Understanding reproduction and development benefits enormously from a comparative approach. The CRISPR/Cas9 system of targeted DNA modification opens many additional experimental opportunities for studying animal function in otherwise non-traditional model systems and an important goal is to widely spread the application quickly. Sea urchins have been an important organism for many studies on, e.g. cell fate determination, morphogenesis, gene regulatory networks, and recently it was reported that CRISPR/Cas9 can inform how signaling networks lead to left-right asymmetry (Lin and Su, 2016). Here we add to this approach by streamlining the gRNA construction step, by documenting a simple visual readout of functional gene disruption, and by targeting a non-essential gene that enables quick assessment of successful CRISPR/Cas9 function in the embryo.
Echinoderms make a variety of pigments referred to as echinochromes, and key elements of their synthetic pathway were recently documented (Barsi et al., 2015; Calestani et al., 2003). Essential to the final pigment product is the enzyme polyketide synthase-1 (PKS1) and knock down of this enzyme by morpholino antisense oligonucleotides (MASO) resulted in albino embryos, though transiently by virtue of the MASO life time. Here we targeted PKS1 by CRISPR-Cas9. We focused on a rapid throughput of gRNA construction and found the CRISPR-scan approach of identifying quality gRNA sites and of synthesis of the gRNAs to be well suited [http://www.crisprscan.org/; (Moreno-Mateos et al., 2015)]. The construction of gRNAs is the rate limiting step for widespread utilization of CRISPR-directed gene inactivation and the CRISPR-scan approach is unique in that no cloning is necessary. The tail sequence of the CRISPR gRNA is that region required to form the dsRNA hairpins recognized and bound by Cas9 and is constant for each gRNA of this type II CRISPR/Cas9 system (Jinek et al., 2012). The targeting site, however, is variable and shorter (18–20 nucleotides). By designing targeting sites with a constant sequence to anneal to the complementary tail sequence, and by adding the sequence for the T7 promoter to the 5’end of the targeting sequence, one can efficiently make the DNA template for a guide RNA for less than $20US in a matter of hours. In contrast, our efforts to make gRNA DNA templates by using complementary primers and cloning into a T7 vector takes over a week at much greater cost in terms of money and time. Further, the CRISPRscan website provides an effective algorithm for gRNA targeting with an output that includes the T7 site for direct transcription of the gRNA and the annealing site for the tail DNA primer [see also Help section CRISPRscan.org].
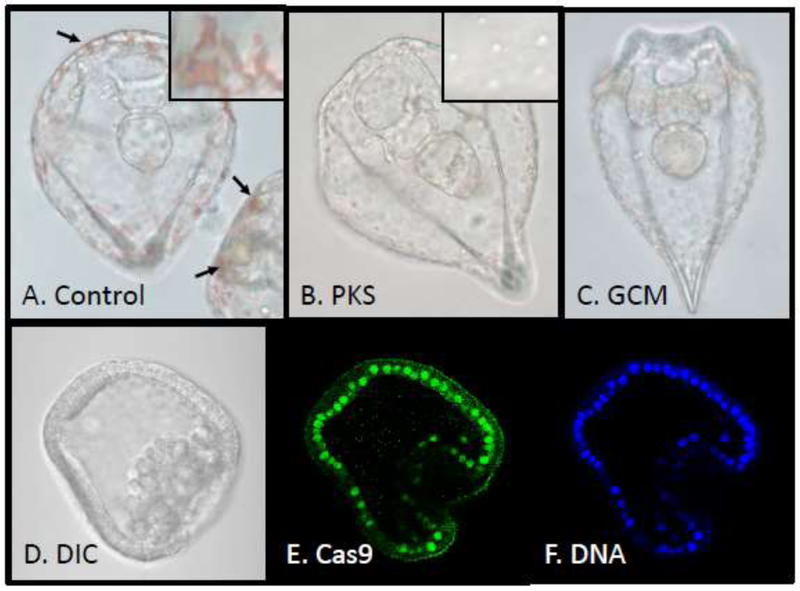
Here we tested three gRNAs to PKS and from DNA ordering to analysis of animal phenotypes, it took less than 1 week time. Following injection of the RNA constructs into egg (Cheers and Ettensohn, 2004) we analyzed Cas9 entry into the nucleus by using mRNA synthesized from the plasmid pCS2–3xFLAG-NLS-SpCas9-NLS (a gift from Yonglong Chen (Addgene plasmid #51307), which encodes Cas9 (codon optimized for mammalian cells) along with two nuclear localization sequences (NLS) (one N-terminal and one C-terminal of the Cas9 sequence) and three FLAG-tags (at the N-terminus; (Guo et al., 2014)). All cells appear to direct Cas9 into nuclei (Figure) but have no discernable disruptive phenotype for long term culture of the embryos. Adding a combination of targeting gRNAs for PKS1 (see Figure and Supplemental Figure) also resulted in no change in the development of the embryo but the resulting larvae lacked pigment and phenocopied larvae from an antisense oligonucleotide approach to knock-down the PKS1 mRNA translaiton (Calestani et al., 2003). We found the approach highly penetrant and efficient; of the 121 gRNA embryos analyzed, 119 were devoid of any pigment, whereas 1 had two pigmented cells, and another was indistinguishable from controls. Pigment cells are progeny of the veg2 tier of cells formed at the ~64 cell stage: in order for larvae to be albino, either PKS1 was disrupted in all cells at that stage, or earlier in their precursor cells. From the low variation of mutated genomic DNA sequences (see Supplemental Figure) we surmise that the targeted gene disruption occurred in all cells of the embryo between the 2–4 cell stage.
CRISPR/Cas9 directed inactivation of genes involved in synthesis of pigment and assayed by albinism. Over 100 pigment cells decorate the larva (arrows in A, inset is magnification of the pigment (red) from control injected embryos). CRISPR/Cas9 targeted gene inactivation of the enzyme responsible for the final synthetic step of pigment (B. polyketide synthase, PKS1, (Calestani et al., 2003); inset shows lack of pigment) or the transcription factor responsible for PKS1 transcription (C. Glial cells missing (GCM), (Ransick and Davidson, 2012)) both yield albinism in otherwise healthy, normal developing animals. D-F show Cas9 enrichment in nuclei of early embryos.
As a further test of the efficiency of the approach, a second gene essential for pigment formation was targeted. The transcription factor Glial Cells Missing (GCM) is essential for transcribing PKS1 (Ransick and Davidson, 2012). The same procedure was followed for gRNA design and synthesis, with similar efficiency (over 100 embryos analyzed; Figure). Further, the resulting larvae perfectly phenocopied the MASO morphants (Ransick and Davidson, 2012).
In conclusion, the CRISPR/Cas9 system is highly effective in early sea urchin embryos. By use of protocols for simple, rapid, and cost-efficient gRNA construction and easy analysis of a DNA disruption phenotype – that does not alter the developmental program or cell fate in the embryo – one now has a quick and easy visual marker for use in combination with other gRNAs for gene disruption. Investigators can co-introduce gRNAs for their target gene of interest, along with the gRNA for pigment synthesis used here (see Supplemental Figures), and assess which embryos to analyze for their experimental gene disruption (CRISPR/Cas9 positive) based on albinism.
Supplementary Material
Acknowledgements:
The authors thank Yi-Hsien Su for help and encouragement, Isabelle Peter for stimulating the concept, and the National Institutes of Health for support (NIH 2R01HD028152).
Grant Support: National Institutes of Health (NIH 2R01HD028152).
References:
- Barsi JC, Tu Q, Calestani C, Davidson EH, 2015. Genome-wide assessment of differential effector gene use in embryogenesis. Development 142, 3892–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH, 2003. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 130, 4587–4596. [DOI] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA, 2004. Rapid microinjection of fertilized eggs. Methods in cell biology 74, 287–310. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y, 2014. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development 141, 707–714. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Su YH, 2016. Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Developmental biology 409, 420–428. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ, 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Davidson EH, 2012. Cis-regulatory logic driving glial cells missing: self-sustaining circuitry in later embryogenesis. Developmental biology 364, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.