Abstract
Human induced pluripotent stem cells (hiPSCs) have received enormous attention because of their ability to differentiate into multiple cell types that demonstrate the patient’s original phenotype. The use of hiPSCs is particularly valuable to the study of cardiac biology, as human cardiomyocytes are difficult to isolate and culture and have a limited proliferative potential. By deriving iPSCs from patients with heart disease, and subsequently differentiating these hiPSCs to cardiomyocytes, it is feasible to study cardiac biology in vitro and model cardiac diseases. While there are many different methods for deriving hiPSCs, clinical use of these hiPSCs will require derivation by methods that do not involve modification of the original genome (non-integrative) or incorporate xeno-derived products (such as bovine serum albumin) which may contain xeno-agents. Ideally, this derivation would be carried out under chemically-defined conditions to prevent lot-to-lot variability and enhance reproducibility. Additionally, derivation from cell types such as fibroblasts requires extended culture (4–6 weeks), greatly increasing the time required to progress from biopsy to hiPSC. Herein, we outline a method of culturing peripheral blood mononuclear cells (PBMCs) and reprogramming PBMCs into hiPSCs using a non-integrative Sendai virus.
Keywords: hiPSC, cardiac disease modeling, Sendai virus, cardiomyocytes
1. Introduction
Stem cells have the ability to differentiate into many different cellular lineages. The use of stem cells is particularly important in regenerative medicine because stem cells can be induced to differentiate into most cell types and can further be used to repair damaged tissue. While there are ethical concerns regarding the use of human embryonic derived stem cells (ESCs) 1, human induced pluripotent stem cells (hiPSCs) can be created from differentiated adult and mature cell types. Initially, mouse iPSCs were created by exogenously expressing only four genes POU5F1 (OCT4), SOC2, KLF4, and MYC (c-MYC) in mouse embryonic fibroblasts (MEF) 2. Since this discovery, hiPSCs have been successfully derived by expressing these four genes, and other similar combinations, in human adult fibroblasts 3, 4, keratinocytes 5, 6, blood 7, 8, adipose stromal cells9, and multiple other cell types.
For hiPSCs to be efficiently created, reprogramming factors must be expressed in the proper stoichiometry 10. In addition, for clinical use of hiPSCs, any exogenous genes should not be carried over and expressed within the hiPSCs after creation. Viral delivery of OCT4, SOC2, KLF4, and MYC reprogramming factors is commonly used to created hiPSCs. Lentiviral and retroviral methods of delivery require integration of exogenous genes within the host genome and subsequent epigenetic downregulation of expression; however, leaky expression of OCT4, SOX2, KLF4, and MYC can be found in hiPSCs even after prolonged culture 11. Therefore, it is important to deliver OCT4, SOX2, KLF4, and MYC by a non-integrating method such as episomal plasmid 12, minicircle plasmid 13, mRNA 14, miRNA 15, or more recently Sendai virus 16. It will also be important to develop chemically-defined culture conditions because they are more cost-effective and elimination of xeno-proteins can prevent activation of the host immune response 17.
Acquiring patient cells may be difficult because some patients are reluctant to donate biological material, such as punch biopsies. It is therefore imperative to derive strategies to create hiPSCs by non-invasive procedures. A mildly, non-invasive procedure is drawing blood from a patient. hiPSCs can be created by isolating the nucleated cells contained in blood (peripheral blood mononuclear cells (PBMCs)) and expressing OCT4, SOX2, KLF4, and MYC into these cells.
In the following text, we have outlined a method of isolating PBMCs from patient blood by Percoll centrifugation and described the culture conditions necessary for PBMC expansion and survival. In addition, we have provided a protocol for reprogramming PBMCs into hiPSCs using non-integrative Sendai virus expressing the reprogramming factors OCT4, SOX2, KLF4, and MYC. Cardiomyocytes derived in this manner will be a valuable resource in studying various cardiac diseases and conditions.
2. Materials
2.1. Blood Media Components [Based on Chou et al., 2011 and modified to be xeno-free)]
-
–
IMDM with glutamine (Life Technologies 12440–053)
-
–
F12 with glutamine (Life Technologies 11765–054)
-
–
Human serum albumin (HSA) (Sigma-Aldrich A1653)
-
–
Chemically defined lipid concentrate (Life Technologies 11905–031)
-
–
Recombinant human insulin (Life Technologies 12585–014)
-
–
Recombinant human transferrin (Sigma T3705)
-
–
Sodium selenite (Sigma-Aldrich S5261)
-
–
Ascorbic acid-2-phosphate (Sigma-Aldrich A8960)
-
–
1-Thioglycerol (Sigma-Aldrich M6145)
-
–
Recombinant human SCF (Peprotech AF-300–07)
-
–
Recombinant human IL3 (Peprotech AF-200–03)
-
–
Recombinant human EPO (R&D 287-TC-500)
-
–
Recombinant human IGF1 (R&D 291-G1–01M)
-
–
Dexamethasone (Sigma-Aldrich D8893)
2.2. iPSC Media
-
–
Essential 8 (Life Technologies A14625DJ and A146265A), combine supplement with base media.
-
–
Essential 7 (Life Technologies), combine supplement with base media.
2.3. Isolation of PBMCs
-
–
Ficoll-Paque PLUS at Room Temperature (GE Healthcare 17–1440-20)
-
–
PBS (Life Technologies 14190–144)
-
–
EDTA (Life Technologies 15575–020)
-
–
Patient blood: At least 10 mL in EDTA preservatives (lavender tubes)
2.4. Reprogramming Factors
-
–
CytoTune®-iPS Sendai Reprogramming Kit (Life Technologies A1378001)
2.5. Additional Reagents
-
–
Matrigel - Growth factor reduced, hESC-qualified
-
–
DMEM/F12 (Life Technologies 11330–032)
-
–
TrypLE Express (Life Technologies 12560–036), store at RT, there is no need to warm to 37°C before use
-
–
Y27632 (ROCK inhibitor, Tocris 1254), 10 mM stocks in Ultrapure water, store at −20 °C
-
–
Ultrapure water (Life Technologies 10977–015)
-
–
Sodium butyrate (NaB) (Sigma-Aldrich B5587)
2.6. Equipment
-
–
6-well cell culture plates (surface area = 9.8 cm2) (Greiner cat. no. 657160) coat with 2 mL Matrigel
-
–
12-well cell culture plates (surface area = 3.8 cm2) (Greiner cat. no. 665180) coat with 1 mL Matrigel
-
–
10 cm tissue culture plates (surface area = 58.95 cm2) (BD Biosciences, cat. no. 353003) coat with 12 mL Matrigel
-
–
15 and 50 mL polypropylene conical tubes (BD Biosciences, cat. no. 2097, 2098)
-
–
2 mL plastic aspiration pipettes (BD Biosciences)
-
–
5, 10, 25 and 50 mL plastic pipettes (BD Biosciences, cat no. 357543, 357551, 357525, 357550)
-
–
250 and 500 mL PES media filters (Corning or Millipore, cat no. SCGPU02RE, SCGPU05RE, SCGPU10RE)
-
–
Countess Cell Counter, slides and trypan blue (Life Technologies)
-
–
Tissue culture incubator capable of 37 ºC, 5% CO2, and 85% relative humidity (such as New Brunswick Galaxy 170R)
-
–
Dual gas tissue culture incubator capable of 37 ºC, 10% CO2, 5% O2, and 85% relative humidity with split inner door (such as New Brunswick Galaxy 170R)
-
–
Centrifuge (such as Beckman Coulter Allegra X-12 or Sorvall)
-
–
Inverted tissue culture microscope (such as Nikon Ti) with heated stage (such as Tokai-Hit)
3. Methods
3.1. Prepare the Blood Media
-
–
Dilute the following components into the outlined stock concentrations, combine each component and filter sterilize.
| Blood Media Composition | 100 mL | 200 mL |
|---|---|---|
| IMDM | 49 mL | 98 mL |
| F12 | 49 mL | 98 mL |
| 5 mg/mL HSA | 500 mg | 1 g |
| 1 x Lipid Concentrate (100x) | 1 mL | 2 mL |
| 10 ug/mL Insulin (4 mg/mL) | 250 μL | 500 μL |
| 100 μg/mL Transferrin (50 mg/mL) | 200 μL | 400 μL |
| 14 ng/mL Sodium selenite | 20 μL | 40 μL |
| 64 μg/mL L-Ascorbic acid 2-phosphate (64 mg/mL) | 100 μL | 200 μL |
| 450 uM 1-Thioglycerol (11.5 M) | 3.5 μL | 7 μL |
| 50 ng/mL SCF (100 ng/uL) | 100 μL | 200 μL |
| 10 ng/mL IL3 (100 ng/uL) | 10 μL | 20 μL |
| 2U/mL EPO (1U/uL) | 200 μL | 400 μL |
| 40 ng/mL IGF1 (100 ng/uL) | 40 μL | 80 μL |
| 1 uM Dexamethasone (1 mM) | 100 μL | 200 μL |
3.2. PBMC Isolation from Blood by Percoll Separation
Prepare a PBS buffer mix solution containing phosphate-buffered saline (PBS), pH 7.2, and 2 mM EDTA. Filter sterilize and keep buffer cold (2–8 °C).
Dilute blood cells in 2–4x the volume of buffer (the more dilute the blood sample, the better the purity of the mononuclear cells).
Carefully layer 35 mL of diluted cell suspension over 15 mL of Ficoll-Paque 15 mL (ρ = 1.077 g/mL) in a 50 mL conical tube (Figure 1 A).
Centrifuge at 400×g for 30–40 minutes at 20°C in a swinging-bucket rotor without brake.
Aspirate the upper layer leaving the mononuclear cell layer (lymphocytes, monocytes, and thrombocytes) (Figure 1 B) undisturbed at the interphase.
Carefully transfer the mononuclear cell layer to a new 50 mL conical tube.
Fill the conical tube with PBS buffer, mix, and centrifuge at 300×g for 10 minutes at 20°C. Carefully remove the supernatant completely to remove the Percoll.
Resuspend PBMC in 50 mL of PBS buffer mix and centrifuge at 300×g for 10 minutes at 20°C.
Aspirate PBS buffer mix and resuspend PBMC in Blood medium to 2–3 million cells per mL.
Plate out PBMC in a standard TC coated 24-well plate, change media by removal, centrifugation, and resuspension every 2–3 days for 6–8 days (Figure 1 C).
Figure 1.
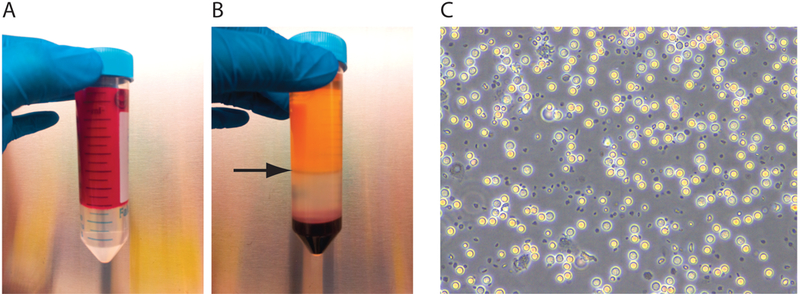
Purification of PBMCs. (A) Slowly layer 35 mL of blood onto 15 mL of Percoll. (B) After centrifugation, remove the thin layer of PBMCs (arrow denotes layer of PBMCs). (C) After multiple rounds of washing, PMBCs will appear as rounded spheres in suspension.
3.3. Infect PBMCs with Sendai virus Reprogramming Factors
Prepare Sendai virus cocktail: 10 uL each of OCT4, SOX2, KLF4, and MYC as per the manufacturer’s instructions. Keep on ice.
Plate 1×105 - 5×105 PBMCs into 200 uL of Blood media in a 24-well plate.
Add combined Sendai virus reprogramming cocktail (40 uL) to PBMCs.
The following day, remove the virus by centrifugation (300×g for 6 minutes). Note: Some PBMCs may be left behind so additional washes of the well may be required. Resuspend the PBMCs into 500 uL of blood media and add to 1 well of a 12-well plate.
Allow PBMCs to grow for three days in suspension in the Blood media + 0.5 mM NaB.
3.4. Prepare Matrigel Plates
Thaw stock bottle of Matrigel at 4°C overnight.
Keep all supplies on ice. Make aliquots of recommended size (270–300 uL, see product insert), and store at −20°C.
Thaw 1 Matrigel aliquot at 4°C.
Add one aliquot of Matrigel (270–300 uL) to 50 mL of cold DMEM/F12 media. Add 2 mL to each well of a 6-well plate.
5. Place the 6-well plates in the incubator and allow the Matrigel to set overnight.
3.5. PBMC Reprogramming
Three days after Sendai virus infection, remove the Blood media by centrifugation and resuspend the cells in 2 mL of E7 media + 0.5 mM NaB.
Add the PBMCs to one well of a 6-well plate coated with Matrigel.
Monitor the cells over the next three days. Suspended cells should settle and adhere to the bottom of the Matrigel-coated wells (Figure 2 A). Once cells have become adherent, aspirate E7 media and feed cells daily with 2 mL of E7.
Continue to monitor the cells for proliferation and morphology change.
At day 15 after Sendai virus infection, replace E7 media with E8 media.
Colonies will appear around day 20 (Figure 2 B) and should be picked (using a stem cell knife or p20 pipette tip) onto 1 well of a fresh Matrigel coated 6-well plate into E8 + 10 uM Y27632 [passage 1 (=p1)].
Figure 2.
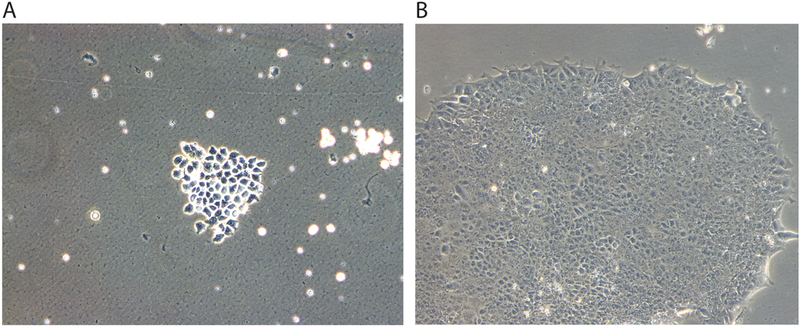
Reprograming of PBMCs. (A) After Sendai virus infection, PMBCs will be adherent to Matrigel coated plates. (B) Twenty days after Sendai virus infection, iPSC colonies can be seen.
3.6. Colony Purification and Expansion
After 7–10 days colonies will have grown out and become dense. Cut up colonies into 10–20 pieces with a p20 pipette tip and transfer into 1 well of a Matrigel coated 6-well plate into Essential 8 + 10 uM Y27632 (=p2).
After 7–10 days, colonies will have grown out and become dense, add 1 mL TrypLE and passage into 1 well of a Matrigel coated 6-well plate into Essential 8 + 10 uM Y27632 (=p3).
After 4–5 days, colonies will have grown out and become dense, add 1 mL TrypLE and passage into 3 wells of a Matrigel coated 6-well plate into Essential 8 + 10 uM Y27632 (=p4).
Freeze down 5 vials from the 3 wells and continue to grow at a 1:6 split ratio, freezing vials every 5–10 passages.
Grow cells to at least passage 20 for efficient differentiation.
Figure 3.

Timeline of blood reprograming. The timeline outlines cell culture conditions and media requirements for the different stages of reprogramming PBMCs to iPSCs.
References
- 1.Lebacqz K Stumbling on status: Abortion, stem cells, and faulty reasoning. Theor Med Bioeth. 2012;33:75–82 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676 [DOI] [PubMed] [Google Scholar]
- 3.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long qt syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229 [DOI] [PubMed] [Google Scholar]
- 4.Lai WH, Ho JC, Lee YK, Ng KM, Au KW, Chan YC, Lau CP, Tse HF, Siu CW. Rock inhibition facilitates the generation of human-induced pluripotent stem cells in a defined, feeder-, and serum-free system. Cell Reprogram. 2010;12:641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2011;108:8797–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raya A, Rodriguez-Piza I, Navarro S, Richaud-Patin Y, Guenechea G, Sanchez-Danes A, Consiglio A, Bueren J, Izpisua Belmonte JC. A protocol describing the genetic correction of somatic human cells and subsequent generation of ips cells. Nat Protoc. 2010;5:647–660 [DOI] [PubMed] [Google Scholar]
- 7.Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L. Efficient human ips cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT, Wu JC. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106:15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiemann U, Sgodda M, Warlich E, Ballmaier M, Scholer HR, Schambach A, Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–435 [DOI] [PubMed] [Google Scholar]
- 11.Hanley J, Rastegarlari G, Nathwani AC. An introduction to induced pluripotent stem cells. Br J Haematol. 2010;151:16–24 [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human ipsc derivation and culture. Nat Methods. 2011;8:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human ips cells. Nat Methods. 2010;7:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mrna. Cell Stem Cell. 2010;7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient mirna-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on sendai virus, an rna virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MJ, Muotri A, Gage F, Varki A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 2005;11:228–232 [DOI] [PubMed] [Google Scholar]


