Abstract
Objective:
Poly(ADP-ribose) polymerase (PARP) inhibitors have shown substantial activity in homologous recombination- (HR-) deficient ovarian cancer and are undergoing testing in other HR-deficient tumors. For reasons that are incompletely understood, not all patients with HR-deficient cancers respond to these agents. Preclinical studies have demonstrated that changes in alternative DNA repair pathways affect PARP inhibitor (PARPi) sensitivity in ovarian cancer models. This has not previously been assessed in the clinical setting.
Methods:
Clonogenic and plasmid-based HR repair assays were performed to compare BRCA1-mutant COV362 ovarian cancer cells with or without 53BP1 gene deletion. Archival biopsies from ovarian cancer patients in the phase I, open-label clinical trial of PARPi ABT-767 were stained for PARP1, RAD51, 53BP1 and multiple components of the nonhomologous end-joining (NHEJ) DNA repair pathway. Modified histochemistry- (H-) scores were determined for each repair protein in each sample. HRD score was determined from tumor DNA.
Results:
53BP1 deletion increased HR in BRCA1-mutant COV362 cells and decreased PARPi sensitivity in vitro. In 36 women with relapsed ovarian cancer, responses to the PARPi ABT-767 were observed exclusively in cancers with HR deficiency. In this subset, 7 of 18 patients (39%) had objective responses. The actual HRD score did not further correlate with change from baseline tumor volume (r = 0.050; p = 0.87). However, in the HR-deficient subset, decreased 53BP1 H-score was associated with decreased antitumor efficacy of ABT-767 (r = −0.69, p = 0.004).
Conclusion:
Differences in complementary repair pathways, particularly 53BP1, correlate with PARPi response of HR-deficient ovarian cancers.
Keywords: 53BP1, PARP inhibitors, ovarian cancer, DNA damage, HR-deficiency
INTRODUCTION
Multiple poly(ADP-ribose) polymerase (PARP) inhibitors (PARPis) have recently received regulatory approval for the treatment of ovarian cancer and are undergoing extensive clinical testing in additional homologous recombination (HR) deficient cancers, including triple-negative breast, prostate, and pancreatic cancer [1–4]. Among recurrent high grade serous or endometrioid ovarian carcinomas, the response rate to PARPi therapy is highest in BRCA1- or BRCA2-mutant carcinomas, intermediate in BRCA1/2-wildtype carcinomas that show evidence of HR deficiency as manifested by high loss of heterozygosity, and lowest in HR-proficient carcinomas [5]. Given the toxicities of PARPi therapy, including prolonged nausea, myelosuppression and possible risk of myleodysplastic syndrome and acute myeloid leukemia, identifying carcinomas that are unlikely to respond could spare a subset of ovarian cancer patients unnecessary side effects. How to distinguish HR-deficient ovarian carcinomas that will or will not respond to PARPis remains incompletely understood.
Preclinical studies have suggested several pathways that might modulate sensitivity of HR-deficient cancers to PARPis [6–8]. In particular, the PARP trapping model [9, 10] suggests that cancers with diminished PARP1 expression might be less sensitive to PARPis. Studies in ovarian cancer cell lines [11, 12] and BRCA1 mutation-associated murine breast cancers [13] have also indicated that downregulation of components of the nonhomologous end-joining (NHEJ) DNA repair pathway, including KU70, KU80 and Artemis, or diminished levels of the 53BP1 protein that regulates engagement of the NHEJ pathway are associated with PARPi resistance. In the case of 53BP1 loss, this PARPi resistance has been attributed to restoration of HR despite the continued absence of BRCA1 [14–16]. The pertinence of these findings to clinical PARPi responses is currently unknown.
ABT-767 is a potent orally bioavailable small molecule inhibitor of PARP1 and PARP2 (Ki = 0.47 and 0.85 nM, respectively) that demonstrated anticancer activity in preclinical models [17]. A recent phase I study (NCT01339650) evaluated ABT-767 in subjects with advanced solid tumors harboring deleterious BRCA1 or BRCA2 mutations or subjects with recurrent ovarian, fallopian tube, or peritoneal cancer [17]. In the present study we examined the relationship between HRD score, BRCA1 and BRCA2 mutation status, expression of repair proteins, and response of ovarian cancers treated with ABT-767 on this trial.
METHODS
Patient population and study design
NCT01339650, a Phase I, open-label, multicenter study of the PARPi ABT-767, included dose escalation and safety expansion cohorts [17]. ABT-767 was administered orally on Days 1–28 of 28-day cycles until patients experienced progressive disease (PD) or unacceptable toxicity. From an initial dose level of 20 mg once daily, ABT-767 was escalated to 500 mg twice daily (BID) using a 3+3 trial design. At the recommended phase 2 dose of 400 mg BID, an expansion cohort with BRCA1/BRCA2-mutated advanced solid tumors and another cohort with advanced ovarian, fallopian tube or primary peritoneal carcinoma (hereafter collectively called “ovarian cancer”) were enrolled. The present analysis focused exclusively on ovarian cancer patients enrolled in this trial. When available, archival tissue was submitted for biomarker analysis. Objective response rate (ORR: confirmed complete response [CR] and partial response [PR]) was measured by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 in patients who had measurable disease at baseline [18]. As defined by RECIST, CR indicates disappearance of all target lesions with pathologic lymph nodes reduced to <10 mm on the short axis, PR required ≥30% decrease in sum of the diameters of target lesions, and non-responders included both progressive disease (PD, ≥20% increase in sum of diameters of target lesions) and stable disease (SD). CA-125 response was measured by Gynecologic Cancer Intergroup criteria [19].
Homologous Recombination Deficiency (HRD) Score and BROCA analysis
HRD score, which is a weighted sum of LOH, telomeric allelic imbalance and large scale transitions, was assayed at Myriad Genetics as described by Telli et al [20]. Samples were considered HR-deficient if the HRD score was ≥42. Tumor mutation status of BRCA1 and BRCA2 was simultaneously determined at Myriad Genetics. Mutations were considered deleterious only if they were nonsense mutations or missense mutations known previously to be associated with altered function or strongly correlated with disease penetrance [21]. In the sample set, HR deficiency was defined as an HRD score ≥42 and/or the presence of a deleterious BRCA1 or BRCA2 mutation.
To search for additional HR gene mutations, DNA from HR-deficient cases that lacked deleterious BRCA1 or BRCA2 mutations was isolated from FFPE slides by laser capture microdissection and assayed for mutations in genes involved in DNA repair (Table S1) by BROCA-HR DNA sequencing as previously described [22]. Mutations were considered deleterious if they were truncating or were missense mutations with evidence of functional compromise. Sanger sequencing was used to confirm deleterious mutations.
Methylation Analysis
As previously reported [5, 23], DNA was bisulfite converted (EZ Methylation Direct kit, Zymo Research, Irvine, CA) and evaluated with methylation sensitive PCR for BRCA1 and RAD51C. In vitro methylated DNA and known unmethylated DNA (Zymo Research #D5014) were used as positive and negative controls, respectively, for bisulfite conversion and validation of amplicon size. Water (H2O) was substituted for DNA to rule out cross-contamination of samples.
Cell lines
The identity of all cell lines was confirmed by short tandem repeat analysis in the Mayo Clinic Cytogenetics Core. To assess the impact of 53BP1 on PARP inhibitor sensitivity, BRCA1-mutant COV362 cells ([24], kind gift from Robert van Waardenburg, University of Alabama Birmingham) were utilized. To provide positive and negative controls for IHC, the following cell lines were utilized: OVCAR8 (kind gift from Dominic Scudiero, NCI Frederick) as well as OVCAR8 cells transiently transfected with siRNA targeting the RAD51, KU80, KU70, XRCC4, or 53BP1 mRNAs; parental and PARP1−/− HCT116 cells ([25], a kind gift from Eric Hendrickson, University of Minnesota); or parental MO59J cells (lacking DNA-PKCS) and MO59K cells expressing DNA-PKCS ([26], kind gift from Jann Sarkaria, Mayo Clinic, Rochester, MN). HR-proficient OV90 ([24], kind gift from Robert van Waardenburg) and HR-deficient, BRCA2-mutant PE01 cells [11, 23] served as positive and negative controls, respectively, for the plasmid reactivation assays.
COV362, OV90 and OVCAR8 cell lines were grown in RPMI medium 1640 supplemented with 10% (vol/vol) FCS (medium A); HCT116 cell lines were grown in McCoy’s 5A medium supplemented with 10% (vol/vol) FCS; PE01 cells were grown as previously described [12]]; and MO59J and MO59K cell lines were grown in DMEM/F-12 (1:1) medium supplemented with 15% (vol/vol) FCS and 1 mM sodium pyruvate. All media contained 50 units/mL penicillin G, 50 μg/mL streptomycin, and 1 mM glutamine.
To generate 53BP1 knockout cells, the oligonucleotides (5′-TTGATCTCACTTGTGATTCG −3′) guiding to human 53BP1 2023–2042 (accession number: AF078776.1) were synthesized, annealed, and cloned into the BsmBI site of lentiCRISPR-v2 plasmid (Addgene, Cambridge, MA). 53BP1 targeting virus and empty vector were packaged by transfecting HEK293T cells with the packaging vector psPAX3, envelope vector pMD2.G, and lentiCRISPR-v2–53BP1 2023–2042 or empty vector using Lipofectamine 2000 (ThermoFisher, Waltham, MA). Two days after viral transduction, COV362 cells were selected with 3 μg/ml puromycin. Pooled cells were utilized for the assays described below. 53BP1 knockout was verified by immunoblotting.
The following siRNA constructs were purchased from Dharmacon (Lafayette, CO): RAD51 (3M-003530–04, SMARTpool Human), KU80 (J-010491–07, ON-TARGETplus siRNA), and XRCC4 (5’-AUAUGUUGGUGAACUGAGATT-3’)[27]; or from Ambion (Austin, TX), USA): KU70 (s5457, 5’-GACAUAUCCUUGUUCUACA-3’), 53BP1 (s14313, 5’-GAAGGACGGAGUACUAAUA-3’), and Negative Control No. 1 (cat. No. 4390884). Oligonucleotides were resuspended according to the suppliers’ instructions. Cells suspended in medium A were subjected to electroporation using a BTX830 square wave electroporator (Harvard Apparatus, Holliston, MA, USA) delivering two 10-ms pulses at 280 V. After a 48-h incubation, 90% of the cells were embedded to serve as immunohistochemistry (IHC) controls. Whole cell lysates were prepared from the remaining cells to confirm knockdown of siRNA targets by immunoblotting.
Clonogenic assays
Aliquots containing 500 COV362 EV or COV362 53BP1−/− cells in medium A containing 3 μg/ml puromycin, were plated in 35 mm plates, allowed to adhere for 14–18 hours, and treated with varying concentrations of veliparib (added from 1000X stocks in DMSO). Cells were incubated for 10 days to allow colony formation. PE01 and OV90 cells were assayed similarly except that 400 and 600 cells, respectively, were plated and cells were grown in their usual growth media. Survival was calculated as the ratio of colonies per well treated with drug compared with diluent.
Immunoblotting
Aliquots of control and knockout or knockdown cell lines prepared as IHC controls were harvested, washed in calcium- and magnesium-free Dulbecco’s phosphate buffered saline (PBS), sonicated in buffered 6 M guanidine hydrochloride under reducing conditions, and prepared for electrophoresis as described [28]. Samples containing 50 μg of protein were separated on SDS-polyacrylamide gels containing 8% (wt/vol) acrylamide, transferred to nitrocellulose and probed with antibodies [28].
HR assays.
The previously described [29] reporter plasmid DR-GFR (provided by L. Karnitz, Mayo Clinic, Rochester MN) was transfected, along with pCherry and plasmid encoding I-SceI or empty vector, by electroporation using a BTX 830 square-wave electroporator. Conditions used for various cell lines were: COV362 parental, COV362 53BP1−/− and OV90, 260 V for two 10 msec pulses; PE01, 240 V for two 10 msec pulses. After incubation for 48 h, cells were trypsinized, washed in PBS, and fixed in 4% (w/v) formaldehyde in PBS. Flow cytometry was performed on a Becton Dickinson LSR II flow cytometer (BD Biosciences; Franklin Lakes, NJ) as described previously [12]. Results were expressed as a ratio of double-positive cells (EGFP+mCherry+) to the total number of mCherry+ cells to normalize for transfection efficiency, and then normalized to 1.0 for the value in COV362 cells in each experiment. Whisker plots summarize 4 independent experiments.
Immunohistochemistry (IHC)
After formalin fixed, paraffin embedded archival cancer specimens were sectioned at 5 microns, antigens were detected using automated IHC staining. Antigen retrieval was performed for 20 min using Epitope Retrieval 1 (Citrate; Leica). Antibodies to DNA-PKCS (1:600; IHC00044, Bethyl Labs), KU80 (1:2000, clone EPR3467, Abcam), KU70 (1:500, clone N3H10, Abcam), XRCC4 (1:8000, clone 4/XRCC4, BD Biosciences), PARP1 (1:300, clone E102, Abcam), RAD51 (1:2000, clone EPR4030(3), Abcam), and 53BP1 (mouse monoclonal [30], a kind gift from Thanos Halazonetis, University of Geneva, Switzerland) were diluted in Background Reducing Diluent (Dako) and incubated for 15 min.
A Leica Polymer Refine Detection System was used according to the supplier’s instructions. Slides were rinsed between steps with 1X Bond Wash Buffer (Leica). Immunostaining was visualized by incubating slides for 10 minutes in 3,3’diaminobenzidine (DAB) and DAB buffer (1:19 mixture) from the Bond Polymer Refine Detection System. Slides were counterstained for 5 min using Schmidt hematoxylin and molecular biology grade water (1:1 mixture) followed by several rinses in 1X Bond Wash Buffer and distilled water. Once completed, slides were rinsed in tap water for 3 min, dehydrated in increasing concentrations of ethanol, and cleared in 3 changes of xylene prior to permanent coverslipping in xylene-based medium.
OVCAR8 cells transfected with siRNA to DNA repair proteins as described above, parental HCT116 cells, PARP1−/− HCT116 cells, MO59J cells, and MO59K cells served as staining controls. In each case, cells were fixed in PBS containing 4% (w/v) paraformaldehyde at 20–22 °C for 2 h, washed in PBS, sedimented, embedded in paraffin in the Mayo Clinic Histochemistry Core, sectioned, and stained in each IHC run to serve as positive and negative controls.
Slides were scored by investigators (A.E.W.H. and D.V.) blinded to all clinical results. Using consensus estimates of the percentages of cells that were stained weakly, moderately or strongly, a modified H-score was calculated as 1 x % weak + 2 x % moderate + 3 x % strong [31]. For samples that lacked sufficient slides to assess all antigens, 53BP1 and KU80 were prioritized.
Statistical Analysis
Change in baseline tumor was defined as the greatest reduction in tumor size observed for a patient. Responses (CR and PR, as defined above) were assigned according to RECIST version 1.1 as per the parent clinical trial [18]. Non-responders included both SD and PD. Relationships between DNA repair proteins (H-scores) and the change in baseline tumor volume were assessed graphically and via Spearman correlation. Differences in H-scores by response group (non-responders vs. complete/partial responders) were assessed using Wilcoxon rank-sum tests; differences in HRD status by response group were assessed using Fisher’s exact test.
RESULTS
53BP1 loss decreases sensitivity of BRCA1-mutant ovarian cancer cells to PARP inhibitor in vitro
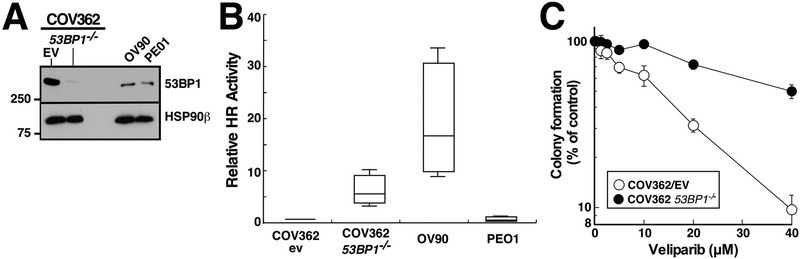
Previous studies have demonstrated that interruption of the 53BP1 gene, which regulates repair pathway choice between HR and NHEJ, is associated with increased HR activity and decreased PARPi sensitivity in BRCA1-mutant breast cancer or immortalized BRCA1−/− retinal pigment epithelial cells [14–16]. To assess whether 53BP1 loss could also potentially impact PARPi sensitivity in human ovarian cancer, we used CRISPR/Cas9 reagents to target the 53BP1 gene in COV362 cells, a high grade serous ovarian cancer line that harbors a truncating BRCA1 mutation [24]. As indicated in Fig. 1A, 53BP1 was markedly diminished in the pool of cells that grew back after puromycin selection, indicating success of the knockout in the majority of cells. 53BP1 gene disruption in this cell line was accompanied by increased HR as assessed using a plasmid-based repair assay (Figs. 1B and S1). Consistent with these results, 53BP1−/− COV362 cells were more resistant to the PARPi veliparib (ABT-888) than COV362 cells transduced with empty vector (Fig. 1C), with an increase in IC50 of 2.9 ± 0.4-fold (mean ± SEM, n = 5). Thus, PARPi sensitivity in human ovarian cancer cells can be modulated by 53BP1 loss as previously reported in BRCA1-mutant breast cancer and retinal pigment epithelium models.
Figure 1. 53BP1 loss and PARP inhibitor sensitivity in HR-deficient ovarian cancer cells.
A, expression of 53BP1 of was assessed by immunoblotting in whole cell lysates of COV362 transduced with empty vector (EV) or sgRNA targeting 53BP1−/− as well as OV90 and PE01 cells. B, homologous recombination repair was assayed using the DR-GFP plasmid as described in the Methods and illustrated in Fig. S1. OV90 (HR proficient—ref. 24) and PE01 cells (HR deficient–refs. 11 and 23) served as controls for this assay. Error bars, summary of 4 independent assays. PARPi sensitivity of the positive and negative controls is shown in Fig. S1C. C, COV362 empty vector (EV) and COV362 53BP1−/− cells were continuously exposed to increasing concentrations of PARP inhibitor veliparib in a clonogenic assay. Error bars, ± SEM from triplicate plates in a single assay. Across 5 independent assays, the IC50 for veliparib was 2.9 ± 0.4-fold (mean ± SEM) higher in the 53BP1−/− cells.
Development and Validation of DNA Repair Protein IHC Assays
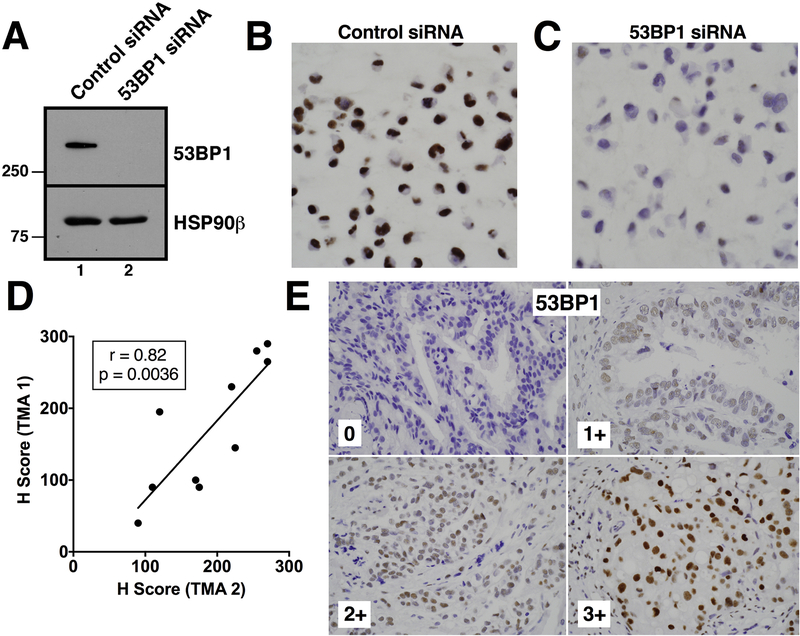
This observation and others in preclinical models suggest that changes in several different alternative DNA repair pathway proteins can modulate PARPi sensitivity in HR-deficient cells [6, 7, 11–15]. To test various hypotheses, we developed assays to assess expression of PARP1, RAD51, 53BP1 and classical NHEJ pathway proteins by IHC. Each assay was validated by showing that the IHC signal was markedly attenuated by gene knockout or highly effective siRNA (Fig. 2A-C and Fig. S2). Staining of 53BP1 for quality control purposes showed a strong correlation (r = 0.82) between the H-Score from two different tissue microarrays (TMAs), each containing different cores from the same ovarian cancers (Fig. 2D-E).
Figure 2. Validation of 53BP1 antibody staining.
OVCAR8 cells transfected with nontargeting siRNA (siRNA control) or siRNA targeting 53BP1 (si53BP1) were subjected to immunoblotting (A) or formalin fixed, embedded in paraffin, and stained to confirm specificity of the anti-53BP1 antibody (B, C). D, cores from the same ovarian cancer included on two different TMAs were independently stained with anti-53BP1 and scored. E, 53BP1 staining of four ovarian cancers to illustrate range of expression in the present study.
Relationship between BRCA1 or BRCA2 mutations, HRD score and ABT-767 response
The PARPi ABT-767 was evaluated in a phase I clinical trial with expansion cohort [17]. Among the 93 patients placed on trial (Fig. S3), 80 had recurrent ovarian cancer, 10 had breast cancer, and 3 had other neoplasms. The present analysis focused on the 36 ovarian cancer patients who were i) treated at the seven highest dose levels, ii) had archival material available for molecular and immunohistochemical analysis, iii) had a known HR status and iv) were evaluable for objective response. Among these 36 ovarian cancers, 15 had deleterious BRCA1 or BRCA2 mutations.
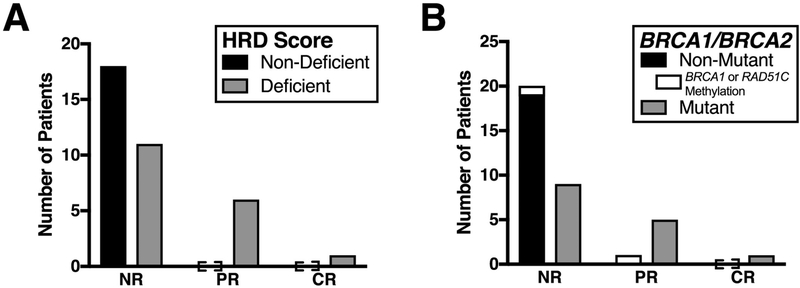
Eighteen of the 36 ovarian cancers had a deficiency in HR, as defined by the presence of a BRCA1/BRCA2 mutation and/or a Myriad HRD assay score of ≥42 [20]. Of the three HR-deficient ovarian cancers lacking demonstrable BRCA1/BRCA2 mutations, RAD51C or BRCA1 promoter hypermethylation was observed in the two samples available for analysis (Fig. S4). In the BROCA analysis of these samples, no clearly deleterious mutations in DNA repair genes other than TP53 were identified despite the HRD score ≥42.
All responses were observed in HR-deficient ovarian cancers. Of the 18 HR-deficient ovarian carcinomas, seven (39%) had objective responses, including six PRs and one CR (Fig. 3A and3B). This ability of HR gene mutation analysis or HRD score to enrich for PARPi responders, but not completely predict PARPi response is similar to recent reports [5, 32, 33]. Among the six ovarian cancers that exhibited PRs, five harbored a clearly deleterious BRCA1 or BRCA2 mutation and one displayed RAD51C promoter hypermethylation (Fig. 3C). The ovarian cancer that achieved a CR lacked hypermethylation of the RAD51C and BRCA1 promoters as well as truncating mutations in the genes listed in Table S1 (except TP53) but had a somatic BRCA2 mutation [BRCA2 c.7753G>A (p.Gly2585Arg)] that was recently classified as deleterious by functional criteria [34]. For the 30 ovarian cancers where clinical data regarding the most recent response to a platinum-containing regimen was available, three of six patients (50%) with a progression-free interval of 6–12 months responded, as compared to three of 24 (12.5%) with a progression-free interval of less than 6 months (Table S2).
Figure 3. HR-Deficiency status, BRCA1/BRCA2 mutation status, and response to ABT-767 among ovarian cancers.
Archival tissue from ovarian cancers in patients treated in the highest seven dose levels was assessed for HR-deficiency (A) and sequenced for BRCA1/BRCA2 mutations (B).
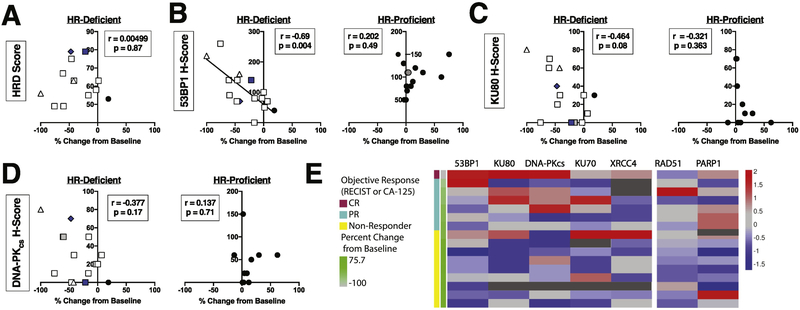
Because all responses occurred in the HR-deficient subset, we assessed whether HRD score correlated with percent change of tumor area from baseline in this subset of patients. No correlation was observed (Fig. 4A, Spearman r = 0.050; p = 0.87). Thus, while a positive HRD test enriched for responders, the actual value of the score provided no additional information regarding the extent of response.
Figure 4. Relationship between H-scores for multiple NHEJ DNA repair pathway proteins and carcinoma shrinkage.
A, relationship between Myriad HRD score and largest percent change in tumor cross sectional area from baseline in target lesions by RECIST [18] in the subset of ovarian cancers for which a numerical HRD score was available (n=13). B-D, IHC staining for 53BP1 (B), KU80 (C), and DNA-PKCS (D) was evaluated to give a modified H-score, which was compared to percent change in tumor cross-sectional area from baseline [18]. HR gene alterations are denoted as mutations (white) or methylation (blue) of BRCA1 (squares), BRCA2 (triangle), or RAD51C (diamond). Symbols that represent two separate patients but are not separable because of overlapping data points are denoted in grey. Ovarian cancers that were stained were sorted into those that were HR deficient (left panels) and those that were not HR deficient (right panels). Because not all ovarian cancers had sufficient sample to assess all antigens, the number of samples analyzed was: 53BP1 (B), HR-deficient n=16, HR-proficient n=16; KU80 and DNA-PKCS (C, D), HR-deficient n=15, HR-proficient n=10. Heat map analysis (E) reflects the H-score for all proteins analyzed, sorted by HR-deficiency and percent change in tumor cross-sectional area from baseline.
Relationship between PARP1 expression and response
In view of preclinical studies described above suggesting that differences in repair pathways can affect PARPi sensitivity, we stained the cancer specimens from this trial for a series of DNA repair proteins. When the relationship between response and staining intensity was examined across the entire study population, there was a statistically significant association between HRD status and response, as anticipated, but no significant correlation between protein H-scores and response (Table 1). Because previous reports have demonstrated the role of NHEJ in vitro and in vivo specifically in the setting of HR deficiencies [11–15], we next performed a preplanned analysis examining the relationship between repair protein levels and response in HR-deficient and HR-proficient carcinomas (as defined by the Myriad HRD assay) separately.
Table 1.
Univariate analysis of response with clinical parameters and H-scores
| Non-Responder (N=29) | Responder (N=7) | p value | |
|---|---|---|---|
| HR Status | 0.0081 | ||
| Deficient | 11 (37.9%) | 7 (100%) | |
| BRCA1/2 Mutant | 9 | 6* | |
| HR-proficient | 18 (62.1%) | 0 (0%) | |
| 53BP1 H-score | 0.122 | ||
| median | 90 | 140 | |
| Q1, Q3 | 70, 110 | 80, 180 | |
| Ku80 H-score | 0.132 | ||
| median | 10 | 40 | |
| Q1, Q3 | 0, 40 | 25, 65 | |
| Ku70 H-score | 0.192 | ||
| median | 0 | 10 | |
| Q1, Q3 | 0, 10 | 5, 30 | |
| PARP1 H-score | 0.242 | ||
| median | 60 | 110 | |
| Q1, Q3 | 25, 105 | 70, 115 | |
| XRCC4 H-score | 0.292 | ||
| median | 30 | 5 | |
| Q1, Q3 | 7.5, 50 | 0, 30 | |
| DNA-PKCS H-score | 0.432 | ||
| median | 20 | 50 | |
| Q1, Q3 | 5, 55 | 20, 60 | |
| RAD51 H-score | 0.972 | ||
| median | 15 | 20 | |
| Q1, Q3 | 5, 40 | 7.5, 25 |
The 7th patient responded had an ovarian cancer harboring a RAD51C promoter methylation
Fisher’s exact test
Kruskal-Wallis rank sum test
When we examined the relationship between response and PARP1, a protein whose loss is associated with poor PARPi response in cell lines [9, 10, 35], no association was observed between PARP1 expression and clinical response (p = 0.203, Fig. S5A). Accordingly, when the relationship between PARP1 H-score and percent change in tumor cross sectional area from baseline was examined specifically in the HR-deficient or HR-proficient subsets, only weak correlations (r = −0.16, p = 0.55 and r = 0.38, p = 0.20, respectively) were observed (Fig. S5B).
Relationship between 53BP1 expression and response
In contrast, in HR-deficient carcinomas, there was a strong negative correlation between 53BP1 H-score and percent change of tumor area from baseline (Fig. 4B, left panel; r = −0.69, p = 0.004), consistent with pre-clinical studies showing that 53BP1 loss in BRCA1-mutant murine cells is associated with PARPi resistance [13–15]. Additionally, there was a trend toward increased shrinkage of tumor from baseline and higher KU80 H-score (Fig. 4C, left panel; r = 0.46, p = 0.08) or higher DNA-PKCS H-score (Fig. 4D, left panel; r = −0.38; p = 0.17). In contrast, no significant correlations were observed between 53BP1, KU80 or DNA-PKCS expression and change in tumor size among the HR-proficient tumors (Fig. 4B-D, right panels). Moreover, no significant correlations were observed in either HR-deficient or HR-proficient carcinomas between KU70, XRCC4, or RAD51 H-scores and percent change of tumor area (Fig. S6).
In view of the fact that several proteins in the same repair pathway either correlated with ABT-767 response (53BP1, Fig. 4B) or showed a trend (KU80, Fig. 4C), we also analyzed the relationship between expression of various proteins in this pathway. As indicated in Fig. 4E, expression levels of 53BP1, KU80, and DNA-PKCS tended to track with each other. Accordingly, multivariate analysis taking into account expression of all three of these proteins was performed. Recursive partitioning did not yield any additional significant findings when looking at change in baseline tumor response with DNA repair proteins (results not shown).
DISCUSSION
Although PARPis are rapidly changing the treatment of HR-deficient ovarian cancer [1–4, 8, 36], objective response rates to these agents in BRCA1/BRCA2-mutant relapsed, platinum sensitive ovarian cancer range from 30–80% [5, 32, 33]. Factors that determine response in the clinical setting remain incompletely understood [8, 36]. Even though several potential mechanisms of PARPi resistance have been identified in tissue culture or animal models [1, 2, 7], reversion mutations in HR genes and demethylation of the BRCA1 promoter are the only changes previously shown to correlate with poor objective responses to PARPis in the clinical setting [5, 37]. In preclinical models, changes in complementary DNA repair pathways have also been reported to modulate PARPi sensitivity, as summarized in Fig. 5. In particular, loss of 53BP1 has been reported to restore HR in BRCA1-mutant cells [13–15]; and downregulation of KU80 or DNA-PKCS has been reported to induce PARPi resistance in BRCA2- or BRCA1-mutant cell lines [11, 12]. These observations prompted us to examine the relationship between selected repair proteins and PARPi sensitivity in the context of a single-agent PARPi trial.
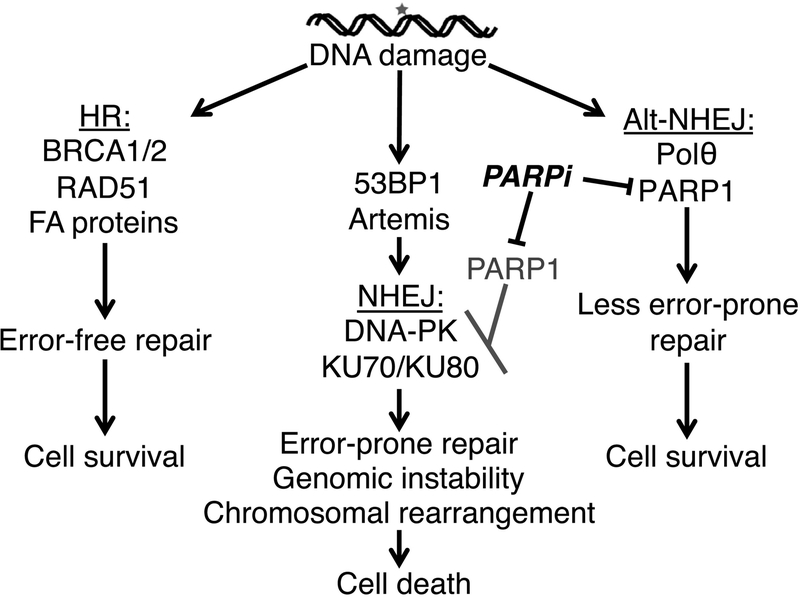
Figure 5. Complementary pathways in DNA double strand (ds) break repair and PARP inhibitor response.
In the setting of a DNA ds break, HR preferentially restores DNA integrity in an error-free fashion (left column). In the setting of HR-deficiency, however, the error-prone NHEJ pathway and the less error-prone alternative NHEJ (alt-NHEJ) pathways (middle and right columns, respectively) play a larger role in DNA ds break repair. PARP inhibitors inhibit alt-NHEJ and simultaneously facilitate NHEJ, leading to increased chromosomal rearrangements, genetic instability, and cell death. In the setting of 53BP1 downregulation, an inhibitory influence on HR is removed [16] and HR is restored. In the setting of NHEJ protein down-regulation, other DNA repair pathways with higher fidelity repair also appear to be utilized [11, 12], although the mechanistic details are incompletely understood.
The PARP trapping hypothesis suggests that ovarian cancers with low PARP1 expression might be less sensitive to PARPis. In the present study, however, we did not observe any correlation between PARP1 expression and response (Fig. S5). These observations suggest that low PARP1 expression is not an explanation for poor response to single-agent PARPi therapy in this trial.
A previous study has suggested that the PARPi niraparib shows benefit even in patients with an HRD score <42 [38]. Importantly, that result was observed in platinum responsive relapsed ovarian cancer treated with maintenance niraparib; and the number of weeks of improved progression-free survival was much lower in HR-proficient cancers than in ovarian cancers with a positive HRD score. In contrast, in the present study where the endpoint was tumor shrinkage, responses to ABT-767 were only observed in HR-deficient cancers (Fig. 3A). Even so, the numerical value of the HRD score did not further correlate with percent change in tumor area within the HR-deficient subset (Fig. 4A), suggesting that factors beyond HRD score must be considered to identify ovarian cancers most likely to respond to PARPi therapy.
Considering only the HR-deficient ovarian cancers, there was a strong correlation in the present study between low 53BP1 levels and poor carcinoma response (Fig. 4B), consistent with studies in model systems indicating that 53BP1 loss restores HR and confers diminished PARPi sensitivity mechanisms in BRCA1-mutant cells [Fig. 5 and refs. 13, 14, 15]. Trends were also observed between low KU80 or low DNA-PKCS and poor response (Figs. 4C and 4D), although this association did not reach statistical significance in the present small sample set. Strikingly, these proteins were all expressed at somewhat higher levels in ovarian cancers that respond to ABT-767 (Fig. 4E).
The present analysis was performed on cancer samples harvested at the time of initial diagnosis. While secondary mutations that restore expression of HR proteins can occur between diagnosis and subsequent treatment [5, 37, 39], it is unclear how frequently expression of repair proteins encoded by nonmutated, nonmethylated genes changes over the course of therapy. Analyses that compare archival versus pretreatment biopsies are required to further address this issue, as well as clarify the potential role of low 53BP1 expression in intrinsic versus acquired PARPi resistance in ovarian cancer. Additional studies are also needed to determine whether low 53BP1 expression correlates with poorer PARPi response in other cancers that are being treated with PARPis, including HR-deficient breast and prostate cancers.
In summary, the present study suggests a possible role of repair proteins in complementary pathways in the response to the PARPi ABT-767 as a single agent in the clinical setting. Among HR-deficient ovarian cancers, 53BP1 H-score demonstrated a strong correlation with the percent change of tumor volume. These results not only extend previous preclinical studies [11–15] into the clinical setting, but also point toward a strategy to identify patients most likely to respond to PARPi therapy. Accordingly, studies to assess the relationship between 53BP1 as well as possibly other repair proteins and response to other PARPis should be considered, as this type of analysis may add value to the HRD score in predicting response. In view of correlation between PARPi sensitivity and platinum sensitivity [36, 40], studies to assess the relationship between platinum response and expression of these same proteins also appear warranted.
Supplementary Material
Highlights.
Not all cancers with defects in homologous recombination (HR) demonstrate clinical responses to PARP inhibitors.
In a BRCA1-mutant ovarian cancer cell line, 53BP1 loss restores HR and decreases PARP inhibitor sensitivity.
Samples from a phase I clinical trial of the PARP inhibitor ABT-767 were stained with validated IHC assays for DNA repair proteins.
In HR-deficient ovarian cancer, 53BP1 expression negatively correlated with percent change in tumor size from baseline.
Assessment of proteins that modulate DNA repair pathway choice might inform PARP inhibitor response in ovarian cancer.
Acknowledgements
The work was supported in part by NIH grants P50 CA136393 (S.H.K., A.E.W.H., and D.W.V.) and R01 CA190423 (S.H.K., A.E.W.H., E.M.S., and D.W.V.), and DOD Translational Synergy Grant W81XWH-13–1-0485 (to E.M.S. and S.H.K.) as well as a fellowship (to R.M.H.) and support (S.H.K.) from the Mayo Foundation for Education and Research. A.E.W.H., E.M.S., and S.H.K. were supported by Stand Up To Cancer – Ovarian Cancer Research Fund Alliance – National Ovarian Cancer Coalition Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT16–15). Stand Up to Cancer is a program of the Entertainment Industry Foundation; research grants are administered by the American Association for Cancer Research, a scientific partner of SU2C.
Footnotes
Conflict of Interest Statement
P.A. and M.D. are employed and hold stock and intellectual property from Abbvie. S.H.K. receives royalties from Millipore and Topogen for unrelated technology and is an inventor on a patent held by Mayo Clinic regarding NHEJ proteins in ovarian cancer. S.P.S. holds stock through and was previously employed at Abbvie. The Phase I trial was funded by Abbvie, but no funding was provided for the follow-up biomarker study presented here.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5:1137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) Polymerase Inhibitors: Recent Advances and Future Development. Journal of Clinical Oncology. 2015;33:1397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miller RE, Ledermann JA. The status of poly(adenosine diphosphate-ribose) polymerase (PARP) inhibitors in ovarian cancer, part 1: olaparib. Clin Adv Hematol Oncol. 2016;14:619–27. [PubMed] [Google Scholar]
- [4].del Rivero J, Kohn EC. PARP Inhibitors: The Cornerstone of DNA Repair-Targeted Therapies. Oncology (Williston Park). 2017;31:265–73. [PubMed] [Google Scholar]
- [5].Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- [6].Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381–8. [DOI] [PubMed] [Google Scholar]
- [7].Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res. 2014;20:540–7. [DOI] [PubMed] [Google Scholar]
- [8].Stover EH, Konstantinopoulos PA, Matulonis UA, Swisher EM. Biomarkers of Response and Resistance to DNA Repair Targeted Therapies. Clin Cancer Res. 2016;22:5651–60. [DOI] [PubMed] [Google Scholar]
- [9].Satoh MS, Lindahl T. Role of Poly (ADP-ribose) Formation in DNA Repair. Nature. 1992;356:356–8. [DOI] [PubMed] [Google Scholar]
- [10].Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. [DOI] [PubMed] [Google Scholar]
- [11].Patel A, Sarkaria J, Kaufmann SH. Nonhomologous end-joining drives PARP inhibitor synthetic lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Choi YE, Meghani K, Brault ME, Leclerc L, He YJ, Day TA, et al. Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer. Cell Rep. 2016;14:429–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013;3:68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Noordermeer SM, Adam S, Setiaputra D, Barazas M, Pettitt SJ, Ling AK, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van der Biessen DAJ, Gietema JA, de Jonge MJA, Desar IME, den Hollander MW, Dudley M, et al. A phase 1 study of PARP-inhibitor ABT-767 in advanced solid tumors with BRCA1/2 mutations and high-grade serous ovarian, fallopian tube, or primary peritoneal cancer. Invest New Drugs. 2018;36:828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- [19].Rustin GJ, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG). Int J Gynecol Cancer. 2011;21:419–23. [DOI] [PubMed] [Google Scholar]
- [20].Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res. 2016;22:3764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Norquist BM, Brady MF, Harrell MI, Walsh T, Lee MK, Gulsuner S, et al. Mutations in Homologous Recombination Genes and Outcomes in Ovarian Carcinoma Patients in GOG 218: An NRG Oncology/Gynecologic Oncology Group Study. Clin Cancer Res. 2018;24:777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].AlHilli MM, Becker MA, Weroha SJ, Flatten KS, Hurley RM, Harrell MI, et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol. 2016;143:379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alotaibi M, Sharma K, Saleh T, Povirk LF, Hendrickson EA, Gewirtz DA. Radiosensitization by PARP Inhibition in DNA Repair Proficient and Deficient Tumor Cells: Proliferative Recovery in Senescent Cells. Radiation Res. 2016;185:229–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Allalunis-Turner MJ, Barron GM, Day RS 3rd,, Dobler KD, Mirzayans R. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiation Res. 1993;134:349–54. [PubMed] [Google Scholar]
- [27].Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. [DOI] [PubMed] [Google Scholar]
- [28].Kaufmann SH. Reutilization of Immunoblots After Chemiluminescent Detection. Analytical Biochemistry. 2001;296:283–6. [DOI] [PubMed] [Google Scholar]
- [29].Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hirsch FR, Varella-Garcia M, Bunn PA Jr., Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–807. [DOI] [PubMed] [Google Scholar]
- [32].Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–61. [DOI] [PubMed] [Google Scholar]
- [33].Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 2013;14:882–92. [DOI] [PubMed] [Google Scholar]
- [34].Guidugli L, Shimelis H, Masica DL, Pankratz VS, Lipton GB, Singh N, et al. Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. Am J Hum Genet. 2018;102:233–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase inhibitors reflects poisoning of both enzymes. J Biol Chem. 2012;287:4198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Evans T, Matulonis U. PARP inhibitors in ovarian cancer: evidence, experience and clinical potential. Ther Adv Med Oncol 2017;9:253–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI, et al. Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2017;7:984–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–64. [DOI] [PubMed] [Google Scholar]
- [39].Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229:422–9. [DOI] [PubMed] [Google Scholar]
- [40].Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.