Abstract
New strategies are urgently needed to target MRSA, a major global health problem and the leading cause of mortality from antibiotic-resistant infections in many countries. Here, we report a general approach to this problem exemplified by the design and synthesis of a vancomycin–D-octaarginine conjugate (V–r8) and investigation of its efficacy in addressing antibiotic-insensitive bacterial populations. V–r8 eradicated MRSA biofilm and persister cells in vitro, outperforming vancomycin by orders of magnitude. It also eliminated 97% of biofilm-associated MRSA in a murine wound infection model and displayed no acute dermal toxicity. This new dual-function conjugate displays enhanced cellular accumulation and membrane perturbation as compared to vancomycin. Based on its rapid and potent activity against biofilm and persister cells, V–r8 is a promising agent against clinical MRSA infections.
Graphical Abstract
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is the leading cause of mortality from antibiotic-resistant infections in the United States and is estimated to be responsible for >50% of all hospital-associated infections in Asia and North and South America.1–3 MRSA predominantly manifests as skin and soft tissue infections (SSTIs) and can progress to life-threatening invasive diseases.4 Its propensity to form biofilms and persister cells is linked to recurrent and difficult-to-treat infections including intravenous catheter-related bacteremia, endocarditis, and osteomyelitis.5,6 Such biofilms consist of slow-growing bacteria surrounded by a self-produced protective extracellular matrix which blocks entry of many drugs, while persister cells are dormant and highly antibiotic-tolerant cells that can exist in planktonic or biofilm state within or external to mammalian cells.7,8 The emergence of recurrent MRSA infections in both hospital and community settings, coupled to a 90% decline in FDA approvals for new antibiotics over the last 30 years,9 renders treatment of MRSA an immediate and formidable challenge.
Vancomycin is a glycopeptide antibiotic produced by Amycolatopsis orientalis. In use since 1958, it is generally regarded as a first-line therapy for hospitalized patients with MRSA SSTIs.10,11 The antibiotic inhibits bacterial growth by binding to the D-Ala-D-Ala termini of un-cross-linked peptidoglycan precursors at the cell surface and cell septum, preventing cell wall assembly.12–14 However, the bactericidal activity of vancomycin in vivo is slow15 and often associated with inefficient clearance of infection16 and, in some cases, clinical failure.17 To achieve an effective concentration at the site of infection, vancomycin is commonly administered intravenously with high and frequent doses, increasing the risk of side effects such as nephrotoxicity and ototoxicity.18 The slow bactericidal activity of vancomycin also promotes the emergence of resistant and/or dormant bacterial survivors.19 Additionally, the penetration of vancomycin into tissues is not optimal and can greatly affect its pharmacokinetics and therapeutic window, especially in diabetic patients who are particularly susceptible to bacterial infections.20
Promising vancomycin derivatives have been introduced to enhance efficacy and to attempt to overcome vancomycin resistance mechanisms.21,22 Oritavancin is one of the most remarkable semisynthetic derivatives, approved in 2014 by the FDA for the treatment of SSTIs.23 Oritavancin differs from vancomycin by the presence of the 4-epi-vancosamine sugar and a p-chlorophenylbenzyl substituent. Oritavancin is more effective than vancomycin, exhibits activity against vancomycin-resistant organisms, and can sterilize vancomycin-insensitive S. aureus biofilms.24 Telavancin, dalbavancin, and teicoplanin are FDA-approved semisynthetic vancomycin derivatives, each distinguished by a long lipophilic tail that is thought to enhance association with bacterial membranes and thus promote inhibition of cell-wall synthesis.22 Bacterial membrane integrity is additionally compromised by telavancin treatment.25 Other vancomycin derivatives incorporate alternative cationic lipophilic and/or antimicrobial peptide components and exhibit notably improved antibacterial activity and synergistic modes of action, including perturbation of membrane integrity.22,26–29 Moreover, a recent study directed at the design and synthesis of vancomycin derivatives creatively combined the chlorobiphenyl functionality of oritavancin and a C-terminal lipophilic cationic ammonium group together with the replacement of the residue 4 amide carbonyl with a methylene to improve the binding affinity to peptidoglycan with D-Ala-D-Lac peptide stem termini produced by vancomycin-resistant organisms.27,30–33 Notwithstanding these noteworthy advances, the challenge posed by the difficult-to-treat persistent and chronic infections involving biofilms, persister cells, and intracellular colonization still represents an unmet therapeutic need.
We hypothesized that the biological activity and therapeutic performance of vancomycin, as well as those of many other antibiotics, could be enhanced by their attachment to cellassociating or cell-penetrating molecular transporters (MoTrs) that would enable stronger association with cell surface anions, thereby weakening the cell membrane and cell wall while positioning cell surface acting antibiotics such as vancomycin to better arrest cell-wall synthesis. Alternatively or concurrently, antibiotic–MoTr conjugates could benefit from enabled or enhanced cell uptake, thereby accessing previously inaccessible intracellular drug binding targets. In either case, such dual function systems could provide for synergistic enhancements of antimicrobial activities.
To explore these strategies, we designed, synthesized, and evaluated a new vancomycin derivative conjugated to a cellpenetrating guanidinium-rich molecular transporter (GR–MoTr), specifically D-octaarginine (r8). Our selection of “r8” was based on our early identification of cell-penetrating oligoarginines and more generally designed GR–MoTrs, inspired by the observation that HIV-1 Tat, a protein essential for viral transcription, penetrates mammalian cells due to its highly basic nine amino acid domain (RKKRRQRRR).34–37 Although the passage of such a highly polar, polycationic agent across nonpolar membranes appeared at first to be counterintuitive, our extensive chemical and biophysical studies indicate that this transport process is driven by the number and spatial array of guanidinium groups and their initial association with the negatively charged cell surface carboxylic acids, sulfates, and phosphates.38 Our group and others have greatly expanded on this strategy for drug delivery, reporting a range of cell-penetrating MoTrs, including arginine-rich contiguous and spaced peptides, and guanidinium-rich peptoids and nonpeptidic agents,38–42 that deliver a variety of cargos, including antimicrobials,43–47 chemotherapeutics,48,49 peptides,50–52 proteins,53 inositol polyphosphates,54 and oligonucleotides (siRNA, DNA, pDNA)55–59 into cells in vitro and in vivo. GR–MoTr conjugates successfully cross a number of mammalian barriers including in vivo delivery into human skin.50 They have also been conjugated to antimicrobial agents, producing conjugates with varying efficacy depending on the antimicrobial agent, transporter, and pathogen.43,46,47,60–63 Additionally, GR–MoTrs have delivered fluorescent cargos into nonmammalian cells, including parasites,43,44 algae,64 and bacteria.65
Previously, we proposed that conjugation of a GR–MoTr to an antibiotic would enhance its delivery into bacterial cells, although the ability of such delivery systems to penetrate biofilm barriers was not explored.66 Herein, we show that conjugation of a GR–MoTr to an antibiotic produces a dualfunction conjugate with improved and new activities arising from enhanced access to and association with bacterial cellenvelope constituents, penetration of biofilm barriers, and membrane disruption. An attractive aspect of this strategy is the step and time economy achieved by taking a known agent and augmenting its function or creating new function by attaching it to a cell associating or penetrating transporter.67 Given the urgent clinical need for new antibacterial strategies and the potential generality of enhancing or creating new antibiotic activity through GR–MoTr conjugation to antibiotics, we focused our initial investigation on the design and evaluation of a vancomycin–Gr–MoTr conjugate as a prelude to the broader investigation of such conjugates for clinical applications.
RESULTS AND DISCUSSION
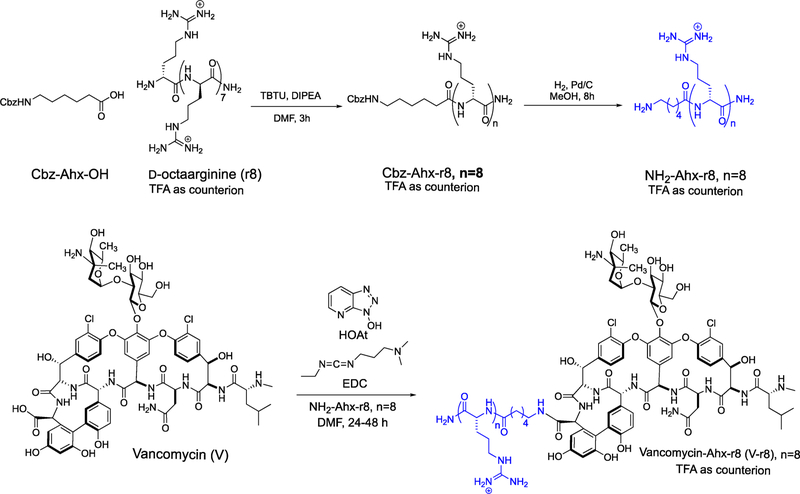
Design and Synthesis of V–r8.
For the synthesis of the vancomycin–GR–MoTr conjugate, we attached a cellpenetrating GR–MoTr to the C-terminus of vancomycin at the derivatizable carboxylic acid group that is not involved in vancomycin’s mode of action.14 Based on our previous work with oligo-D- and oligo-L-arginine transporters, unnatural Damino acids were chosen because of the increased stability of the resulting oligomer to proteolysis.37 Upon consideration of optimal uptake efficiency and step-economical synthesis, an eight-residue peptide was selected for study.37 D-Octaarginine (r8) was first coupled to an N-protected aminohexanoic (Ahx) acid, followed by deprotective hydrogenolysis to yield NH2-Ahx–r8 (Scheme 1).68 NH2-Ahx–-r8 was conjugated to the Cterminus of vancomycin to produce V–r8.69 V–r8 was purified as a trifluoroacetic acid (TFA) salt by reversed-phase high-performance liquid chromatography (RP-HPLC) and characterized by high-resolution mass spectrometry (HRMS) and 1H NMR. The corresponding HCl salt of V–r8 was also generated,70 as HCl salts of vancomycin derivatives have been reported to exhibit improved antibacterial activity and superior mammalian cell compatibility compared to TFA salts.71,72 A vancomycin-linker–peptide conjugate with four Darginines (V–r4) was also synthesized to evaluate the influence of peptide length on the conjugate’s activity. Finally, fluorescein-tagged derivatives of vancomycin (Fl-V) and V–r8 (Fl-V–r8) were synthesized to optically evaluate uptake and localization (Scheme SI-1).
Scheme 1.
Synthesis of Vancomycin Conjugate V–r8
Evaluation of V–r8 in Targeting Biofilms, Persister Cells, and Planktonic Cell Populations.
Vancomycin is effective at inhibiting the growth of planktonic S. aureus, including MRSA, but it is not effective in eradicating S. aureus biofilms.54–56 To investigate whether attachment of a GR–MoTr to vancomycin would produce a conjugate with improved delivery and efficacy against S. aureus biofilms, the activities of V–r8 and vancomycin (V) were compared in biofilm eradication assays using the Calgary Biofilm Device with a reference methicillin-susceptible S. aureus (MSSA) strain and two clinically relevant MRSA strains (USA300 LAC and USA400 MW2).73,74 Significantly, V–r8 (TFA) was an order of magnitude more effective than vancomycin in nutrient-rich media (tryptic soy broth, TSB), yielding median minimum biofilm eradication concentrations (MBECs) of 9.5–20 μM (Table 1). V–r8 (HCl) showed antibiofilm activity comparable to that of V–r8 (TFA) (Table SI-1), and in all subsequent in vitro experiments, V–r8(TFA) was used, whereas V–r8 was used for in vivo experiments. The MBECs of oritavancin and dalbavancin, two recently developed glycopeptides, were also evaluated for comparison to V–r8. Oritavancin and dalbavancin yielded MBEC ranges of 12–32 and 8–20 μM, respectively, comparable to that of V–r8 (Table SI-1). Furthermore, the shorter V–r4 was not as effective as V–r8 in biofilm assays, indicating, in line with prior work, that the number of arginine residues is important (Table SI-1).
Table 1.
Minimum Biofilm Eradication Concentrations (MBECs, μM)a and Minimum Inhibitory Concentrations (MICs, μM)b
| MBECs |
MICs |
|||||||
|---|---|---|---|---|---|---|---|---|
| strain | V | V–r8 (TFA) | r8 | V + r8 | V | V–r8 (TFA) | r8 | V + r8 |
| MISSA (29213) | ≥500 | 20 (16–25) | >80 | 80 (63–125) | 0.50 (0.50–0.63) | 1.8 (1.0–2.0) | 60 (40–80) | 0.50 |
| MRSA (USA400 MW2) | ≥500 | 10 (6–26) | >80 | >20 (20–>80) | 0.50 (0.31–0.63) | 0.94 (0.63–1.0) | 20 | 0.50 |
| MRSA (USA300 LAC) | ≥400 | 9.5 (3–16) | >80 | 16 (10–40) | 0.50 (0.31–0.50) | 2.0 (1.8–2.0) | 40 | 0.50 |
Median MBEC values from two to six independent experiments, where ranges are given in parentheses. For V and r8, MBEC values often exceeded the highest concentrations tested.
Median MIC values from two to seven independent experiments, where ranges are given in parentheses.
The activities of vancomycin and V–r8 in planktonic bacteria were also compared using minimum inhibitory concentration (MIC) assays. V–r8 exhibited a 2–4-fold reduction in activity as compared to vancomycin in S. aureus (Table 1). Thus, V–r8 does not display enhanced potency in targeting planktonic exponentially growing cells, indicating that the remarkably enhanced antibiofilm activity of the conjugate involves differences of cells in the biofilm, considered further below. We also examined the antibiotic activity of V–r8 against a panel of Gram-positive organisms, including a vancomycin-intermediate S. aureus (VISA) strain and three vancomycin-resistant Enterococci (VRE) strains. VISA strains produce thicker cell walls and were similarly affected by vancomycin and V–r8 as vancomycin-susceptible S. aureus strains, with a 2–4 fold increase in the observed MIC for V–r8 as compared to vancomycin (Table SI-2). V–r8 and vancomycin also yielded comparable MICs in vancomycin-susceptible Enterococci (VSE) strains. However, V–r8 exhibited enhanced potency as compare to vancomycin against VRE strains that can produce cell walls with altered D-Ala-D-Lac stem termini, including a >100-fold improvement for vancomycin-resistant E. faecium (Table SI-2). This activity presents a similarity between V–r8 and oritavancin, wherein a vancomycin conjugate is effective against bacteria with altered peptidoglycan typically characterized by a reduced binding affinity for vancomycin. The activity of V–r8 is further tested and discussed below in terms of the possible molecular basis for its activity.
The potential antibacterial activity of r8 alone was also evaluated. Treatment with r8 alone revealed some antibacterial activity, but with MICs at least 30 times higher than vancomycin and it did not eradicate biofilms in the concentration range tested. (Table 1). A noncovalent equimolar mixture of vancomycin and r8 exhibited identical MICs as vancomycin alone (Table 1), whereas the mixture exhibited enhanced antibiofilm activity compared to vancomycin alone but was less potent than V–r8. Thus, the covalent conjugation of V and r8 affords improved efficacy against biofilms (Table 1).
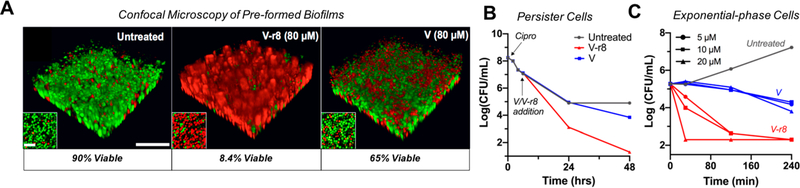
Confocal microscopy imaging of V–r8-treated biofilms was also employed to evaluate the efficacy of V–r8 against biofilms. Biofilms grown on fibronectin-coated glass chamber slides for 48 h were stained with SYTO 9 to label all bacteria (green) and with propidium iodide (PI) to identify dead cells (red) through its access and binding to nucleic acids in nonviable cells. Significantly, after a 5 h treatment with equivalent concentrations of vancomycin and V–r8 (80 μM), only 8% of V–r8 treated biofilm bacteria were viable, while 65% of Vtreated bacteria remained viable (Figure 1). A V–r8 concentration of 32 μM was also effective at eradicating biofilm bacteria, and killing was observed for both concentrations even after a 2 h treatment time (Figure SI-1). Thus, covalent conjugation of r8 to vancomycin produced a conjugate with significantly improved efficacy in killing biofilm-embedded bacteria.
Figure 1.
V–r8 kills biofilm bacteria, persister cell, and exponential-phase bacteria more effectively than vancomycin (V). (A) 3D-reconstructed confocal images of untreated, V–r8-treated and V-treated MRSA USA400 MW2 biofilms in TSB for 5 h. Insets show bacterial cells from the bottom plane of each image. Biofilm bacteria were stained with SYTO 9 (green: live) and PI (red: dead). Quantification of fluorescence showed that significantly more bacteria were killed by V–r8 than by V as compared to the untreated biofilm. Scale bars represent 20 μm in the main panels and 5 μm in the insets. (B) MRSA USA 300 LAC persister cell time-kill curves for compounds with final concentrations of 10 μM introduced 6 h postciprofloxacin treatment (40 μM). (C) Time-kill kinetic analysis of V or V–r8-treated exponential-phase MRSA USA400 MW2 at treatment concentrations of 5, 10, or 20 μM. Data presented in (A)–(C) are from representative experiments.
Given the efficacy of V–r8 in eradicating biofilm bacteria, we next evaluated the conjugate’s ability to kill MRSA persister cells. Persister cells can be generated in the laboratory by treating exponentially growing bacteria with a high concentration (10X MIC) of ciprofloxacin, which kills most cells but leaves the remaining cells in a nonreplicating dormant state.75 V–r8 (10 μM) was >150-fold more efficacious in killing these cells than vancomycin (10 μM) after a 48 h treatment (Figure 1B). Additionally, the noncovalent V + r8 mixture showed some enhanced killing of persister cells, whereas no activity was observed for r8 alone or for V–r4 (Figure SI-2).
To further examine the superior efficacy of V–r8 over vancomycin, and to gain preliminary insight into its mode of action, we compared the rate of killing of exponentially growing cells by V–r8 and vancomycin. V–r8 added to cells in nutrient broth exerted remarkably rapid killing effects after only 30 min and eventually resulted in killing to the limit of detection in an antibiotic sensitivity assay (Figure 1C), showing a faster rate of killing and superior overall efficacy of V–r8 relative to V. This enhanced kill rate and efficacy are consistent with the hypothesis that V–r8 exhibits dual function antimicrobial activity with the vancomycin subunit binding to Lipid II, while the r8 subunit synergistically engages proximate anionic lipids such as wall- or lipo-teichoic acids resulting in local disruption of the membrane barrier. Alternatively or concurrently, the r8 subunit could mediate passage into the cell, enabling the vancomycin subunit to engage intracellular targets that would be inaccessible to vancomycin itself. Thus, we designed a series of experiments to evaluate and compare the modes of action of V and V–r8: (i) the extent of cellassociation using synthetically prepared fluorescent conjugates, (ii) the influence of cell metabolism and cell growth state on compound activity, and (iii) the impact on membrane barrier function and cellular integrity.
Evaluation of Cellular Localization of V–r8 Using Fluorescent Conjugates.
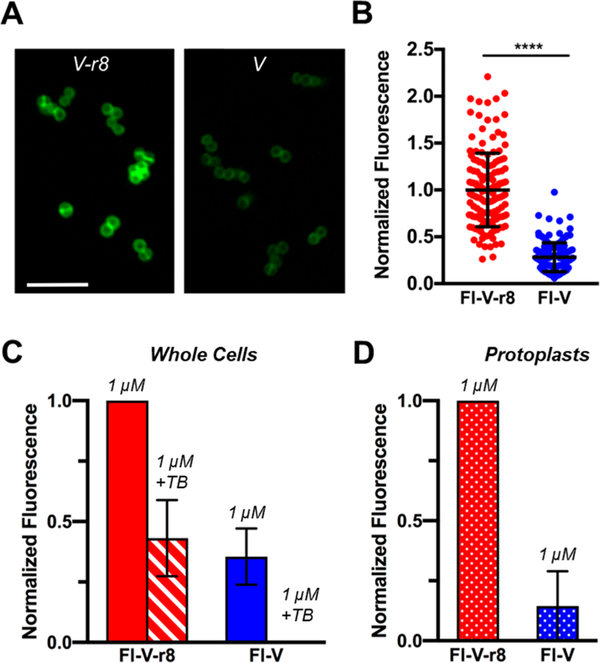
To evaluate the localization and uptake of V–r8 in MRSA, we synthesized fluorescent derivatives of vancomycin (Fl-V) and V–r8 (Fl-V–r8) that retained the antibacterial activity of the corresponding parent compounds (Scheme SI-1) and have equal fluorescence intensities (Figure SI-3).76 Previous studies have shown that fluorescently labeled vancomycin localizes to the cell wall and preferentially to cell septa during cell divsion.77 We treated MRSA with 5 μM Fl-V or Fl-V–r8 and evaluated the uptake and localization of the two compounds. Bacteria treated with Fl-V–r8 exhibited twice as much fluorescence as compared to bacteria treated with 5 μM Fl-V (Figure 2A,B). The difference between Fl-V and Fl-V–r8 uptake observed by confocal microscopy was supported by quantitative analysis using fluorescence-activated cell sorting (FACS, Figure 2C). We also observed concentration-dependent cellular uptake of Fl-V and Fl-V–r8 by FACS (Figure SI-3). The increased fluorescent signal from Fl-V–r8-treated bacteria reveals that V–r8 exhibits stronger cellular association than vancomycin, as expected due to the r8 component. However, it does not specifically identify whether V–r8 could have been transported into the cell, is externally cell membrane associated, or is positioned in both locations.
Figure 2.
MRSA USA400 MW2 bacteria treated with Fl-V–r8 exhibits greater cell-associated and protoplast-associated fluorescence than Fl-V. (A) Confocal microscopy of bacteria treated with 5 μM FlV–r8 for 5 min and (B) analysis of fluorescence intensities of individual bacteria imaged as in (A). Intensity values were normalized to the mean intensity of Fl-V–r8-treated cells. Error bars represent standard deviations. P < 0.0001 determined using a Mann–Whitney test. (C) FACS analysis of MRSA whole cells treated with Fl-V–r8 or Fl-V. (D) FACS analysis of protoplasts prepared from MRSA whole cells treated with Fl-V–r8 and Fl-V as in (C). In (C) and (D), each bar represents median normalized fluorescence values from two experiments, with data normalized to the highest fluorescence value in each experiment. Error bars represent the range of normalized fluorescence values obtained in two experiments.
To examine the spatial localization of V–r8 in MRSA bacteria, we used Trypan Blue (TB) as a quenching agent to eliminate fluorescence from fluorophores located extracellularly.78 After treatment with TB, approximately 45% of the fluorescent signal from Fl-V–r8 treated cells remained, whereas fluorescence from Fl-V treated bacteria was fully quenched, as determined by FACS (Figure 2C). To further confirm the intracellular localization of Fl-V–r8 versus Fl-V biochemically, we treated cells with Fl-V and Fl-V–r8, generated MRSA protoplasts by digesting and removing the majority of the cell wall peptidoglycan with lysostaphin,79 and measured the remaining fluorescence in the protoplasts by FACS.
We observed stronger fluorescence in protoplasts from FlV–r8-treated bacteria compared to the Fl-V treated ones. Specifically, cells treated with 1 μM Fl-V–r8 exhibited a greater than 7-fold stronger fluorescence compared to that of cells treated with Fl-V (Figure 2D). To summarize, Fl-V–r8 treated bacteria exhibit strong fluorescence, about half of which is not accessible or quenched by TB, and the fluorescence signal is maintained in protoplasts after digestion of the mature cell wall. These observations are consistent with the proposed mechanism of tight cellular association with enhanced Lipid II binding at the membrane exoface and/or internalization of the Fl-V–r8, thereby accessing intracellular targets. The enhanced cellular association of V–r8 is likely mediated through polycationic r8’s interaction with the negatively charged groups on the cell surface, such as phosphates from walland/or lipo-teichoic acids or the undecaprenyl pyrophosphate of Lipid II. The strong unquenched fluorescence from Fl-V–r8 also supports participation of an r8 driven internalization process that would allow access and binding to intracellular DAla-D-Ala containing precursors, such as Park’s nucleotides.
Evaluation of V–r8’s Activity on Bacterial Growth, Morphology, and Membrane Integrity in Relation to Metabolic State.
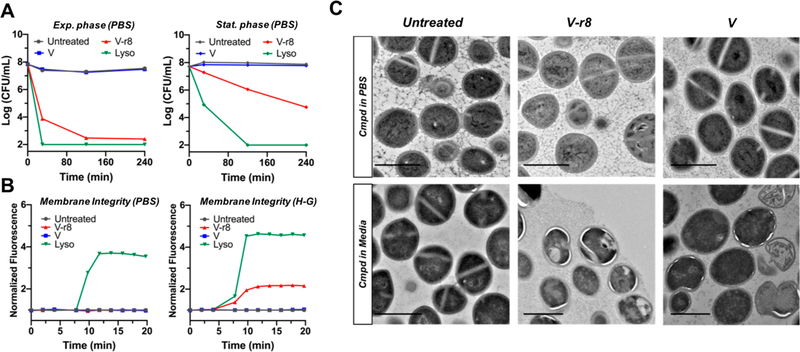
The combined effects of a dual-function antibiotic (target and membrane binding) should enable the compound to remain cell associated and effective even after cells are transferred to antibiotic-free nutrient medium. Thus, we evaluated V and V–r8 in this way and additionally compared efficacy of killing using cells collected from exponential and stationary phase growth states. V or V–r8 was added to harvested cells resuspended in PBS for 1, 2, or 4 h treatment times followed by microdilution plating onto tryptic soy agar. V–r8 exhibits potent inhibition activity for cells in exponential phase and is comparable to that of treating cells with lysostaphin which digests the cell wall, rendering cells immediately compromised and unable to grow on agar. As shown in Figure 3A, V–r8’s activity is growth-state dependent and requires a slightly longer treatment time to exhibit activity on cells in stationary phase than on cells in exponential phase (Figure 3A). Cells in stationary phase have thicker cell walls than cells in exponential phase,80,81 and longer V–r8 treatment times may be needed to access key cell-surface Lipid II and immature peptidoglycan template strands. Additionally, teichoic acid content, a candidate for electrostatic association with the r8 subunit, may be reduced in stationary phase cells.82
Figure 3.
Evaluation of V–r8 activity as a function of bacterial growth phase and treatment condition and its influence on membrane and cellular integrity. (A) Comparative time-kill kinetics of MRSA USA400 MW2 harvested from exponential-phase or stationary phase and resuspended in PBS for treatment followed by CFU enumeration on agar. (B) Evaluation of PI uptake and fluorescence as a reporter for the perturbation of membrane barrier function upon treatment with V–r8, V, or lysostaphin. Experiments were performed with stationary-phase MRSA USA400 MW2 resuspended in either PBS or HEPES–glucose (H–G) buffer. In (A) and (B), V and V–r8 treatments were performed at 10 μM, lysostaphin treatments were performed at 125 μg/mL (all ∼10X MIC), and data are from representative experiments. (C) TEM images of untreated, V–r8, or V-treated stationary-phase MRSA USA400 MW2 in PBS or exponential-phase bacteria in tryptic soy broth. Treatments were performed for 90 min at final concentrations of 8 μM in the PBS experiment (top) or 4 μM in the tryptic soy broth experiment (bottom). Scale bars = 1 μm.
Rapid killing of bacteria is often reported for cationic antimicrobial peptides that interfere with bacterial membrane integrity and barrier function, leading to cell death.83,84 Thus, we sought to investigate the possible influence of V–r8 on MRSA cell membrane integrity using propidium iodide (PI).26 Lysostaphin, a positive control, is known to induce membrane permeability via degradation of the bacterial cell wall. Experiments with MRSA resuspended in PBS and treated with V–r8 or V did not show membrane permeabilization, in sharp contrast to lysostaphin (Figure 3A). Like lysostaphin, peptide antibiotics like nisin also compromise membrane integrity in this assay.85–87 However, experiments with MRSA resuspended in HEPES-glucose (H-G) buffer to preserve bacterial metabolic activity88,89 showed uptake of PI in V–r8-treated MRSA (Figure 3B) at concentrations as low as 1X MIC (Figure SI-4). These results reveal that V–r8 perturbs bacterial membrane barrier function when cells are incubated in the presence of a carbon source, glucose, but exhibits no membrane disruption or immediate killing in PBS in the absence of cell metabolism and growth. Visualization of bacteria cells by transmission electron microscopy (TEM) supported the biochemical observations. When MRSA cells were treated with antimicrobial compounds as suspensions in PBS, neither V–r8 nor V altered bacterial morphology. In contrast, when MRSA cells were treated in nutrient medium encouraging active growth and metabolism, V–r8 and V induced irregular cell division and hyperhydration of cell membranes. As displayed in Figure 3C, V–r8 treated cells also displayed intracellular inclusions near the cellular membranes, indicating antibiotic-induced membrane alteration.90 These results suggested that V–r8 may exert its uniquely rapid bacterial killing in part through membrane disruption during cell growth.
Mechanistic Considerations for V–r8 activity.
The combined antibacterial efficacy data, including activity against preassembled biofilms and persister cells (Figure 1), enhanced cell-association through Fl-V–r8 analysis (Figure 2), the experiments in which cells are treated with V–r8 exclusively in a PBS suspension followed by plating (Figure 3A), and the influence on membrane permeability and cell integrity (Figure 3B and C) are compatible with either model described above in which V–r8 could exhibit enhanced cell-association but remain extracellular and/or it could be internalized. In the first model, the r8 component of one V–r8 molecule might bind to wall teichoic acid(s), while the vancomycin component binds to a D-Ala-D-Ala stem terminus of membrane-associated Lipid II. Then, as cells begin to grow and divide and push new cell wall layers outward, wall teichoic acid would move away from the cell, potentially resulting in a tug-of-war with V–r8 also being pulled away through r8 association with teichoic acid and, at the same time, pulling Lipid II and disrupting membrane permeability (supported by PI uptake for active cells in Figure 3B). A similar scenario could be envisioned for r8 association with lipoteichoic acid(s), which is membrane associated, and vancomycin association with immature cell-wall D-Ala-D-Ala termini close to the membrane but no longer part of Lipid II. Either type of multivalent interaction could explain the rapid and profound local activity to lyse and kill MRSA cells. In an alternative model consistent with the data, V–r8 may be transported into cells where it could bind Lipid II or Park’s nucleotide and/or the very first D-Ala-D-Ala precursors. In this model, cell-wall synthesis would be halted through sequestration of the precursors. The observation of V–r8-induced membrane perturbation, not observed for V, could result from cells attempting to translocate V–r8-associated Lipid II across the membrane, disrupting the membrane through the attempted transport of the large and bulky glycopeptide. Such intracellular activity would be somewhat similar to that of bacitracin, which enters cells and binds the undecaprenyl pyrophosphate component of Lipid II to prevent continued peptidoglycan synthesis.91
Finally, V–r8 is a derivative of one of the most well-studied and well-pursued antibiotics, vancomycin. V–r8’s unique and exciting activity alone motivates future dissection into its exact molecular mechanism, recruiting structural, biochemical, and biophysical analyses. Furthermore, the mechanism of V–r8 in MRSA biofilms is of great interest, as its improved efficacy may result from enhanced penetration of biofilms and/or enhanced potency against slow-growing cells within the biofilm. Given the ability of V–r8 to kill persister cells, we hypothesize that V–r8 is able to better target slow-growing cells within the biofilm than vancomycin. In contrast to other antibiotics, vancomycin also penetrates slowly through biofilms, so it is also possible that addition of r8 improves its ability to penetrate biofilms, enhancing killing.8 Yet, given the tremendous unmet clinical need for new antibiotics, testing the efficacy of V–r8 in an in vivo animal model of infection would significantly propel multidisciplinary efforts to analyze V–r8 and to design new potentially more potent conjugates toward preclinical development. Thus, we sought to evaluate V–r8 in an in vivo model of difficult-to-treat MRSA infections.
Evaluation of V–r8 in a Murine Wound Model of Biofilm Infection.
MRSA is a major cause of both acute and chronic wound and soft tissue infections. Chronic wound infections are difficult to treat due to challenges in eliminating associated biofilms.92,93 MRSA biofilms are also involved in the colonization of catheters, leading to bacteremia, infection of prosthetic implants after surgery, and other infections such as chronic osteomyelitis and endocarditis.94,95 Thus, to begin to address the effects of V–r8 on MRSA biofilms within a complex biochemical in vivo environment, we used a previously characterized excisional mouse wound biofilm model.96 Prior to performing in vivo experiments in animals, an in vitro toxicity assessment was performed where V–r8 exhibited minimal hemolytic activity (HC50 > 320 μM), comparable to vancomycin. Cytotoxicity evaluation with HaCaT skin cells indicated that V–r8 concentrations in excess of 10 times the MIC against MRSA were needed to exert 50% cytotoxicity in HaCaT skin cells (Figure SI-5). While V–r8 exhibited more cytotoxicity in this assay than vancomycin, V–r8 was comparable to a clinically used drug, oritavancin. These results warranted further examination of the efficacy of V–r8 in vivo.
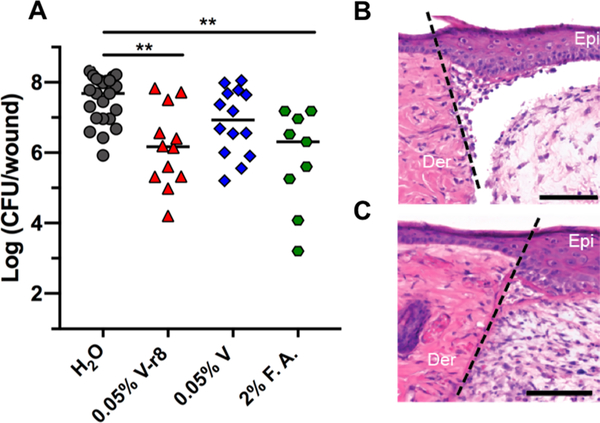
MRSA USA300 LAC biofilms were established within a partial-thickness skin wound for 24 h prior to inoculation of the treatment compounds into the wound for 5 h.96,97 We observed a significant reduction in biofilm load (97% eradication) after topical treatment with a 0.05% solution of V–r8 (HCl) compared to untreated mice (Figure 4A). A 6-fold reduction in median CFU/wound was observed in wounds treated with 0.05% vancomycin as compared to untreated mice, but this difference was not statistically significant. V–r8 was also compared to the parent antibiotic vancomycin and to fusidic acid (F.A.), a drug with activity against S. aureus biofilms that has limited efficacy in the clinic because of emerging resistance.98,99 Significantly, 0.05% V–r8 was as effective as 2% F.A. in reducing the bacterial load in wounds after 5 h, with V–r8 employed at a 40-fold lower percent solution (or a 220-fold lower molar concentration). V–r8 was 6-fold more efficacious than vancomycin when employed at matched percent solution (3-fold lower molar concentration). To determine whether V–r8 has any dermal toxicity on mouse skin, we inoculated uninfected punch-biopsy wounds with a 0.05% solution of V–r8 and performed histological examination 3-days post treatment. This analysis indicated the absence of necrosis and apoptosis (Figure 4B,C). These results demonstrate that V–r8 can effectively reduce biofilm bacterial loads in the in vivo wound environment. To further evaluate the therapeutic potential of V–r8, we examined the evolution of resistance of S. aureus bacteria over a 14 day period. We observed a 4-fold increase in the MIC of V–r8, which matched the observed increase for vancomycin in this assay. The absence of high-level resistance emergence in passaged bacteria treated with V–r8 further supports its therapeutic potential.
Figure 4.
In vivo evaluation of V–r8 in a skin wound biofilm model. (A) Each data point represents Log(CFU/wound) from one mouse, with the median values indicated by bars. Data were compiled from two to three independent experiments containing four to five animals per treatment group. Statistical analysis was performed using the nonparametric Kruskal–Wallis test with Dunn’s post ad hoc test for intergroup comparisons. **P < 0.01. To examine in vivo cytotoxicity, sterile wounds were generated and inoculated with water (B) or 0.05% V–r8 (C) in the absence of bacteria. The epidermis and dermis layers from mice treated with V–r8 appeared similar to those from mice treated with water and showed no signs of necrosis or apoptosis by hematoxylin and eosin staining. Wound healing was comparable between groups indicated by dermal granulation tissue formation and epidermal healing. A small increase in neutrophil infiltration was observed in V–r8-treated mice as compared to untreated mice. Dotted line represents the site of wounding. Epi: skin epidermis; Der: skin dermis. Scale bars = 100 μm. Images are representative of three independent samples.
CONCLUSION
V–r8 exhibits novel activity and clinical potential against difficult-to-treat MRSA populations, including biofilms and persister cell. V–r8 outperformed vancomycin, often by orders of magnitude, in all persister cell and biofilm assays, and demonstrated a faster bactericidal mode of action, tighter cellular association, and enhanced membrane disruption as compared to vancomycin. V–r8 reduced biofilm bacterial loads in vivo, while exhibiting no acute toxicity or damage to skin tissues. The ability of V–r8 to treat slow-growing bacteria makes it a unique candidate for therapeutic use. Moreover, the new activities and enhanced efficacy associated with conjugation of r8 to V could potentially be broadly extended to other antibiotics and antibiotic derivatives, serving thereby as a more general strategy for targeting multidrug resistant bacteria and biofilms.
METHODS
Synthesis of Vancomycin Conjugates.
Full details regarding the synthesis and chemical characterization of V–r8 (TFA), V–r8 (HCl), V–r4 (TFA), Fl-V, and Fl-V–r8 (TFA) are provided in the Supporting Information.
Determination of MBECs.
Minimum biofilm eradication concentrations (MBECs) were determined in accordance with literature methods.100 To cultivate biofilms, 150 μL of inoculum (1 × 107 CFU/mL, prepared in TSB) was added to each well of a 96-well base plate (Nunc 269787), and a plate lid containing 96 pegs (Nunc 445497) was added to the base. The entire apparatus was sealed with Parafilm and placed in a sealed plastic bag lined with moistened paper towels for 48 h growth with shaking (150 rpm) at 35 °C. Prior to treatment, biofilms were rinsed in a 96-well plate containing 200 μL PBS/well for 1 min and then the peg lid was transferred to the treatment plate containing 2-fold serial dilutions of compound in TSB or PBS (final treatment volume: 200 μL/well). The plate was sealed with Parafilm and incubated at 35 °C for 24 h with 150 rpm shaking. Upon 24 h treatment, the peg lid was removed, rinsed in 200 μL PBS buffer twice for 1 min each, then transferred to a recovery plate containing 200 μL of TSB + 1% Tween-20 per well. The apparatus was placed in a steel tray and sonicated in a water bath sonicator (Branson 1510) for 10 min to dislodge biofilms. The peg lid was subsequently removed, and the media-containing plate was incubated for 20 h at 35 °C with 150 rpm shaking to recover biofilms. The MBEC value was recorded as the lowest treatment concentration where no bacterial growth occurred, as determined by OD600 measurements on a microplate reader.
Determination of MICs.
MICs were determined using broth microdilution in accordance with CLSI methods.101 One day prior to each MIC experiment, bacterial strains were streaked for single colonies on TSA plates from frozen glycerol stocks stored at −80 °C. Three to five colonies from each plate were harvested with a disposable inoculating loop and resuspended in 500 μL of PBS. This suspension was diluted in PBS to an OD600 of 0.1 (~ × 108 CFU/ ml), and the OD 0.1 suspension was diluted 1:100 in Mueller–Hinton broth (MHB, Difco 257530) just prior to inoculating the 96-well polypropylene treatment plate (Costar 3879). A 50 μL portion of inoculum was added to a treatment plate containing 2-fold serial dilutions of compound in MHB (50 μL of treatment/well) to lend a final total volume of 100 μL/well and a final inoculum density of ~ × 105 CFU/ml. The completed assay plate was sealed with Parafilm, placed in a lidded plastic tray lined with moistened paper towels, and incubated at 37 °C for 18–20 h. The MIC was read as the lowest treatment concentration where no bacterial growth occurred, as determined by OD600 measurements on a microplate reader.
Evaluation of Antibacterial Activity against Persister Cells.
USA300 LAC persister cells were generated by diluting a stationary phase culture of USA300 bacteria 1:1000 in 4 mL MHB and growing to OD600 0.5 at 37 °C with 200 rpm shaking. The OD600 0.5 culture was treated with 10× MIC of ciprofloxacin (40 μM treatment) for 6 h at 37 °C with 200 rpm shaking, and culture aliquots were taken every 2 h, serially diluted in PBS, and plated on TSA to enumerate CFU/ mL. Upon 6 h treatment with ciprofloxacin, 500 μL culture aliquots were treated with compounds to yield the desired final concentrations. The aliquots were incubated at 37 °C with 200 rpm shaking. CFU/mL were enumerated at indicated time points to monitor treatment efficacy, where the detection limit was 1 log CFU/mL.
Confocal Microscopy of Biofilms.
USA400 MW2 were grown in TSB shaking at 37 °C until the stationary phase was reached. The cultures were adjusted in TSB to a final inoculum density of 1 × 107 CFU/mL. An eight-well chambered coverglass slide was coated with 50 μg/mL of fibronectin for 30 min at room temperature and then washed twice with PBS. A 300 μL portion of inoculum was added to each well, and the biofilms were grown statically in a humidified chamber at 37 °C for 48 h. Biofilms were washed twice with PBS and then stained with 150 μL LIVE/DEAD BacLight bacterial viability kit, with components solubilized in TSB. A 10× solution of vancomycin or V–r8 was added and mixed by gentle pipetting. The biofilms were incubated with the compounds statically at 37 °C and imaged at the designated time points using a Zeiss LSM 700 confocal microscope with an EC Plan-Neofluar 40×/0.75 oil-immersion objective. Images in each experiment were taken with identical laser intensities and processed using Volocity software.
Time-Kill Kinetics Experiments.
For time-kill kinetics experiments performed in MHB, a stationary-phase culture of USA400 MW2 was diluted to 1 × 106 CFU/mL, and 50 μL of this suspension was mixed with 50 μL of MHB containing twice the desired final concentrations of compounds. The plate was incubated at 37 °C with 200 rpm shaking, and CFU/mL were enumerated on TSA at determined time points (detection limit: 2.3 log CFU/mL). For time-kill kinetics experiments with exponential-phase bacteria in PBS, overnight cultures of USA400 MW2 were diluted 1:500 in TSB and grown to OD600 of 0.6. The bacteria were then pelleted, washed with PBS, and resuspended in PBS buffer to an OD600 of 0.3 and treated with compounds at 37 °C with 200 rpm. For time-kill kinetics experiments with stationary-phase bacteria in PBS, overnight cultures of USA400 MW2 grown in TSB were pelleted. The bacteria were then pelleted, washed with PBS, and then subsequently resuspended in PBS to an OD600 of 0.3 and treated with compounds at 37 °C with 200 rpm. At indicated times, aliquots were removed, serially diluted in PBS, and plated on TSA to enumerate CFU/mL, where the detection limit was 2 log CFU/mL.
TEM.
To prepare exponential-phase bacterial TEM samples, an overnight culture of USA400 MW2 (prepared in TSB) was diluted 1:300 in TSB and grown at 37 °C with 200 rpm shaking until an OD600 of 0.6 was reached. Compound treatments were performed (4 μM V or V–r8) for 90 min. To prepare stationary-phase bacterial samples, an overnight culture of USA400 MW2 was pelleted and washed 1× in PBS and then diluted to an OD600 of 1.0 in PBS. Compound treatments were performed at 8 μM for V and V–r8 for 90 min. Upon treatment, all cells were then pelleted at 10000g at 4 °C for 10 min and then resuspended in fixative (2% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate, pH 7.4). Upon three PBS washes to remove excess fixative, the samples were enrobed in gelatin, placed in a cold block, and postfixed with 1% OsO4 in phosphate buffer. Samples were then washed three times with ddH2O, stained with 1% uranyl acetate overnight at 4 °C, and then subsequently dehydrated with 95% EtOH and embedded in Epon. A Leica Ultracut S microtome was used to prepare thin sections, which were viewed on a JEOL JEM-1400.
Evaluation of Membrane Integrity with Propidium Iodide.
The propidium iodide assay was adapted from the literature.27 For experiments with stationary-phase bacteria, overnight cultures of USA400 MW2 grown in TSB were pelleted, washed three times in PBS, and then subsequently resuspended in either PBS or HEPES–glucose buffer (5 mM HEPES, 5 mM glucose, pH 7.2) to an OD600 of 0.3. For experiments with exponential-phase bacteria, overnight cultures of USA400 MW2 were diluted 1:500 in TSB and grown to OD600 of 0.6. The bacteria were then pelleted and resuspended in HEPES–glucose buffer to an OD600 of 0.3. Cultures were treated with 10 μM propidium iodide solution, and 130 μL of bacterial suspension was added to a 96-well, black-walled, clear-bottom plate (Costar 3603). The plate was incubated 5 min at 37 °C with shaking in a fluorescent microplate reader (SpectraMax M5, Molecular Devices), and the fluorescence signal was measured (excitation 535, emission 617). Twenty microliters of compounds at desired concentrations was added (prepared in either HEPES–glucose or PBS to match bacterial buffer), and changes in fluorescence post-treatment were monitored. Lysostaphin was used as a positive control, where concentrations of lysostaphin tested were based on MICs determined in the absence of bovine serum albumin (BSA).
Confocal Microscopy for Planktonic Bacteria.
Stationaryphase cultures of USA400 MW2 in TSB were diluted 1:20 and then spotted onto poly-L-lysine-coated slides for 15 min. Nonadherent bacteria were aspirated, and adherent bacteria were washed with PBS. Adherent bacteria were incubated with 5 μM treatment in PBS at room temperature for 5 min in the dark and then washed with PBS. Fluorescence of Fl-V and Fl-V–r8 stock solutions was measured before the experiment on a fluorescent plate reader to ensure equivalent fluorescent signals from treatment solutions at matched concentrations. All samples were imaged on a Zeiss LSM 700 confocal microscope with an EC Plan-Neofluar 40×/0.75 oil-immersion objective (excitation and emission wavelengths of 495 and 525 nm, respectively). Images in each experiment were taken with identical laser intensities and processed in ImageJ with the same settings. For quantification, we used a circular region of interest slightly larger than an individual coccus to select and capture the integrated fluorescence intensity in ImageJ. These raw values were divided by the mean fluorescence intensity of the Fl-V–r8 population and plotted as a scatter graph.
Flow Cytometry Experiments.
OD 0.5 pellets were prepared from stationary-phase USA400 MW2 cultures via centrifugation of cultures at 8000 rpm for 5 min followed by aspiration of supernatant. Pellets were resuspended in PBS buffer, and desired concentrations of fluorescent compound were added for a final treatment volume of 500 μL. Samples were incubated for 5 min at 37 °C in the dark. Upon completion of treatment, the samples were immediately centrifuged at 8000 rpm for 5 min to remove unbound compound, and the supernatant in each sample was aspirated. The pelleted bacteria were resuspended in 400 μL of PBS and transferred into round-bottom polystyrene sample tubes (Falcon, Cat. No.352058). Flow cytometry experiments were performed with 1500–2500 cells on a FACScan flow cytometer at 488 nm. For Trypan Blue quenching experiments, bacteria were treated with 0.2% Trypan Blue (2.3 mM final concentration) and incubated for 5–10 min before analysis. This concentration was selected based on similar Trypan Blue quenching experiments in the literature102 and its ability to fully quench cellassociated Fl-V over a range of treatment concentrations (1–10 μM) as measured on a fluorescent microplate reader. For protoplast FACS experiments, treated cell pellets were resuspended in PBS buffer supplemented with 1 M sucrose and 75 μg/mL lysostaphin for 1 h at 37 °C, 200 rpm to generate protoplasts.79 The resultant protoplast suspension was analyzed by FACS.
In Vitro Toxicity Assessment.
To assess the hemolytic activity of V–r8, a 1 mL aliquot of single-donor human red blood cells (IPLAWB3–22136, Innovative Research, Inc.) was centrifuged at 1000g for 10 min. The supernatant was aspirated, and the resulting pellet of erythrocytes was resuspended in PBS and centrifuged again at 1000g for 10 min. Upon two additional PBS washes and centrifugation cycles, the final erythrocyte pellet was resuspended in PBS to yield a 1% volume/volume suspension. A 100 μL portion of 1% erythrocyte solution was added to the wells of a V-bottomed 96 well microtiter plate. Treatments (100 μL) treatments were added to yield the desired final compound concentrations. Blank PBS was used as a negative control and 1% TX-100 as a positive control. The plate was incubated statically for 1 h at 37 °C and subsequently centrifuged for 5 min at 1500g at room temperature. A 100 μL portion of the resulting supernatant was transferred to a flat-bottomed microtiter plate and analyzed on a microplate reader via absorbance measurements at 450 nm. Percent hemolysis was determined by dividing background-corrected absorbance measurements by background-corrected measurements for 1% TX-100.
Cellular cytotoxicity of V–r8 was assessed using an MTT percent viability assay. Briefly, HaCaT cells were plated at 20000 cells/well in 96 well plates for 24 h at 37 °C in DMEM media containing serum. Cells were washed with 200 μL of PBS two times prior to treatment. In a separate 96-well plate, compounds were serially diluted in serumfree DMEM over a range of 160–5 μM. The second plate was transferred to the plate containing cells and incubated at 37 °C for 2 h, at which point the media was removed. The cells were rinsed with 200 μL of PBS two times, 200 μL of serum-containing DMEM was added to each well, and the cells incubated for an additional 18 h. Ten microliters of a 5 mg/mL solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in DMEM was added to each well. After incubation for 2 h, 100 μL of solubilizing solution (10% Triton-X-100, 90% 0.1 N HCl in 2-propanol) was added to each well, and colorimetry data were obtained on a plate reader at 690 and 570 nm. Percent viability was determined by dividing the average colorimetric value obtained for a treated sample by the average colorimetric value obtained for untreated cells.
Monitoring of Antimicrobials in Murine Skin Wound Excisional Model.
Wound infection procedures were adopted from previously described protocols, with minor modifications.97 Male wild-type C57BL/6 mice (7–8 weeks old, 22 to 25g, InVivos, Singapore) were anesthetized in an induction chamber supplied with 3% isoflurane. Anesthetization was maintained with a nose cone throughout the procedure. Dorsal hair of the mice was removed by trimming and applying hair removal cream (Nair cream, Church and Dwight Co.). The trimmed hair was then gently removed using a scalpel blade. Skin was disinfected with 70% ethanol and excised with a 6 mm biopsy punch (Integra Miltex, New York). A 10 μL drop of PBS containing 7 × 104 CFU of USA300 LAC was inoculated directly onto the open wound surface. The inoculum was allowed to dry before applying an 8 mm Finn Chamber (SmartPractice, Phoenix, AZ) over it and sealed with an additional transparent dressing (Tegaderm 3M, St. Paul, MN). Mice were monitored for signs of pain and distress, and analgesics were administered as necessary. After 24 h of infection, the wound dressing was removed without physically affecting the skin layer. Ten microliters of PBS-containing vancomycin, V–r8 (HCl), fusidic acid (Sigma-Aldrich)), or water (negative control) was added to infected wounds at the indicated concentrations, followed by reapplication of Tegaderm. After 5 h, an area of 1 cm × 1 cm of skin surrounding the wound site was excised and collected in sterile PBS. Skin samples were homogenized, serially diluted, and plated on TSA plates containing 1.5% agar and MRSASelect II agar plates (Bio-Rad, USA) for selective viable USA300 enumeration to determine the efficacy of bacterial clearance post-treatment. Wounds contaminated with non-USA300 bacteria were monitored by growth on TSB agar plates containing no antibiotics for selection and were excluded from analysis.
In Vivo Histological Cytotoxicity Assessment.
To assess the toxicity of V–r8 (HCl), skin wounds were created in anesthetized male wild-type C57BL/6 mice (7–8 weeks old, 22 to 25g, InVivos, Singapore) in an induction chamber supplied with 3% isoflurane (same as described above). Uninfected wounds were administered with 10 μL of water or 0.05% of V–r8 immediately after wounding and excised after 3 days as described above. Samples were fixed in 4% paraformaldehyde in PBS and incubated in 20% sucrose for 24 h. Tissues were then embedded into Optical Cutting Temperature (OCT) compound (Sakura, CA) and snap frozen in liquid nitrogen. Twelve micrometer sections were obtained on a Leica CM1860 UV cryotome (Leica Biosystems, Ernst-Leitz Strasse, Germany), stained with hematoxylin and eosin (H&E), and visualized using an Axio Scan.Z1 slide scanner (Carl Zeiss, Göttingen, Germany) under 20x/0.8 Apochrome objective as previously described.97
Statistical Analysis.
Statistical analysis of animal data between treated and nontreated groups was performed using the nonparametric Kruskal–Wallis test with Dunn’s post ad hoc test for intergroup comparisons.
Resistance Evolution Experiment.
To evaluate the propensity of V–r8 to induce resistance, an MIC assay using the previously described protocol was performed each day for 14 days using strain 29213, where each assay was incubated 24 h at 37 °C. Bacteria from the 1/2 MIC well the prior day were used to generate the inoculum for that day’s assay.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grants R01GM117278 (L.C.) and NIH-CA031845 (P.A.W.) and the Singapore National Medical Research Council NMRC/CBRG/0086/2015 grant (to K.A.K.). Support of this work through fellowships from the Stanford Center for Molecular Analysis and Design (A.S.A.) and the National Science Foundation (M.A.H.) are acknowledged. We gratefully acknowledge the Stanford University NMR Facility, Stanford University Mass Spectrometry, Stanford Cell Sciences Imaging Facility, and the Stanford Shared FACS Facility for instrumentation. We thank Sam Keyser and Prof. Carolyn Bertozzi’s laboratory for the use of their HPLC, Prof. Steve Boxer’s laboratory for the use of their NanoDrop instrument, and John Perrino for assistance with TEM imaging. We thank Milton Kwek for technical assistance with histology.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b08711. Synthetic methods and supplementary data (PDF)
REFERENCES
- (1).Klein EY; Mojica N; Jiang W; Cosgrove SE; Septimus E; Morgan DJ; Laxminarayan R Trends in Methicillin-Resistant Staphylococcus Aureus Hospitalizations in the United States, 2010–2014. Clin. Infect. Dis 2017, 65, 1921–1923. [DOI] [PubMed] [Google Scholar]
- (2).Stefani S; Chung DR; Lindsay JA; Friedrich AW; Kearns AM; Westh H; MacKenzie FM Meticillin-Resistant Staphylococcus Aureus (MRSA): Global Epidemiology and Harmonisation of Typing Methods. Int. J. Antimicrob. Agents 2012, 39, 273–282. [DOI] [PubMed] [Google Scholar]
- (3).Hassoun A; Linden PK; Friedman B Incidence, Prevalence, and Management of MRSA Bacteremia across Patient Populations-a Review of Recent Developments in MRSA Management and Treatment. Crit. Care London Engl 2017, 21, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tattevin P; Schwartz BS; Graber CJ; Volinski J; Bhukhen A; Bhukhen A; Mai TT; Vo NH; Dang DN; Phan TH; Basuino L; Perdreau-Remington F; Chambers HF; Diep BA Concurrent Epidemics of Skin and Soft Tissue Infection and Bloodstream Infection Due to Community-Associated Methicillin-Resistant Staphylococcus Aureus. Clin. Infect. Dis 2012, 55, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fisher RA; Gollan B; Helaine S Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol 2017, 15, 453–464. [DOI] [PubMed] [Google Scholar]
- (6).Bhattacharya M; Wozniak DJ; Stoodley P; Hall-Stoodley L Prevention and Treatment of Staphylococcus Aureus Biofilms. Expert Rev. Anti-Infect. Ther 2015, 13, 1499–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Waters EM; Rowe SE; O’Gara JP; Conlon BP Convergence of Staphylococcus Aureus Persister and Biofilm Research: Can Biofilms Be Defined as Communities of Adherent Persister Cells? PLoS Pathog. 2016, 12, e1006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Singh R; Ray P; Das A; Sharma M Penetration of Antibiotics through Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms. J. Antimicrob. Chemother 2010, 65, 1955–1958. [DOI] [PubMed] [Google Scholar]
- (9).Spellberg B The Future of Antibiotics. Crit. Care 2014, 18, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Liu C; Bayer A; Cosgrove SE; Daum RS; Fridkin SK; Gorwitz RJ; Kaplan SL; Karchmer AW; Levine DP; Murray BE; J Rybak M; Talan DA; Chambers HF Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus Aureus Infections in Adults and Children: Executive Summary. Clin. Infect. Dis 2011, 52, 285–292. [DOI] [PubMed] [Google Scholar]
- (11).Levine DP Vancomycin: A History. Clin. Infect. Dis 2006, 42 (Suppl. 1), S5–S12. [DOI] [PubMed] [Google Scholar]
- (12).Reynolds PE Structure, Biochemistry and Mechanism of Action of Glycopeptide Antibiotics. Eur. J. Clin. Microbiol. Infect. Dis 1989, 8, 943–950. [DOI] [PubMed] [Google Scholar]
- (13).Barna JC; Williams DH The Structure and Mode of Action of Glycopeptide Antibiotics of the Vancomycin Group. Annu. Rev. Microbiol 1984, 38, 339–357. [DOI] [PubMed] [Google Scholar]
- (14).Walsh CT; Fisher SL; Park I-S; Prahalad M; Wu Z Bacterial Resistance to Vancomycin: Five Genes and One Missing Hydrogen Bond Tell the Story. Chem. Biol 1996, 3, 21–28. [DOI] [PubMed] [Google Scholar]
- (15).Cantoni L; Glauser MP; Bille J Comparative Efficacy of Daptomycin, Vancomycin, and Cloxacillin for the Treatment of Staphylococcus Aureus Endocarditis in Rats and Role of Test Conditions in This Determination. Antimicrob. Agents Chemother 1990, 34, 2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Levine DP; Fromm BS; Reddy BR Slow Response to Vancomycin or Vancomycin plus Rifampin in Methicillin-Resistant Staphylococcus Aureus Endocarditis. Ann. Intern. Med 1991, 115, 674–680. [DOI] [PubMed] [Google Scholar]
- (17).Fusco DN; Alexander EL; Weisenberg SA; Mediavilla JR; Kreiswirth BN; Schuetz AN; Jenkins SG; Rhee KY Clinical Failure of Vancomycin in a Dialysis Patient with Methicillin-Susceptible Vancomycin-Heteroresistant S. Diagn. Microbiol. Infect. Dis 2009, 65, 180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bosso JA; Nappi J; Rudisill C; Wellein M; Bookstaver PB; Swindler J; Mauldin PD Relationship between Vancomycin Trough Concentrations and Nephrotoxicity: A Prospective Multicenter Trial. Antimicrob. Agents Chemother 2011, 55, 5475–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hall CW; Mah T-F Molecular Mechanisms of Biofilm-Based Antibiotic Resistance and Tolerance in Pathogenic Bacteria. FEMS Microbiol. Rev 2017, 41, 276–301. [DOI] [PubMed] [Google Scholar]
- (20).Rybak M; Lomaestro B; Rotschafer JC; Moellering R; Craig W; Billeter M; Dalovisio JR; Levine DP Therapeutic Monitoring of Vancomycin in Adult Patients: A Consensus Review of the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists. Am. J. Health-Syst. Pharm 2009, 66, 82–98. [DOI] [PubMed] [Google Scholar]
- (21).Okano A; Isley NA; Boger DL Total Syntheses of Vancomycin-Related Glycopeptide Antibiotics and Key Analogues. Chem. Rev 2017, 117, 11952–11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Blaskovich MAT; Hansford KA; Butler MS; Jia Z; Mark AE; Cooper MA Developments in Glycopeptide Antibiotics. ACS Infect. Dis 2018, 4, 715–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Brade KD; Rybak JM; Rybak MJ Oritavancin: A New Lipoglycopeptide Antibiotic in the Treatment of Gram-Positive Infections. Infect. Dis. Ther 2016, 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Belley A; Neesham-Grenon E; McKay G; Arhin FF; Harris R; Beveridge T; Parr TR; Moeck G Oritavancin Kills Stationary-Phase and Biofilm Staphylococcus Aureus Cells In Vitro. Antimicrob. Agents Chemother 2009, 53, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Higgins DL; Chang R; Debabov DV; Leung J; Wu T; Krause KM; Sandvik E; Hubbard JM; Kaniga K; Schmidt DE; Gao Q; Cass RT; Karr DE; Benton BM; Humphrey PE Telavancin, a Multifunctional Lipoglycopeptide, Disrupts Both Cell Wall Synthesis and Cell Membrane Integrity in Methicillin-Resistant Staphylococcus Aureus. Antimicrob. Agents Chemother 2005, 49, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yarlagadda V; Akkapeddi P; Manjunath GB; Haldar J Membrane Active Vancomycin Analogues: A Strategy to Combat Bacterial Resistance. J. Med. Chem 2014, 57, 4558–4568. [DOI] [PubMed] [Google Scholar]
- (27).Okano A; Isley NA; Boger DL Peripheral Modifications of [Ψ[CH2NH]Tpg4]Vancomycin with Added Synergistic Mechanisms of Action Provide Durable and Potent Antibiotics. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E5052–E5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Blaskovich MAT; Hansford KA; Gong Y; Butler MS; Muldoon C; Huang JX; Ramu S; Silva AB; Cheng M; Kavanagh AM; Ziora Z; Premraj R; Lindahl F; Bradford TA; Lee JC; Karoli T; Pelingon R; Edwards DJ; Amado M; Elliott AG; Phetsang W; Daud NH; Deecke JE; Sidjabat HE; Ramaologa S; Zuegg J; Betley JR; Beevers APG; Smith RAG; Roberts JA; Paterson DL; Cooper MA Protein-Inspired Antibiotics Active against Vancomycin- and Daptomycin-Resistant Bacteria. Nat. Commun 2018, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Arnusch CJ; Pieters RJ; Breukink E Enhanced Membrane Pore Formation through High-Affinity Targeted Antimicrobial Peptides. PLoS One 2012, 7, e39768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Leung SSF; Tirado-Rives J; Jorgensen WL Vancomycin Analogs: Seeking Improved Binding of D-Ala-D-Ala and D-Ala-D-Lac Peptides by Side-Chain and Backbone Modifications. Bioorg. Med. Chem 2009, 17, 5874–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).McComas CC; Crowley BM; Boger DL Partitioning the Loss in Vancomycin Binding Affinity for D-Ala-D-Lac into Lost HBond and Repulsive Lone Pair Contributions. J. Am. Chem. Soc 2003, 125, 9314–9315. [DOI] [PubMed] [Google Scholar]
- (32).Crowley BM; Boger DL Total Synthesis and Evaluation of [Ψ[CH2NH]Tpg4]Vancomycin Aglycon: Reengineering Vancomycin for Dual d-Ala-d-Ala and d-Ala-d-Lac Binding. J. Am. Chem. Soc 2006, 128, 2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Okano A; Nakayama A; Schammel AW; Boger DL Total Synthesis of [Ψ[C(=NH)NH]Tpg4]Vancomycin and Its (4-Chlorobiphenyl)Methyl Derivative: Impact of Peripheral Modifications on Vancomycin Analogues Redesigned for Dual d-Ala-d-Ala and d-Ala-d-Lac Binding. J. Am. Chem. Soc 2014, 136, 13522–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Frankel AD; Pabo CO Cellular Uptake of the Tat Protein from Human Immunodeficiency Virus. Cell 1988, 55, 1189–1193. [DOI] [PubMed] [Google Scholar]
- (35).Green M; Loewenstein PM Autonomous Functional Domains of Chemically Synthesized Human Immunodeficiency Virus Tat Trans-Activator Protein. Cell 1988, 55, 1179–1188. [DOI] [PubMed] [Google Scholar]
- (36).Vives E; Brodin P; Lebleu B A Truncated HIV-1 Tat̀ Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem 1997, 272, 16010–16017. [DOI] [PubMed] [Google Scholar]
- (37).Wender PA; Mitchell DJ; Pattabiraman K; Pelkey ET; Steinman L; Rothbard JB The Design, Synthesis, and Evaluation of Molecules That Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc. Natl. Acad. Sci. U. S. A 2000, 97, 13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Stanzl EG; Trantow BM; Vargas JR; Wender PA Fifteen Years of Cell-Penetrating, Guanidinium-Rich Molecular Transporters: Basic Science, Research Tools, and Clinical Applications. Acc. Chem. Res 2013, 46, 2944–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bechara C; Sagan S Cell-Penetrating Peptides: 20 Years Later, Where Do We Stand? FEBS Lett. 2013, 587, 1693–1702. [DOI] [PubMed] [Google Scholar]
- (40).Torchilin VP Tat Peptide-Mediated Intracellular Delivery of Pharmaceutical Nanocarriers. Adv. Drug Delivery Rev 2008, 60, 548–558. [DOI] [PubMed] [Google Scholar]
- (41).Nakase I; Takeuchi T; Futaki S Cell Penetrating Peptides for Chemical Biological Studies. In Cell-Penetrating Peptides; Methods in Molecular Biology; Humana Press, New York, NY, 2015; pp 387–396. [DOI] [PubMed] [Google Scholar]
- (42).Madani F; Lindberg S; Langel Ü; Futaki S; Graslund A Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. J. Biophys 2011, 2011, No. 414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Samuel BU; Hearn B; Mack D; Wender P; Rothbard J; Kirisits MJ; Mui E; Wernimont S; Roberts CW; Muench SP; Rice DW; Prigge ST; Law AB; McLeod R Delivery of Antimicrobials into Parasites. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 14281–14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sparr C; Purkayastha N; Kolesinska B; Gengenbacher M; Amulic B; Matuschewski K; Seebach D; Kamena F Improved Efficacy of Fosmidomycin against Plasmodium and Mycobacterium Species by Combination with the Cell-Penetrating Peptide Octaarginine. Antimicrob. Agents Chemother 2013, 57, 4689–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lei EK; Pereira MP; Kelley SO Tuning the Intracellular Bacterial Targeting of Peptidic Vectors. Angew. Chem., Int. Ed 2013, 52, 9660–9663. [DOI] [PubMed] [Google Scholar]
- (46).Purkayastha N; Capone S; Beck AK; Seebach D; Leeds J; Thompson K; Moser HE Antibacterial Activity of Enrofloxacin and Ciprofloxacin Derivatives of β-Octaarginine. Chem. Biodiversity 2015, 12, 179–193. [DOI] [PubMed] [Google Scholar]
- (47).Luedtke NW; Carmichael P; Tor Y Cellular Uptake of Aminoglycosides, Guanidinoglycosides, and Poly-Arginine. J. Am. Chem. Soc 2003, 125, 12374–12375. [DOI] [PubMed] [Google Scholar]
- (48).Dubikovskaya EA; Thorne SH; Pillow TH; Contag CH; Wender PA Overcoming Multidrug Resistance of Small-Molecule Therapeutics through Conjugation with Releasable Octaarginine Transporters. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 12128–12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wender PA; Galliher WC; Bhat NM; Pillow TH; Bieber MM; Teng NNH Taxol-Oligoarginine Conjugates Overcome Drug Resistance in-Vitro in Human Ovarian Carcinoma. Gynecol. Oncol 2012, 126, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Rothbard JB; Garlington S; Lin Q; Kirschberg T; Kreider E; McGrane PL; Wender PA; Khavari PA Conjugation of Arginine Oligomers to Cyclosporin A Facilitates Topical Delivery and Inhibition of Inflammation. Nat. Med 2000, 6, 1253–1257. [DOI] [PubMed] [Google Scholar]
- (51).Chen L; Wright LR; Chen C-H; Oliver SF; Wender PA; Mochly-Rosen D Molecular Transporters for Peptides: Delivery of a Cardioprotective ΕPKC Agonist Peptide into Cells and Intact Ischemic Heart Using a Transport System, R7. Chem. Biol 2001, 8, 1123–1129. [DOI] [PubMed] [Google Scholar]
- (52).Jameson KL; Mazur PK; Zehnder AM; Zhang J; Zarnegar B; Sage J; Khavari PA IQGAP1 Scaffold-Kinase Interaction Blockade Selectively Targets RAS-MAP Kinase–Driven Tumors. Nat. Med 2013, 19, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Benner NL; Zang X; Buehler DC; Kickhoefer VA; Rome ME; Rome LH; Wender PA Vault Nanoparticles: Chemical Modifications for Imaging and Enhanced Delivery. ACS Nano 2017, 11, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Pavlovic I; Thakor DT; Vargas JR; McKinlay CJ; Hauke S; Anstaett P; Camuña RC; Bigler L; Gasser G; Schultz C; Wender PA; Jessen HJ Cellular Delivery and Photochemical Release of a Caged Inositol-Pyrophosphate Induces PH-Domain Translocation in Cellulo. Nat. Commun 2016, 7, 10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Geihe EI; Cooley CB; Simon JR; Kiesewetter MK; Edward JA; Hickerson RP; Kaspar RL; Hedrick JL; Waymouth RM; Wender PA Designed Guanidinium-Rich Amphipathic Oligocarbonate Molecular Transporters Complex, Deliver and Release SiRNA in Cells. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 13171–13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wender PA; Huttner MA; Staveness D; Vargas JR; Xu AF Guanidinium-Rich, Glycerol-Derived Oligocarbonates: A New Class of Cell-Penetrating Molecular Transporters That Complex, Deliver, and Release SiRNA. Mol. Pharmaceutics 2015, 12, 742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).McKinlay CJ; Vargas JR; Blake TR; Hardy JW; Kanada M; Contag CH; Wender PA; Waymouth RM Charge-Altering Releasable Transporters (CARTs) for the Delivery and Release of MRNA in Living Animals. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E448–E456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Benner NL; Near KE; Bachmann MH; Contag CH; Waymouth RM; Wender PA Functional DNA Delivery Enabled by Lipid-Modified Charge-Altering Releasable Transporters (CARTs). Biomacromolecules 2018, 19, 2812–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).McKinlay CJ; Benner NL; Haabeth OA; Waymouth RM; Wender PA Enhanced MRNA Delivery into Lymphocytes Enabled by Lipid-Varied Libraries of Charge-Altering Releasable Transporters. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E5859–E5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Brezden A; Mohamed MF; Nepal M; Harwood JS; Kuriakose J; Seleem MN; Chmielewski J Dual Targeting of Intracellular Pathogenic Bacteria with a Cleavable Conjugate of Kanamycin and an Antibacterial Cell-Penetrating Peptide. J. Am. Chem. Soc 2016, 138, 10945–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wesolowski D; Alonso D; Altman S Combined Effect of a Peptide-Morpholino Oligonucleotide Conjugate and a Cell-Penetrating Peptide as an Antibiotic. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 8686–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Defaus S; Gallo M; Abengozar MA; Rivas L; Andreu D´ A. Synthetic Strategy for Conjugation of Paromomycin to Cell-Penetrating Tat(48–60) for Delivery and Visualization into Leishmania Parasites. Int. J. Pept 2017, 2017, No. 4213037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Abushahba MF; Mohammad H; Seleem MN Targeting Multidrug-Resistant Staphylococci with an Anti-RpoA Peptide Nucleic Acid Conjugated to the HIV-1 TAT Cell Penetrating Peptide. Mol. Ther.–Nucleic Acids 2016, 5, e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Hyman JM; Geihe EI; Trantow BM; Parvin B; Wender PA A Molecular Method for the Delivery of Small Molecules and Proteins across the Cell Wall of Algae Using Molecular Transporters. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Geueke B; Namoto K; Agarkova I; Perriard J-C; Kohler H-PE; Seebach D Bacterial Cell Penetration by Β3-Oligohomoarginines: Indications for Passive Transfer through the Lipid Bilayer. ChemBioChem 2005, 6, 982–985. [DOI] [PubMed] [Google Scholar]
- (66).Rothbard J; Wender P Method and Composition for Enhancing Transport across Biological Membranes. US20030162719 A1, August 28, 2003.
- (67).Wender PA; Quiroz RV; Stevens MC Function through Synthesis-Informed Design. Acc. Chem. Res 2015, 48, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wang T; Zhang Z; Han Y; Yin Z; Scola PM Guanidine Derivatives for the Treatment of Hepatitis C. US9655902B2, August 1, 2014.
- (69).Mishra NM; Briers Y; Lamberigts C; Steenackers H; Robijns S; Landuyt B; Vanderleyden J; Schoofs L; Lavigne R; Luyten W; Vanderleyden J; Schoofs L; Lavigne R; Luyten W; Van der Eycken EV Evaluation of the Antibacterial and Antibiofilm Activities of Novel CRAMP-Vancomycin Conjugates with Diverse Linkers. Org. Biomol. Chem 2015, 13, 7477–7486. [DOI] [PubMed] [Google Scholar]
- (70).Andrushchenko VV; Vogel HJ; Prenner EJ Optimization of the Hydrochloric Acid Concentration Used for Trifluoroacetate Removal from Synthetic Peptides. J. Pept. Sci 2007, 13, 37–43. [DOI] [PubMed] [Google Scholar]
- (71).Lu J; Yoshida O; Hayashi S; Arimoto H Synthesis of Rigidly-Linked Vancomycin Dimers and Their in vivo Efficacy against Resistant Bacteria. Chem. Commun. 2007, 3, 251–253. [DOI] [PubMed] [Google Scholar]
- (72).Cornish J; Callon KE; Lin CQ-X; Xiao CL; Mulvey TB; Cooper GJS; Reid IR Trifluoroacetate, a Contaminant in Purified Proteins, Inhibits Proliferation of Osteoblasts and Chondrocytes. Am. J. Physiol. - Endocrinol. Metab 1999, 277, E779–E783. [DOI] [PubMed] [Google Scholar]
- (73).Olson ME; Ceri H; Morck DW; Buret AG; Read RR Biofilm Bacteria: Formation and Comparative Susceptibility to Antibiotics. Can. J. Vet. Res 2002, 66, 86–92. [PMC free article] [PubMed] [Google Scholar]
- (74).Ceri H; Olson ME; Stremick C; Read RR; Morck D; Buret A The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol 1999, 37, 1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lechner S; Lewis K; Bertram R Staphylococcus Aureus Persisters Tolerant to Bactericidal Antibiotics. J. Mol. Microbiol. Biotechnol 2012, 22, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Xu R; Ruan L; Ge M; Zhang Y Synthesis of Fluorescein Isothiocyanate-Labeled Vancomycin and Its Bacterial Detection by Fluorescence, Synthesis of Fluorescein Isothiocyanate-Labeled Vancomycin and Its Bacterial Detection by Fluorescence. Chin. J. Appl. Chem 2013, 31, 220–224. [Google Scholar]
- (77).Daniel RA; Errington J Control of Cell Morphogenesis in Bacteria: Two Distinct Ways to Make a Rod-Shaped Cell. Cell 2003, 113, 767–776. [DOI] [PubMed] [Google Scholar]
- (78).Avelar-Freitas BA; Almeida VG; Pinto MCX; Mourao F. a. G.; Massensini AR; Martins-Filho OA; Rocha-Vieira E; Brito-Melo GE. Trypan Blue Exclusion Assay by Flow Cytometry. Braz. J. Med. Biol. Res 2014, 47, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Nygaard R; Romaniuk JAH; Rice DM; Cegelski L Spectral Snapshots of Bacterial Cell-Wall Composition and the Influence of Antibiotics by Whole-Cell NMR. Biophys. J. 2015, 108, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Williams I; Paul F; Lloyd D; Jepras R; Critchley I; Newman M; Warrack J; Giokarini T; Hayes AJ; Randerson PF; Venables WA Flow Cytometry and Other Techniques Show That Staphylococcus Aureus Undergoes Significant Physiological Changes in the Early Stages of Surface-Attached Culture. Microbiology 1999, 145, 1325–1333. [DOI] [PubMed] [Google Scholar]
- (81).Zhou X; Cegelski L Nutrient-Dependent Structural Changes in S. Aureus Peptidoglycan Revealed by Solid-State NMR Spectroscopy. Biochemistry 2012, 51, 8143–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Romaniuk JAH; Cegelski L Peptidoglycan and Teichoic Acid Levels and Alterations in Staphylococcus Aureus by Cell-Wall and Whole-Cell Nuclear Magnetic Resonance. Biochemistry 2018, 57, 3966–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Glukhov E; Stark M; Burrows LL; Deber CM Basis for Selectivity of Cationic Antimicrobial Peptides for Bacterial Versus Mammalian Membranes. J. Biol. Chem 2005, 280, 33960–33967. [DOI] [PubMed] [Google Scholar]
- (84).Deslouches B; Steckbeck JD; Craigo JK; Doi Y; Mietzner TA; Montelaro RC Rational Design of Engineered Cationic Antimicrobial Peptides Consisting Exclusively of Arginine and Tryptophan, and Their Activity against Multidrug-Resistant Pathogens. Antimicrob. Agents Chemother 2013, 57, 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Wu JA; Kusuma C; Mond JJ; Kokai-Kun JF Lysostaphin Disrupts Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms on Artificial Surfaces. Antimicrob. Agents Chemother 2003, 47, 3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Ceotto-Vigoder H; Marques SLS; Santos INS; Alves MDB; Barrias ES; Potter A; Alviano DS; Bastos MCF Nisin and Lysostaphin Activity against Preformed Biofilm of Staphylococcus Aureus Involved in Bovine Mastitis. J. Appl. Microbiol. 2016, 121, 101–114. [DOI] [PubMed] [Google Scholar]
- (87).Kim W; Conery AL; Rajamuthiah R; Fuchs BB; Ausubel FM; Mylonakis E Identification of an Antimicrobial Agent Effective against Methicillin-Resistant Staphylococcus Aureus Persisters Using a Fluorescence-Based Screening Strategy. PLoS One 2015, 10, e0127640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Peng B; Su Y; Li H; Han Y; Guo C; Tian Y; Peng X Exogenous Alanine and/or Glucose plus Kanamycin Kills Antibiotic-Resistant Bacteria. Cell Metab. 2015, 21, 249–262. [DOI] [PubMed] [Google Scholar]
- (89).Allison KR; Brynildsen MP; Collins JJ Metabolite-Enabled Eradication of Bacterial Persisters by Aminoglycosides. Nature 2011, 473, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Rabanal F; Grau-Campistany A; Vila-Farres X; Gonzalez-´ Linares J; Borras M; Vila J; Manresa A; Cajal Y. A Bioinspired̀ Peptide Scaffold with High Antibiotic Activity and Low in Vivo Toxicity. Sci. Rep. 2015, 5, 10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Economou NJ; Cocklin S; Loll PJ High-Resolution Crystal Structure Reveals Molecular Details of Target Recognition by Bacitracin. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 14207–14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Percival SL; Suleman L; Vuotto C; Donelli G Healthcare-Associated Infections, Medical Devices and Biofilms: Risk, Tolerance and Control. J. Med. Microbiol 2015, 64, 323–334. [DOI] [PubMed] [Google Scholar]
- (93).Omar A; Wright JB; Schultz G; Burrell R; Nadworny P Microbial Biofilms and Chronic Wounds. Microorganisms 2017, 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Patel DA; Stephens JM; Gao X; Verheggen BG; Shelbaya A; Haider S Economic Burden of Inpatient and Outpatient Antibiotic Treatment for Methicillin-Resistant Staphylococcus Aureus Complicated Skin and Soft-Tissue Infections: A Comparison of Linezolid, Vancomycin, and Daptomycin. Clin. Outcomes Res. CEOR 2013, 5, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Abdelhady W; Bayer AS; Seidl K; Nast CC; Kiedrowski MR; Horswill AR; Yeaman MR; Xiong YQ Reduced Vancomycin Susceptibility in an In Vitro Catheter-Related Biofilm Model Correlates with Poor Therapeutic Outcomes in Experimental Endocarditis Due to Methicillin-Resistant Staphylococcus Aureus. Antimicrob. Agents Chemother 2013, 57, 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Roche ED; Renick PJ; Tetens SP; Carson DL A Model for Evaluating Topical Antimicrobial Efficacy against Methicillin-Resistant Staphylococcus Aureus Biofilms in Superficial Murine Wounds. Antimicrob. Agents Chemother. 2012, 56, 4508–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Chong KKL; Tay WH; Janela B; Yong AMH; Liew TH; Madden L; Keogh D; Barkham TMS; Ginhoux F; Becker DL; Kline KA Enterococcus Faecalis Modulates Immune Activation and Slows Healing During Wound Infection. J. Infect. Dis 2017, 216, 1644–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).O’Neill AJ; Larsen AR; Skov R; Henriksen AS; Chopra I Characterization of the Epidemic European Fusidic Acid-Resistant Impetigo Clone of Staphylococcus Aureus. J. Clin. Microbiol 2007, 45, 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Siala W; Rodriguez-Villalobos H; Fernandes P; Tulkens PM; Bambeke FV Activity of Combinations of Antistaphylococcal Antibiotics with Fusidic Acid against Staphylococcal Biofilms in in Vitro Static and Dynamic Models. Antimicrob. Agents Chemother. 2018, No. AAC.00598-18, DOI: 10.1128/AAC.00598-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Harrison JJ; Stremick CA; Turner RJ; Allan ND; Olson ME; Ceri H Microtiter Susceptibility Testing of Microbes Growing on Peg Lids: A Miniaturized Biofilm Model for High-Throughput Screening. Nat. Protoc 2010, 5, 1236–1254. [DOI] [PubMed] [Google Scholar]
- (101).Wiegand I; Hilpert K; Hancock REW Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc 2008, 3, 163–175. [DOI] [PubMed] [Google Scholar]
- (102).Benincasa M; Barriere Q; Runti G; PIERRE O; Bourge M; Scocchi M; Mergaert P. Single Cell Flow Cytometry Assay for Peptide Uptake by Bacteria. BIO-PROTOC 2016, 6, e2038. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.