Understanding the effect of sphingolipid metabolism on viral pathogenesis provide important insights into the development of therapeutic strategies against microbial infections. In this study, we demonstrate a critical role of ceramide during influenza A virus infection. We demonstrate that ceramide produced through de novo biosynthesis possess an antiviral role. These observations unlock new opportunities for the development of novel antiviral therapies against influenza.
KEYWORDS: influenza virus, ceramide, ceramide analogue, ceramide synthase, de novo pathway, serine palmitoyltransferase, sphingolipids
ABSTRACT
Annual influenza outbreaks are associated with significant morbidity and mortality worldwide despite the availability of seasonal vaccines. Influenza pathogenesis depends on the manipulation of host cell signaling to promote virus replication. Ceramide is a sphingosine-derived lipid that regulates diverse cellular processes. Studies highlighted the differential role of ceramide de novo biosynthesis on the propagation of various viruses. Whether ceramide plays, a role in influenza virus replication is not known. In this study, we assessed the potential interplay between the influenza A (IAV) and ceramide biosynthesis pathways. The accumulation of ceramide in human lung epithelial cells infected with influenza A/H1N1 virus strains was evaluated using thin-layer chromatography and/or confocal microscopy. Virus replication was assessed upon the regulation of the de novo ceramide biosynthesis pathway. A significant increase in ceramide accumulation was observed in cells infected with IAV in a dose- and time-dependent manner. Inoculating the cells with UV-inactivated IAV did not result in ceramide accumulation in the cells, suggesting that the induction of ceramide required an active virus replication. Inhibiting de novo ceramide significantly decreased ceramide accumulation and enhanced virus replication. The addition of exogenous C6-ceramide prior to infection mediated an increase in cellular ceramide levels and significantly attenuated IAV replication and reduced viral titers (≈1 log10 PFU/ml unit). Therefore, our data demonstrate that ceramide accumulation through de novo biosynthesis pathway plays a protective and antiviral role against IAV infection. These findings propose new avenues for development of antiviral molecules and strategies.
IMPORTANCE Understanding the effect of sphingolipid metabolism on viral pathogenesis provide important insights into the development of therapeutic strategies against microbial infections. In this study, we demonstrate a critical role of ceramide during influenza A virus infection. We demonstrate that ceramide produced through de novo biosynthesis possess an antiviral role. These observations unlock new opportunities for the development of novel antiviral therapies against influenza.
INTRODUCTION
Influenza A virus (IAV) is a major etiologic agent of acute respiratory tract infections. Influenza infections are associated with a significant morbidity and mortality burden worldwide. The Centers for Disease Control and Prevention (CDC) recently estimated that influenza results in 650,000 deaths worldwide annually (1). IAV belongs to the Orthomyxoviridae family, and it is divided into subtypes based on the surface proteins: hemagglutinin (HA) and the neuraminidase (NA) (2, 3). Currently, H1N1 and H3N2 are the circulating influenza A virus subtypes among humans (4). Avian IAVs (e.g., H5N1 and H7N9) remain a public health concern due to their pandemic potential (2, 3, 5, 6). Antiviral drugs such as neuraminidase inhibitors are highly effective against influenza; however, the propensity of the virus to mutate and develop resistant mutations undermines the efficacy of these drugs (7–11). This highlights the urgent need for new therapeutic targets that are not influenced by IAV genetic diversity and yet capable of inhibiting its replication (12). Understanding the cellular mechanisms underlying virus replication can identify such host targets. Infection by IAV is initiated through its binding to sialic acid receptors on the host membrane. After endocytosis, the viral and endosomal membranes fuse and the viral genome is released into the host cytoplasm (13). This genome consists of viral RNA bound to RNA-dependent RNA polymerase to form viral ribonucleoprotein (vRNP). The middle region of the vRNP is associated with an oligomeric nucleoprotein (NP) (5, 6, 14, 15). Viral RNA (vRNA) is transported from the cytoplasm into the nucleus, where it undergoes transcription into viral mRNA. This process is primer dependent and is orchestrated by the viral RNA polymerase that adds a short-capped primer derived from host pre-mRNA transcripts in a process called cap-snatching. Then, it sutures a poly(U) stretch near the 5′ end, resulting in autopolyadenylation (16, 17). The newly formed viral transcripts are exported to the cytoplasm, where they are processed by the host machinery to produce new viral proteins (18). In contrast, vRNA synthesis is an unprimed process mediated by reverse transcription from viral cRNA that acts as a template. The newly synthesized vRNA couples with nucleoprotein to form vRNP that is exported from the nucleus to the cell membrane, where the assembly of new virions occurs (13, 17, 18).
Sphingolipids are key lipid constituents of the plasma membranes of eukaryotes. This lipid family consists of bioactive molecules that act as second messengers and regulate a myriad of intracellular signals (19). Ceramide is a central intermediate in sphingolipid metabolism. It is responsible for maintaining membrane dynamics, fluidity, and internal membrane transport (20). Also, it plays a pivotal role in regulating cell viability, senescence, differentiation, cell cycle, and stress response through a number of signaling cascades (21–27). Ceramide is produced through three distinct metabolic pathways: de novo biosynthesis, sphingomyelin (SM) hydrolysis, and salvage pathway. De novo biosynthesis takes place in the endoplasmic reticulum (ER) and ER-associated membranes. The process starts with the condensation of serine and palmitoyl coenzyme A, a rate-limiting step catalyzed by serine palmitoyltransferase (SPT), to produce 3-keto-dihydrosphingosine. The product is reduced to dihydrosphingosine (sphinganine) and then acylated by ceramide synthase (CerS) to form dihydroceramide. Finally, the desaturase inserts a double bond between carbons 4 and 5 of the sphingoid backbone to produce ceramide, which is transported from the ER to the Golgi apparatus to be further metabolized into complex sphingolipids (28). Ceramide can be also produced by hydrolysis of SM, the most abundant membrane sphingolipid, which is catalyzed by acid or neutral sphingomyelinase (SMase) enzymes within the plasma membrane or lysosomes. In the salvage pathway, the ceramide pool produced from the degradation of sphingolipids and glycolipids is hydrolyzed by ceramidases into sphingosine and free fatty acid. Long-chain sphingoid bases can be recycled back into ceramide by CerS or phosphorylated by sphingosine kinases to form sphingosine-1-phosphate (S1P), which has potent bioactivities generally opposite to those of ceramide (28–30). The spatial separation of the enzymes contributing to ceramide production permits a differential regulation of these pathways.
Ceramide is associated with different pathobiological disorders such as cancer, liver disease, diabetes, cardiovascular disease, and lung inflammation (31–35). Previous studies demonstrated that drugs capable of inducing ceramide accumulation are potential anticancer treatments (23–25, 29, 36–38). Ceramide can also enhance or inhibit viral replication suggesting that clinical manipulation of ceramide metabolism might be a potential target against viral infections (39–48). Some studies highlighted the role of S1P in modulating IAV infection and replication (12, 49). However, the interplay between ceramide and IAV has not been addressed. Intriguingly, pathways that are regulated by IAV and ceramide are interconnected, which raises the question about a potential role for ceramide during IAV infection (25, 26, 50–53). In the present study, we sought to determine the effect of ceramide produced through the de novo and salvage pathways on IAV replication.
RESULTS
IAV infection triggers ceramide accumulation in a dose- and time-dependent manner.
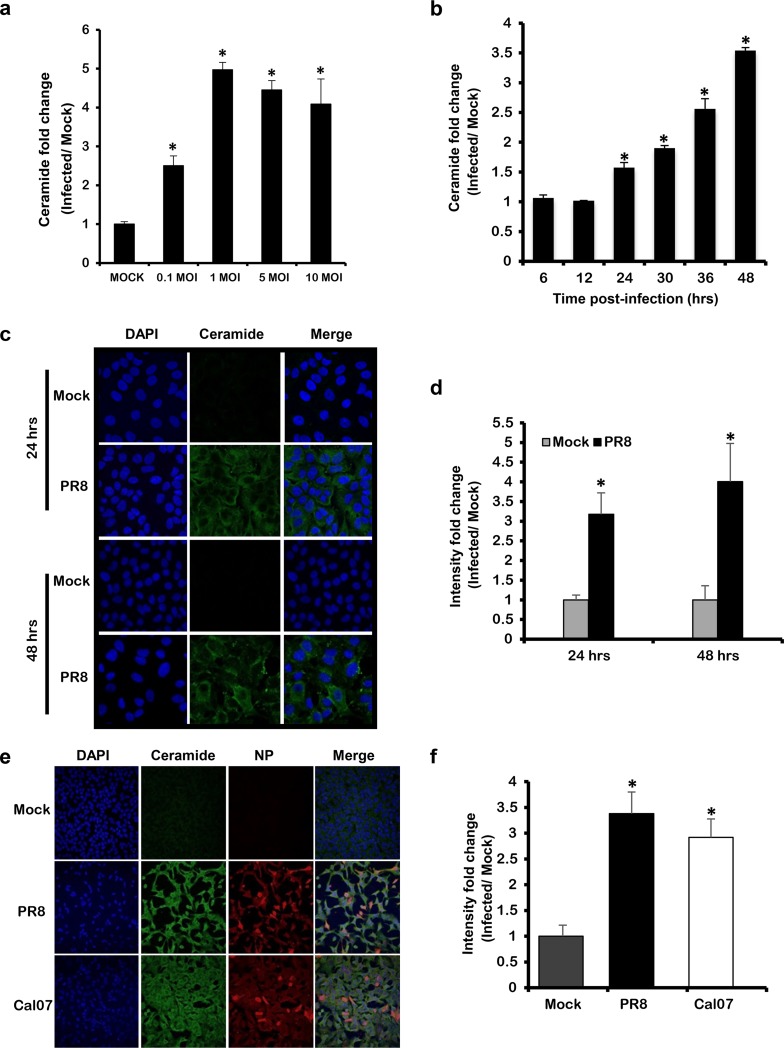
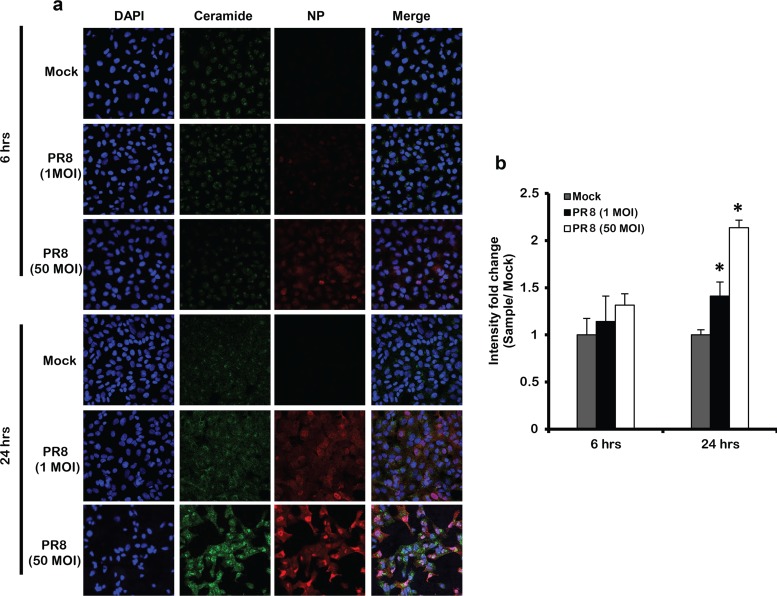
First, we assessed whether IAV can trigger ceramide accumulation in infected cells. A549 cells were inoculated with IAV (PR8) at increasing multiplicities of infection (MOIs; 0.1, 1, 5, and 10). For noninfected controls (Mock), the cells were inoculated with virus infection medium (VIM) only. After 48 h, the cells were trypsinized and washed with phosphate-buffered saline (PBS). Lipid extraction was performed followed by ceramide quantification using the DGK assay (Fig. 1a). Ceramide accumulation was normalized with respect to total sample phosphate and calculated in terms of fold change relative to Mock. Cells infected with as low as 0.1 MOI displayed a significant increase in ceramide compared to Mock. The level of ceramide significantly increased in a dose-dependent manner to reach its maximum with 1 MOI and plateaued at higher MOIs. We next assessed the time at which ceramide starts to accumulate in infected cells. A549 cells were infected with 1 MOI, and the cells were harvested at selected time points (6, 12, 24, 30, 36, and 48 h). At each time point, the fold change in ceramide level in infected cells was calculated relative to the time matching Mock (Fig. 1b). A significant increase in ceramide level was first observed at 24 h postinfection (hpi) and continued to rise until 48 hpi. These findings were confirmed by using anti-ceramide antibody and confocal microscopy (Fig. 1c and d). In order to verify that the ceramide increase in response to IAV infection was not a specific response to the PR8 strain, we assessed ceramide accumulation upon infecting cells with an influenza A/H1N1pdm09 virus (Cal07) using confocal microscopy (Fig. 1e). Ceramide level was increasing in IAV-infected cells whereby Cal07 induced a 3-fold increase in ceramide level compared to Mock that was similar to that produced in PR8-infected cells (Fig. 1f). In order to investigate the involvement of ceramide during the early replication cycle of IAV, which could have been missed with the used 1 MOI, we infected the cells with a high virus inoculum (50 MOI) and assessed the ceramide levels at 6 and 24 hpi (Fig. 2a and b). The ceramide levels could not be assessed at 48 hpi in the cells infected with 50 MOI due to excessive cell death. Cells infected with 50 MOI displayed similar viability to Mock and 1 MOI at 6 hpi; however, their viability was largely reduced (40% versus 75% with 1 MOI) at 24 hpi. No detectable change in ceramide was noted at the early time point (6 hpi) in cells infected with 50 MOI relative to Mock (Fig. 2a and b). At 24 hpi, cells infected with 50 MOI possessed higher levels of ceramide compared to 1 MOI (Fig. 2a and b), probably because at the lower MOI a sufficient number of cells should be infected in order to reach detectable variation in ceramide level. These findings collectively suggest the minimal involvement of ceramide during the early replication cycles of IAV and that several replication cycles are required to trigger the intracellular accumulation of ceramide.
FIG 1.
Ceramide accumulates in response to IAV infection in a dose- and time-dependent manner. (a) A549 cells were infected with increasing virus titer of IAV (PR8) for 48 h, or (b, c, and d) with 1 MOI for the indicated time points. (e and f) A549 cells were infected with Cal07 or PR8 at 1 MOI for 48 h. (a and b) Ceramide accumulation was determined using (DGK) assay. For each condition, ceramide accumulation was normalized to total phosphate, and the ceramide fold change was computed with respect to time matching noninfected controls (Mock). (c and e) Ceramide accumulation was detected by confocal microscopy using fluorescent anti-ceramide antibody (green), nuclei were counterstained with DAPI (blue), and IAV was visualized using anti-NP antibody (red) at ×63 (c) and ×40 magnifications. The intensity fold change in ceramide levels between infected cells and time matching Mock was assessed in panels d and f. Statistical significance was determined using the t test (*, P < 0.05) relative to time matching Mock.
FIG 2.
IAV infection does not induce ceramide accumulation at early viral replication cycles. (a) Ceramide accumulation was detected by confocal microscopy in cells infected by IAV (PR8) using 1 and 50 MOI at the indicated time points (6 and 24 h) or those left noninfected (Mock). Immunostaining was done using anti-ceramide antibody (green), nuclear stain DAPI (blue), and anti-NP antibody (red) at ×40 magnification. (b) The intensity fold change in ceramide levels was assessed between infected cells and time matching Mock, Statistical significance was determined using the t test (*, P < 0.05) relative to time matching Mock.
Ceramide accumulates in response to an active IAV infection.
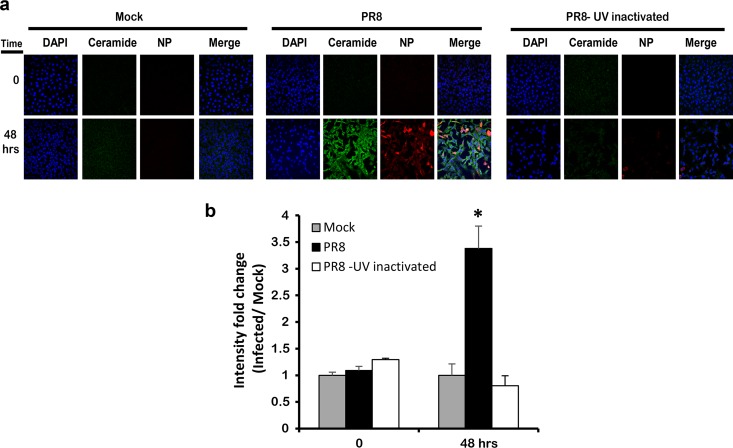
To verify that ceramide accumulation depends on active virus replication, we assessed the level of ceramide in cells infected with 1 MOI of UV-inactivated PR8. In this experimental setup, IAV would be able to internalize but without further replication, and this was confirmed by plaque assay (data not shown). After 48 h, we assessed ceramide accumulation by immunostaining and the variation ceramide levels were assessed as intensity fold change relative to the noninfected control (Mock). The cells that were infected with UV-inactivated PR8 did not show an elevation of endogenous ceramide levels at 48 hpi (Fig. 3a and b). These data show that ceramide accumulation is a specific response to active IAV infection, which requires IAV binding and replication.
FIG 3.
Ceramide accumulation requires an active IAV infection. UV-inactivated virus (PR8) at 1 MOI was used to inoculate A549 cells. After 48 hpi, the cells were immunostained using fluorescent anti-ceramide antibody (green) and anti-NP (IAV) antibody (red), and the nucleus was stained in blue using DAPI. The slides were visualized by confocal microscopy at ×40 (a), and the variation in intensity fold change between different conditions was calculated (b). Statistical significance was assessed using the t test (*, P < 0.05) relative to time matching noninfected cells (Mock).
Ceramide accumulation during IAV infection via the de novo biosynthesis.
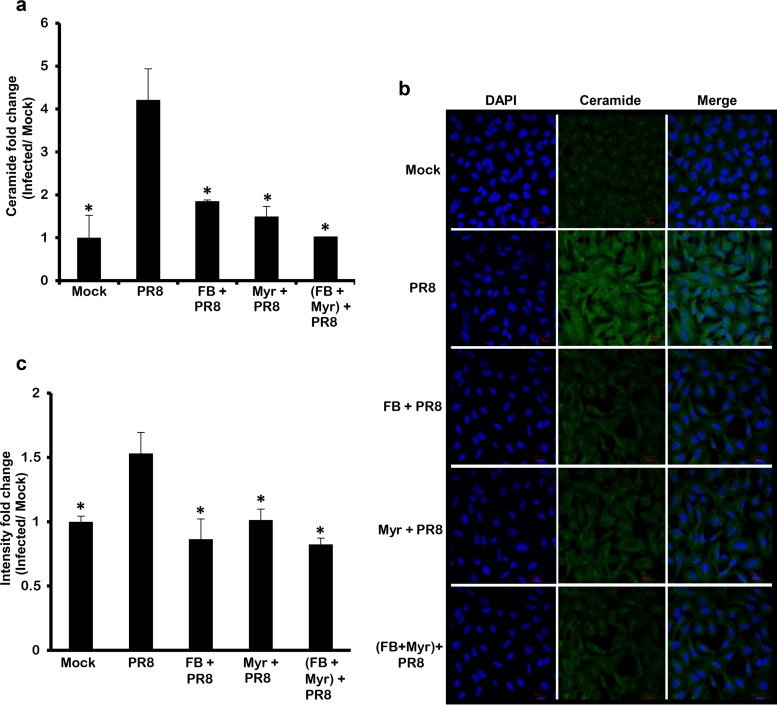
Intracellular ceramide biosynthesis occurs through two pathways: the de novo and salvage pathways (20). Many studies highlighted the role of these two pathways in modulating the life cycle and replication of some viruses (40–42, 44, 48, 54–56). In our study, ceramide levels significantly increased at later time points (starting 24 hpi) post-IAV infection. Previous studies showed that the late accumulation of ceramide in response to adenovirus infection or TNF-α treatment occurs through the de novo biosynthesis pathway (48, 57). To verify whether the IAV-induced ceramide accumulation is through the de novo pathway, we evaluated ceramide accumulation in infected cells treated with de novo ceramide inhibitors myriocin (Myr), a selective inhibitor for SPT, and fumonisin B1 (FB), a potent inhibitor of CerS (30). In this experimental setup, A549 cells were treated with subcytotoxic concentrations of FB and/or Myr, as confirmed using an MTT assay (data not shown). After treating the cells with the selected drug or vehicle for 2 h, the cells were infected with IAV (PR8) at 1 MOI. The variation in ceramide levels in this experimental setup was assessed in terms of fold change relative to the vehicle-treated noninfected control (Mock) (Fig. 4a). Blocking the first and rate-limiting step of the de novo pathway using Myr resulted in a potent reduction in ceramide accumulation compared to the vehicle-treated infected cells reaching baseline level indicated by the Mock. The levels of ceramide in cells treated with FB or both inhibitors were similar to Myr-treated cells. A similar pattern was observed by using confocal microscopy (Fig. 4b and c). These findings suggested that the accumulation of cellular ceramide in response to IAV infection is mainly driven via the de novo biosynthesis pathway.
FIG 4.
IAV infection mediates ceramide biosynthesis through the de novo pathway. A549 cells were treated with 50 μM FB and/or 0.1 μM Myr or vehicle for 2 h. The cells were infected with IAV (PR8) at 1 MOI in the presence or absence of the inhibitor(s) or left noninfected. After 48 h, ceramide accumulation was determined by two methods: DGK assay where ceramide fold change was assessed with respect to time matching noninfected controls (Mock) (a) or by confocal microscopy (b and c). Immunostaining was done using anti-ceramide antibody (green) and nuclear stain DAPI (blue) at ×40 for panel b. (c) The intensity fold change between different experimental setups and Mock was measured. Statistical significance was assessed using the t test (*, P < 0.05) relative to IAV-infected, nontreated cells.
Inhibiting the de novo ceramide pathway promotes IAV replication and production.
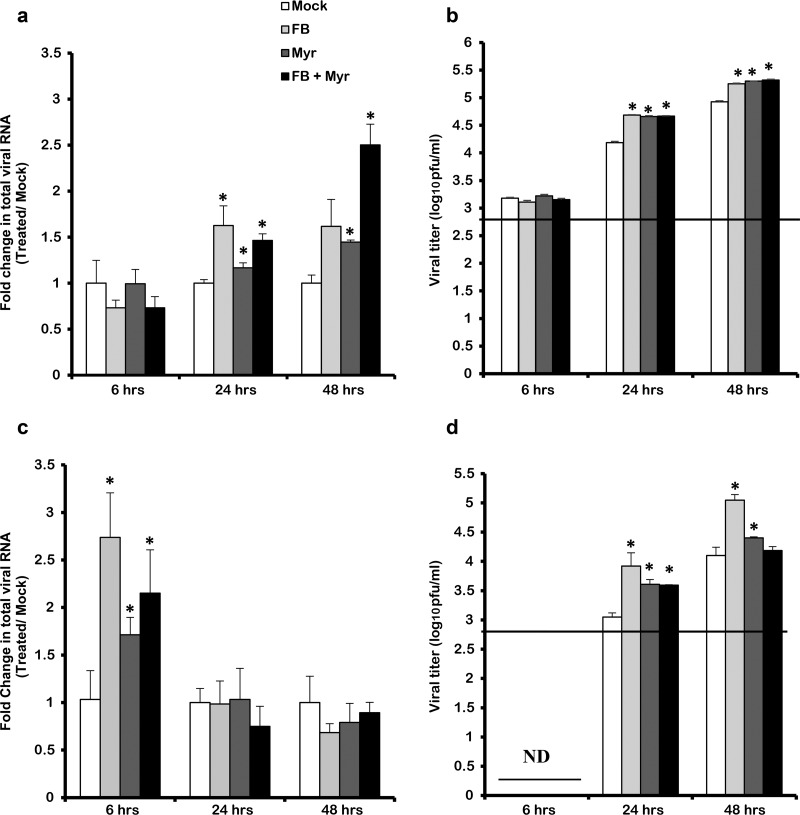
The accumulation of ceramide during IAV infection suggested that it was either required for virus replication or it was produced by the host as an antiviral response. In order to address this question, we investigated the effect of inhibiting of de novo ceramide pathway on virus replication at the level of RNA synthesis and virus production. The inhibition of the de novo ceramide pathway was associated with a significant increase in total viral RNA (Fig. 5a and c). This increase was observed as early as 6 hpi in cells infected with 0.01 MOI (Fig. 5c). On the other hand, in the cells infected with an MOI of 1, the increase in viral RNA was observed at later time points (24 and 48 hpi) (Fig. 5a). This increase in total viral RNA upon inhibition of ceramide synthesis resulted in an equivalent increase in virus production. A slight increase in viral progeny was detected in cells infected with 1 MOI (Fig. 5b), and yet a more prominent increase was obtained from the cells that were infected with the lower MOI (≈1 log10 PFU/ml unit) (Fig. 5d). Therefore, our data revealed that IAV replication and production is enhanced upon blocking de novo ceramide biosynthesis, suggesting an antiviral role for ceramide.
FIG 5.
IAV replication and production are enhanced upon inhibiting the de novo pathway. A549 cells were treated with 50 μM FB and/or 0.1 μM Myr or vehicle for 2 h. Cells were then infected with different viral titers of IAV (PR8) at the indicated time points: 1 MOI (a and b) or 0.01 MOI (c and d). Replication of viral RNA was determined by qRT-PCR using gene-specific primers and probes (a and c). Variation in the viral titer in response to treatment was measured by using a plaque assay (b and d). The line represents the lowest detection limit indicated at 2.69 log10 PFU/ml, and ND represents lack of virus detection in the tested samples. Statistical significance was assessed using the t test (*, P < 0.05) relative to vehicle-treated IAV-infected cells (Mock).
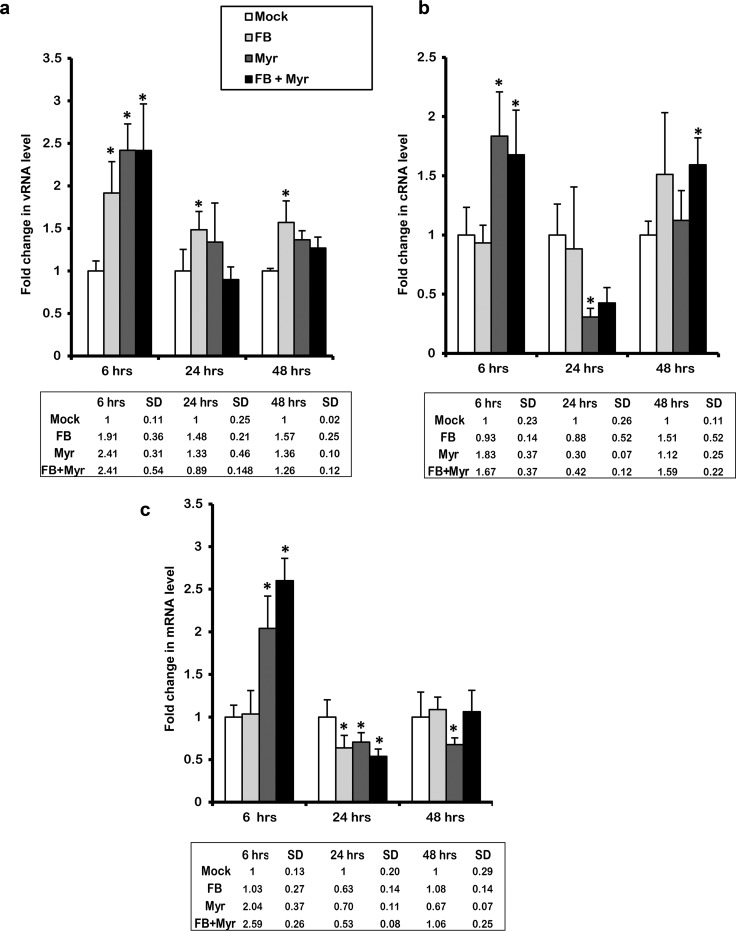
To understand the regulation of IAV replication upon inhibition of de novo ceramide, we decided to measure the levels of the various viral RNA species in the presence of the inhibitors. In order to distinguish between the different viral RNA species (vRNA, cRNA, and mRNA), cDNA was synthesized by reverse transcription (RT) using tagged primers specific to each of the RNA species (Table 1). The hot start method was used in order to eliminate nonspecific overlap between the three different RNA species. Then, qRT-PCR was performed with one primer complementary to the tag sequence of RT primers and the other primer complementary to viral RNA sequence (58). De novo pathway inhibition using FB and/or Myr resulted in a significant increase in viral RNA species at 6 hpi in cells infected with 0.01 MOI (Fig. 6a to c). This was similar to the changes observed at the level of total viral RNA (Fig. 5c). However, ceramide inhibition using FB mediated a differential change in the levels of RNA species. The vRNA levels in the infected cells that were pretreated with Myr or combined inhibitors increased by 6 hpi only. On the other hand, the infected cells that were pretreated by FB alone demonstrated an increase in the vRNA levels at 6 hpi and at later time points as well, albeit to a lesser extent compared to the early time point. In the case of mRNA, a slight but significant drop in transcript levels was observed in cells treated with either one or both inhibitors at 24 hpi (Fig. 6c). Overall, the data indicate that inhibition of the de novo biosynthesis pathway promotes IAV replication early during infection; however, the effect on virus production is not observed until later time points (24 hpi), supporting the antiviral role of ceramide.
TABLE 1.
Primers and probes used in this study
| Target | Primer or probe | Sequence (5′–3′) or sourcea |
|---|---|---|
| M gene | Forward | BEI Resources NR-15594 |
| Reverse | BEI Resources NR-15595 | |
| Probe | BEI Resources NR-15590 | |
| RNase P | Forward | BEI Resources NR-15612 |
| Reverse | BEI Resources NR-15613 | |
| Probe | BEI Resources NR-15611 | |
| vRNA | Reverse transcription | GGCCGTCATGGTGGCGAATGAATGGACGGAGAACAAGGATTC |
| Forward | GGCCGTCATGGTGGCGAAT | |
| Reverse | CTCAATATGAGTGCAGACCGTGC | |
| cRNA | Reverse transcription | GCTAGCTTCAGCTAGGCATCAGTAGAAACAAGGGTATTTTTCTTT |
| Forward | GCTAGCTTCAGCTAGGCATC | |
| Reverse | CGATCGTGCCCTCCTTTG | |
| mRNA | Reverse transcription | CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTCTTTAATTGTC |
| Forward | CCAGATCGTTCGAGTCGT | |
| Reverse | CGATCGTGCCCTCCTTTG | |
| β-Actin | Reverse transcription | Oligo(dT) |
| Forward | ATTGGCAATGAGCGGTTCC | |
| Reverse | GGTAGTTTCGTGGATGCCACA |
Tagged sequences are indicated in boldface.
FIG 6.
Inhibiting de novo ceramide biosynthesis affects virus replication, transcription, and translation. The de novo ceramide pathway was inhibited using 50 μM FB and/or 0.1 μM Myr or vehicle for 2 h followed by infection with IAV (PR8) at an MOI of 0.01. Variation in viral RNA species in response to treatment was measured using qRT-PCR after 6, 24, and 48 hpi. The levels of vRNA (a), cRNA (b), and mRNA (c) were normalized relative to host β-actin. Fold change was quantified relative to vehicle-treated infected cells (Mock). Statistical significance was assessed using the t test (*, P < 0.05) relative to Mock.
Exogenous C6-ceramide suppresses influenza virus replication.
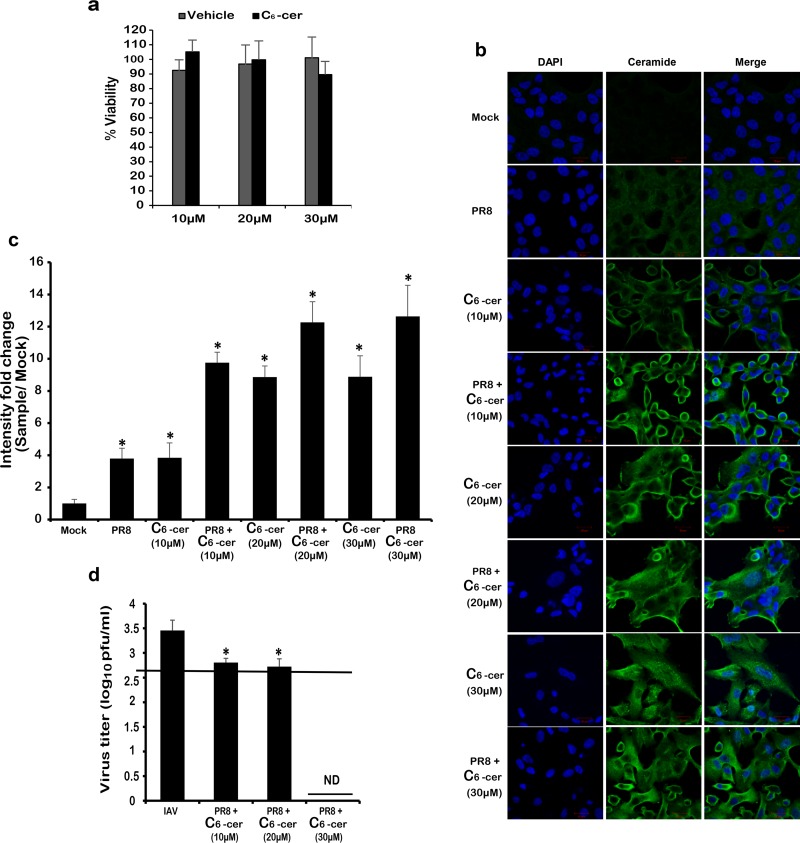
The increase in virus replication upon inhibition of the de novo pathway suggested a potential antiviral role for ceramide. To better assess this role of ceramide, we determined the effect of treating the cells with C6-ceramide, a synthetic cell-permeable ceramide analogue, on IAV production. According to previous studies, exogenous C6-ceramide activates de novo ceramide biosynthesis in A549 cells. In this process, the C6-ceramide gets deacylated then its sphingosine base gets reacylated by CerS to produce “endogenous” long-chain ceramides (59). In this experimental setup, A549 cells were treated with increasing concentrations of exogenous C6-ceramide (10, 20, and 30 μM) for 24 h, followed by infection with IAV (PR8) at 0.01 MOI. An MTT assay confirmed that none of the used concentrations was toxic in A549 cells (Fig. 7a). After 24 hpi, ceramide levels in the treated-infected cells were compared to the control groups (treated/noninfected or vehicle-treated/noninfected [Mock]) using immunofluorescent ceramide staining (Fig. 7b and c). First, a dose-dependent increase in ceramide levels was observed in the noninfected cells that were treated with increasing concentrations of C6-ceramide. These results confirmed the successful uptake and utilization of the exogenous ceramide analogue. At high concentrations (20 and 30 μM), we did not detect a large difference in fluorescence because the signal became saturated. Second, the cells that were treated with exogenous ceramide and infected with IAV displayed a higher increase in intercellular ceramide levels. Next, we investigated the effect of boosting intracellular ceramide levels on virus production by means of plaque assay (Fig. 7d). Treating the cells with 30 μM C6-ceramide suppressed virus replication to below the detection limit (2.69 log10 PFU/ml). Lower C6-ceramide concentrations (10 and 20 μM) resulted in a less prominent but significant reduction in virus titer (≈0.7 log10 PFU/ml unit). These results confirmed the antiviral role of ceramide on IAV production.
FIG 7.
Treatment with exogenous C6-ceramide induces ceramide accumulation and inhibits IAV propagation. Cells were treated with the indicated concentrations of C6-ceramide for 48 h. (a) Cell viability was assessed using an MTT assay and is expressed as the percent viability relative to untreated cells. A549 cells were treated with the indicated concentrations of C6-ceramide or with the vehicle for 24 h prior to infection with 0.01 MOI of IAV (PR8). (b) Twenty-four hours after C6-ceramide treatment and/or virus inoculation, the cells were stained with a fluorescent anti-ceramide antibody (green) and nuclear stain DAPI (blue) and then visualized by confocal microscopy (×63). (c) The variation in intensity fold change between the different experimental setups and the vehicle-treated noninfected control (Mock) was assessed. (d) Variation in the viral titer in response to treatment was determined using plaque assay. The line represents the lowest detection limit indicated at 2.69 log10 PFU/ml. “ND” indicates the lack of virus detection in the tested samples. Statistical significance was assessed using the t test (*, P < 0.05) relative to Mock.
DISCUSSION
Ceramide determines the curvature and fluidity of cell membranes and plays an important role as a second messenger in diverse cell signaling pathways, including immune responses (45, 60). Like other viral pathogens, IAV lacks the machinery required for lipid biosynthesis. As it hijacks the host cellular machinery for its replication (17, 61), newly assembled IAV acquires its envelope from the host membrane, where sphingolipids are key constituents (62). Previous studies have shown that IAV requires lipid rafts and intact sphingolipid biosynthesis for replication and morphogenesis (62–66). Nonetheless, the full extent of the regulation of sphingolipid biosynthesis during infection and its effect on virus replication remained unclear. In this study, we demonstrated that endogenous ceramide levels increase in response to IAV infection in a dose- and time-dependent manner. We showed for the first time that ceramide exerts an antiviral role during IAV replication. This was evident by the enhanced IAV replication upon inhibiting the de novo biosynthesis pathway, while the addition of exogenous ceramide suppressed virus production.
Endogenous ceramide biosynthesis occurs through the de novo biosynthesis and salvage pathways (20). Several studies highlighted the role of these pathways in viral infections and the disease manifestations, making them potential therapeutic targets. It has been shown that pharmacological upregulation of ceramide using fenretinide inhibited the human immunodeficiency virus (HIV) infection in TZM-bl cells (41). This antiviral effect was mediated through the reduction of HIV entry and the perturbation of host membrane structure leading to the production of noninfective viral progeny (41). In a similar approach, Myr treatment successfully suppressed the hepatitis C virus (HCV) replication (54). Also, adenovirus utilizes de novo ceramide biosynthesis to regulate host SR proteins that are crucial for its pathogenesis (48). Recently, a differential role for de novo ceramide was revealed in the life cycles of two flaviviruses, the West Nile and dengue viruses, both shown to trigger ceramide biosynthesis (60, 67). Ceramide was found to be required for the efficient replication and production of West Nile virus, whereas it exerts an antiviral function against dengue virus. Colocalization between ceramide with viral protein was also observed in case of West Nile but not dengue virus, suggesting that the former virus required ceramide for its replication, whereas the latter probably utilized downstream sphingolipid by-products. In the present study, inhibiting SPT and/or CerS, two key enzymes in the de novo biosynthesis pathway, in IAV-infected cells reduced ceramide levels to baseline level detected in noninfected cells. These findings strongly indicate that IAV induces ceramide accumulation mainly through the de novo pathway.
Two previous studies used prolonged (3 days) Myr or FB treatments to block complex sphingolipid biosynthesis prior to IAV infection (68, 69). The first study, by Tafesse et al., showed that the surface expression of IAV proteins (HA and NA) were significantly reduced in cells that genetically lack SM synthase or those treated with Myr, an SPT inhibitor, for 72 h. The prolonged treatment of the cells with Myr inhibited SM synthesis by reducing the availability of its precursor “ceramide” leading to a decreased SM synthase1 activity. Their results showed that SM is important for IAV maturation beyond the trans-Golgi network (68). In the second study, prolonged treatment of MDCK cells with FB was done to suppress glycosphingolipids (GSLs) and sphingomyelin (SM) levels. This led to an altered distribution of the HA protein and suppressed its incorporation into the plasma membrane, resulting in reduced virus production. Their study highlighted the indispensable role of GSLs in IAV life cycle whereby it contributes to the viral infection more potently than SM (69). In the present study, we decided to investigate the direct role of ceramide produced through de novo biosynthesis on IAV replication without manipulating SM and GSL levels. Therefore, we suppressed ceramide biosynthesis by treating the cells with FB and/or Myr for only 2 h prior to infection. This resulted in increased replication of the virus genome (vRNA and cRNA) and synthesis of mRNA transcripts (particularly in Myr-treated cells) at 6 hpi. This upregulation of viral RNA species translated to increased production of IAV at later time points. The early suppression of IAV genome replication upon ceramide inhibition is surprising given that significant accumulation of ceramide is only observed at later time points. We speculate that subtle yet undetectable changes in ceramide levels during the early replication cycle could be sufficient and responsible for triggering the early effect observed on virus replication. The exact mechanism through which each of the used drugs modulates viral RNA transcription and replication is not yet understood and requires further investigation. Further studies are warranted to fully understand the regulation of various stages of virus life cycle due to ceramide production in infected cells.
The enhancement of virus propagation upon inhibiting ceramide biosynthesis suggested that it plays an antiviral role during IAV infection. Therefore, we tested the effect of treating the cells with exogenous C6-ceramide on IAV production. Exogenous C6-ceramide enhanced the cellular ceramide levels and inhibited IAV replication by orders of magnitude. These findings confirmed the antiviral role of ceramide during IAV infection. Ceramide and sphingosine-1-phosphate (S1P) are intermediates in sphingolipid biosynthesis. Ceramide can be converted to sphingosine through the action of ceramidase enzyme. Sphingosine undergoes phosphorylation into S1P in a reversible reaction by sphingosine kinase (SK), and the opposite reaction is catalyzed by sphingosine lyase. Ceramide and S1P have opposing biological functions, and the balance between the two interconvertible lipids is known as the sphingolipid rheostat (70). Seo et al. previously showed that overexpression of S1P lyase, the enzyme responsible for irreversible degradation of S1P, inhibited IAV propagation, whereas overproduction of S1P through upregulation of sphingosine kinase 1 enhanced IAV replication (49, 71). Also, it was shown in mice that inhibiting SK protects the mice from lethal IAV infection (72). S1P has been shown to act as a noncompetitive inhibitor of CerS2, and thus its overexpression could dampen the cellular levels of ceramide (73). Moreover, de novo synthesis of ceramide was found to be upregulated in cells overexpressing S1P lyase (74). Therefore, our results complement the previous findings since S1P formation will reduce the cellular ceramide pool and thus increasing IAV replication (49, 71, 72). Ceramide analogues and mimetics have been proposed as potential immunotherapeutic remedies against viral infections (45). Furthermore, exogenous short-chained ceramides were shown to induce death in Kaposi’s sarcoma-associated herpesvirus and human T-cell leukemia-lymphoma virus-infected cells (40, 44, 55, 56), enhanced maturation and activation of DCs in response to lymphocytic choriomeningitis virus and IAV infections, and inhibited HIV and HCV viral replication (45). Therefore, targeting the sphingolipid pathway in a way to enhance ceramide production or the use of ceramide analogues could be a promising antiviral approach against influenza infections.
Our data, as well as those previously published (49, 71, 72), suggest that ceramide and downstream products likely interferes with the IAV life cycle at various stages, including replication, protein transport, and assembly. Confirming the biological significance of our results in vivo is something that we are planning to investigate in future work. However, minor differences in viral titers can result in a significant biological effect in vivo. For instance, Zaraket et al. showed that a mutation in HA of pathogenic H5N1 mediates a 1- to 1.5-log10 PFU/ml unit decrease in virus titer in A549 cells but significantly reduce virus replication in the lungs of ferrets (75). Further investigations are required for better understanding of the exact cellular processes taking place downstream ceramide in response to IAV infection.
MATERIALS AND METHODS
Reagents and antibodies.
Fumonisin B1 (Enzo), myriocin (Sigma), C6-ceramide (Sigma), anti-ceramide antibody (Sigma), anti-NP antibody (Thermo Fisher Scientific), Alexa Fluor 488 goat anti-mouse IgG antibody (Thermo Fisher Scientific), and Alexa Fluor 594 donkey anti-rabbit IgG antibody (Thermo Fisher Scientific) were used in this study.
Cell culture.
Madin-Darby canine kidney (MDCK; ATCC) and human lung adenocarcinoma epithelial cell line (A549; ATCC) were cultured in Dulbecco modified Eagle medium-high glucose (Sigma) supplemented with 10% fetal bovine serum and 1% of 100 U/ml penicillin-streptomycin and maintained in 5% CO2 at 37°C.
Virus propagation and titration.
MDCK cells were infected by IAV A/Puerto Rico/8/34 (PR8) or A/California/07/2009 (Cal07) prepared in VIM composed of minimal essential media (MEM) supplemented with bovine serum albumin (BSA; 0.3%), penicillin-streptomycin (5%), MEM vitamin (5%), glutamine (2 mM), and gentamicin (0.04 mg/ml). Infection was done using an MOI equivalent to 0.01 PFU/cell. Adsorption was allowed for 1 h accompanied by gentle shaking every 15 min. The infected cells were incubated in VIM containing 1 μg/ml TPCK-trypsin (Sigma) for 48 to 72 h postinfection (hpi). After observing the cytopathic effect, the supernatant containing the virus was collected and stored at –80°C. Virus titration was performed using a plaque assay. Briefly, MDCK cells were seeded at 7 × 105 cells per well in 6-well plates to form a homogeneously confluent monolayer. The cells were then washed with PBS with calcium and magnesium, and 10-fold serial dilutions of the virus were prepared. Virus dilutions (200 μl) were added to each well and incubated at 37°C for 1 h (shaking every 15 min). The cells were covered with 3 ml of freshly prepared agarose (0.5%) nutritive overlay. The dishes were kept at room temperature for 10 min to allow their solidification and then incubated at 37°C. After 72 h, the agarose overlay was removed, and the cells were fixed/stained with crystal violet solution.
Inhibitors and exogenous ceramide.
A dosing regimen was selected upon testing the cytotoxic effect of the drugs on A549 cells. Fumonisin B1 (FB, 50 μM), myriocin (Myr, 0.01 μM), or combination of both drugs were used to inhibit de novo ceramide biosynthesis. The cells were incubated with the inhibitor(s) for 2 h, followed by infection at 0.01 or 1 MOI. The inhibitors were maintained in the media for the duration of the experiment.
A549 cells were treated with increasing concentrations (10 to 30 μM) of exogenous C6-ceramide for 24 h, followed by infection with IAV at 0.01 MOI. The short-chained ceramide analogue was maintained in the culture media throughout the experiment. The variation in viral titers between the different experimental conditions was measured by using the plaque assay.
The effect of C6-ceramide on the viability of A549 cells was assessed using an MTT assay. Briefly, cells were cultured in 96-well plates at a seeding density of 2 × 104 cell per well. Then, the cells were treated with C6-ceramide (10, 20, and 30 μM). After 48 h, 20 μl of the tetrazolium dye (MTT) was added, and the cells were incubated at 37°C for 2 h. During this time the viable cells would reduce MTT into an insoluble formazan, which is solubilized using 100 μl of isopropanol. The absorption was measured using spectrophotometer at a wavelength of 595 nm.
Lipid extraction and ceramide quantification.
Cell pellets were lysed in an organic solvent (chloroform-methanol), and lipids were extracted according to the Bligh and Dyer method (76). Two aliquots were dried by speed vacuum and used for ceramide and phosphate measurements, respectively. Ceramide levels were assessed using the diacylglycerol kinase (DGK) assay as described previously (25, 26, 77). In short, lipids were incubated with 20 μl of β-octylglucoside/dioleoyl-PG micelles (7.5%), 70 μl of the reaction mixture (1 mM dithiothreitol, 5 μg DGK) and 10 μl of ATP mixture (2.5 mM ATP and 0.13 μCi/μl adenosine 5′-triphosphate [γ-32P]ATP) for 30 min at room temperature. After several steps of adding organic solvents and water followed by centrifugation, the lower phase was removed and dried by speed vacuum. The dried lipids from the samples and standards were resuspended in 50 μl of 9:1 chloroform-methanol, and the lipid species were separated using thin-layer chromatography plates (Whatman). Radioactive ceramide species were detected using X-ray films by autoradiography. The area of interest was scraped and its radioactivity was measured by liquid scintillation. Ceramide levels were quantified according to the standard curve.
Reverse transcription and quantitative RT-PCR.
Total RNA was extracted using a PureLink Viral RNA/DNA minikit (Invitrogen) according to the manufacturer’s specifications. Quantitative reverse transcription-PCR (RT-PCR) was done using primers and probes targeting the M gene (IAV) and host RNase P as a control housekeeping gene (Table 1 ). One-Step RT-PCR AgPath-ID (Thermo Fisher Scientific) was used to amplify the target according to the following protocol: 45°C for 10 min, 95°C for 10 min, and then 45 cycles of 95°C for 15 s and 55°C for 1 min. The level of total viral RNA was normalized with respect to RNase P.
To quantify the individual viral RNA species, total RNA was extracted using TRIzol (Sigma) according to the manufacturer’s protocol. The RNA concentration was measured using DeNovix DS-11 spectrophotometer. cDNA complementary to the vRNA, cRNA, and mRNA species were synthesized from 1 μg of RNA by using a RevertAid First Strand cDNA synthesis kit (Thermo Fisher). Hot-start reverse transcription was performed in the presence of 5 μM tagged primers specifically targeting the individual RNA species of PR8 (modified from Kawakami et al.) (Table 1) (58). The mixture was heated to 65°C for 10 min, chilled for 5 min, and heated again at 60°C for 5 min. Reverse transcription was initiated by heating at 42°C for 1 h, followed by enzymatic inactivation at 70°C for 5 min. qRT-PCR was performed using the tagged primers listed in Table 1 and SYBR Green JumpStart Taq ReadyMix (Sigma) on a CFX96 real-time system (Bio-Rad, Hercules, CA). The PCR conditions were as follows: 94°C for 10 min, 40 cycles of 94°C for 15 s, 60°C for 1 min (68°C for vRNA), and 72°C for 1 min. Host β-actin was used as a housekeeping gene to normalize the levels of the viral RNA species (78). Primer efficiency was tested for all used primers, and quantification was done according to the Livak method (79). The fold change in viral RNA was assessed with respect to nontreated infected cells.
Immunocytochemistry.
A549 cells grown on 18 mm coverslips were infected with IAV at 80% confluence. The cells were fixed at the required time points using 4% paraformaldehyde for 15 min and permeabilized (0.2% Triton X-100 in PBS) for 20 min. The blocking solution (1% BSA and 0.1% Triton X-100 in PBS) was applied for 1 h, followed by overnight incubation with monoclonal antibody (1:100) in blocking buffer. The cells were then washed with PBS-Tween (0.1%) and incubated with the fluorescent secondary antibody (1:250) for 1 h. Finally, the cells were mounted with 200 μl of mounting medium and visualized using a laser scanning confocal microscopy (Zeiss LSM710) 40× and 63× oil immersion objective lenses. The pinhole was maintained at 1 Airy unit (Au), and multitrack acquisition setting was set to avoid interchannel cross talk. Intensities were quantified using 6 to 10 images from each experimental condition using ZEN 2 (blue edition).
Statistical analysis.
Statistical analysis was performed using the Student t test equipped with Excel 2016 (Microsoft, Redmond, WA). The results are presented as means ± the standard deviations of duplicates from two to three independent experiments.
ACKNOWLEDGMENTS
This research was supported by intramural funding through the Medical Practice Plan and the University Research Board of the American University of Beirut.
We thank Mona Diab-Assaf for her helpful suggestions and support.
REFERENCES
- 1.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, Asten L, van Silva SP, da Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Proschle CVS, Mallegas NV, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Herrera Betsy. A, Llanés MJ, Fischer TK, Krause TG, Mølbak K, Nielsen J, Trebbien R, Bruno A, Ojeda J, Ramos H, Heiden M, An der Signor L, del CC, Serrano CE, Bhardwaj R, Chadha M, Narayan V, Kosen S, Bromberg M, Glatman-Freedman A, Kaufman Z, Arima Y, Oishi K, Chaves S, Nyawanda B, Al-Jarallah RA, Kuri-Morales PA, Matus CR, Corona MEJ, Burmaa A, Darmaa O, Obtel M, Cherkaoui I, Wijngaard CC, van den Hoek W, van der Baker M, Bandaranayake D, Bissielo A, Huang S, Lopez L, Newbern C, Flem E, Grøneng GM, Hauge S, Cosío FG, de Moltó Y, de Castillo LM, Cabello MA, Horoch M, von Osis JM, Machado A, Nunes B, Rodrigues AP, Rodrigues E, Calomfirescu C, Lupulescu E, Popescu R, Popovici O, Bogdanovic D, Kostic M, Lazarevic K, Milosevic Z, Tiodorovic B, Chen M, Cutter J, Lee V, Lin R, Ma S, Cohen AL, Treurnicht F, Kim WJ, Delgado-Sanz C, Ontañón S, de Mateo Larrauri A, León IL, Vallejo F, Born R, Junker C, Koch D, Chuang J-H, Huang W-T, Kuo H-W, Tsai Y-C, Bundhamcharoen K, Chittaganpitch M, Green HK, Pebody R, Goñi N, Chiparelli H, Brammer L, Mustaquim D. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26:D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2017. Influenza type A viruses: avian influenza (flu). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 5.Fodor E. 2013. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol 57:113–122. [DOI] [PubMed] [Google Scholar]
- 6.Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza RNA synthesis machine. RNA Biol 8:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaraket H, Saito R, Suzuki Y, Baranovich T, Dapat C, Caperig-Dapat I, Suzuki H. 2010. Genetic makeup of amantadine-resistant and oseltamivir-resistant human influenza A/H1N1 viruses. J Clin Microbiol 48:1085–1092. doi: 10.1128/JCM.01532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2018. Influenza antiviral drug resistance: seasonal influenza (flu). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 9.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li TCM, Chan MCW, Lee N. 2015. Clinical implications of antiviral resistance in influenza. Viruses 7:4929–4944. doi: 10.3390/v7092850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaraket H, Kondo H, Hibino A, Yagami R, Odagiri T, Takemae N, Tsunekuni R, Saito T, Myint YY, Kyaw Y, Oo KY, Tin HH, Lin N, Anh NP, Hang NLK, Mai LQ, Hassan MR, Shobugawa Y, Tang J, Dbaibo G, Saito R. 2016. Full genome characterization of human influenza A/H3N2 isolates from Asian countries reveals a rare amantadine resistance-conferring mutation and novel PB1-F2 polymorphisms. Front Microbiol 7:262. doi: 10.3389/fmicb.2016.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo Y-J, Pritzl CJ, Vijayan M, Bomb K, McClain ME, Alexander S, Hahm B. 2013. Sphingosine kinase 1 serves as a pro-viral factor by regulating viral RNA synthesis and nuclear export of viral ribonucleoprotein complex upon influenza virus infection. PLoS One 8:e75005. doi: 10.1371/journal.pone.0075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Te Velthuis AJW, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arranz R, Coloma R, Chichón FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortín J, Martín-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 15.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RWH. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 17.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crépin T, Hart D, Lunardi T, Nanao M, Ruigrok RWH, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martínez-Alonso M, Fodor E. 2014. Conserved and host-specific features of influenza virion architecture. Nat Commun 5:4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolesnick R. 2002. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest 110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro BM, Prieto M, Silva LC. 2014. Ceramide: a simple sphingolipid with unique biophysical properties. Prog Lipid Res 54:53–67. doi: 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Holthuis JC, Pomorski T, Raggers RJ, Sprong H, Van Meer G. 2001. The organizing potential of sphingolipids in intracellular membrane transport. Physiol Rev 81:1689–1723. doi: 10.1152/physrev.2001.81.4.1689. [DOI] [PubMed] [Google Scholar]
- 22.Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA. 2002. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells dependence on protein phosphatase-1. J Biol Chem 277:12587–12595. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 23.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell 82:405–414. [DOI] [PubMed] [Google Scholar]
- 24.Perry DK, Carton J, Shah AK, Meredith F, Uhlinger DJ, Hannun YA. 2000. Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J Biol Chem 275:9078–9084. [DOI] [PubMed] [Google Scholar]
- 25.Dbaibo GS, Pushkareva MY, Rachid RA, Alter N, Smyth MJ, Obeid LM, Hannun YA. 1998. p53-dependent ceramide response to genotoxic stress. J Clin Invest 102:329–339. doi: 10.1172/JCI1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hage-Sleiman R, Esmerian MO, Kobeissy H, Dbaibo G. 2013. p53 and ceramide as collaborators in the stress response. Int J Mol Sci 14:4982–5012. doi: 10.3390/ijms14034982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husari AW, Dbaibo GS, Bitar H, Khayat A, Panjarian S, Nasser M, Bitar FF, El-Sabban M, Zaatari G, Mroueh SM. 2006. Apoptosis and the activity of ceramide, Bax and Bcl-2 in the lungs of neonatal rats exposed to limited and prolonged hyperoxia. Respir Res 7:100. doi: 10.1186/1465-9921-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannun YA, Obeid LM. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 29.Young SA, Mina JG, Denny PW, Smith TK. 2012. Sphingolipid and Ceramide Homeostasis: Potential Therapeutic Targets. Biochem Res Int Res Article 2012:1. doi: 10.1155/2012/248135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitatani K, Idkowiak-Baldys J, Hannun YA. 2008. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal 20:1010–1018. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitatani K, Taniguchi M, Okazaki T. 2015. Role of sphingolipids and metabolizing enzymes in hematological malignancies. Mol Cells 38:482–495. doi: 10.14348/molcells.2015.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasumov T, Li L, Li M, Gulshan K, Kirwan JP, Liu X, Previs S, Willard B, Smith JD, McCullough A. 2015. Ceramide as a mediator of non-alcoholic fatty liver disease and associated atherosclerosis. PLoS One 10:e0126910. doi: 10.1371/journal.pone.0126910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D, Suoniemi M, Hurme R, März W, Scharnagl H, Stojakovic T, Vlachopoulou E, Lokki M-L, Nieminen MS, Klingenberg R, Matter CM, Hornemann T, Jüni P, Rodondi N, Räber L, Windecker S, Gencer B, Pedersen ER, Tell GS, Nygård O, Mach F, Sinisalo J, Lüscher TF. 2016. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 37:1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galadari S, Rahman A, Pallichankandy S, Galadari A, Thayyullathil F. 2013. Role of ceramide in diabetes mellitus: evidence and mechanisms. Lipids Health Dis 12:98. doi: 10.1186/1476-511X-12-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker KA, Riethmüller J, Zhang Y, Gulbins E. 2010. The role of sphingolipids and ceramide in pulmonary inflammation in cystic fibrosis. Open Respir Med J 4:39–47. doi: 10.2174/1874306401004020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogretmen B. 2006. Sphingolipids in cancer: regulation of pathogenesis and therapy. FEBS Lett 580:5467–5476. doi: 10.1016/j.febslet.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 37.Barth BM, Cabot MC, Kester M. 2011. Ceramide-based therapeutics for the treatment of cancer. Anticancer Agents Med Chem 11:911–919. [DOI] [PubMed] [Google Scholar]
- 38.Bieberich E. 2008. Ceramide signaling in cancer and stem cells. Future Lipidol 3:273–300. doi: 10.2217/17460875.3.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caretti A, Torelli R, Perdoni F, Falleni M, Tosi D, Zulueta A, Casas J, Sanguinetti M, Ghidoni R, Borghi E, Signorelli P. 2016. Inhibition of ceramide de novo synthesis by myriocin produces the double effect of reducing pathological inflammation and exerting antifungal activity against A. fumigatus airways infection. Biochim Biophys Acta 1860:1089–1097. doi: 10.1016/j.bbagen.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Dai L, Trillo-Tinoco J, Bai A, Chen Y, Bielawski J, Del Valle L, Smith CD, Ochoa AC, Qin Z, Parsons C. 2015. Ceramides promote apoptosis for virus-infected lymphoma cells through induction of ceramide synthases and viral lytic gene expression. Oncotarget 6:24246–24260. doi: 10.18632/oncotarget.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finnegan CM, Rawat SS, Puri A, Wang JM, Ruscetti FW, Blumenthal R. 2004. Ceramide, a target for antiretroviral therapy. Proc Natl Acad Sci U S A 101:15452–15457. doi: 10.1073/pnas.0402874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aktepe TE, Pham H, Mackenzie JM. 2015. Differential utilization of ceramide during replication of the flaviviruses West Nile and dengue virus. Virology 484:241–250. doi: 10.1016/j.virol.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 43.Hinkovska-Galcheva V, Boxer L, Mansfield PJ, Schreiber AD, Shayman JA. 2003. Enhanced phagocytosis through inhibition of de novo ceramide synthesis. J Biol Chem 278:974–982. doi: 10.1074/jbc.M206199200. [DOI] [PubMed] [Google Scholar]
- 44.Darwiche N, Abou-Lteif G, Najdi T, Kozhaya L, Tayoun AA, Abou Tayyoun A, Bazarbachi A, Dbaibo GS. 2005. Human T-cell lymphotropic virus type I-transformed T cells have a partial defect in ceramide synthesis in response to N-(4-hydroxyphenyl)retinamide. Biochem J 392:231–239. doi: 10.1042/BJ20050578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pritzl CJ, Seo Y-J, Xia C, Vijayan M, Stokes ZD, Hahm B. 2015. A ceramide analogue stimulates dendritic cells to promote T cell responses upon virus infections. Ji 194:4339–4349. doi: 10.4049/jimmunol.1402672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grassmé H, Riehle A, Wilker B, Gulbins E. 2005. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. J Biol Chem 280:26256–26262. doi: 10.1074/jbc.M500835200. [DOI] [PubMed] [Google Scholar]
- 47.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog 8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanj SS, Dandashi N, El-Hed A, Harik H, Maalouf M, Kozhaya L, Mousallem T, Tollefson AE, Wold WS, Chalfant CE, Dbaibo GS. 2006. Ceramide regulates SR protein phosphorylation during adenoviral infection. Virology 345:280–289. doi: 10.1016/j.virol.2005.09.060. [DOI] [PubMed] [Google Scholar]
- 49.Seo Y-J, Blake C, Alexander S, Hahm B. 2010. Sphingosine 1-phosphate-metabolizing enzymes control influenza virus propagation and viral cytopathogenicity. J Virol 84:8124–8131. doi: 10.1128/JVI.00510-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nailwal H, Sharma S, Mayank AK, Lal SK. 2015. The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43. Cell Death Dis 6:e1768. doi: 10.1038/cddis.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodcock J. 2006. Sphingosine and ceramide signalling in apoptosis. IUBMB Life 58:462–466. doi: 10.1080/15216540600871118. [DOI] [PubMed] [Google Scholar]
- 52.Gaur P, Munjhal A, Lal SK. 2011. Influenza virus and cell signaling pathways. Med Sci Monit 17:RA148–RA154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demarchi F, Bertoli C, Greer PA, Schneider C. 2005. Ceramide triggers an NF-κB-dependent survival pathway through calpain. Cell Death Differ 12:512–522. doi: 10.1038/sj.cdd.4401592. [DOI] [PubMed] [Google Scholar]
- 54.Tatematsu K, Tanaka Y, Sugiyama M, Sudoh M, Mizokami M. 2011. Host sphingolipid biosynthesis is a promising therapeutic target for the inhibition of hepatitis B virus replication. J Med Virol 83:587–593. doi: 10.1002/jmv.21970. [DOI] [PubMed] [Google Scholar]
- 55.Dai L, Plaisance-Bonstaff K, Voelkel-Johnson C, Smith CD, Ogretmen B, Qin Z, Parsons C. 2014. Sphingosine kinase-2 maintains viral latency and survival for KSHV-infected endothelial cells. PLoS One 9:e102314. doi: 10.1371/journal.pone.0102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Z, Dai L, Trillo-Tinoco J, Senkal C, Wang W, Reske T, Bonstaff K, Del Valle L, Rodriguez P, Flemington E, Voelkel-Johnson C, Smith CD, Ogretmen B, Parsons C. 2014. Targeting sphingosine kinase induces apoptosis and tumor regression for KSHV-associated primary effusion lymphoma. Mol Cancer Ther 13:154–164. doi: 10.1158/1535-7163.MCT-13-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dbaibo GS, El-Assaad W, Krikorian A, Liu B, Diab K, Idriss NZ, El-Sabban M, Driscoll TA, Perry DK, Hannun YA. 2001. Ceramide generation by two distinct pathways in tumor necrosis factor alpha-induced cell death. FEBS Lett 503:7–12. [DOI] [PubMed] [Google Scholar]
- 58.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, Bielawska A, Obeid LM, Hannun YA. 2002. Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line: role for endogenous ceramide in mediating the action of exogenous ceramide. J Biol Chem 277:12960–12969. doi: 10.1074/jbc.M110699200. [DOI] [PubMed] [Google Scholar]
- 60.Martín-Acebes MA, Blázquez A-B, Jiménez de Oya N, Escribano-Romero E, Saiz J-C. 2011. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One 6:e24970. doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondal A, Dawson AR, Potts GK, Freiberger EC, Baker SF, Moser LA, Bernard KA, Coon JJ, Mehle A. 2017. Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. Elife 6:e26910. doi: 10.7554/eLife.26910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanner LB, Chng C, Guan XL, Lei Z, Rozen SG, Wenk MR. 2014. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J Lipid Res 55:1357–1365. doi: 10.1194/jlr.M049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheiffele P, Roth MG, Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J 16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parton DL, Tek A, Baaden M, Sansom MSP. 2013. Formation of raft-like assemblies within clusters of influenza hemagglutinin observed by MD simulations. PLoS Comput Biol 9:e1003034. doi: 10.1371/journal.pcbi.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Razinkov VI, Cohen FS. 2000. Sterols and sphingolipids strongly affect the growth of fusion pores induced by the hemagglutinin of influenza virus. Biochemistry (Mosc) 39:13462–13468. doi: 10.1021/bi0012078. [DOI] [PubMed] [Google Scholar]
- 66.Heung LJ, Luberto C, Del Poeta M. 2006. Role of sphingolipids in microbial pathogenesis. Infect Immun 74:28–39. doi: 10.1128/IAI.74.1.28-39.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog 8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tafesse FG, Sanyal S, Ashour J, Guimaraes CP, Hermansson M, Somerharju P, Ploegh HL. 2013. Intact sphingomyelin biosynthetic pathway is essential for intracellular transport of influenza virus glycoproteins. Proc Natl Acad Sci U S A 110:6406–6411. doi: 10.1073/pnas.1219909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hidari KIPJ, Suzuki Y, Suzuki T. 2006. Suppression of the biosynthesis of cellular sphingolipids results in the inhibition of the maturation of influenza virus particles in MDCK cells. Biol Pharm Bull 29:1575–1579. doi: 10.1248/bpb.29.1575. [DOI] [PubMed] [Google Scholar]
- 70.Van Brocklyn JR, Williams JB. 2012. The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: oxidative stress and the seesaw of cell survival and death. Comp Biochem Physiol B Biochem Mol Biol 163:26–36. doi: 10.1016/j.cbpb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 71.Seo Y-J, Alexander S, Hahm B. 2011. Does cytokine signaling link sphingolipid metabolism to host defense and immunity against virus infections? Cytokine Growth Factor Rev 22:55–61. doi: 10.1016/j.cytogfr.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xia C, Seo Y-J, Studstill CJ, Vijayan M, Wolf JJ, Hahm B. 2018. Transient inhibition of sphingosine kinases confers protection to influenza A virus infected mice. Antiviral Res 158:171–177. doi: 10.1016/j.antiviral.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Futerman AH. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem 283:5677–5684. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 74.Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, Kelly S, Wang E, Merrill AH, Saba JD. 2004. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem 279:1281–1290. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- 75.Zaraket H, Bridges OA, Duan S, Baranovich T, Yoon S-W, Reed ML, Salomon R, Webby RJ, Webster RG, Russell CJ. 2013. Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J Virol 87:9911–9922. doi: 10.1128/JVI.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 77.Younes A, Kahn DW, Besterman JM, Bittman R, Byun HS, Kolesnick RN. 1992. Ceramide is a competitive inhibitor of diacylglycerol kinase in vitro and in intact human leukemia (HL-60) cells. J Biol Chem 267:842–847. [PubMed] [Google Scholar]
- 78.Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM. 2011. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J Lipid Res 52:68–77. doi: 10.1194/jlr.M009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]