Abstract
Background
Currently, diagnosis of equine coronavirus (ECoV) relies on the exclusion of other infectious causes of enteric disease along with molecular detection of ECoV in feces or tissue. Although this approach is complete, it is costly and may not always be achievable.
Objective
We hypothesized that the overall fecal shedding of ECoV in hospitalized horses is low. Our objective was to determine whether systemically healthy horses and horses with gastrointestinal disorders shed ECoV in their feces at the time of admission to a referral hospital and after 48 hours of stress associated with hospitalization.
Animals
One‐hundred thirty adult horses admitted to the Washington State University Veterinary Teaching Hospital for gastrointestinal disease (n = 65) or for imaging under anesthesia (n = 65) that were hospitalized for 48 hours. Owner consent was obtained before sampling.
Methods
Fecal samples were collected at admission and 48 hours later. Polymerase chain reaction (PCR) for ECoV and electron microscopy (EM) were performed on all samples.
Results
Only 1 of 258 fecal samples was PCR‐positive for ECoV. Electron microscopy identified ECoV‐like particles in 9 of 258 samples, parvovirus‐like particles in 4 of 258 samples, and rotavirus‐like particles in 1 of 258 samples.
Conclusions and Clinical Importance
The presence of ECoV in feces of hospitalized adult horses was low. Thus, fecal samples that are PCR‐positive for ECoV in adult horses that have clinical signs consistent with this viral infection are likely to be of diagnostic relevance. The clinical relevance of the viruses observed using EM remains to be investigated.
Keywords: anesthesia, anorexia, electron microscopy, fever, gastrointestinal disease, lethargy, PCR
Abbreviations
- CT
computed tomography
- DI
deionized
- ECoV
equine coronavirus
- EM
electron microscopy
- MRI
magnetic resonance imaging
- PCR
polymerase chain reaction
- RNA
ribonucleic acid
- VTH
Veterinary Teaching Hospital
- WSU
Washington State University
1. INTRODUCTION
Equine coronavirus (ECoV), a member of the Coronaviridae family, is a single‐stranded, positive sense, enveloped ribonucleic acid (RNA) virus associated with outbreaks of enteric disease in adult horses.1, 2 Infection with ECoV reportedly causes fever, lethargy, colic, or diarrhea of variable severity in adult horses.1, 3 Hyperammonemic encephalopathy also has been reported.3 Morbidity and mortality range from 10% to 83%1, 2, 3, 4 and from 7% to 27%,1, 2, 3, 4 respectively. Historically, electron microscopy (EM) was considered the gold standard for diagnosis of coronavirus in feces.5, 6 Because of the expertise and equipment required for this technique and its inability to characterize viruses beyond the family level, EM has been augmented in clinical settings by real‐time polymerase chain reaction (PCR) as this test provides a sensitive and specific diagnostic tool to document the presence of ECoV in feces of horses.1
Epidemiological information regarding ECoV is limited,7 and as a result, definitive diagnosis of ECoV in adult horses remains challenging. It is well established that clinically affected horses shed ECoV in feces for 2‐5 weeks after infection, 3, 7, 8 although longer periods have been described.8 The virus also is demonstrable in feces of unaffected horses during outbreaks, suggesting that inapparent carrier horses or transient shedding of ECoV may occur.1, 3 Coronaviral‐induced disease is associated with stress in other adult large animals.9, 10, 11, 12 Subclinical carriers and fecal viral load appeared to be minimal in other continents,13, 14 and seroprevalence in the United States was reported to be low (<10%).5 However; the incidence of fecal viral shedding in healthy, unexposed horses in the United States remains unknown. As a result, ECoV‐positive fecal samples are difficult to interpret on their own. Thus, the diagnosis of ECoV infection currently relies on the exclusion of other infectious causes of enteric disease in horses with clinical signs of ECoV along with molecular detection of ECoV in feces or tissue.3, 7 Although this approach is complete, it poses an additional financial burden to the horse's owner. In addition, with this approach, diagnosis may not be achievable in places where testing for multiple equine enteric pathogens is not readily available. Thus, our aim was to determine whether systemically healthy horses and horses with gastrointestinal disorders shed ECoV in their feces at the time of admission to a referral hospital and after 48 hours of stress associated with hospitalization. Based on the fact that ECoV outbreaks have not been reported in referral hospitals, we hypothesized that the overall fecal shedding of ECoV in hospitalized horses would be low. Fecal samples from horses presented to the Washington State University Veterinary Teaching Hospital (WSU‐VTH) for evaluation of gastrointestinal diseases or those that were anesthetized for imaging were collected for this prospective study.
2. MATERIALS AND METHODS
2.1. Study design
Prospective, Descriptive study.
2.2. Animals
Adult horses (>1 year of age) that were presented to the WSU‐VTH with clinical signs of gastrointestinal tract disease localized aboral to the stomach and hospitalized for a minimum of 48 hours were included in the Gastrointestinal Disease group (n = 65). Similarly, adult horses (without clinical signs of gastrointestinal tract disease) that presented to the WSU‐VTH for diagnostic imaging under anesthesia and were hospitalized for a minimum of 48 hours were included in the Anesthesia group (n = 65). Client consent was obtained from owners before fecal samples were collected. The experimental protocol was reviewed and approved by the Animal Care and Use Committee at WSU‐VTH (ASAF# 04793‐001).
2.3. Samples
Sample collection was performed from January 2016 through July 2018. Two fecal samples were collected from each horse. The first sample was collected as soon as a sample was available after admission to the VTH and the second sample 48 hours after collection of the first sample. Fecal samples were collected from the stall floor shortly after each horse defecated, were placed in a sterile container and immediately stored at 4°C until shipped to the Real‐time PCR Research and Diagnostic Core facilities at the University of California, Davis, California, for ECoV RT‐PCR or to the North Dakota State University Veterinary Diagnostic Laboratory, Fargo, North Dakota, for electron microscopic (EM) analysis. All samples were refrigerated (ice packs) and shipped overnight to the respective laboratories. The maximum storage time of samples at 4°C was 6 days. Samples were collected at 2 different times to improve chances of viral identification and to evaluate the possible effect of the stress of hospitalization and anesthesia on viral shedding in feces.7
2.4. Polymerase chain reaction
Fecal samples were processed at the Real‐time PCR Research and Diagnostic Core facilities at the University of California, Davis, California. A previously described ECoV PCR technique routinely performed at the laboratory was used 1, 6 Briefly, 200 μg of feces and 2 mL of phosphate buffer solution were vortexed and centrifuged. Nucleic acid purification from 200 μL of supernatant was performed using a semi‐automated nucleic acid extraction system (QIAamp 96 DNA QIAcube HT Kit; QIAcube, Qiagen Valencia, California) according to the manufacturer's instructions and eluted in 100 μL of diethylpyrocarbonate‐treated water. Following the manufacturer's recommendations, RNA was converted to complementary DNA as previously described.6 Equine coronavirus PCR was performed based on a specific 142 base‐pair product of the N gene of ECoV (GenBank accession number EF446615).1 All samples then were amplified in a combined thermocycler/fluorometer (7900 HT Fast; Applied Biosystems, Foster City, California) using a standard thermal cycling protocol.1 A real‐time PCR assay targeting a universal sequence of the bacterial 16S rRNA gene was used as quality control and as an indicator of fecal inhibition.1, 15 A biological positive control (extracted feces from a horse with ECoV infection confirmed by immunohistochemistry and histology) as well as a negative control (sterile water) were included in each run.
2.5. Electron microscopy
Electron microscopic evaluation was performed at the North Dakota State University Veterinary Diagnostic Laboratory, Fargo, North Dakota, as previously described.16 Briefly, fecal material was mixed with 3‐9 times its volume of deionized (DI) water to obtain the desired consistency. The suspension was allowed to settle and cleared by low‐speed and then high‐speed centrifugation. Pellets were resuspended in DI water, placed on 400‐mesh, carbon‐stabilized grids and negatively stained for 30‐60 seconds with 2% phosphotungstic acid solution. The grids were examined on a JEOL JEM‐100CX II transmission electron microscope and evaluated by an experienced electron microscopist.
2.6. Statistical analysis
Data were analyzed using commercial software (SigmaPlot; SPSS, Chicago, Illinois). Normality of the data and equality of variances were assessed using the Shapiro‐Wilk and Levene's tests, respectively. Differences in breed and age by group were assessed using 2‐way repeated measures analysis of variance; multiple comparisons were performed using the Holm‐Sidak method. Significance was set at P < .05. Clinical problem and season was summarized in a contingency table.
3. RESULTS
During the scheduled 2‐year collection period (January 2016 through December 2017), 1847 cases were seen at the WSU‐VTH and 65 horses met the inclusion criteria for the Gastrointestinal Disease group. In order to collect the same number of horses in the Anesthesia group, the collection period had to be extended through May 2018 for this second group. The total number of horses seen by the VTH in that 2.5‐year period was 2174.
The most common horse breeds in the study were Quarter Horses (54.6%), Warmblood breeds (14%), and Thoroughbred horses (8.4%). Other less commonly represented breeds included pony and draft breeds, Arabians and miniature horses. There were no significant differences in the number of horses of each breed by group but the number of Quarter Horses was significantly higher than the rest of the other breeds (P < .001) in both groups. The age of hospitalized horses included in the study ranged from 13 months to 24 years. Horses in the Anesthesia group were significantly (P = .001) younger (median, 8 years; range, 13 months to19 years) than horses in the Gastrointestinal Disease group (median, 10 years; range, 2‐24 years). There were no significant differences in the sex of horses admitted into each group. Overall, geldings accounted for 42% of the total admissions and were significantly more common than stallions (P = .006) and mares (P = .03). Mares also were significantly more common than stallions (P = .006) in both groups.
For the total period of study, the largest percentage of horses was admitted during fall followed by spring (Figure 1).
Figure 1.
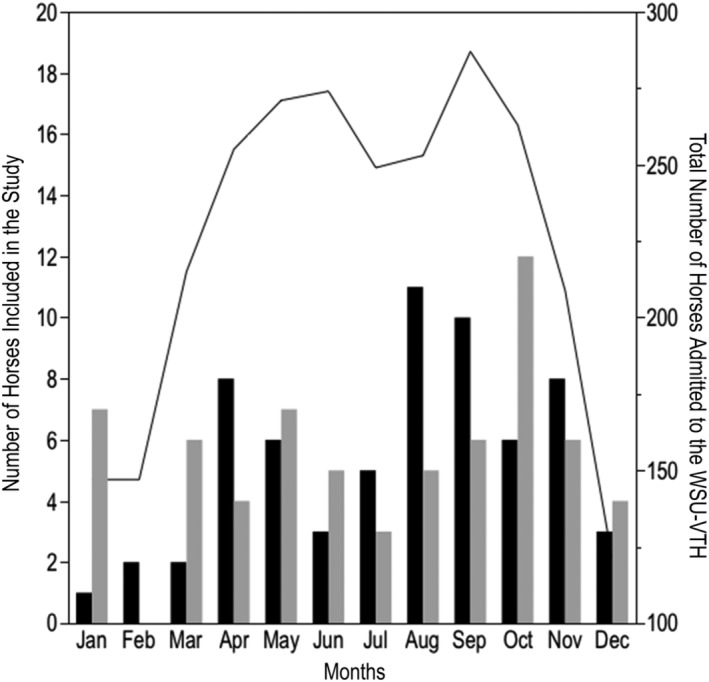
Monthly distribution of the total number of horses admitted to the VTH‐WSU (solid line, right y‐axis) and the number of horses included in the study (bars, left y‐axis) in the Gastrointestinal Disease group (black bars) and in the Anesthesia group (gray bar)
The most common cause of hospitalization for horses in the Gastrointestinal Disease group was colic of intestinal origin. Large colon conditions that required medical (impaction, displacement) or surgical (impaction, displacement, and volvulus) treatment were the most common causes of colic. Whenever the cause of colic could not be confirmed, the case was classified as “Colic‐Unknown.” Colitis (thickened large colon wall on ultrasonographic evaluation, neutropenia, and hypoalbuminemia), diarrhea (marked increase in frequency and volume of manure), and liver disease (increased liver enzyme activities and bile acid concentrations, lethargy, and fever) also were diagnosed several times. Other conditions that were not commonly diagnosed (fever localized to the abdomen, small intestinal or small colon pathology, gastrointestinal neoplasia) were grouped under a single category labeled “Other.” In the Anesthesia group, horses were classified based on the imaging modality used. Thus, horses were classified as having a computed tomography (CT) of the limbs, magnetic resonance imaging (MRI) of the limbs, CT‐MRI of the limbs (limbs imaged using both modalities during the same anesthetic period), or CT of the skull or a myelogram. Clinical diagnoses/lesion localization by group are shown in Table 1.
Table 1.
Clinical diagnosis/lesion localization of horses included in the Gastrointestinal Disease (n = 65) and Anesthesia (n = 65) groups (Total number of horses in each category (n) and percentage (%) of the total group are shown.)
| Gastrointestinal Disease group | n (%) | Anesthesia group | n (%) |
|---|---|---|---|
| Large colon: medical | 26 (40) | Navicular bone/bursa | 10 (15.4) |
| Large colon: surgical | 8 (12.3) | Deep digital tendon | 8 (12.3) |
| Colitis | 8 (12.3) | Third phalanx | 7 (10.8) |
| Liver disease | 2 (3.1) | Suspensory ligament | 5 (7.7) |
| Diarrhea | 2 (3.1) | Sinus | 5 (7.7) |
| Other | 8 (12.3) | Tarsus | 4 (5.4) |
| Colic: unknown | 11 (16.9) | Sesamoidean ligaments | 4 (5.4) |
| Other | 17 (26.2) | ||
| Undiagnosed | 5 (7.7) | ||
3.1. Polymerase chain reaction
Only 1 of the 130 fecal samples collected for the Gastrointestinal Disease group was positive for ECoV. None of the 130 fecal samples collected in the Anesthesia group was positive. The positive sample belonged to an 8‐year‐old Miniature Horse gelding that presented to the WSU‐VTH for fever and anorexia of 6 day's duration. The horse was admitted during summer (end of July) and was discharged after 20 days of hospitalization with a diagnosis of colitis and hepatic lipidosis secondary to prolonged anorexia. The gelding developed diarrhea and hypoproteinemia 3 days after admission, and both resolved within a week of medical treatment. Neutropenia was not observed. The admission fecal sample was negative for ECoV. In contrast, the sample collected at 48 hours was positive. Five fecal samples submitted for Salmonella culture were negative.
3.2. Electron Microscopy
Direct EM investigation of negatively stained samples was performed in all but 1 horse from the Gastrointestinal Disease group (because its fecal samples were accidently left out of the refrigerator for a prolonged period of time). Viral particles with coronavirus‐like characteristics were identified in 9 of 258 fecal samples (3.4%). Four of these samples belonged to horses in the Gastrointestinal Disease group: 2 were from horses that presented with large colon medical conditions and 2 from one horse with liver disease. The first 2 horses had viral particles only in the samples collected 48 hours after admission; the initial fecal samples were negative. The horse with liver disease had viral particles present in the admission sample and 48 hours later. The additional 5 fecal samples with coronavirus‐like particles were admitted for CT of the limbs (2), MRI of the limbs (2), and myelogram (1). Viral particles were observed only in the samples collected at admission in both horses undergoing MRI and only in the 48 hours sample in the remaining horses. In addition, 4 horses (3 in the Gastrointestinal Disease group and 1 in the Anesthesia group) had parvovirus‐like viral particles and 1 horse (Gastrointestinal Disease group) had a rotavirus‐like viral particle. All these additional viral particles were observed only in the fecal samples collected 48 hours after admission.
4. DISCUSSION
Our results confirmed our hypothesis that shedding of ECoV in the selected population of hospitalized horses is low. Although fever, lethargy, and anorexia are the most common clinical signs of ECoV infection,1, 7 these are nonspecific clinical signs that can be present as a result of many different conditions. Thus, we included horses that were hospitalized for gastrointestinal disease because colic, enteritis, and diarrhea have been reported with this infection.3, 4, 17, 18 Horses in the Anesthesia group were included to evaluate the effect that stress (hospitalization, anesthesia) may have had on fecal shedding of ECoV in systemically healthy horses. Only horses that were anesthetized for imaging (CT, MRI, or myelogram) were included in this group to maintain homogeneity of the stressor (hospitalization and anesthesia). Apart from their presenting complaints (eg, lameness, sinus problem, spinal ataxia), all of these horses were systemically healthy.
Fecal samples were selected because ECoV can be shed consistently in feces of affected and exposed horses.2, 7 Samples were collected upon admission and after 48 hours of hospitalization to increase chances of viral detection because shedding may be low during the peracute stages of infection or may be delayed because of gastrointestinal tract stasis.1 In addition, serial sampling was performed to evaluate the role of stress in ECoV fecal shedding, because stress has been shown to affect shedding of other equine pathogens.19, 20, 21 A >48‐hour sampling period between samples was not feasible, because most horses undergoing anesthesia for imaging purposes were hospitalized for ≤2 days.
One hundred thirty horses were included in the study. After the breed distribution of the WSU‐VTH equine admissions, Quarter Horse was the most common breed included in our study. Although they made up for almost half of the sampled horses, many other breeds (Warmblood breeds, Thoroughbreds, Arabians, Draft breeds, Miniature horses, Mustangs, Morgans, and Pony breeds) were included in the study, providing a varied breed representation. The number of stallions was significantly lower than the other sex and reflects the equine population routinely admitted to our hospital. A wide range of ages was included because there appears to be no age predilection for this infection in adult horses.5 Horses in the Anesthesia group were significantly younger than horses in the Gastrointestinal Disease group. This is likely because horses undergoing limb imaging usually are actively performing and therefore are younger. As expected, based on the normal distribution of admissions to our hospital, most horses included in the study were admitted during fall and spring. A 2‐year sample period was used to account for year‐to‐year viral shedding variation that may have occurred and to include a larger number of horses. In addition, this period allowed us to evaluate multiple seasons. Although colder months (October to April) appear to be associated with a higher incidence of infection,7 the only PCR‐positive horse with clinical signs consistent with ECoV was admitted during summer indicating that this disease may occur any time of the year and supporting the sporadic nature of this viral infection reported by others.7
Fecal samples were tested using a previously described ECoV real‐time PCR technique that has been shown to provide a sensitive and specific diagnostic tool to document the presence of ECoV in feces of horses.1, 6 Polymerase chain reaction assays are considered highly sensitive diagnostic techniques, but their sensitivity may be decreased by the presence of PCR amplification inhibitory factors in feces, viral strain differences that affect primer specificity, and lack of optimization or consistency in protocols.15, 16 To account for inhibition, our study included an internal target (a universal bacterial gene). Hence, all negative results were considered truly negative. In addition to PCR, all of the fecal samples collected in the study also were analyzed using EM. Electron microscopy historically has been considered a gold standard for diagnosis of viral presence in feces because of the very low number of false positives associated with this technique.5, 6 Although the use of EM for viral diagnosis does not require organism‐specific reagents, the equipment and expertise for this assay are not widely available. In addition, EM may not be useful if viral particles are not present in sufficient numbers7 and can only identify viruses to the family level.
As we hypothesized, shedding of ECoV in the specific hospitalized population evaluated in our study was very low. Only 1 of 258 fecal samples from 130 horses tested in our study was PCR positive for ECoV. The positive horse, an 8‐year‐old Miniature Horse gelding, was admitted for fever and anorexia of 6 days' duration, and ultimately was diagnosed with colitis. These clinical signs are consistent with ECoV infection.7 Salmonella infection was ruled out based on serial fecal culture. Tests for Clostridia (spp. difficile and perfringens) were not performed.
Coronavirus‐like viral particles were identified by EM in 9 of 258 fecal samples from 129 horses. Interestingly, none of these horses was PCR‐positive for ECoV, suggesting the presence of a novel ECoV/coronavirus‐like virus. Further identification of these organisms using deep sequencing would be of interest, but was beyond the objectives of our study. Electron microscopy also identified parvovirus‐like viral particles in the feces of 4 horses (3 admitted for colic and 1 for CT of the limbs) and rotavirus‐like viral particles in an additional horse admitted for liver disease. The finding of additional viruses was not unexpected. Parvovirus‐like particles are not uncommon in adult horses.22 Interestingly, all of these viral particles were identified only in the samples obtained 48 hours after hospital admission. Stress has been shown to affect the shedding of other horse pathogens.19, 20, 21 Therefore, it is possible that the stress of hospitalization increased the shedding of these viruses because the initial samples were negative. The clinical relevance of these viruses remains to be determined.
Although our results support the overall low prevalence of ECoV in healthy horses reported by others,5, 7, 13, 14 all horses included in the study were from the Pacific Northwest of the United States. Thus, these results may not be readily extrapolated to equine populations in other geographic areas. Moreover, our results should not be extrapolated to foals, where shedding of ECoV infection appears to be more common.23
In summary, we evaluated the prevalence of ECoV in the feces of clinically healthy adult horses hospitalized for diagnostic imaging or in horses hospitalized for gastrointestinal disease and evaluated the effect of the stress of hospitalization on fecal viral shedding of ECoV. The prevalence of ECoV in feces of the included adult horses was low and supports the findings by others.5, 7 Thus, fecal samples that are PCR‐positive for ECoV in adult horses that have clinical signs consistent with this viral infection (fever, anorexia, colic, diarrhea)2 are likely to be of diagnostic relevance. Although additional testing of other enteric pathogens provides useful information about potential coinfections, our data along with previous work5 suggests that such tests may not always be required to obtain a definitive diagnosis of ECoV infection.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
The experimental protocol was reviewed and approved by the Animal Care and Use Committee at WSU (ASAF# 04793‐001). Owner consent was obtained prior to sampling.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
ACKNOWLEDGMENT
This study was funded by the Boehringer Ingelheim Advancement in Equine Research Award.
Sanz MG, Kwon SY, Pusterla N, Gold JR, Bain F, Evermann J. Evaluation of equine coronavirus fecal shedding among hospitalized horses. J Vet Intern Med. 2019;33:918–922. 10.1111/jvim.15449
Present address Dr Bain, Merck Animal Health, 2 Giralda Farms, Madison, NJ 07940. Email: fairfield.bain@merck.com
Funding information Boehringer Ingelheim
REFERENCES
- 1. Pusterla N, Mapes S, Wademan C, et al. Emerging outbreaks associated with equine coronavirus in adult horses. Vet Microbiol. 2013;162:228‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oue Y, Morita Y, Kondo T, et al. Epidemic of equine coronavirus at Obihiro Racecourse, Hokkaido, Japan in 2012. J Vet Med Sci. 2013;75:1261‐1265. [DOI] [PubMed] [Google Scholar]
- 3. Fielding CL, Higgins JK, Higgins JC, et al. Disease associated with equine coronavirus infection and high case fatality rate. J Vet Intern Med. 2015;29:307‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oue Y, Ishihara R, Edamatsu H, et al. Isolation of an equine coronavirus from adult horses with pyrogenic and enteric disease and its antigenic and genomic characterization in comparison with the NC99 strain. Vet Microbiol. 2011;150:41‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kooijman LJ, James K, Mapes SM, Theelen MJP, Pusterla N. Seroprevalence and risk factors for infection with equine coronavirus in healthy horses in the USA. Vet J. 2017;220:91‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pusterla N, Kass PH, Mapes S, et al. Surveillance programme for important equine infectious respiratory pathogens in the USA. Vet Rec. 2011;169:12. [DOI] [PubMed] [Google Scholar]
- 7. Pusterla N, Vin R, Leutengegger C, et al. Equine coronavirus: an emerging enteric virus of adult horses. Equine Vet Educ. 2016;28:216‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodrich EL, Mittel LD, Glaser A, Ness SL, Radcliffe RM, Divers TJ. Novel findings from a beta coronavirus outbreak on an American Miniature Horse breeding farm in upstate New York. Equine Vet Educ. 10.1111/eve.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract. 2010;26:123‐146. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Genova SG, Streeter RN, Simpson KM, Kapil S. Detection of an antigenic group 2 coronavirus in an adult alpaca with enteritis. Clin Vaccine Immunol. 2008;15:1629‐1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kapil S, Yeary T, Evermann JF. Viral diseases of new world camelids. Vet Clin North Am Food Anim Pract. 2009;25:323‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins JK, Riegel CA, Olson JD, Fountain A. Shedding of enteric coronavirus in adult cattle. Am J Vet Res. 1987;48:361‐365. [PubMed] [Google Scholar]
- 13. Hemida MG, Chu DKW, Perera R, et al. Coronavirus infections in horses in Saudi Arabia and Oman. Transbound Emerg Dis. 2017;64:2093‐2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miszczak F, Tesson V, Kin N, et al. First detection of equine coronavirus (ECoV) in Europe. Vet Microbiol. 2014;171:206‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mapes S, Rhodes DM, Wilson WD, Leutenegger CM, Pusterla N. Comparison of five real‐time PCR assays for detecting virulence genes in isolates of Escherichia coli from septicaemic neonatal foals. Vet Rec. 2007;161:716‐718. [DOI] [PubMed] [Google Scholar]
- 16. Hazelton PR, Gelderblom HR. Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg Infect Dis. 2003;9:294‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giannitti F, Diab S, Mete A, et al. Necrotizing enteritis and hyperammonemic encephalopathy associated with equine coronavirus infection in equids. Vet Pathol. 2015;52:1148‐1156. [DOI] [PubMed] [Google Scholar]
- 18. Huang JC, Wright SL, Shipley WD. Isolation of coronavirus‐like agent from horses suffering from acute equine diarrhoea syndrome. Vet Rec. 1983;113:262‐263. [DOI] [PubMed] [Google Scholar]
- 19. Traub‐Dargatz JL, Salman MD, Jones RL. Epidemiologic study of salmonellae shedding in the feces of horses and potential risk factors for development of the infection in hospitalized horses. J Am Vet Med Assoc. 1990;196:1617‐1622. [PubMed] [Google Scholar]
- 20. Ward MP, Alinovi CA, Couetil LL, et al. Evaluation of a PCR to detect Salmonella in fecal samples of horses admitted to a veterinary teaching hospital. J Vet Diagn Invest. 2005;17:118‐123. [DOI] [PubMed] [Google Scholar]
- 21. Badenhorst M, Page P, Ganswindt A, Laver P, Guthrie A, Schulman M. Detection of equine herpesvirus‐4 and physiological stress patterns in young Thoroughbreds consigned to a South African auction sale. BMC Vet Res. 2015;11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Divers TJ, Tennant BC, Kumar A, et al. New parvovirus associated with serum hepatitis in horses after inoculation of common biological product. Emerg Infect Dis. 2018;24:303‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slovis NM, Elam J, Estrada M, Leutenegger CM. Infectious agents associated with diarrhoea in neonatal foals in central Kentucky: a comprehensive molecular study. Equine Vet J. 2014;46:311‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]


