Abstract
Background: Tripartite motif 13 (TRIM13) plays a significant role in various biological processes including cell growth, apoptosis, transcriptional regulation, and carcinogenesis. However, the prognostic significance of TRIM13 gene in breast cancer treatment remains largely unclear. Methods: We performed a bioinformatics analysis of the clinical parameters and survival data as it relates to TRIM13 in breast cancer patients using several online databases including Oncomine, bcGenExMiner, PrognoScan, and UCSC Xena. Results: We found that TRIM13 was lower-expressed in different subtypes of breast cancer with respect to normal tissues. Estrogen receptor and progesterone receptor status were positively correlated with TRIM13 level; whereas, the Scarff–Bloom–Richardson grade, Nottingham prognostic index, nodal status, basal-like status, and triple-negative status were negatively related to TRIM13 expression in breast cancer patients with respect to normal individuals. Lower TRIM13 expression correlated with worse distant metastasis free survival, relapse free survival, disease specific survival, and metastatic relapse free survival. We also confirmed a positive correlation between TRIM13 and RAB11FIP2 gene expression. Conclusion: Bioinformatics analysis revealed that TRIM13 may be adopted as a promising predictive biomarker for prognosis of breast cancer. More in-depth experiments and clinical trials are needed to validate the value of TRIM13 in breast cancer treatment.
Keywords: Breast cancer, Biomarker, Prognosis, TRIM13
Introduction
Breast cancer remains the most common malignant tumor and a leading cause of cancer-related mortality in women worldwide [1]. Breast cancer treatment includes locoregional surgery, systemic chemotherapy, precision radiotherapy, endocrine therapy, biological targeting agents, and a combination of the above, mainly depending on clinical, pathological, and molecular features. Biomarkers are served as surrogates of these features for establishing prognostics and predicting outcomes. Finding more effective, sensitive, and specific biomarkers for the prognosis of patients is therefore urgently needed in breast cancer research [2].
The family of tripartite motif (TRIM) proteins (http://trimbase.tigem.it/) is one of the subfamilies of the RING type E3 ubiquitin ligases and contains more than 70 members in human chromosomes [3]. Accumulated evidence has suggested that some members of TRIM are involved in various biological processes including cell growth, apoptosis, transcriptional regulation, and carcinogenesis [4]. For example, knockdown of TRIM66 inhibited malignant behavior and epithelial–mesenchymal transition in non-small-cell lung cancer, suggesting that TRIM66 was an oncogene that promoted lung cancer progression [5]. TRIM36 played a tumor suppressive role by reducing cell proliferation and migration as well as promoting apoptosis in prostate cancer [6]. Researchers and clinicians are increasingly regarding TRIM as important factor in cancer development [7,8].
TRIM13, a tumor suppressor gene in the 13q14 chromosome region, was frequently lost in various malignancies [9–13]. It has been reported to play a role in endoplasmic reticulum-associated degradation and to regulate autophagy caused by endoplasmic stress [14–16]. To date, caspase-8, Akt, L-type channels, and Nur77 have been shown to be substrates for TRIM13-mediated ubiquitination [16–19]. TRIM13 over-expression caused stabilization of p53 and decrease of Akt kinase activity followed by induction of apoptosis [17]. Taken together, these findings suggest that TRIM13 may not only function as a tumor suppressor, but also as a potential predictive biomarker for prognosis of cancer.
In the present study, we performed a bioinformatics analysis of the clinical parameters and survival data as it relates to TRIM13 in breast cancer patients using several large online databases in order to evaluate the prognostic significance of TRIM13 gene in breast cancer treatment.
Materials and methods
Oncomine
The Oncomine (http://www.oncomine.org) is a web-based data mining platform aimed at facilitating new discovery from genome-wide expression analysis [20]. TRIM13 gene was queried in the database and the results were filtered by selecting breast cancer and Cancer versus Normal Analysis. The threshold included fold change ≥2 in expression between cancers and normal tissues, P-value ≤1E-4, and gene rank ≥ top 10%. The comparisons of mRNA levels of TRIM13 in breast cancer and normal tissues in each individual dataset were analyzed using the Student’s t test. Gene co-expressed with TRIM13 was also analyzed.
bcGenExMiner
The Breast Cancer Gene-Expression Miner v4.1 (bcGenExMiner v4.1, http://bcgenex.centregauducheau.fr/BC-GEM), a mining tool of 36 published annotated genomics data (total of 5861 patients), was used to analyse TRIM13 gene with clinical parameters such as age, Scarff–Bloom–Richardson (SBR) grade, Nottingham prognostic index (NPI), estrogen receptor (ER), progesterone receptor (PR), epidermal growth factor receptor-2 (HER-2), and nodal status using the expression module [21,22]. Relevance of TRIM13 and metastatic relapse event was calculated using the prognostic module, and correlation of TRIM13 and RAB11FIP2 was assessed using the correlation module. Data were last updated on December 2017.
PrognoScan
The PrognoScan (http://www.prognoscan.org/) is a popular online database for evaluating the biological relationship between gene expression and survival data including distant metastasis free survival, relapse free survival, and disease specific survival in breast cancer patients across a large collection of publicly available cancer microarray datasets [23]. This tool could automatically calculate P-value, HR, and 95% confidence intervals based on a certain gene expression. Data were last updated on March 2013.
UCSC Xena
The UCSC Xena (http://xena.ucsc.edu/) is a functional genomics browser that provides visualization and integration for analyzing and viewing the public data hubs. The heat map was generated by data mining in The Cancer Genome Atlas (TCGA) Breast Cancer using the UCSC Xena browser.
Results
Reduced expression of TRIM13 gene in breast cancer patients
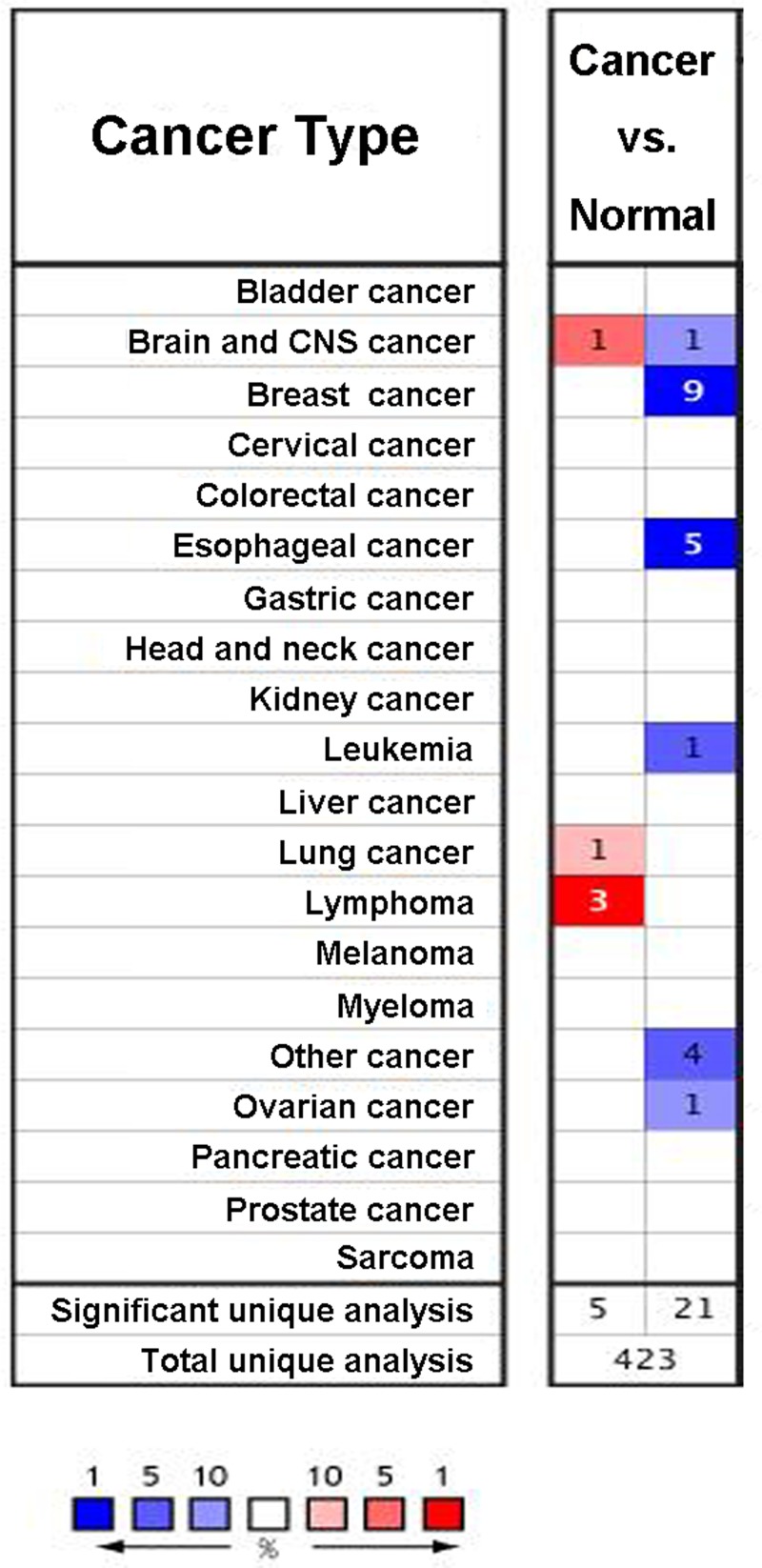
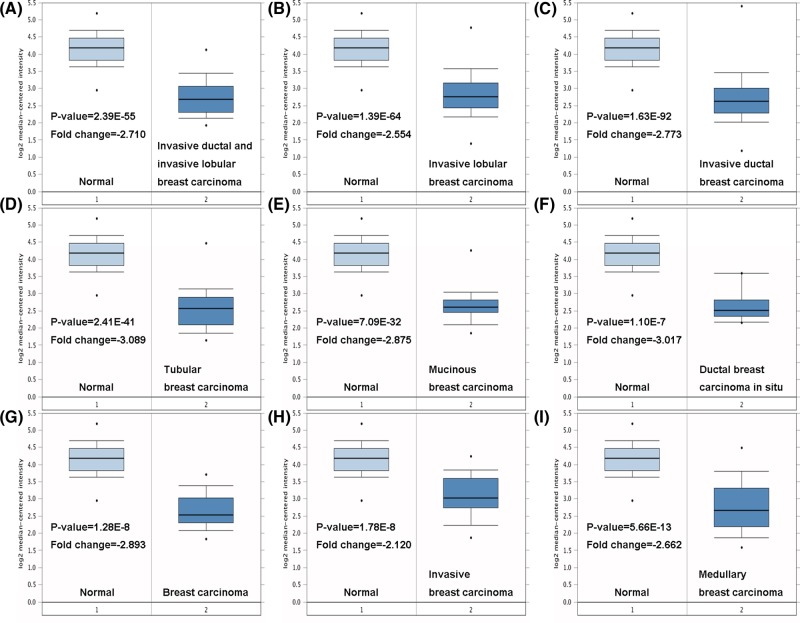
We first measured the expression of TRIM13 gene in 20 common types of cancer and compared its level to normal tissues using the Oncomine online database. Increased level of TRIM13 (red) was found in brain cancer, lung cancer, and lymphoma; whereas, decreased level of TRIM13 (blue) was observed in esophageal cancer, leukemia, ovarian cancer, and especially breast cancer (Figure 1). Oncomine analysis also revealed that TRIM13 was significantly lower-expressed in invasive ductal and invasive lobular breast carcinoma, invasive lobular breast carcinoma, invasive ductal breast carcinoma, tubular breast carcinoma, mucinous carcinoma, ductal breast carcinoma in situ, breast carcinoma, invasive breast carcinoma, and medullary carcinoma with respect to normal individuals (Figure 2A–I, Table 1).
Figure 1. Expression of TRIM13 gene in 20 common cancers versus paired normal tissues using the Oncomine database with the threshold of fold change ≥2, P-value ≤1E-4, and gene rank ≥ top 10%.
Red and blue respectively stand for the numbers of datasets with statistically significant (P<0.05) increased and decreased levels of TRIM13 gene.
Figure 2. Box plot comparing TRIM13 expression in normal individuals and breast cancer patients obtained from the Oncomine database.
Analysis is shown for invasive ductal and invasive lobular breast carcinoma (A), invasive lobular breast carcinoma (B), invasive ductal breast carcinoma (C), tubular breast carcinoma (D), mucinous carcinoma (E), ductal breast carcinoma in situ (F), breast carcinoma (G), invasive breast carcinoma (H), and medullary carcinoma (I).
Table 1. TRIM13 expression among different subtypes of breast cancer and normal individuals using the Oncomine database.
| Breast cancer subtype | P-value | t test | Fold change | Sample |
|---|---|---|---|---|
| Invasive ductal and invasive lobular breast carcinoma | 2.39E-55 | −23.164 | −2.710 | 90 |
| Invasive lobular breast carcinoma | 1.39E-64 | −22.594 | −2.554 | 148 |
| Invasive ductal breast carcinoma | 1.63E-92 | −37.680 | −2.773 | 1556 |
| Tubular breast carcinoma | 2.41E-41 | −21.724 | −3.089 | 67 |
| Mucinous breast carcinoma | 7.09E-32 | −20.140 | −2.875 | 46 |
| Ductal breast carcinoma in situ | 1.10E-7 | −11.852 | −3.017 | 10 |
| Breast carcinoma | 1.28E-8 | −10.636 | −2.893 | 14 |
| Invasive breast carcinoma | 1.78E-8 | −8.060 | −2.120 | 21 |
| Medullary breast carcinoma | 5.66E-13 | −10.639 | −2.662 | 32 |
TRIM13 expression and clinical parameters of breast cancer patients
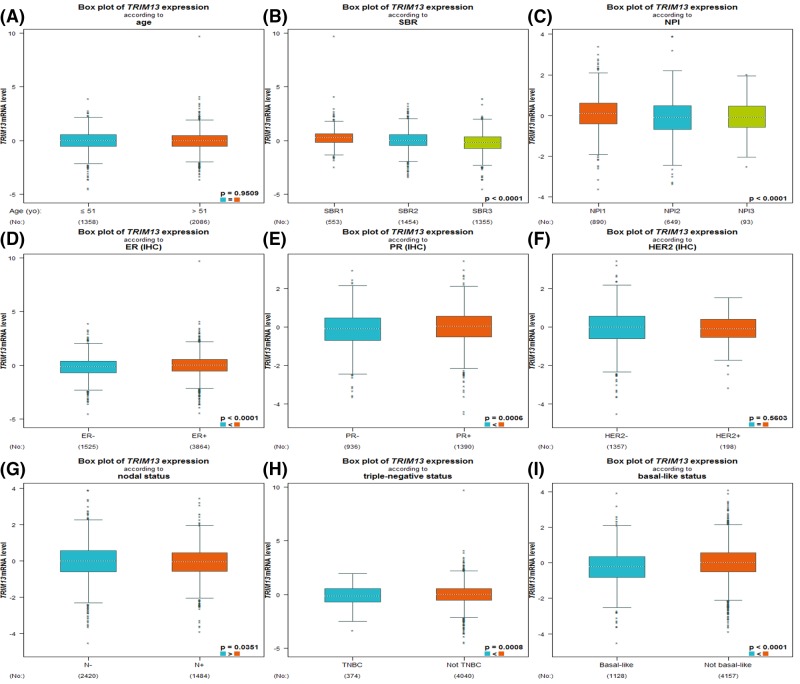
We next evaluated TRIM13 expression among different groups of patients based on several clinical parameters using the bc-GenExMiner software. For age criteria, no significant difference was found between ≤51 year and >51 year group (Figure 3A, Table 2). The SBR is a histological grade that evaluates tubule formation, nuclear characteristics of pleomorphism, and mitotic index. Based on tumor size, lymph node stage, and tumor grade, the NPI has been validated to stratify patients into additional prognostic groups. The SBR grade and NPI index are two accepted prognostic factors for breast cancer [24,25]; more advanced SBR grade and NPI index were associated with lower TRIM13 level (Figure 3B,C). ER-positive or PR-positive breast cancer patients tended to express higher TRIM13 gene compared with ER-negative or PR-negative patients (Figure 3D,E; Table 2). Regarding HER-2, there was no significant difference between positive and negative group (Figure 3F, Table 2). Patients with nodal-positive status showed reduced expression of TRIM13 than nodal-negative patients (Figure 3G, Table 2). Moreover, TRIM13 was significantly elevated in non-triple-negative and non-basal-like breast cancer patients with respect to triple-negative and basal-like breast cancer patients (Figure 3H,I; Table 2).
Figure 3. Box plot evaluating TRIM13 expression among different groups of patients based on clinical parameters using the bc-GenExMiner software.
Analysis is shown for age (A), SBR grade (B), NPI index (C), ER (D), PR (E), HER-2 (F), nodal status (G), triple-negative status (H), and basal-like status (I).
Table 2. Relationship between TRIM13 expression and clinical parameters of breast cancer patients using the bc-GenExMiner database.
| Variables | No. of patients | TRIM13 mRNA | P-value |
|---|---|---|---|
| Age | 0.9509 | ||
| ≤51 | 1358 | - | |
| >51 | 2086 | - | |
| ER | <0.0001 | ||
| Negative | 1525 | - | |
| Positive | 3864 | Increased | |
| PR | 0.0006 | ||
| Negative | 936 | - | |
| Positive | 1390 | Increased | |
| HER-2 | 0.5603 | ||
| Negative | 1357 | - | |
| Positive | 198 | - | |
| Nodal status | 0.0351 | ||
| Negative | 2420 | Increased | |
| Positive | 1484 | - | |
| Triple-negative status | 0.0008 | ||
| Non-triple-negative | 4040 | Increased | |
| Triple-negative | 374 | - | |
| Basal-like status | <0.0001 | ||
| Non-basal-like | 4157 | Increased | |
| Basal-like | 1128 | - |
TRIM13 expression and survival data of breast cancer patients
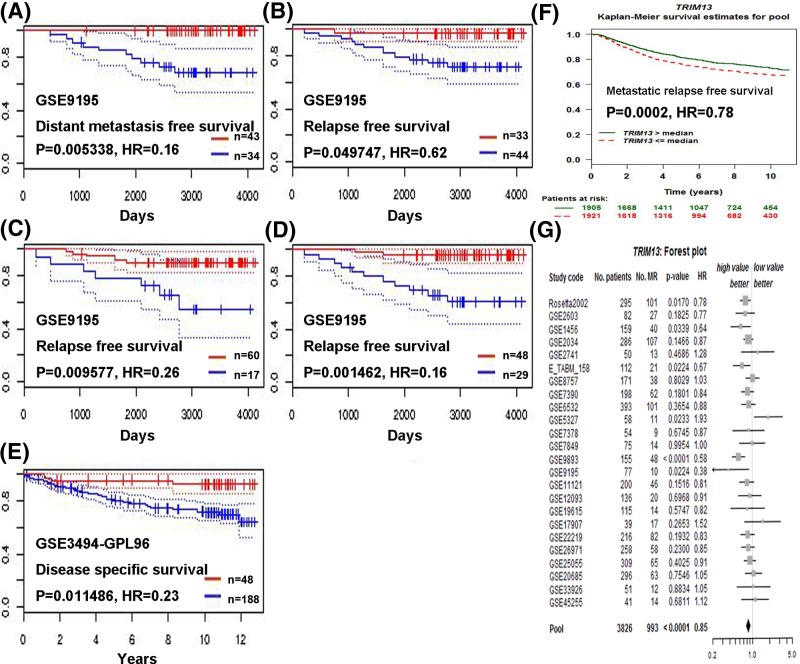
We then investigated the prognostic value of TRIM13 gene using the PrognoScan database. Higher expression of TRIM13 (red) was significantly related to preferable distant metastasis free survival (Figure 4A, Table 3). Breast cancer patients with increased TRIM13 level (red) presented better relapse free survival (Figure 4B–D, Table 3). Conversely, down-regulated TRIM13 gene (blue) was strongly associated with worse disease specific survival (Figure 4E, Table 3). To further investigate the role of TRIM13 in breast cancer prognosis, we used the bc-GenExMiner software to verify our findings. TRIM13 was observed to be positively correlated with metastatic relapse free survival, as suggested by the survival curve and forest plot (Figure 4F–G, Table 3).
Figure 4. Survive curve and forest plot evaluating the prognostic value of TRIM13 using the PrognoScan and bc-GenExMiner database.
Analysis is shown for distant metastasis free survival (A), relapse free survival for different probe (B–D), disease specific survival (E), metastatic relapse free survival (F), and forest plot (G). For A–E, red and blue stand for high and low expression of TRIM13 gene, respectively.
Table 3. TRIM13 expression and survival data of breast cancer patients using the PrognoScan database.
| Dataset | Probe ID | End point | No. | Cox P-value | HR |
|---|---|---|---|---|---|
| GSE9195 | 203659_s_at | Distant metastasis free survival | 77 | 0.005338 | 0.16 [0.04–0.58] |
| GSE9195 | 229943_at | Relapse free survival | 77 | 0.049747 | 0.62 [0.38–1.00] |
| GSE9195 | 230192_at | Relapse free survival | 77 | 0.009577 | 0.26 [0.09–0.72] |
| GSE9195 | 203659_s_at | Relapse free survival | 77 | 0.001462 | 0.16 [0.05–0.49] |
| GSE3494–GPL96 | 203659_s_at | Disease specific survival | 236 | 0.011486 | 0.23 [0.08–0.72] |
Co-expression of TRIM13 gene
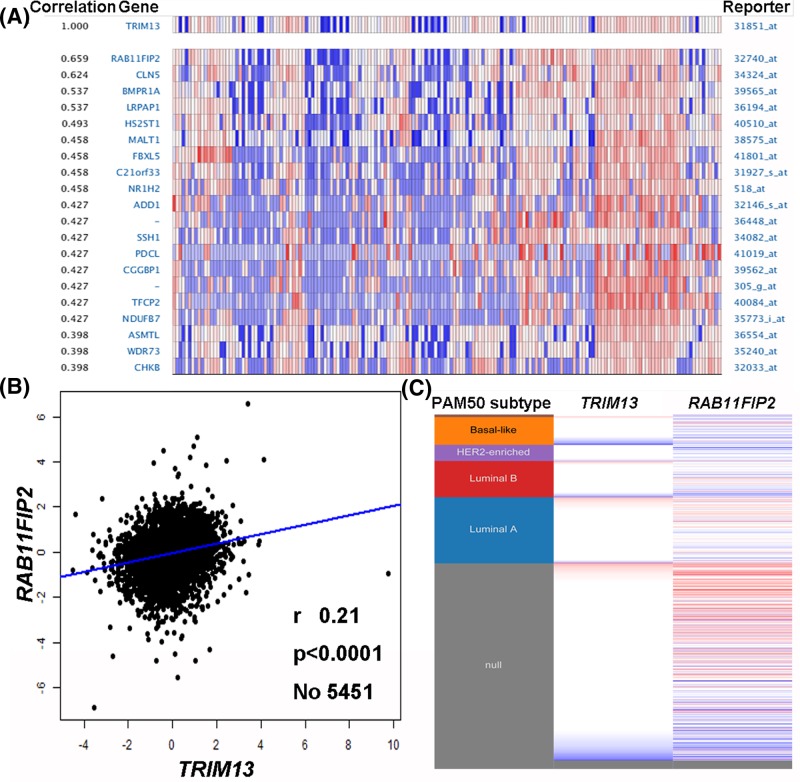
We finally checked the co-expression of TRIM13 gene using the Oncomine database. The co-expression profile of TRIM13 was identified with a large cluster of 8603 genes across 174 cancer samples including 26 breast carcinomas (Figure 5A). Data mining using the bc-GenExMiner software revealed a positive correlation between TRIM13 and RAB11FIP2 expression (Figure 5B). After analyzing breast cancer patient data in the TCGA database using the UCSC Xena web-based tool, we also confirmed a positive correlation between TRIM13 and RAB11FIP2 expression, as shown in the heat map (Figure 5C).
Figure 5. Co-expression of TRIM13 gene.
(A) TRIM13 co-expression of genes identified using the Oncomine database. (B) Correlation between TRIM13 and RAB11FIP2 expression in breast cancer analyzed using the bc-GenExMiner software. (C) Heat map of TRIM13 and RAB11FIP2 expression across a 50-gene qPCR assay (PAM50) breast cancer subtypes in the TCGA database generated using the UCSC Xena web-based tool.
Discussion
TRIM13, a member of the TRIM family, has been reported to play a role in diverse cellular functions such as cell growth, immunity, and tumorigenesis including lipoma, leukemia, and myeloma [9–13,26,27]. TRIM13 over-expression caused increased stability of p53 and decreased activity of Akt kinase, resulting in induction of apoptosis [17]. However, the significance of TRIM13 expression in the development and prognosis of breast cancer remains largely unclear. To the best of our knowledge, this is the first study to identify TRIM13 as a potential predictive biomarker for prognosis of breast cancer.
In the present work, we performed a bioinformatics analysis of the clinical parameters and survival data as it relates to TRIM13 in breast cancer patients by pooling and analyzing several online tools. Oncomine database revealed that TRIM13 was lower-expressed in invasive ductal and invasive lobular breast carcinoma, invasive lobular breast carcinoma, invasive ductal breast carcinoma, tubular breast carcinoma, mucinous carcinoma, ductal breast carcinoma in situ, breast carcinoma, invasive breast carcinoma, and medullary carcinoma with respect to normal tissues. For HER-2 criterion, there was no significant difference between positive and negative group. ER and PR status were positively correlated with TRIM13 level. Conversely, SBR grade, NPI index, nodal status, basal-like status, and triple-negative status were negatively related to TRIM13 expression in breast cancer patients in respect to normal individuals. Generally, breast cancer patients with ER or PR positive, nodal negative, non-basal-like or non-triple-negative status have a preferable outcome. Therefore, these results indicated that high expression of TRIM13 may predict a better prognosis.
We further investigated the prognostic value of TRIM13 in breast cancer using the PrognoScan and bc-GenExMiner software. The pooled results showed that lower TRIM13 expression correlated with worse distant metastasis free survival, relapse free survival, disease specific survival, and metastatic relapse free survival. Such findings are in agreement with the notion of TRIM13 as a tumor suppressor gene and a useful predictive biomarker for prognosis of breast cancer. We finally checked the co-expression of TRIM13 gene using the Oncomine, bc-GenExMiner, and UCSC Xena web-based tools and found that RAB11FIP2 was positively correlated with TRIM13 expression. RAB11FIP2, a member of the RAB11 family of interacting proteins, exhibited potential tumor suppressor function since blocking of the miR-192/215-RAB11FIP2 axis could inhibit gastric cancer progression [28]. This observation, along with our findings of TRIM13 in survival information, provides evidence that TRIM13 gene might inhibit tumor migration and invasion associated with RAB11FIP2 expression.
In conclusion, the present work suggests that TRIM13 was lower-expressed in different subtypes of breast cancer compared with normal tissues and was associated with several clinical parameters. TRIM13 could be adopted as a promising predictive biomarker for prognosis of breast cancer with co-expressed RAB11FIP2 gene. More in-depth experiments and clinical trials are needed to validate the value of TRIM13 in breast cancer treatment.
Abbreviations
- ER
estrogen receptor
- HER2
epidermal growth factor receptor-2
- NPI
Nottingham prognostic index
- PR
progesterone receptor
- SBR
Scarff–Bloom–Richardson
- TRIM
Tripartite motif 13
Funding
This work was supported by grants from the Natural Science Foundation of China [grant number 81702591], and the Natural Science Foundation of Jiangsu Province [grant number BK20170294].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
W.C. and L.C. conceived and designed the experiments. W.C. and L.X. analyzed the data. Q.Q. and Y.Z. contributed analysis tools. W.C. wrote the paper.
References
- 1.Fan L., Strasser-Weippl K., Li J.J., St Louis J., Finkelstein D.M. and Yu K.D. (2014) Breast cancer in China. Lancet Oncol. 15, e279–e289 10.1016/S1470-2045(13)70567-9 [DOI] [PubMed] [Google Scholar]
- 2.Duffy M.J., Walsh S., McDermott E.W. and Crown J. (2015) Biomarkers in breast cancer: where are we and where are we going? Adv. Clin. Chem. 71, 1–23 10.1016/bs.acc.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 3.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S. and Luzi L. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatakeyama S. (2011) TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804 10.1038/nrc3139 [DOI] [PubMed] [Google Scholar]
- 5.Dai H.Y., Ma Y., Da Z. and Hou X.M. (2018) Knockdown of TRIM66 inhibits malignant behavior and epithelial-mesenchymal transition in non-small cell lung cancer. Pathol. Res. Pract. 214, 1130–1135 10.1016/j.prp.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 6.Kimura N., Yamada Y., Takayama K.I., Fujimura T., Takahashi S. and Kume H. (2018) Androgen-responsive TRIM36 enhances tumor-suppressive effect by regulating apoptosis-related pathway in prostate cancer. Cancer Sci. 12, 3840–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatakeyama S. (2017) TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 42, 297–311 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 8.Crawford L.J., Johnston C.K. and Irvine A.E. (2018) TRIM proteins in blood cancers. J. Cell Commun. Signaling 12, 21–29 10.1007/s12079-017-0423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapanadze B., Kashuba V., Baranova A., Rasool O., van Everdink W. and Liu Y. (1998) A cosmid and cDNA fine physical map of a human chromosome 13q14 region frequently lost in B-cell chronic lymphocytic leukemia and identification of a new putative tumor suppressor gene, Leu5. FEBS Lett. 426, 266–270 10.1016/S0014-5793(98)00357-3 [DOI] [PubMed] [Google Scholar]
- 10.Baranova A., Hammarsund M., Ivanov D., Skoblov M., Sangfelt O. and Corcoran M. (2003) Distinct organization of the candidate tumor suppressor gene RFP2 in human and mouse: multiple mRNA isoforms in both species- and human-specific antisense transcript RFP2OS. Gene 321, 103–112 10.1016/j.gene.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 11.Tyybakinoja A., Vilpo J. and Knuutila S. (2007) High-resolution oligonucleotide array-CGH pinpoints genes involved in cryptic losses in chronic lymphocytic leukemia. Cytogenet. Genome Res. 118, 8–12 10.1159/000106435 [DOI] [PubMed] [Google Scholar]
- 12.Panagopoulos I., Gorunova L., Lobmaier I., Andersen H.K., Bjerkehagen B. and Heim S. (2017) Cytogenetic analysis of a pseudoangiomatous pleomorphic/spindle cell lipoma. Anticancer Res. 37, 2219–2223 10.21873/anticanres.11557 [DOI] [PubMed] [Google Scholar]
- 13.Gatt M.E., Takada K., Mani M., Lerner M., Pick M. and Hideshima T. (2013) TRIM13 (RFP2) downregulation decreases tumour cell growth in multiple myeloma through inhibition of NF Kappa B pathway and proteasome activity. Br. J. Haematol. 162, 210–220 10.1111/bjh.12365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lerner M., Corcoran M., Cepeda D., Nielsen M.L., Zubarev R. and Ponten F. (2007) The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol. Biol. Cell 18, 1670–1682 10.1091/mbc.e06-03-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomar D., Singh R., Singh A.K., Pandya C.D. and Singh R. (2012) TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim. Biophys. Acta 1823, 316–326 10.1016/j.bbamcr.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 16.Tomar D., Prajapati P., Sripada L., Singh K., Singh R. and Singh A.K. (2013) TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim. Biophys. Acta. 1833, 3134–3144 10.1016/j.bbamcr.2013.08.021 [DOI] [PubMed] [Google Scholar]
- 17.Joo H.M., Kim J.Y., Jeong J.B., Seong K.M., Nam S.Y. and Yang K.H. (2011) Ret finger protein 2 enhances ionizing radiation-induced apoptosis via degradation of AKT and MDM2. Eur. J. Cell Biol. 90, 420–431 10.1016/j.ejcb.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 18.Altier C., Garcia-Caballero A., Simms B., You H., Chen L. and Walcher J. (2011) The Cavbeta subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat. Neurosci. 14, 173–180 10.1038/nn.2712 [DOI] [PubMed] [Google Scholar]
- 19.Huang B., Pei H.Z., Chang H.W. and Baek S.H. (2018) The E3 ubiquitin ligase Trim13 regulates Nur77 stability via casein kinase 2alpha. Sci. Rep. 8, 13895 10.1038/s41598-018-32391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R. and Ghosh D. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jezequel P., Campone M., Gouraud W., Guerin-Charbonnel C., Leux C. and Ricolleau G. (2012) bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 131, 765–775 10.1007/s10549-011-1457-7 [DOI] [PubMed] [Google Scholar]
- 22.Jezequel P., Frenel J.S., Campion L., Guerin-Charbonnel C., Gouraud W. and Ricolleau G. (2013) bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database 2013, bas060 10.1093/database/bas060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno H., Kitada K., Nakai K. and Sarai A. (2009) PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genet. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Doussal V., Tubiana-Hulin M., Friedman S., Hacene K., Spyratos F. and Brunet M. (1989) Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer 64, 1914–1921 [DOI] [PubMed] [Google Scholar]
- 25.Haybittle J.L., Blamey R.W., Elston C.W., Johnson J., Doyle P.J. and Campbell F.C. (1982) A prognostic index in primary breast cancer. Br. J. Cancer 45, 361–366 10.1038/bjc.1982.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B. and Baek S.H. (2017) Trim13 potentiates toll-like receptor 2-mediated nuclear factor kappaB activation via K29-linked polyubiquitination of tumor necrosis factor receptor-associated factor 6. Mol. Pharmacol. 91, 307–316 10.1124/mol.116.106716 [DOI] [PubMed] [Google Scholar]
- 27.Hatchi E.M., Poalas K., Cordeiro N., N’Debi M., Gavard J. and Bidere N. (2014) Participation of the E3-ligase TRIM13 in NF-kappaB p65 activation and NFAT-dependent activation of c-Rel upon T-cell receptor engagement. Int. J. Biochem. Cell Biol. 54, 217–222 10.1016/j.biocel.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Peng Y., Huang Y., Deng S., Feng X. and Hou G. (2018) Inhibition of the miR-192/215-Rab11-FIP2 axis suppresses human gastric cancer progression. Cell Death Dis. 9, 778 10.1038/s41419-018-0785-5 [DOI] [PMC free article] [PubMed] [Google Scholar]