Abstract
Type III-A CRISPR-Cas systems employ the Cas10-Csm complex to destroy bacteriophages and plasmids, using a guide RNA to locate complementary RNA molecules from the invader and trigger an immune response that eliminates the infecting DNA. In addition, these systems possess the non-specific RNase Csm6 which provides further protection for the host. While the role of Csm6 in immunity during phage infection was previously determined, how this RNase is used against plasmids is unclear. Here we show that S. epidermidis Csm6 is required for immunity when transcription across the plasmid target is infrequent, leading to impaired target recognition and inefficient DNA degradation by the Cas10-Csm complex. In these conditions Csm6 causes a growth arrest in the host and prevents further plasmid replication through the indiscriminate degradation of host and plasmid transcripts. In contrast, when plasmid target sequences are efficiently transcribed, Csm6 is dispensable and DNA degradation by Cas10 is sufficient for anti-plasmid immunity. Csm6 therefore provides robustness to the type III-A CRISPR-Cas immune response against difficult targets for the Cas10-Csm complex.
Introduction
In prokaryotes, clustered, regularly interspaced, short, palindromic repeats (CRISPR) loci provide defence against parasitic phages1 and plasmids2. Defence is mediated by the acquisition of short “spacer” sequences (30–40 nt) from the invading phage or plasmid during infection, which are inserted in between the CRISPR repeats. Spacers are then transcribed and processed into CRISPR RNAs (crRNAs), which guide CRISPR-associated (Cas) nucleases to the invader’s target (known as the protospacer) through complementary base pairing3–6 and trigger its destruction.
To date, six major types of CRISPR systems have been identified, which vary in their cas gene composition and mechanism of action7. Type III systems are uniquely able to degrade both the DNA and RNA of the invader8. The Cas10-Csm (type III-A) and Cas10-Cmr (type III-B) complexes use crRNA guides to detect and anneal to transcripts harbouring a complementary sequence4,9–11. This base pair interaction unleashes the single-stranded DNase activity of Cas1012–14, and the sequence-specific RNase activity of Csm39,10,15 or Cmr44,11. DNA degradation by Cas10 is transient, and cleavage of the protospacer RNA by Csm3/Cmr4 results in Cas10 inactivation and the dissociation of the effector complex from its target12–14.
In addition to the Cas10 complexes, type III systems also use an accessory RNase, Csm6 for type III-A and Csx1 for type III-B16. Csm6 is an endoribonuclease17 whose activity is modulated by cyclic oligoadenylate (cOA), a second messenger synthesised by the Palm domain of Cas10 upon target recognition by the crRNA18–20. The de-activation of Csm6 is the consequence of two events: the lack of synthesis of new cOA molecules by the Palm domain after cleavage of the target transcript by Csm3/Cmr419–21, and the degradation of the existing cOA, presumably by specific ring nucleases22.
The function of Csm6 has been studied during the type III-A CRISPR-Cas immune response against lambda-like dsDNA phages, where Csm6 RNase activity is required only when the target is transcribed late in the phage life cycle23. Because late targeting cannot prevent phage replication, it is hypothesized that Csm6 degradation of viral transcripts prevents the completion of the lytic cycle and allows Cas10 DNase activity to clear the phage genomes that accumulated before the transcription of the target. Csm6 has also been shown to be required for the prevention of plasmid conjugation and plasmid transformation by type III-A CRISPR-Cas systems24,25; however how Csm6 contributes to plasmid clearance is still not understood. Here we show that low levels of target transcription are sufficient to activate Csm6 and trigger non-specific degradation of both host and plasmid transcripts. This accelerates plasmid clearance by the Cas10-Csm complex, presumably through the depletion of transcripts required for efficient plasmid replication and maintenance. Simultaneously, the destruction of host transcripts produces a growth arrest, as was previously proposed19,20,26. Since plasmid DNA degradation leads to the disappearance of the targets that activate Csm6, the growth arrest is short lived and the cells resume normal growth following plasmid clearance. Our study furthers our understanding of the mechanisms by which type III-A CRISPR-Cas systems employ a two-pronged combination of DNase and RNase activities to provide robust immunity against foreign genetic parasites.
Results
Csm6 RNase activity is required for immunity against poorly transcribed targets in pG0400
Spc1 in the CRISPR-cas locus of S. epidermidis RP62A (Fig. 1a) targets the nickase (nes) gene of the conjugative plasmid pG0400, preventing pG0400 transfer2. However, Northern analysis of the crRNAs derived from spc1 indicated that the CRISPR locus is transcribed unidirectionally27 and that the spc1 crRNA has the same, not the complementary, sequence as the putative transcript of the nes gene. Given the requirement of direct binding between the crRNA guide and a target RNA for type III-A CRISPR-Cas immunity28,29, we performed RNA-seq analysis of pG0400 transcripts to look for the presence of nes antisense transcripts that could elicit the spc1-mediated anti-plasmid response. We found very low levels of reads corresponding to transcripts derived from the nes non-template strand, which we hypothesised would result in infrequent activation of the DNase activity of the spc1-crRNA/Cas10-Csm complex. In turn, inefficient clearance of the pG0400 genome would lead to the Csm6 requirement for anti-plasmid immunity that we reported previously25.
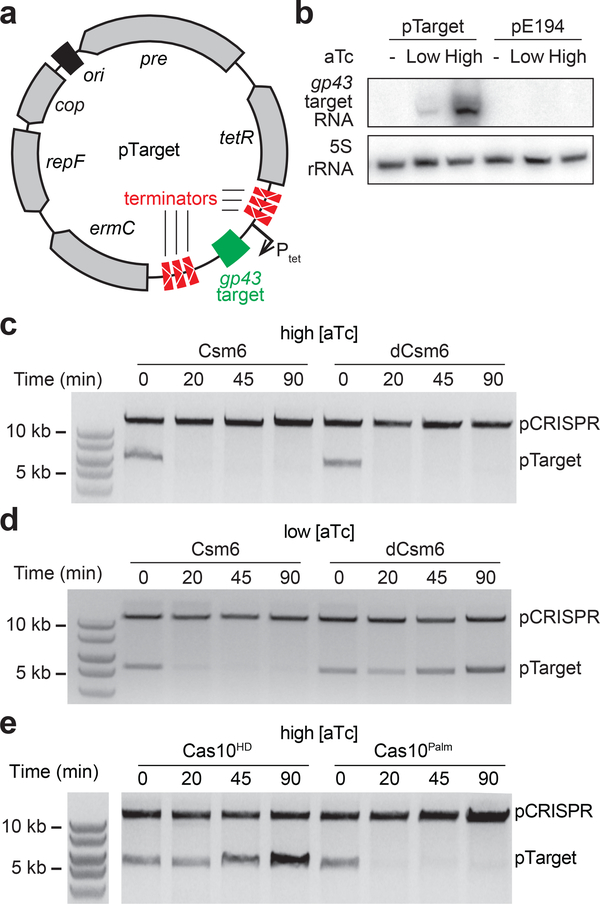
Figure 1 – Csm6 is required for interference against pG0400 when the target is weakly transcribed.
a, The S. epidermidis RP62A type III-A CRISPR, with the catalytic residues of Cas10 and Csm6 highlighted. The black boxes, representing repeats, flank the coloured boxes, symbolising spacers. Spc1 matches the nes antisense transcript of the conjugative plasmid pG0400. b, The architecture around the nes protospacer in pG0400, with RNA-seq traces representing read depth. The annealing positions for spc1 and spc1-flip are indicated. RPM, reads per million. c, Conjugation of pG0400 or pG0420 into S. aureus cells containing pCRISPRs with the indicated csm6 variant and spacer, after filter mating. Each bar represents the mean of three biological replicates ±s.e.m. d, Like b, but for plasmid pG0420, with the inserted transcriptional promoter shown as a black arrow.
To test this hypothesis, we generated a construct harbouring a reversed spc1 (spc1-flip) that would produce a crRNA complementary to the highly abundant nes transcript (Fig. 1b). This was achieved by using Staphylococcus aureus RN422030 harbouring an S. epidermidis type III-A CRISPR-Cas locus on the chloramphenicol-resistant pC194 staphylococcal plasmid, pCRISPR25, with modified spacer sequences. We found that, in contrast to spc1 where targeting requires csm625 (Fig. 1c), spc1-flip blocks pG0400 conjugation in recipients expressing a catalytically dead (R364A, H369A) csm6 version (dcsm6)17,23 (Fig. 1c). We also constructed a pG0400 derivative (pG0420) in which we inserted a strong promoter that drives high transcription of the nes non-template strand (Fig. 1d). Next we tested the ability of spc1 to mediate immunity against pG0420 in the presence or absence of Csm6 RNase activity and found that, similarly to spc1-flip, the increased transcription of the target RNA eliminated the Csm6 requirement observed for pG0400 (Fig. 1c). Altogether, these results support our hypothesis that Csm6 RNA degradation is not required for plasmid clearance in conditions of high target transcription. We also determined that the other type III-A RNase, Csm3 within the Cas10-Csm complex, is not required for anti-plasmid immunity under low transcription conditions (Fig. S1).
Csm6 RNase activity accelerates plasmid clearance in conditions of low target transcription
To determine how Csm6 affects anti-plasmid immunity in the presence of different levels of target transcription, we placed the gp43 protospacer23 (from phage фNM1γ629) under the control of a tight and tuneable anhydrotetracycline (aTc)-inducible promoter31, flanked by strong transcriptional terminators (pTarget, Fig. 2a). We performed Northern blots to validate these useful features of pTarget (Fig. 2b); we did not detect a target transcript in the absence of aTc, and found that target transcription levels correlate with the aTc concentration. We first investigated the effect of Csm6 on the fate of plasmid DNA. We generated S. aureus cultures harbouring both pTarget and pCRISPR, added different concentrations of aTc, purified the plasmids, and visualized them using agarose gel electrophoresis. We found that at high levels of aTc, i.e. high target transcription, plasmid loss was equally efficient in wild-type and dcsm6 backgrounds (Fig. 2c), even shortly after induction (Fig. S2). This result corroborates our previous result that type III-A immunity is independent of Csm6 at high levels of target transcription, and that this ribonuclease is not necessary for DNA degradation per se. In contrast, at low levels of aTc, pTarget remained stable in hosts carrying the dcsm6 allele (Fig. 2d), demonstrating that the RNase activity of Csm6, which is essential for immunity in these conditions, leads to an accelerated rate of plasmid DNA loss.
Figure 2 – Csm6 accelerates plasmid clearance when interfering against a weakly transcribed protospacer.
a, Overview of the pTarget plasmid, with salient features labelled. Upon the addition of the inducer anhydrotetracycline (aTc), transcription is initiated from promoter Ptet, and the gp43 protospacer is transcribed. This transcription allows interference against pTarget by the III-A CRISPR system (pCRISPR). The transcriptional terminators prevent non-specific background transcription across the gp43 target. b, Northern blot analysis of pTarget (performed once), detecting the gp43 target transcript prior to, or two minutes after, addition of either low or high levels of aTc. The empty plasmid pE194 is used as a control. c, pTarget plasmid curing assay, where plasmid DNA is extracted from cells containing pTarget and a pCRISPR with the specified csm6 allele, before or at the specified time after adding high levels of aTc, and visualised by gel electrophoresis. Gel image is representative of three independent experiments. d, Like c, but with induction of protospacer transcription by low levels of aTc. Gel image is representative of three independent experiments. e, Like c, but with mutations in the Cas10 Palm or HD domains, both in a dcsm6 background. Gel image is representative of three independent experiments.
Csm6 activation leads to non-specific degradation of host and plasmid transcripts
How can an RNase impact plasmid DNA stability? We investigated if Csm6 was responsible for the degradation of host and/or plasmid transcripts important for plasmid replication/maintenance using RNA-seq. Because plasmid degradation carried out by the DNA cleavage activity of the type III-A system would lead to differences in transcript abundance due to different plasmid template levels (not only due to Csm6 RNase activity), we performed RNA-seq in a cas10 mutant genetic background lacking DNase activity.
Previously, we reported that the Palm domain of the Cas10 subunit of the type III-A S. epidermidis complex is implicated in DNA cleavage15. However, recent work on other type III systems showed that Cas10 nuclease activity relies on its HD domain12–14,32 and that the Palm domain is involved in the synthesis of Csm6’s inducer, cOA14,20,21. To clarify this discrepancy, we repeated the experiment of Figure 2c in a genetic background harbouring HD or Palm domain mutations [H14A, D15A (Cas10HD) or D586A, D587A (Cas10Palm)] (Fig. 1a). We tested conditions of high target transcription, in a dCsm6 background, to avoid the interference of RNA degradation in the interpretation of the results. We found that the HD, but not the Palm, domain of Cas10 is required for the clearance of pTarget (Fig. 2e), and therefore used a pCRISPR-cas10HD mutant for our RNA-seq experiment.
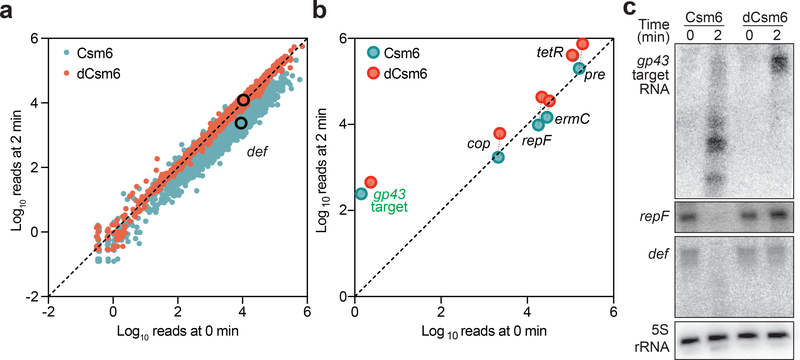
For the RNA-seq experiment, wild-type or dcsm6 cells were harvested in duplicate either before or two minutes after induction with a low concentration of aTc. RNA was extracted, sequenced, and the average transcript abundance of every gene at 0 or 2 minutes after induction was plotted. Host transcripts exhibited a marked decrease in global transcript abundance in the presence of Csm6, i.e. most genes falling below the identity line (Fig. 3a). In the absence of the RNase activity of Csm6, however, this reduction was not detected. pTarget transcripts showed a similar difference in abundance (Fig. 3b). To corroborate the RNA-seq results, Northern blot analysis was performed to detect a subset of transcripts (Fig. 3c). The transcripts corresponding to the pTarget protospacer, the plasmid replication repF gene, and the chromosomal gene def (peptide deformylase) were all rapidly degraded in the presence of Csm6 RNase activity. Together, these experiments demonstrate that Csm6 activation during the type III-A CRISPR-Cas immune response results in significant depletion of both host and plasmid transcripts.
Figure 3 – Csm6 activation results in non-specific degradation of host and plasmid transcripts.
a, RNA-seq was performed with S. aureus cells harbouring a pCRISPR with wild-type (red) or mutated csm6 (blue), extracting RNA before or after inducing target transcription by adding low levels of aTc. L. seeligeri spike-in RNA was added to allow absolute normalisation of read depth. Chromosomal genes falling on the identity line are unchanged from 0 to 2 minutes, while genes falling below the line are depleted after 2 minutes. Both are done in a Cas10HD background. The def gene (peptide deformylase) is highlighted. The average of two replicates for each condition is shown. b, Like a, but showing pTarget genes. c, Northern blot analysis of RNA from cells with pTarget and pCRISPRs containing either csm6 or dcsm6, before or after inducing pTarget transcription with low levels of aTc (performed once). Probes detecting either the gp43 target transcript, the repF transcript of pTarget, or the chromosomal transcript def are used.
Degradation of host transcripts induces a growth arrest
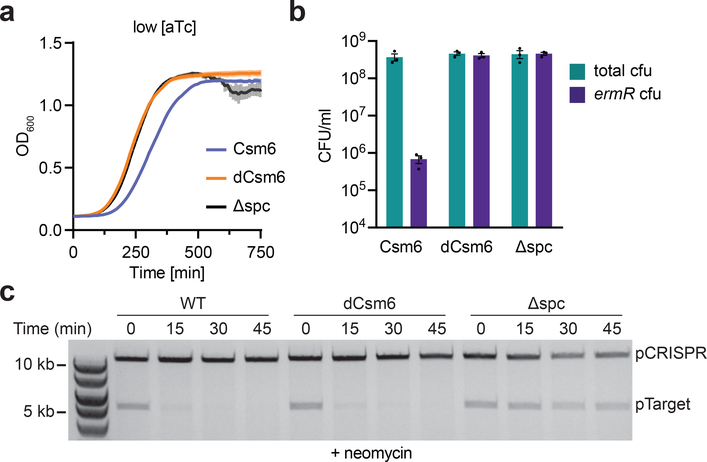
The results described above suggest two hypotheses: (i) that the general destruction of host transcripts should result in host cell toxicity during type III-A CRISPR-Cas immunity, and (ii) that the degradation of transcripts important for replication and/or maintenance should prevent the replenishment of plasmid DNA and therefore facilitate plasmid clearance by the DNase activity of the Cas10-Csm complex. We tested the first hypothesis by looking at the effect of CRISPR immunity against pTarget on host growth. In the presence of erythromycin, cell survival requires the presence of pTarget to provide resistance against the antibiotic and the growth of the culture correlates with the rate of plasmid loss observed in Figures 2c and 2d (Fig. S3a-b). Therefore, to determine if Csm6 activation results in collateral RNA degradation we measured the effect of type III-A targeting on culture growth in the absence of erythromycin, i.e. without plasmid selection. Under low target transcription conditions that require Csm6 for rapid plasmid loss (Figs. 2d and S1b), we detected a significant growth arrest that was dependent on the presence of Csm6’s RNase activity (Fig. 4a). To measure plasmid destruction we enumerated the cells that still contained pTarget at the end of the growth experiment by counting the erythromycin-resistant colony forming units (cfu) that remain after induction (Fig. 4b). We found a csm6-dependent decrease in cfu that reflects extensive plasmid clearance in the culture. These results show that Csm6 activation, which is required for plasmid loss under low target transcription conditions, is also responsible for generating a growth defect of the host. Interestingly, this growth defect is triggered by Csm6’s enzymatic activity even in conditions of high transcription, when this RNase is not required for plasmid clearance (Fig. S3c-d).
Figure 4 – Prevention of expression of genes important for plasmid replication accelerates plasmid clearance.
a, Growth of staphylococci containing pTarget and pCRISPRs with either csm6 or dcsm6, upon inducing transcription across the target with low levels of aTc, in the absence of erythromycin. Δspc is a no spacer control. OD600 measurements are taken every 10 minutes, each data point representing the mean of three biological replicates ±s.e.m. b, Cells at the end of the experiment in a are spotted onto TSB plates in the presence or absence of erythromycin, selecting for the presence of pTarget, and enumerated. Each bar represents the mean of three biological replicates ±s.e.m. c, Plasmid curing assay, where plasmid DNA is extracted from cells harbouring the specified pCRISPR and pTarget, before or at different times after induction of protospacer transcription by the addition of low levels of aTc, and visualised by gel electrophoresis. The cells are grown in the presence of the translational inhibitor neomycin. Gel image is representative of three independent experiments.
Non-specific degradation of host and plasmid transcripts facilitates plasmid clearance
Next, we investigated the second hypothesis; whether the prevention of expression of genes important for plasmid replication could accelerate plasmid clearance. pTarget genes such as repF and cop are important for plasmid replication and maintenance, and our RNA-seq analysis determined that these transcripts are targeted by Csm6. In addition, Csm6-mediated depletion of host transcripts required for plasmid replication, such as DNA polymerase and single-stranded DNA binding protein33, could also impair plasmid stability and facilitate clearance by the type III-A CRISPR-Cas system. To test if a global reduction of gene expression, similar to that caused by Csm6 activation, can accelerate plasmid loss during the type III-A CRISPR-Cas immune response, we used neomycin, an inhibitor of the 30S ribosomal subunit, and measured pTarget clearance after induction with low levels of aTc. In contrast to Figure 2d results, repeating this experiment in the presence of neomycin led to rapid pTarget clearance also in the dCsm6 background (Fig. 4c), comparable to that of cells expressing wild-type Csm6. Importantly, neomycin did not affect pTarget stability in non-targeting hosts (Fig. 4c, Δspc) or in cells without Cas10 DNase activity (Fig. S4). These results support the idea that reduction in expression of both plasmid and host genes, either by the non-specific mRNA degradation by Csm6 or by neomycin-induced inhibition of translation, can facilitate the type III-A CRISPR-Cas immunity against plasmids with low rates of protospacer transcription.
Complete plasmid clearance requires DNA cleavage by Cas10
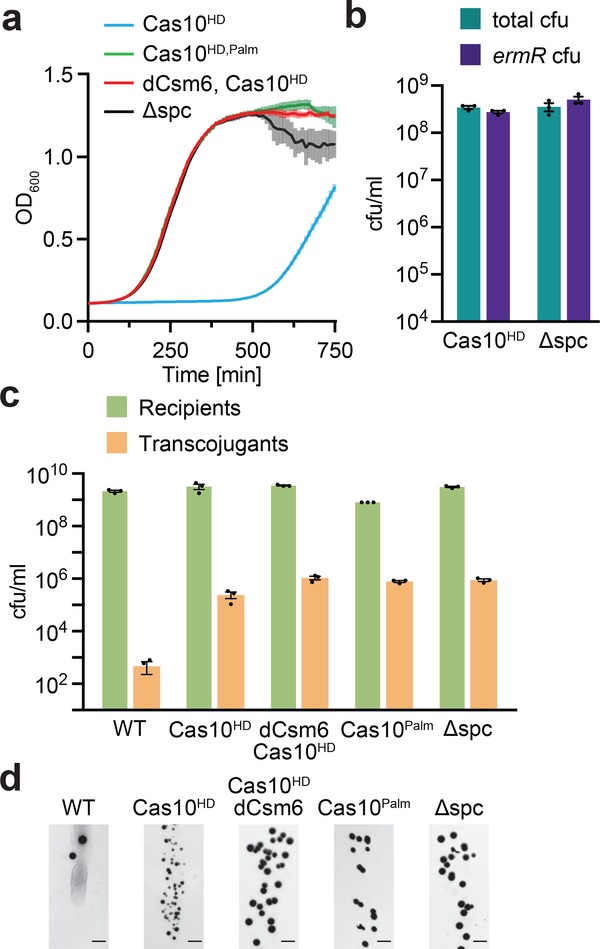
Finally, we examined whether transcript degradation by Csm6 alone is sufficient for anti-plasmid type III-A CRISPR-Cas immunity. We measured the growth of cells carrying the cas10HD mutation after induction of target transcription in the absence of erythromycin selection. We found a growth arrest that was much more severe than detected with wild-type Cas10 (Fig. 4a), likely due to the absence of DNA cleavage and the persistence of pTarget, leading to continuous activation of Csm6 (Fig. 5a). This growth arrest was dependent on the ability of the Cas10 Palm domain to make cOA, and on the RNase activity of Csm6. In addition, all the cells that recovered at the end of the experiment were able to grow on erythromycin and therefore retained pTarget (Fig. 5b), escaping arrest through the accumulation of mutations that abrogate type III-A immunity and the activation of Csm6 (Fig. S5). Altogether these results demonstrate that the Csm6-mediated non-specific degradation of host and plasmid transcripts that affect plasmid replication is not sufficient to mount an efficient type III-A CRISPR-Cas immune response, which also requires the destruction of plasmid DNA by the Cas10 nuclease.
Figure 5 – The Cas10 HD domain is required for efficient plasmid clearance during type III-A immunity.
a, The OD600 of staphylococci harbouring pTarget and the specified pCRISPR is measured every 10 minutes after induction of protospacer transcription by the addition of high levels of aTc, in the absence of erythromycin. Each data point representing the mean of three biological replicates ±s.e.m. b, Cells at the end of the experiment in a are spotted onto TSB plates in the presence or absence of erythromycin, selecting for the presence of pTarget, and enumerated. Each bar represents the mean of three biological replicates ±s.e.m. c, Conjugation of pG0400 into S. aureus cells carrying the specified pCRISPRs, after filter mating. Each bar represents the mean of three biological replicates ±s.e.m. d, Representative plate images from the conjugation experiment in panel c, performed once. Scale bar, 5 mm.
We also investigated the effect of Csm6-mediated growth arrest on the type III-A immune response against pG0400, the natural plasmid target of the S. epidermidis CRISPR system. Previously we showed that mutations in csm6 and cas10 Palm domain, but not in its HD domain, led to an increase in the number of pG0400 transconjugants, i.e. lack of type III-A immunity25. Similar results were recently obtained by analyzing this system in an Escherichia coli heterologous host24. Given our findings with pTarget, we thought that we might have failed to see pG0400 transconjugants in the cas10HD background if the Csm6-mediated growth arrest affected colony growth. Therefore we performed conjugation assays and incubated the plates seeded with transconjugants for a longer time (24 hours, as opposed to the 16 hours used in previous assays). We found that indeed cas10HD hosts do produce a large number of transconjugants, indicating a loss of CRISPR immunity (Figs. 5c-d). However, transconjugant colonies are markedly small (Fig. 5d). As we hypothesized, Csm6 is responsible for this phenotype, which is eliminated by the addition of the dcsm6 mutation (Cas10HD vs Cas10HD/dCsm6, Fig. 5d). Altogether, these results with both pTarget and pG0400 suggest that Csm6-mediated growth arrest is only temporary and not essential for type III-A immunity against plasmids. Cas10-mediated DNA cleavage, on the other hand, is absolutely necessary to fully eliminate the plasmid target and turn off Csm6’s non-specific RNase activity.
Discussion
Here we show that Csm6 RNase activity is only required for S. epidermidis type III-A CRISPR-Cas immunity under conditions of low transcription of the plasmid target sequence. We found that activated Csm6 results in the non-specific degradation of host and plasmid transcripts and promotes the fast clearance of the target plasmid by reducing the expression of genes important for its replication and/or maintenance. In addition, the global depletion of host transcripts leads to a growth arrest. Csm6 RNase activity is not sufficient for plasmid clearance and the slow degradation of plasmid DNA by the Cas10 nuclease is also required. Upon complete plasmid loss promoted by both activities, the target transcript is eliminated and cOA production ceases (and is eventually degraded), resulting to Csm6 deactivation and the return of normal growth. Although our studies only addressed type III-A CRISPR-Cas systems, type III-B systems have a similar targeting mechanism, and it is therefore likely that their accessory RNase, Csx134, has a similar function to that described here for Csm6.
We believe that when target transcription is low there are few recruitment events of the Cas10-Csm complex to the invading plasmid, and DNA cleavage is inefficient. Under such conditions a “two-hit punch” is required for immunity, where Csm6 prevents the plasmid from replicating while the Cas10-Csm complex slowly degrades the plasmid. This is in line with studies of the type III-A anti-phage response23, where it was found that Csm6 is required when the target is expressed late in the phage lytic cycle. In this case, the accumulation of many phage genomes before the activation of type III-A immunity would overwhelm the DNA cleavage capacity of the Cas10-Csm complex. Csm6-dependent degradation of both host and phage transcripts would hinder phage replication and allow time for the gradual degradation of phage genomes by Cas10. Also, Csm6 was determined to be essential when the phage target contains multiple mismatches with the crRNA guide. It is conceivable that such mismatches will specifically inhibit activation of the HD domain but not the Palm domain, leading to low DNA cleavage but normal Csm6-mediated RNA degradation. Therefore Csm6 appears to be an accessory RNase that rescues type III-A immunity when DNA targets are difficult to detect or eliminate.
One fundamental difference in the Csm6-mediated response against phages and plasmids is the observation of a host growth defect only in the latter. We speculate that as result of both the high concentration of phage genomes and the preferential transcription of these genomes during infection, the host cell contains a much higher proportion of phage transcripts23. Therefore, assuming that the cOA degradation enzymes prevent accumulation of the inducer in most of the bacterial cytoplasm except around its site of production where there is a local concentration of active Csm6 molecules, during the anti-viral type III-A CRISPR response this volume will be occupied mostly by phage RNA, and the impact on host transcripts (and thus growth) is less noticeable.
Interestingly, the indiscriminate RNA degradation of Csm6 has similarities to ribonuclease toxins. Indeed, HEPN domains are found in a wide range of predicted toxin-antitoxin (TA) modules and abortive infection systems in all three domains of life35. In contrast to the function sometimes ascribed to these systems, our data suggests that the role of Csm6 in immunity is not through the induction of host cell death, but instead leads to a temporary growth arrest. Although not found in our experimental system, there are possible scenarios where Csm6 activation could trigger cell death. For example, at very high phage concentrations the Cas10-Csm complex might be unable to clear the virus, prolonging Csm6 activity to degrade total cellular RNA. This could kill the host, but would also prevent the release of functional viral particles, similar to programmed cell death pathways of eukaryotic cells36 and abortive infection defense systems of prokaryotes37. Further, there might be circumstances where the DNase activity of Cas10 is completely compromised, such as base modifications in phage DNA that affect cleavage, or anti-CRISPR proteins that could specifically inhibit the HD domain of Cas10. Future studies will determine if Csm6 global degradation of host transcripts can mediate abortive infection-like immunity.
Methods
Bacterial strains and growth conditions
S. aureus RN422030 was grown in tryptic soy broth (TSB) medium at 37°C, supplemented with 10 μg/ml of chloramphenicol or erythromycin for maintenance of plasmids pC19438 or pE19439 respectively.
Molecular cloning
The plasmids with type III CRISPR systems (pCRISPR) and harbouring either a gp43 spacer (pWJ191, pWJ241) or no spacer (pGG-BsaI-R) have been described elsewhere23,29. The sequences of the oligonucleotides used in this study are provided in supplemental table 1. The plasmids used in this study are shown in supplemental table 2. The plasmid cloning strategies are showed in supplemental table 3. All pCRISPR plasmids contain one spacer flanked by two repeats.
Conjugation
Conjugation was performed using filter mating as previously described2 into recipients containing the specified pCRISPRs (pGG25, pGG-BsaI-R, pJTR111, pJTR135, pJTR138, pJTR175, or pJTR177). Pictures of colonies for Figure 5d were obtained by imaging the plates with the Axygen Scientific GD1000 gel documentation system, 24 hours after plating.
Plasmid curing assay
Overnight RN4220 cultures carrying pTarget (pJTR162) and the specified pCRISPRs (pWJ191, pWJ241, pJTR125, pJTR147, pGG-BsaI-R) were diluted to an OD of exactly 0.15 in TSB containing 10 μg/ml of chloramphenicol. Where relevant, 200 μg/ml of neomycin was added. aTc was added to a final concentration of either 7.5 ng/ml (“low aTc condition”) or 50 ng/ml (“high aTc condition”), and cells were isolated and plasmid extracted at the specified timepoints. 300 ng of total plasmid was then linearised with the common single cutter BamHI-HF (NEB), and imaged by gel electrophoresis.
Growth curves
Triplicate overnight RN4220 cultures carrying pTarget and the specified pCRISPRs (pWJ191, pWJ241, pGG-BsaI-R, pJTR109, pJTR121, pJTR125) were diluted 1:100 in fresh TSB with 10 μg/ml of chloramphenicol, and 10 μg/ml of erythromycin where specified, and outgrown for an hour. Cells were then diluted and normalised for OD, moved to a 96-well plate in triplicate, and aTc was added to a final concentration of either 2.5 ng/ml (“low aTc condition”) or 12.5 ng/ml (“high aTc condition”). OD600 readings were then taken every 10 minutes by a microplate reader (TECAN Infinite 200 PRO). For measuring colony forming units from each well, plateau phase cells from the end of the experiment were resuspended, serial diluted, and spotted on TSB agar plates. Plates contained 10 μg/ml of chloramphenicol when selecting for pCRISPR only, or 10 μg/ml of chloramphenicol and 10 μg/ml of erythromycin when also selecting for pTarget. For the Cas10 dHD targeting escaper growth curves, cells that recovered from the end of the experiment in 5a (Cas10HD cells grown in high aTc conditions) were streaked out, and single colonies were picked for a new growth experiment, in the presence of high levels of aTc. To analyse these escapers, DNA was isolated, subjected by PCR using primers JTR390 and W1022, with the products being visualised by gel electrophoresis. PCR products (escapers 1–3) or isolated plasmid (escaper 4) were sent for Sanger sequencing to confirm the observed deletions.
RNA purification
For isolating RNA for pG0400/pG0420 RNA-seq, 5ml of S. aureus RN4220 cells at OD 0.6 containing the relevant plasmid were spun down. For isolating RNA for Csm6 targeting RNA-seq, 20 ml of S. aureus RN4220 cells at OD 0.15 were spun down at 0 min or 2 min after aTc addition. For both RNA-seq runs, cells were lysed in PBS with treatment with 1 mg/ml lysostaphin and 2 mg/ml lysozyme for five minutes, followed by addition of 1% sarcosyl. For the Csm6 targeting RNA-seq, 2.5 μg of Listeria seeligeri RNA was added at this stage. RNA was then purified using Quick-RNA Miniprep Plus Kit (Zymo research). For Northern blot analysis, 80 ml of OD 0.15 S. aureus RN4220 cells were spun down and lysed as above. The RNA was then isolated by resuspending the lysed cells in Trizol (Thermo Fisher Scientific), and following the Trizol manufacturer’s protocol.
RNA-seq of pG0400/pG0420
RNA was isolated from cells harbouring pG0400/pG0420 as described above, DNase treated (Invitrogen TURBO DNA-free kit according to the manufacturer’s protocol), and rRNase depleted (Illumina Ribo-Zero rRNA Removal Kit (Bacteria) according to the manufacturer’s protocol). Library preparation was done using TruSeq Stranded mRNA kit (Illumina), and the sequencing was performed by Illumina MiSeq. Reads were aligned using STAR40 version 2.5 to either pG040041 (Genebank reference KT780705) or S. aureus NCTC 8325 (Genebank reference CP000253), without normalisation.
RNA-seq of pTarget using a spike RNA
In duplicate, overnight cultures of S. aureus RN4220 carrying pTarget and pCRISPRs containing Cas10HD and either Csm6 or dCsm6 (pJTR109 or pJTR125) were diluted to an OD600 of exactly 0.15 in TSB. Cells were then harvested for the 0 minute timepoint. Then, aTc was added to a final concentration of 7.5 ng/ml (similar to “low” concentration for plasmid curing), and cells were harvested after 2 minutes, quenching the reaction with cold TSB. RNA was then isolated as described above. At the cell lysis stage (after adding the TRI reagent of the Quick-RNA Miniprep Plus Kit (Zymo research)), an equal amount of purified Listeria seeligeri RNA was added to each sample, to be carried through the purification to allow absolute comparisons between the samples. Library preparation was then carried out like for pG0400/pG0420 RNA-seq, and the samples were submitted to NextSeq (Illumina) sequencing at the Rockefeller University Genomics core (New York, USA). For the analysis, inspired by42, the normalisation protocol relied on the number of spike reads mapping to the Listeria seeligeri genome (NC_013891.1). In the first round of read mapping, the reads for each sample were mapped to S. aureus using STAR aligner, with standard parameters except for allowing maximum one mismatch. The unmapped reads were then aligned to the L. seeligeri genome. Since each sample initially had the same number of L. seeligeri spike reads at the lysis stage, a scaling factor was calculated to make number of L. seeligeri reads identical between the samples. This scaling factor is later used to normalise the reads mapping to the S. aureus chromosome or pTarget between samples. Then, for the second round of mapping, all reads are first aligned to L. seeligeri (to eliminate spike reads), and the remaining reads were mapped to either S. aureus or pTarget. The total assigned reads per gene was determined using featureCounts43 with largestOverlap set to TRUE. The number of assigned reads to each gene was then normalised by multiplying with the previously calculated scaling factor, thus allowing absolute comparison between the number of assigned reads to a gene between different samples.
Northern blot
Overnight RN4220 cultures carrying pTarget and the specified pCRISPRs (pWJ191 or pWJ241) were diluted to an OD of exactly 0.15 in TSB containing 10 μg/ml of chloramphenicol. 80 ml of cells were harvested for the 0 minute timepoint. Then aTc was added to a final concentration of 7.5 ng/ml, and 80 ml of cells were again harvested after 2 minutes. RNA was then isolated described as above. The RNA was separated on a 6% PAGE gel by electrophoresis, and blotted onto nylon filters (Invitrogen BrightStar Plus) using a semi-dry blotting apparatus. The oligonucleotide probes were radiolabelled with γ−32P-labelled ATP using PNK (NEB), and incubated with the nylon membranes overnight at 42 °C in the presence of 0.1 mg/ml salmon sperm DNA. The membranes were then visualised by phosphorimaging (Typhoon FLA 7000, GE Life Sciences). For assaying pTarget induction at “low” or “high” aTc concentrations, cells containing either pJTR162 or pE194 were isolated before or 2 minutes after adding 7.5 ng/ml or 50 ng/ml, respectively. The rest of the protocol was done as described above.
Supplementary Material
Acknowledgements.
We would like to thank Alexander Meeske and Charlie Mo for critical reading of the manuscript. We thank Gregory Goldberg for plasmid pGG25; Claire Kenney and Wenyan Jiang for sharing their insights on spc1-flip conjugation; the Rockefeller University Genomics Resource Center for performing the Csm6 targeting Nextseq RNA-seq experiment; and Thomas Carroll of the Rockefeller University Bioinformatics Resource Center, and Elitsa Stoyanova, for helpful discussions on the bioinformatical analysis. J.T.R. was supported by a Boehringer Ingelheim Fonds PhD fellowship. L.A.M. is supported by a Burroughs Wellcome Fund PATH Award and an NIH Director’s Pioneer Award (DP1GM128184).
Footnotes
Competing interests. L.A.M. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics and a cofounder of Eligo Biosciences.
Data availability. The data from this study is available from the authors on request. The raw data for the RNA-seq experiments can be found through the Sequence Read Archive (NIH) through accession code PRJNA506073. Original gels pictures and Northern blots are in Fig. S6
References
- 1.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712, (2007). [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA & Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasiunas G, Barrangou R, Horvath P & Siksnys V Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A 109, E2579–2586, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hale CR et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westra ER et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS & Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 37, 67–78, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pyenson NC & Marraffini LA Type III CRISPR-Cas systems: when DNA cleavage just isn’t enough. Curr. Opin. Microbiol 37, 150–154, (2017). [DOI] [PubMed] [Google Scholar]
- 9.Staals RH et al. RNA Targeting by the Type III-A CRISPR-Cas Csm Complex of Thermus thermophilus. Mol. Cell 56, 518–530, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamulaitis G et al. Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol. Cell 56, 506–517, (2014). [DOI] [PubMed] [Google Scholar]
- 11.Zhang J et al. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. Mol. Cell 45, 303–313, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elmore JR et al. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 30, 447–459, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrella MA, Kuo FT & Bailey S RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 30, 460–470, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C & Siksnys V Spatiotemporal Control of Type III-A CRISPR-Cas Immunity: Coupling DNA Degradation with the Target RNA Recognition. Mol. Cell 62, 295–306, (2016). [DOI] [PubMed] [Google Scholar]
- 15.Samai P et al. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewoehner O & Jinek M Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burroughs AM, Zhang D, Schaffer DE, Iyer LM & Aravind L Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G & Siksnys V A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609, (2017). [DOI] [PubMed] [Google Scholar]
- 20.Niewoehner O et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548, (2017). [DOI] [PubMed] [Google Scholar]
- 21.Rouillon C, Athukoralage JS, Graham S, Gruschow S & White MF Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athukoralage JS, Rouillon C, Graham S, Gruschow S & White MF Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Samai P & Marraffini LA Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164, 710–721, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster K, Kalter J, Woodside W, Terns RM & Terns MP The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatoum-Aslan A, Maniv I, Samai P & Marraffini LA Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. J. Bacteriol 196, 310–317, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anantharaman V, Iyer LM & Aravind L Presence of a classical RRM-fold palm domain in Thg1-type 3’- 5’nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol Direct 5, 43, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marraffini LA & Sontheimer EJ Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L, Garrett RA, Shah SA, Peng X & She Q A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol 87, 1088–1099, (2013). [DOI] [PubMed] [Google Scholar]
- 29.Goldberg GW, Jiang W, Bikard D & Marraffini LA Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreiswirth BN et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712, (1983). [DOI] [PubMed] [Google Scholar]
- 31.Helle L et al. Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157, 3314–3323, (2011). [DOI] [PubMed] [Google Scholar]
- 32.Liu TY, Iavarone AT & Doudna JA RNA and DNA Targeting by a Reconstituted Thermus thermophilus Type III-A CRISPR-Cas System. PLoS One 12, e0170552, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Maso JA et al. Plasmid Rolling-Circle Replication. Microbiol Spectr 3, PLAS-0035–2014, (2015). [DOI] [PubMed] [Google Scholar]
- 34.Sheppard NF, Glover CV 3rd, Terns RM & Terns MP The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22, 216–224, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV & Aravind L Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 15, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upton JW & Chan FK Staying alive: cell death in antiviral immunity. Mol. Cell 54, 273–280, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrie SJ, Samson JE & Moineau S Bacteriophage resistance mechanisms. Nat. Rev. Microbiol 8, 317–327, (2010). [DOI] [PubMed] [Google Scholar]
- 38.Horinouchi S & Weisblum B Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol 150, 804–814, (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horinouchi S & Weisblum B Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol 150, 815–825, (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray MD, Boundy S & Archer GL Transfer of the methicillin resistance genomic island among staphylococci by conjugation. Mol. Microbiol 100, 675–685, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamberte LE et al. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat Microbiol 2, 16249, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.