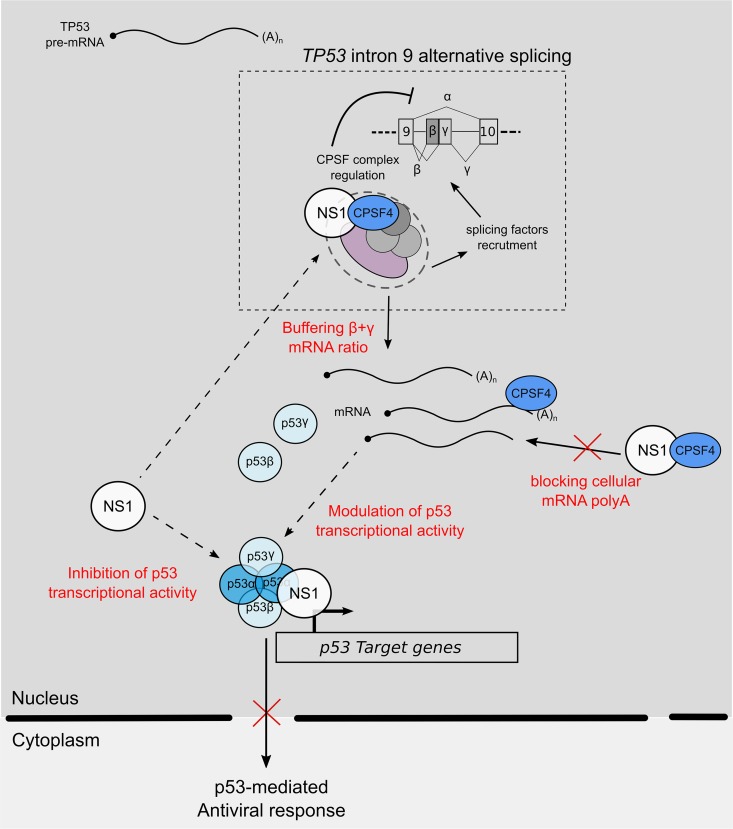
FIG 7.
Working model of interplay between IAV NS1 protein, cellular factor CPSF4, and TP53 splicing. During IAV infection, IAV NS1 inhibits p53 transcriptional activity via its interaction with p53 but also via the modulation of TP53 splicing by “buffering” the function of CPSF4 in mRNA maturation and splicing. As a result, the spliced p53 isoform modulation of p53 transcriptional activity, and notably p53-mediated antiviral responses, coupled to the cellular impact of CPSF4 blockade positively influences viral production. When NS1 is mutated, preventing its binding to CPSF4, this regulation loop is impaired, and the antiviral response is increased, limiting viral production.
All together, our combination of different experimental approaches revealed strong regulatory connections between NS1, CPSF4, and p53. To make a complete picture of this virus-host trio, we focused on TP53 splicing and p53 transcriptional activity. First, we demonstrated that CPSF4 modulates both TP53 splicing and p53 transcriptional activity (Fig. S2). These two events are likely directly correlated, as we previously demonstrated that p53 spliced isoforms regulate p53 transcriptional activity (45, 46). In parallel, we also demonstrated that NS1 modulates the alternative splicing of TP53-i9, in favor of p53β and p53γ isoforms, and that the CPSF4 binding domain of NS1 was involved in this modulation (Fig. 1). This NS1-mediated modulation of TP53 splicing was correlated with a modulation of p53 transcriptional activity, the latter being in agreement with previously published works (37, 42). In addition, several studies, including ours, performed in different physiological and pathological contexts, clearly indicate that alteration or disruption of the splicing machinery has a marked effect on p53 transcriptional activity and p53-mediated cellular responses in a promoter-dependent manner, suggesting a role for p53 spliced isoforms in these processes (47, 58). Our results obtained in the case of IAV infection, or in the experimental contexts of NS1 transient expression/silencing of CPSF4, confirmed these observations, as we described a marked deregulation of TP53 splicing associated with a modulation of p53 transcriptional activity in a promoter dependent-manner (Fig. 1 and 3).
Finally, our results focused on viral production and p53-mediated regulation of IFN-I to complete our understanding of the underlying biological significance of the NS1/CPSF4/p53 interplay. Using specific si-RNAs, we first demonstrated that p53β and p53γ isoforms have a proviral effect (Fig. 4), in line with our previous study (38). In parallel, we have also observed that silencing of CPSF4, which modulates TP53 splicing, significantly affects viral production in a p53-dependent manner (Fig. 5). This experimental silencing of CPSF4 is comparable to that of the early stages of IAV infection, in which NS1 hampers CPSF4 functions. We hypothesize that the role of CPFS4 in IAV infection may rely not only on the maturation of host pre-mRNA (21–23) but also on the modulation of TP53 splicing and p53-mediated responses by NS1. Indeed, little is known about the impact of NS1 on the CPSF complex at the level of splicing regulation. Our hypothesis is that NS1, via its binding with CPSF4, alters the CPSF complex and consecutively modifies its recruitment as a cofactor for splicing regulators. In addition, a converging point between known functions of CPSF4 and those of p53 is the regulation of IFN-I responses. Indeed, CPSF4 is involved in the NS1-dependent regulation of several host genes and particularly in the inhibition of the IFN-I response (24–27). In addition, p53 is known to constitute an amplification loop in the innate immune response, TP53 being part of the family of IFN-stimulated genes (ISGs), and p53 regulates the expression of several genes involved in IFN-I response (reviewed in reference 33). Our results confirmed these observations (Fig. 6) and suggested that spliced p53β and p53γ isoforms are a major contributor to the global p53-mediated regulation of type I IFN and that CPSF4 might be involved in the same regulatory loop.
On the basis of our results, we can propose the following model (depicted in Fig. 7). During IAV infection, NS1 inhibits p53 transcriptional activity via its interaction with p53 but also via the modulation of TP53 splicing by “buffering” the function of CPSF4 in mRNA maturation and splicing (Fig. 7). As a consequence, the spliced p53 isoform modulation of p53 transcriptional activity, and notably p53-mediated antiviral responses, coupled to the cellular impact of CPSF4 blockade, positively influences viral production. When NS1 is mutated, preventing its binding to CPSF4, this regulation loop is impaired, and the antiviral response is increased, limiting viral production (Fig. 7). In future studies, it will be interesting to investigate the presence of naturally occurring mutations in NS1 of IAV circulating strains that could interfere in this NS1/CPSF4/p53 trio, to evaluate a possible impact on strain pathogenesis and virulence.
In conclusion, our results have aided the understanding of the complex mechanisms of IAV hijacking of the host p53 pathway during infection, but they have also highlighted a particular virus-host trio, NS1/CPSF4/p53, that is crucial for viral replication via mRNA maturation and splicing and p53-mediated antiviral responses.
MATERIALS AND METHODS
Cells lines, influenza viruses, and infection.
Human lung epithelial A549 (wild-type p53; ATCC CCL-185) and H1299 (p53 null; ATCC CRL-5803) cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% (or 5% for H1299 cells) of heat-inactivated fetal calf serum (Dutscher), 2 mM l-glutamine (Sigma-Aldrich), penicillin (100 U/ml) and streptomycin (100 μg/ml) (Lonza), under a 5% CO2 atmosphere.
Influenza viruses A/Moscow/10/99 (H3N2) and A/Puerto Rico/8/34 (H1N1), obtained from the French national influenza monitoring network GROG (Groupes Régionaux d’Observation de la Grippe, Lyon, France), were produced in Madin-Darby canine kidney (MDCK) cells (ATCC CCL-34) in Eagle’s minimum essential medium (EMEM; Life Technologies). Viruses were titrated on confluent layers of MDCK cells in 96-well plates to determine the 50% tissue culture infectious dose by endpoint titration (TCID50/ml), which was determined using the Reed and Muench statistical method (59). A549 and H1299 cells were infected at a multiplicity of infection (MOI) of 0.1 or 4 TCID50/cell. After 1 h of viral adsorption, cells were overlaid with DMEM supplemented with 2 mM l-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), and 0.5 μg/ml TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Roche Diagnostics) and incubated at 37°C.
For viral growth kinetic assays, MDCK cells were infected at an MOI of 10−1 or 10−2 TCID50/cell. After a 1-h viral adsorption period, cells were overlaid with Eagle's minimum elementary medium (Lonza) supplemented with 1 μg/ml trypsin (Roche Diagnostics) and further incubated at 34°C. Harvested supernatants were centrifuged at 1,500 × g for 10 min and stored at −80°C until analysis.
Plasmids and the TP53-i9 minigene.
For transient expression of NS1, A549 and H1299 cells at 40% of confluence were transfected using TransIT-LT1 reagent (Mirus), in accordance with the manufacturer’s instructions, with a panel of pCI plasmids empty or carrying wild-type or mutated NS1 sequences derived from the H3N2 strain. Wild-type pCI-NS1 (NS1 wt) was a kind gift from Nadia Naffakh, Institut Pasteur, France. Three pCI-NS1 mutants (NS1-Y89F, NS1-R38A/K41A, and NS1-CPSF4b, mutated from amino acids 184 to 188, sequence RFLRY in place of GLEWN) were obtained by site-directed mutagenesis. Each plasmid was validated by sequencing.
For the TP53 splicing assay, the pcDNA3-ASAi9 p53 minigene plasmid (named the TP53-i9 minigene) was used, the design and protocol of which were previously described (47). The pcDNA3-ASAi9 p53 minigene contains a portion of the TP53 gene that includes the 3′ end of exon 9, the entire intron 9, and the beginning of exon 10, as pictured in Fig. 1A, as well as the neomycin resistance gene used to assess transfection efficiency. In infection experiments, cells were infected as described, 24 to 48 h after TP53-i9 minigene transfection. In cotransfection experiments, the TP53-i9 minigene was transfected simultaneously with pCI-NS1 wt-expressing plasmid (quantities indicated in figure legends).
RNA analysis and real-time quantitative PCR.
Cells lysates were harvested at 8 h postinfection (hpi) (MOI of 4) or at 24 hpi (MOI of 0.1), and the total RNA was extracted using the RNeasy minikit (Qiagen), according to the supplier’s protocol. Reverse transcription was performed on 1 μg of total RNA with SuperScriptII reverse transcriptase (Invitrogen) at 42°C, followed by a quantitative PCR using SYBR green qPCR master mix (Agilent, Santa Clara, CA, USA). In the TP53-i9 minigene experiment, specific primers for each p53 spliced RNA or neomycin resistance gene were used, as previously described (47). For quantitation of endogenous p53 mRNA levels, qPCR was carried out using TaqMan 2× Universal PCR Master Mix No AmpErase UNG (Applied Biosystem, Carlsbad, CA, USA) with specific sets of primers and probes: E8/9 primers for amplification of total p53 isoforms, p53β primers for amplification of β forms, and actin primers for normalization, as previously described (47). The use of actin as a normalization control was validated in control experiments, indicating that IAV infection and/or si-CPSF4 did not affect actin CT by RT-qPCR. A complete list of primers and probes used in this study is available in Table S1 in the supplemental material. The ΔΔCT method was used to determine the fold change of mRNA levels using neomycin resistance or the actin expression level as a reference. Relative mRNA levels were quantified in triplicate, in at least three independent experiments, to perform statistical analyses. The proportion of β+γ mRNA variants relative to total expression was measured by dividing the sum of β and γ relative quantities by the sum of α, β, and γ quantities, all forms being individually normalized against neomycin. This proportion of β+γ variants is used an indicator of the extent of splicing for comparison purposes.
Western blotting.
Total protein was extracted by scraping and syringing cells in 1× NuPAGE LDS buffer (Invitrogen). Approximately 15 to 30 μg of protein was loaded on 10% or 12% SDS gels and analyzed by immunoblotting using the following antibodies: sheep polyclonal antibody anti-total p53 isoforms (SAPU, Dundee, United Kingdom); the rabbit polyclonal antibodies anti-NS1 (30F/31F, kind gift of Daniel Marc, INRA Tours, France), anti-NP (CDC/IVPS, 30AUG01), and anti-phosphorylated (Tyr701) STAT1 (number 9167; Cell Signaling Technology); the mouse monoclonal antibodies anti-p53 TA isoforms (DO-1, Dundee, United Kingdom), anti-NS1 (Santa Cruz, sc-130568), anti-CPSF4 antibody (Santa Cruz, sc-393316), anti-STAT1 (number 9176; Cell Signaling Technology), and anti-Ku80 (AbCam), which was used as a loading control.
Transactivation assay.
A549 cells were transfected with 1 μg of firefly luciferase reporter vectors corresponding to different p53-responsive elements. Cells were cotransfected with empty or NS1-expressing pCI plasmids and harvested at 48 hpi. Transfection efficiency was normalized using 100 ng of Renilla luciferase plasmid. Luciferase activity was measured in whole-cell extracts, in triplicate in two independent experiments, using the Dual-Luciferase reporter assay system (Promega), according to the manufacturer’s instructions. The different reporter vectors used were pG13-Luc with the luciferase gene under the control of 13 copies of the p53-binding consensus sequence (5′-CCAGGCAAGTCCAGGCAGG-3′) and p21-Luc, Mdm2-Luc, and Bax-Luc with the luciferase gene under the control of the complete (p21) or partial (Mdm2 and Bax) promoter sequence of the corresponding genes (60–62).
Recombinant viruses.
Recombinant PR8/NS1 wt and PR8/NS1-CPSF4b viruses were generated by reverse genetics approaches, as previously described (63). Briefly, a pHW2000 plasmid containing NS sequence from H3N2 (49) was mutated to generate a NS1-CPSF4b mutant (from amino acids 184 to 188, sequence RFLRY in place of GLEWN). Recombinant virus generation was performed by transfection of 293T cells (ATCC CRL-3216) using a set of pHW2000 plasmids from the PR8 system and the pHW2000 plasmid containing NS wt or NS1-CPSF4b sequences from H3N2 coding. At 48 hpi, viruses in the culture supernatant were harvested and used to infect a layer of MDCK cells. Viral stocks were prepared after three passages on MDCK cells, and titers were measured using standard TCID50 methods. Full genomes of recombinant viruses were validated by sequencing.
si-RNA treatment.
A549 or H1299 cells (cell confluence, 50%), seeded in antibiotic-free medium, were transfected twice at 24-h intervals with either 50 nM nonspecific si-RNA (si-ctrl, catalog number 0R-0030-Neg05; Eurogentec) or a pool of si-RNA CPSF4 (On-Targetplus human CPSF4 number 10898 siRNA Smartpool; Dharmacon), specific si-RNAs targeting global p53 isoforms (si-P53tot), or spliced p53β and p53γ isoforms (siP53-i9) (Fig. 4A) (Eurogentec based on sequences in reference 47), using oligofectamine (Life Technologies) according to the manufacturer’s instructions. Cells were then infected or treated 24 h after the last si-RNA transfection. The efficiency of si-RNA-mediated knockdown after treatment was evaluated by RT-qPCR and/or Western blotting.
IFN-α/β ELISA.
Twenty-four hours after si-RNA transfection, A549 cells were infected with H3N2 at an MOI of 4 or alternatively treated with 50 μg/ml of extracellular poly(I·C) (Sigma-Aldrich). Forty-eight hours after transfection, IFN-α and IFN-β levels were quantified in supernatants using the VeriKine human interferon alpha multi-subtype serum ELISA kit or the VeriKine-HSTM human interferon beta serum ELISA kit (PBL Assay Science, NJ, USA), respectively, in accordance with the manufacturer’s protocol.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Université Claude Bernard Lyon 1, Lyon, France. J.D. received support from the Région Auvergne-Rhône-Alpes (grant CMIRA ExploRA’DOC) and the Consulat Général de France à Québec (Program Frontenac).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02168-18.
REFERENCES
- 1.Paules C, Subbarao K. 2017. Influenza. Lancet 390:697–708. [DOI] [PubMed] [Google Scholar]
- 2.Neumann G, Kawaoka Y. 2011. The first influenza pandemic of the new millennium. Influenza Other Respir Viruses 5:157–166. doi: 10.1111/j.1750-2659.2011.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palese PSM. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley P (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois J, Terrier O, Rosa-Calatrava M. 2014. Influenza viruses and mRNA splicing: doing more with less. mBio 5:e00070. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marc D. 2014. Influenza virus non-structural protein NS1: interferon antagonism and beyond. J Gen Virol 95:2594–2611. doi: 10.1099/vir.0.069542-0. [DOI] [PubMed] [Google Scholar]
- 7.Krug RM. 2015. Functions of the influenza A virus NS1 protein in antiviral defense. Curr Opin Virol 12:1–6. doi: 10.1016/j.coviro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayllon J, Garcia-Sastre A. 2015. The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- 9.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 10.Ramos I, Carnero E, Bernal-Rubio D, Seibert CW, Westera L, García-Sastre A, Fernandez-Sesma A. 2013. Contribution of double-stranded RNA and CPSF30 binding domains of influenza virus NS1 to the inhibition of type I interferon production and activation of human dendritic cells. J Virol 87:2430–2440. doi: 10.1128/JVI.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira CF, Read EKC, Wise HM, Amorim MJ, Digard P. 2017. Influenza A virus NS1 protein promotes efficient nuclear export of unspliced viral M1 mRNA. J Virol 91:e00528-17. doi: 10.1128/JVI.00528-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mor A, White A, Zhang K, Thompson M, Esparza M, Munoz-Moreno R, Koide K, Lynch KW, Garcia-Sastre A, Fontoura BM. 2016. Influenza virus mRNA trafficking through host nuclear speckles. Nat Microbiol 1:16069. doi: 10.1038/nmicrobiol.2016.69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panthu B, Terrier O, Carron C, Traversier A, Corbin A, Balvay L, Lina B, Rosa-Calatrava M, Ohlmann T. 2017. The NS1 protein from influenza virus stimulates translation initiation by enhancing ribosome recruitment to mRNAs. J Mol Biol 429:3334–3352. doi: 10.1016/j.jmb.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 14.de la Luna S, Fortes P, Beloso A, Ortín J. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol 69:2427–2433.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatada E, Fukuda R. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol 73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A, Wong SM, Yuan YA. 2009. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res 19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- 17.Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ, Randall RE, Russell RJ. 2010. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc Natl Acad Sci U S A 107:1954–1959. doi: 10.1073/pnas.0910715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol 36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 19.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol 9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 20.Mibayashi M, Martínez-Sobrido L, Loo Y-M, Cárdenas WB, Gale M, García-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1:991–1000. [DOI] [PubMed] [Google Scholar]
- 22.Bienroth S, Keller W, Wahle E. 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J 12:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11:1703–1716.] [DOI] [PubMed] [Google Scholar]
- 24.Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, Krug RM, Montelione GT. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A 105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dankar SK, Miranda E, Forbes NE, Pelchat M, Tavassoli A, Selman M, Ping J, Jia J, Brown EG. 2013. Influenza A/Hong Kong/156/1997(H5N1) virus NS1 gene mutations F103L and M106I both increase IFN antagonism, virulence and cytoplasmic localization but differ in binding to RIG-I and CPSF30. Virol J 10:243. doi: 10.1186/1743-422X-10-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. 2014. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 88:12146–12151. doi: 10.1128/JVI.01567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2017. NS1 protein amino acid changes D189N and V194I affect interferon responses, thermosensitivity, and virulence of circulating H3N2 human influenza A viruses. J Virol 91:e01930-16. doi: 10.1128/JVI.01930-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra A, Ou J, Zhu LJ, Green MR. 2015. Global analysis of CPSF2-mediated alternative splicing: integration of global iCLIP and transcriptome profiling data. Genom Data 6:217–221. doi: 10.1016/j.gdata.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misra A, Green MR. 2016. From polyadenylation to splicing: dual role for mRNA 3′ end formation factors. RNA Biol 13:259–264. doi: 10.1080/15476286.2015.1112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinson HG. 2011. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA 2:459–470. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 31.Kruiswijk F, Labuschagne CF, Vousden KH. 2015. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 32.Lane D, Levine A. 2010. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz-Fontela C, Mandinova A, Aaronson SA, Lee SW. 2016. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol 16:741–750. doi: 10.1038/nri.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato Y, Tsurumi T. 2013. Genome guardian p53 and viral infections. Rev Med Virol 23:213–220. doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- 35.Yan W, Wei J, Deng X, Shi Z, Zhu Z, Shao D, Li B, Wang S, Tong G, Ma Z. 2015. Transcriptional analysis of immune-related gene expression in p53-deficient mice with increased susceptibility to influenza A virus infection. BMC Med Genomics 8:52. doi: 10.1186/s12920-015-0127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nailwal H, Sharma S, Mayank AK, Lal SK. 2015. The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43. Cell Death Dis 6:e1768. doi: 10.1038/cddis.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terrier O, Diederichs A, Dubois J, Cartet G, Lina B, Bourdon JC, Rosa-Calatrava M. 2013. Influenza NS1 interacts with p53 and alters its binding to p53-responsive genes, in a promoter-dependent manner. FEBS Lett 587:2965–2971. doi: 10.1016/j.febslet.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Terrier O, Marcel V, Cartet G, Lane DP, Lina B, Rosa-Calatrava M, Bourdon JC. 2012. Influenza A viruses control expression of proviral human p53 isoforms p53beta and Delta133p53alpha. J Virol 86:8452–8460. doi: 10.1128/JVI.07143-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz-Fontela C, Pazos M, Delgado I, Murk W, Mungamuri SK, Lee SW, Garcia-Sastre A, Moran TM, Aaronson SA. 2011. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J Immunol 187:6428–6436. doi: 10.4049/jimmunol.1101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Deng X, Yan W, Zhu Z, Shen Y, Qiu Y, Shi Z, Shao D, Wei J, Xia X, Ma Z. 2012. Stabilization of p53 in influenza A virus-infected cells is associated with compromised MDM2-mediated ubiquitination of p53. J Biol Chem 287:18366–18375. doi: 10.1074/jbc.M111.335422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terrier O, Josset L, Textoris J, Marcel V, Cartet G, Ferraris O, N'Guyen C, Lina B, Diaz JJ, Bourdon JC, Rosa-Calatrava M. 2011. Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway. Virol J 8:285. doi: 10.1186/1743-422X-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Shen Y, Qiu Y, Shi Z, Shao D, Chen P, Tong G, Ma Z. 2010. The non-structural (NS1) protein of influenza A virus associates with p53 and inhibits p53-mediated transcriptional activity and apoptosis. Biochem Biophys Res Commun 395:141–145. doi: 10.1016/j.bbrc.2010.03.160. [DOI] [PubMed] [Google Scholar]
- 43.Turpin E, Luke K, Jones J, Tumpey T, Konan K, Schultz-Cherry S. 2005. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J Virol 79:8802–8811. doi: 10.1128/JVI.79.14.8802-8811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khoury MP, Bourdon JC. 2010. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol 2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joruiz SM, Bourdon JC. 2016. p53 isoforms: key regulators of the cell fate decision. Cold Spring Harb Perspect Med 6:a026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. 2014. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ 21:1377–1387. doi: 10.1038/cdd.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noah DL, Twu KY, Krug RM. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386–395. [DOI] [PubMed] [Google Scholar]
- 49.Terrier O, Moules V, Carron C, Cartet G, Frobert E, Yver M, Traversier A, Wolff T, Riteau B, Naffakh N, Lina B, Diaz JJ, Rosa-Calatrava M. 2012. The influenza fingerprints: NS1 and M1 proteins contribute to specific host cell ultrastructure signatures upon infection by different influenza A viruses. Virology 432:204–218. doi: 10.1016/j.virol.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 50.Wang BX, Brown EG, Fish EN. 2017. Residues F103 and M106 within the influenza A virus NS1 CPSF4-binding region regulate interferon-stimulated gene translation initiation. Virology 508:170–179. doi: 10.1016/j.virol.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 52.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 53.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T, Kawaoka Y. 2015. Influenza virus-host interactomes as a basis for antiviral drug development. Curr Opin Virol 14:71–78. doi: 10.1016/j.coviro.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazo PA, Santos CR. 2011. Interference with p53 functions in human viral infections, a target for novel antiviral strategies? Rev Med Virol 21:285–300. doi: 10.1002/rmv.696. [DOI] [PubMed] [Google Scholar]
- 56.Terrier O, Bourdon JC, Rosa-Calatrava M. 2013. p53 protein isoforms: key regulators in the front line of pathogen infections? PLoS Pathog 9:e1003246. doi: 10.1371/journal.ppat.1003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allende-Vega N, Dayal S, Agarwala U, Sparks A, Bourdon JC, Saville MK. 2013. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 32:1–14. doi: 10.1038/onc.2012.38. [DOI] [PubMed] [Google Scholar]
- 59.Moules V, Ferraris O, Terrier O, Giudice E, Yver M, Rolland JP, Bouscambert-Duchamp M, Bergeron C, Ottmann M, Fournier E, Traversier A, Boule C, Rivoire A, Lin Y, Hay A, Valette M, Marquet R, Rosa-Calatrava M, Naffakh N, Schoehn G, Thomas D, Lina B. 2010. In vitro characterization of naturally occurring influenza H3NA- viruses lacking the NA gene segment: toward a new mechanism of viral resistance? Virology 404:215–224. doi: 10.1016/j.virol.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 60.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. [DOI] [PubMed] [Google Scholar]
- 61.Miyashita T, Reed JC. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293–299. [DOI] [PubMed] [Google Scholar]
- 62.Juven T, Barak Y, Zauberman A, George DL, Oren M. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8:3411–3416. [PubMed] [Google Scholar]
- 63.Essere B, Yver M, Gavazzi C, Terrier O, Isel C, Fournier E, Giroux F, Textoris J, Julien T, Socratous C, Rosa-Calatrava M, Lina B, Marquet R, Moules V. 2013. Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proc Natl Acad Sci U S A 110:E3840–E3848. doi: 10.1073/pnas.1308649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.