Abstract
Background
Ponies are highly susceptible to metabolic derangements including hyperinsulinemia, insulin resistance, and adiposity.
Hypothesis/Objectives
Genetic loci affecting height in ponies have pleiotropic effects on metabolic pathways and increase the susceptibility to equine metabolic syndrome (EMS).
Animals
Two hundred ninety‐four Welsh ponies and 529 horses.
Methods
Retrospective study of horses phenotyped for metabolic traits. Correlations between height and metabolic traits were assessed by Pearson's correlation coefficients. Complementary genome‐wide analysis methods were used to identify a region of interest (ROI) for height and metabolic traits, determine the fraction of heritability contributed by the ROI, and identify candidate genes.
Results
There was an inverse relationship between height and baseline insulin (−0.26) in ponies. Genomic signature of selection and association analyses for both height and insulin identified the same ~1.3 megabase region on chromosome 6 that contained a shared ancestral haplotype between these traits. The ROI contributed ~40% of the heritability for height and ~20% of the heritability for insulin. High‐mobility group AT‐hook 2 was identified as a candidate gene, and Sanger sequencing detected a c.83G>A (p.G28E) variant associated with height in Shetland ponies. In our cohort of ponies, the A allele had a frequency of 0.76, was strongly correlated with height (−0.75), and was low to moderately correlated with metabolic traits including: insulin (0.32), insulin after an oral sugar test (0.25), non‐esterified fatty acids (0.19), and triglyceride (0.22) concentrations.
Conclusions and Clinical Importance
These data have important implications for identifying individuals at risk for EMS.
Keywords: equine metabolic syndrome, genetics, horses, insulin dysregulation
Abbreviations
- AIC
Akaike information criterion
- ANOVA
analysis of variance
- BF
Bayes factor
- bp
base pairs
- EMS
equine metabolic syndrome
- ECA6
equine chromosome 6
- GLU‐OST
glucose after an oral sugar test
- h2SNP
SNP chip heritability
- HMGA2
high‐mobility group AT‐hook 2
- INS‐OST
insulin after an oral sugar test
- IRAK3
interleukin 2 receptor associated kinase 3
- kb
kilobase
- LD
linkage disequilibrium
- Mb
megabase
- MetS
metabolic syndrome in humans
- NCBI
National Center for Biotechnology Information
- NEFA
non‐esterified fatty acids
- OST
oral sugar test
- QC
quality control
- ROI
region of interest
- SNP
single nucleotide polymorphism
1. INTRODUCTION
Equine metabolic syndrome (EMS) describes a clustering of metabolic disturbances including insulin dysregulation (hyperinsulinemia and insulin resistance) and dyslipidemia (elevated triglyceride low‐density lipoprotein concentrations), and generalized obesity and/or regional adiposity (eg, nuchal ligament, tail head).1, 2 Equine metabolic syndrome is an important health concern as affected horses and ponies are predisposed to laminitis.
Ponies (individuals with a wither height less than 58 in.) are more insulin insensitive then large breed horses,3 and metabolic comparisons across breeds have consistently found ponies to be among the more insulin‐resistant groups.4, 5 Unlike many large breed horses, after domestication ponies have maintained a metabolically thrifty phenotype with seasonally adaptive changes including suppressed metabolic rates and excessive fat storage.6 However, the mechanisms underlying ponies' unique metabolic profiles and greater EMS susceptibility have not been identified.
A relationship between individuals of short stature and an increased risk of chronic disease has been well described in humans.7, 8, 9 In particular, there are significant associations between height and the risk of developing Type 2 diabetes or metabolic syndrome (MetS),10, 11, 12, 13, 14, 15 with measured metabolic abnormalities more severe in shorter individuals.10, 11, 16, 17 Many negative correlations between height and specific derangements of the endocrine system include: obesity,16, 18, 19 regional adiposity,14 elevated triglycerides,11, 20 impaired glucose tolerance post oral sugar test (OST),17, 21 and insulin resistance.10, 11, 12, 16 Several underlying mechanisms for these associations have been proposed, including a poor uterine environment, impaired nutrition, adverse social circumstances, and genetic factors.10, 22, 23, 24, 25 The role of genetic factors is supported by the identification of pleiotropic effect between variants within the promoter of the GAD2 gene and low birth weight, decreased length, impaired insulin secretion, and early onset obesity,26 as well as associations between single nucleotide polymorphisms (SNPs) in the LMNA gene with short stature and elevated triglycerides, and obesity and increased waist circumference.27
We hypothesize that loci affecting height could also have pleiotropic effects on metabolic pathways in horses and ponies and increase the risk for EMS. Here we use genomic tools to identify a chromosomal locus associated with both height and fasting insulin concentrations in Welsh ponies and demonstrate that a probable functional mutation in the high mobility group AT‐hook 2 (HMGA2) gene is contributing to both height and metabolic traits.
2. MATERIALS AND METHODS
2.1. Samples
Two hundred ninety‐four Welsh ponies (213 females and 81 males) from 32 farms within the United States were included in the study, with ages ranging from 2 to 33 years (mean age of 11.7 years). As a breed, Welsh ponies are divided into 6 sections based on pedigree and height (see Supporting Information Table 1), which were represented in our cohort as follows: section A (n = 74), section B (n = 146), section C (n = 3), section D (n = 15), section H (n = 26), and unregistered Welsh ponies (n = 10). A total of 529 individuals from 4 large‐breed horses, Quarter horses (n = 59), Arabians (n = 64), Tennessee Walking Horses (n = 48), and Morgan horses (n = 293), as well as 65 horses of other pure or mixed breeds, were also collected. These samples were obtained from farms throughout North America and represented 300 females and 229 males with an age range of 2‐33 years old (mean age of 13 years).
2.2. Phenotype data
Signalment, medical history, height at the withers, and biochemical measurements at baseline and after an OST were collected on all individuals. Baseline measurements and assays included glucose (YSI 2300 STAT Plus glucose and lactate analyzer), insulin (Siemen's TKIN1 Insulin Coat‐A‐Count Kit), ACTH (Siemen's LKAC1 ACTH kits), leptin (Millipore Sigma's XL‐85 K Multi‐Species Leptin RIA), adiponectin (Millipore Sigma's EZHMWA‐64 K Human High Molecular Weight Adiponectin ELISA), triglycerides (Millipore Sigma's TR0100 Serum Triglyceride Determination kit), and non‐esterified fatty acids (NEFA; Wako Diagnostics' HR Series NEFA kit). Oral sugar test measurements comprised insulin (INS‐OST) and glucose (GLU‐OST) levels 75 minutes after oral administration of 0.15 mg/kg Karo light corn syrup.
2.3. Genotype data
Genomic DNA was isolated from whole blood or hair roots per manufacturer recommendations (Puregene Blood Core Kit, Qiagen, Germantown, MD, USA). Welsh ponies were genotyped with either the Axiom Equine MCEc670 (n = 220 Welsh ponies) or MCEc2M (n = 44 Welsh ponies) genotyping arrays, containing 670 805 SNP markers and 2 011 826 SNP markers,28 respectively. For the Welsh ponies not genotyped on the MCEc2M array, Beagle software29, 30 was used to perform genotype imputation and haplotype phasing, using an across‐breed reference population of 516 horses of 14 different breeds, yielding a total of 1 931 311 SNPs.
Quality control (QC) measures were performed on the genotyping data using the PLINK software package.31 This included SNP and individual missingness and genotyping rates, discordant sex information, and abnormally high heterozygosity (≥3 SDs from the mean). All individuals passed QC and were kept in the study cohort. Individual SNPs with a genotyping success rate <90%, minor allele frequency <1.0%, or outside Hardy Weinberg equilibrium were pruned, leaving a total of 1 511 302 SNPs for subsequent analysis.
2.4. FST‐based statistic
Genomic regions of breed‐specific population differentiation were identified in the Welsh ponies using SNP data from the 44 individuals genotyped on the MCEc2M. Calculation of the d i statistic was performed using nonoverlapping 10 kilobase (kb) windows across the 31 equine autosomes with a custom Python script (https://github.com/schae234/PonyTools) based on work previously described.32, 33 The d i statistic detects locus‐specific deviation in allele frequencies for the test population relative to the genome‐wide average of pairwise FST summed across populations. The background population contained 463 individuals from 16 different breeds (Supporting Information Table S2). Significant d i windows were those corresponding to the top 0.1% of the empirical distribution and were considered regions of interest (ROIs) for putative signatures of selection. Two or more contiguous significant d i windows were considered as a single ROI.
2.5. Association analysis
Association analysis for equine chromosome 6 (ECA6; total of 56 246 SNPs) was performed using imputed SNP genotype data from 264 Welsh ponies. Height and EMS traits were treated as quantitative phenotypes. Association analysis was performed using custom code for a mixed linear regression model that included a random polygenic term determined from a genomic relationship matrix calculated from select trait associated SNPs, random herd effect, and fixed covariates sex and age.34 Analysis utilized a combination of the Bayesian Sparse Linear Mixed Model,35 available in the software program Genome‐wide Efficient Mixed Model Association,36 and a linear mixed model implemented in FaST‐LMM37 (additional description provided in Supporting Information Supplemental Methods).
The threshold for genome wide significance was based on the effective number of independent tests for the entire genome (ie, SNPs, after correction for linkage disequilibrium [LD]), as calculated using the Genetic Type 1 Error Calculator.38 The effective number of independent tests was 841 750 resulting in a Bonferroni‐corrected threshold for genome wide significance of 5.9e‐08.
2.6. Estimation of heritability
SNP chip heritability (h2 SNP) for height in Welsh ponies was calculated from the imputed SNP genotype data (n = 264) with the software program Linkage Disequilibrium Adjusted Kinship (LDAK),39, 40 including age, sex, and section as covariates. Two separate techniques were used to estimate the genetic variance explained by our ROI. First, we used genomic partitioning as previously described.39, 41 The second technique fit the top SNPs from the association analysis as covariates in the analysis using LDAK's top‐predictors function. Random subsetting of the data was performed to test the effect of a few cryptically related individuals on the h2 SNP estimates (see Supporting Information Supplemental Methods).
2.7. Haplotype analysis
Local haplotype sharing within the Welsh ponies used for association analysis (n = 264) was calculated from the hapQTL program (http://www.haplotype.org) with default settings.42 This approach relies on a statistical model for LD to infer ancestral haplotypes and their frequencies at each SNP marker for individuals within a population. For each analysis, 1 expectation maximization run was used with 50 steps (‐w 50), 3 upper clusters (‐C 3), 10 lower clusters (‐c 10), and with a prior LD length of 0.5 cM (‐mg 200). Based on recommendations from Xu and Guan (2014), contiguous SNPs with −log10 Bayes factor (BF) >4 were considered significant ROI, and orphan signals were removed from the analysis. Bayes factor values were calculated for each of the 56 740 SNPs on ECA6 using height and baseline insulin as quantitative phenotypes.
2.8. HMGA2 and IRAK3 reconstruction and sequencing
PCR primers were designed for all exons within 2 candidate genes, HMGA2 and interleukin 2 receptor associated kinase 3 (IRAK3), using the Primer3 software.43 Genomic sequences for primer design were retrieved using the National Center for Biotechnology Information (NCBI) Gene tool (https://www.ncbi.nlm.nih.gov/gene); base pair (bp) position of equine exons were confirmed with NCBI's Nucleotide BLAST tool (https://blast.ncbi.nlm.nih.gov/) against the human genome. In some cases, the newly assembled EquCab3 version of the equine genome was queried using a local BLAST tool to confirm exon sequence identity. Details of all HMGA2 and IRAK3 exons, as well as the PCR primer sequences, are presented in Supporting Information Tables 3 and 4.
Genomic DNA from a panel of 51 individuals from 6 different breeds (6 Morgan horses, 6 Arabian horses, 6 Tennessee Walking horses, 12 Quarter horses, 3 Miniature horses, and 18 Welsh ponies) was amplified by standard PCR. The resulting products were submitted to the University of Minnesota Genomics Center for Sanger sequencing after enzymatic cleanup using the ExoSAP‐IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, Massachusetts). Sequencing results were then analyzed, processed, and aligned using the Sequencher software version 5.1 (Gene Codes Corporation, Ann Arbor, Michigan).
2.9. HMGA2 exon 1 variant genotyping
Two methods were employed to genotype the HMGA2 exon 1 mutation (c.83G>A) identified by Frischknecht et al.44 In the first method, standard PCR primers were designed to flank and Sanger sequence this exon (Supporting Information Table 3) in 438 horses, including 150 ponies and 288 large breed horses. In the second method, a TaqMan SNP genotyping assay using the Bio‐Rad CFX96 Real‐Time System was designed as previously described45 and per manufacturer's recommendations (www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_5279.pdf). Results were analyzed with BioRad's CFX Manager Software version 3.1 (see Supporting Information Supplemental Methods for a full description of this assay). Genotypes for this variant using the second genotyping assay were obtained for an additional 144 Welsh ponies and 241 large breed horses.
2.10. Statistical analyses
Statistics were performed using functions within the software package R.46 Metabolic traits were tested for normality using a normal probability plot and a Shapiro test; traits were log or square root transformed when appropriate. Correlations between height and EMS traits (insulin, INS‐OST, glucose, GLU‐OST, NEFA, triglycerides, leptin, adiponectin) and ACTH were calculated using a Pearson's correlation coefficient. After adjusting for multiple testing using a Bonferroni correction (0.05/9), a P‐value of <.005 was considered significant. Analyses were performed as follows: all horses (n = 824), Welsh ponies (n = 294), all large breed horses (n = 529), Quarter horses (n = 59), Arabian horses (n = 64), Morgan horses (n = 293), and Tennessee Walking horses (n = 48). Correlations among genotype for the HMGA2 c83G>A variant and EMS traits, ACTH, or height were calculated for the Welsh ponies (n = 294) using Pearson's correlation coefficient and a Bonferroni‐corrected P‐value (0.05/10; <.005).
Least‐square means were calculated with EMS traits, ACTH, or height as the outcome variable, genotype as the response variable, and age and sex as predictors. The R statistical software package Linear and Nonlinear Mixed Effects Models (nlme)47 fit the linear model using generalized least squares. The R statistical software package Least‐Square Means (lsmeans)48 was used to calculate the predicted marginal means and pair‐wise comparisons.
Model comparison for modes of inheritance between the HMGA2 c.83G>A variant and traits were performed using an analysis of variance (ANOVA) for an additive, dominant, and recessive model. The P‐values of the F‐statistic were compared across all 3 models. The R statistical software package SNPassoc49 was used to calculate the Akaike information criterion (AIC) and P‐value among additive, recessive, dominant, and codominant models. Model selection was based on the lowest AIC values; however, models with less than 10 unit difference between them were considered indistinguishable.
3. RESULTS
3.1. Correlations among height, EMS traits, and ACTH
Correlation analyses between height and biochemical traits in the entire cohort (n = 823) revealed statistically significant inverse correlations for insulin (−0.12), glucose (−0.11), adiponectin (−0.23), and ACTH (−0.12), whereas positive correlations with height were found for triglycerides (0.14) and leptin (0.12) (Table 1). No statistically significant correlations between any of the traits and height were identified in the large breed horses as a whole (n = 529) or within any individual breed (Table 1). However, within the Welsh pony population (n = 294), a statistically significant inverse correlation with height was identified for insulin (−0.26), with the correlation coefficient between height and insulin higher than in the entire population (Table 1), indicating that the pony population was predominately driving the association observed for this trait in the full cohort.
Table 1.
Correlations between height and biochemical traits across breeds
| Breed | INS | INS‐OST | GLU | GLU‐OST | NEFA | TG | LEPTIN | ADIPON | ACTH |
|---|---|---|---|---|---|---|---|---|---|
| All horses (n = 823) | −0.12 (−0.19 to −0.052) | −0.035 (−0.11 to 0.038) | −0.11 (−0.18 to −0.043) | −0.070 (−0.14 to 0.0028) | −0.068 (−0.14 to 0.0097) | 0.14 (0.071 to 0.21) | 0.12 (0.050 to 0.19) | −0.23 (−0.29 to −0.16) | −0.12 (−0.18 to −0.043) |
| P‐value | <.001 | .34 | .001 | .005 | .05 | <.001 | <.001 | <.001 | .001 |
| Welsh ponies (n = 294) | −0.26 (−0.36 to −0.15) | −0.095 (−0.21 to −0.021) | −0.067 (−0.18 to 0.048) | 0.082 (−0.034 to 0.20) | −0.13 (−0.24 to −0.013) | −0.12 (−0.23 to −0.044) | −0.071 (−0.18 to 0.044) | 0.055 (−0.060 to 0.17) | −0.12 (−0.23 to 0.0048) |
| P‐value | <.001 | .12 | .25 | .17 | .03 | .04 | .23 | .35 | .05 |
| Large breed (n = 529) | −0.023 (−0.11 to 0.062) | −0.070 (−0.16 to 0.024) | 0.016 (−0.070 to 0.10) | −0.0068 (−0.10 to 0.087) | 0.12 (0.031 to 0.20) | −0.0013 (−0.087 to 0.085) | −0.059 (−0.14 to 0.027) | −0.046 (−0.13 to 0.040) | 0.026 (−0.060 to 0.11) |
| P‐value | .59 | .15 | .72 | .89 | .007 | .98 | .18 | .29 | .55 |
| Morgans (n = 293) | −0.11 (−0.22 to 0) | −0.13 (−0.24 to 0.0096) | 0.0015 (−0.11 to 0.12) | −0.095 (−0.21 to 0.024) | 0.068 (−0.047 to 0.18) | −0.063 (−0.18 to 0.052) | −0.059 (−0.17 to 0.057) | −0.0034 (−0.12 to 0.11) | −0.047 (−0.16 to 0.069) |
| P‐value | .05 | .03 | .98 | .12 | .25 | .28 | .32 | .95 | .43 |
| QH (n = 59) | 0.19 (−0.067 to 0.43) | 0.24 (−0.093 to 0.52) | 0.12 (−0.14 to 0.36) | 0.17 (−0.16 to 0.47) | 0.25 (−0.013 to 0.47) | 0.13 (−0.13 to 0.37) | −0.0047 (−0.26 to 0.25) | −0.079 (−0.33 to 0.18) | 0.25 (−0.0065 to 0.48) |
| P‐value | .14 | .16 | .38 | .31 | .06 | .33 | .97 | .55 | .05 |
| TWH (n = 48) | 0.23 (−0.060 to 0.48) | 0.082 (−0.28 to 0.42) | 0.14 (−0.15 to 0.41) | 0.13 (−0.23 to 0.46) | −0.12 (−0.39 to 0.17) | −0.080 (−0.36 to 0.17) | 0.036 (−0.26 to 0.32) | −0.17 (−0.43 to 0.12) | −0.077 (−0.35 to 0.21) |
| P‐value | .12 | .66 | .35 | .47 | .41 | .60 | .81 | .25 | .61 |
| Arabians (n = 64) | −0.31 (−0.51 to −0.062) | −0.25 (−0.049 to 0.024) | −0.19 (−0.42 to 0.060) | 0.0099 (−0.26 to 0.28) | 0.12 (−0.13 to 0.36) | −0.21 (−0.44 to 0.041) | −0.12 (−0.36 to 0.13) | 0.018 (−0.23 to 0.27) | −0.12 (−0.36 to 0.13) |
| P‐value | .01 | .07 | .14 | .94 | .34 | .09 | .36 | .89 | .34 |
Pearson's correlation coefficients, 95% confidence intervals, and P‐values for height, 8 EMS biochemical traits, and ACTH across breeds of horses. All traits were corrected for age and sex before analysis. Significant P‐values (<.005) are in bolded text.
Abbreviations: ADIPON, adiponectin; GLU, glucose; GLU‐OST; glucose after an oral sugar test; INS, insulin; INS‐OST, insulin after an oral sugar test; NEFA, non‐esterified fatty acids; QH, quarter horses; TG, triglycerides; TWH, Tennessee Walking horses.
3.2. FST‐based statistic to detect signatures of selection
A total of 212 208 nonoverlapping, 10 kb windows across all 31 equine autosomes were analyzed in the Welsh pony cohort, with an average of 8.2 (±3.2) SNPs per window. A total of 212 windows were within the top 0.1% of the empirical distribution of di values, which in turn represented 134 ROI. Among the significant di windows, 50 (24%) were located on ECA6 and corresponded to 8 separate ROI (Figure 1). One of these ECA6 ROI comprised 42 (20%) of the total significant di windows and spanned an ~782 kb segment. Based on EquCab2, (the equine reference genome available at the time of this analysis), this segment ranged from bp positions 81 003 617 to 81 785 414 (Supporting Information Figure 1). The other 7 significant ROIs on ECA6 were derived from singleton di windows, located at least 1 megabase (Mb) apart. Of note, 162 other significant di windows were distributed throughout all autosomes, except chromosomes 12, 16, 19, 30, and 31.
Figure 1.
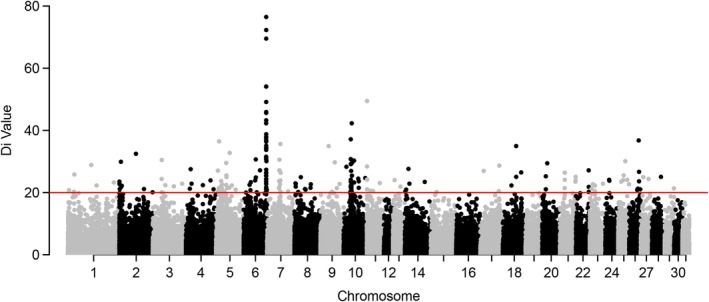
Genome‐wide di values for Welsh ponies. Each d i value is plotted on the y‐axis and each autosome is shown on the x‐axis in alternating colors. Each dot represents a 10 kb window. The red horizontal line represents the top 0.1% of the empirical distribution of d i values. One region of interest on equine chromosome 6 (ECA6) spanned ~782 kb segment, ranging from 81 003 617 to 81 785 414 bp
3.3. Association analysis
For the Welsh pony cohort, P‐values for 142 SNPs on ECA6 associated with height exceeded the threshold for genome‐wide significance (Figure 2A). Based on EquCab2, all 142 SNPs were within the same ~1.3 Mb region and included SNPs from bp position 80 501 273 to 81 808 008. For insulin, P‐values for 58 SNPs on ECA6 exceeded the threshold for genome‐wide significance and included SNPs from bp position 80 639 787 to 81 651 604 (Figure 2B). Significant SNPs within this ROI were not identified for any of the other EMS traits or ACTH.
Figure 2.
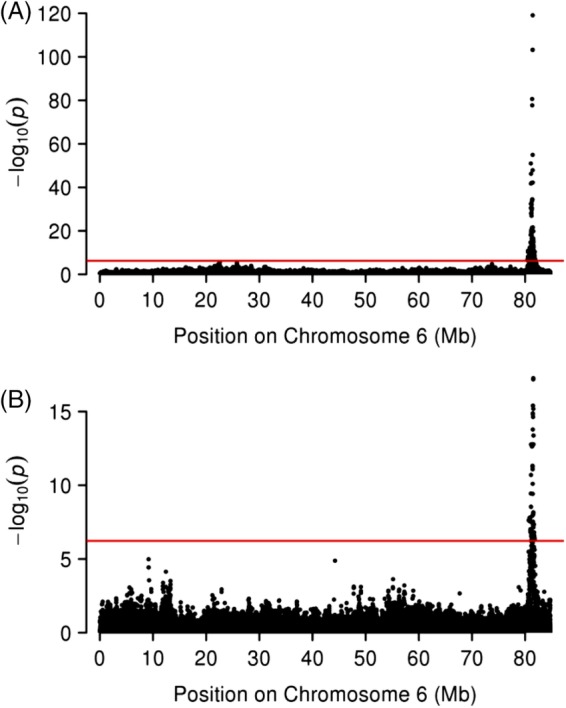
Plot of the association analysis for equine chromosome 6 (ECA6) in 264 Welsh Ponies (WP). The base pair positions for chromosome 6 are plotted along the x‐axis and the −log10 of the P‐values are plotted on the y‐axis. Individual circles represent single nucleotide polymorphisms (SNPs). The red line marks the thresholds for genome‐wide significance. (A) Results obtained in WP for height. Significant associations were noted on ECA6 with SNPs between 80 501 273 and 81 808 008 bp. (B) Results obtained in WP for baseline insulin. Significant associations were noted on ECA6 with SNPs between 80 639 787 and 81 651 604 bp
3.4. Heritability and genetic variation
The h2 SNP for height in the Welsh ponies was 0.87 (SD = 0.084). Using genomic partitioning for height, the percent of the genetic variation contributed by the ROI (SNPs from bp position 80 501 273 to 81 808 008) on ECA6 was 0.34 (SD = 0.083), that is, 39% of the total h2 SNP. The top SNPs from association analysis were included in the h2 SNP model as covariates to estimate the contribution of these SNPs to height in ponies. The 142 SNPs on ECA6 that exceeded the threshold for genome‐wide significance on association analysis were pruned at an LD of >0.8 to avoid over fitting the h2 SNP model, leaving 42 SNPs for analysis. The percent of genetic variation contributed by these 42 SNPs was estimated to be 0.41, that is, 47% of the total h2 SNP. After random subsetting of the data, the resultant mean values for h2 SNP were not significantly different from the original estimates above as follows: 0.89 (SD = 0.087) for the overall h2 SNP estimate of height, 0.38 (SD = 0.087) for genomic partitioning at the ROI, and 0.45 using the top SNPs from association analysis as covariates.
Within this cohort, we previously showed that baseline insulin had an h2 SNP of 0.81 (SD = 0.11), with a mean h2 SNP of 0.82 (mean SE: 0.12) after random subsetting.50 In this analysis, the h2 SNP explained by genomic partitioning was 0.19 (SD = 0.086) or 24% of the total h2 SNP for baseline insulin. Of the 58 significant SNPs found on association analysis, 13 remained in our analysis after pruning for LD. Including these SNPs as top predictors, the percent of genetic variation contributed by these SNPs was 0.13 or 16% of the total h2 SNP. After random subsetting of the data, the mean h2 SNP for genomic partitioning at the ROI was 0.20 (SD = 0.086) and 0.14 using the top SNPs approach.
3.5. Haplotype analyses for height and baseline insulin
Nearly 40% (23058) of all (56740) ECA6 SNPs had a BF value >4 when analyzing height as the trait of interest. A total of 107 SNPs had the highest BF values (>30) and were within the range of bp positions 81 012 766 to 81 782 298 (Figure 3A). Evaluation of all 652 SNPs within and flanking 1 kb of the ROIs identified by association analysis and d i statistic (SNPs from 80 499 826 to 81 809 066 bp), showed that all SNPs exceeded the BF value threshold, with values ranging from 4.17 to 40.12 (Figure 3A). When analyzing haplotypes using baseline insulin as the trait of interest, 290 SNPs on ECA6 had a BF value >4, which included 171 of the 652 SNPs comprising the ROI. The haplotypes consisted of 2 predominant regions where 46 SNPs were within bp positions 81 161 980 to 81 288 528 and 71 SNPs were within bp positions 81 381 221 to 81 583 507 (Figure 3B). The latter region also contained the SNPs with the highest BF values for the entire analysis (maximum BF of 7.5). HapQTL did not identify haplotypes on ECA6 for any of the other traits.
Figure 3.
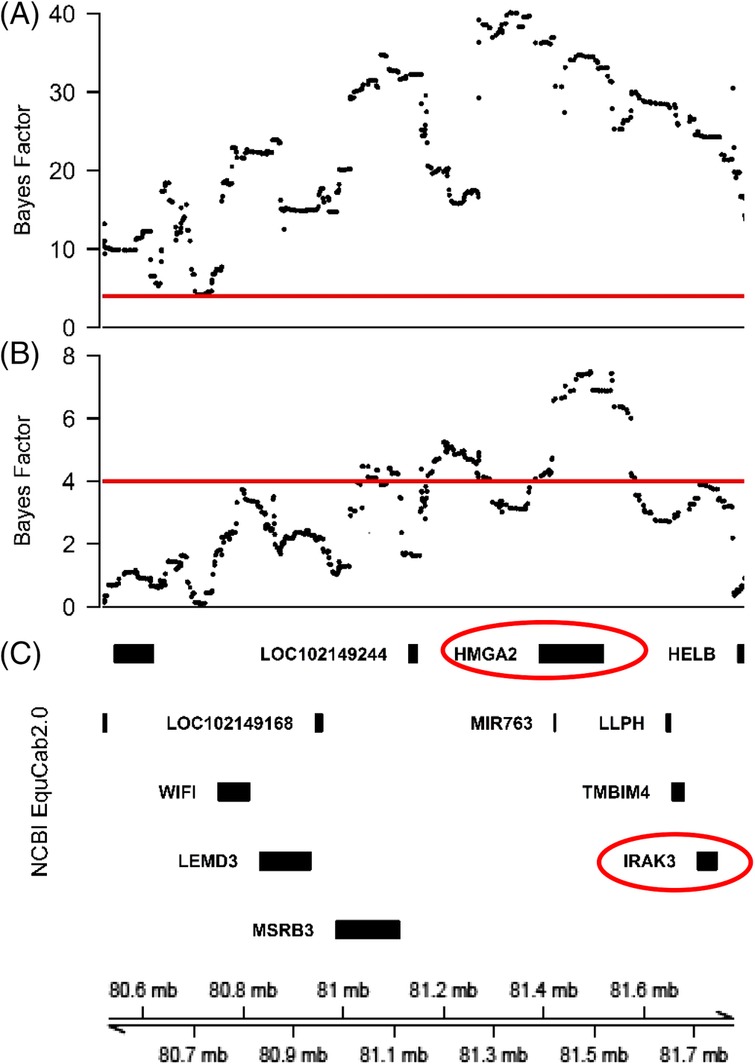
Fine‐scale structure of the region of interest (ROI) on equine chromosome 6 (ECA6). Regions of interest identified from the results of the association analysis and di statistic were used for haplotype analysis for both height (A) and baseline insulin values (B) in Welsh ponies. Bayes Factor values above the red horizontal line are considered significant and represent an ancestral haplotype. Shared ancestral haplotypes between both traits are most predominant from base pair positions 81 161 980 to 81 288 528 and 81 381 221 to 81 583 507. (C) Aligning the NCBI genome browser for the ROI identified HMGA2 (red circle) as a coding gene within the shared haplotype. IRAK3 was also identified as a candidate gene based on proximity and biological data.
3.6. Candidate gene identification, sequencing, and genotyping
The ROI identified in our study from association analysis and d i statistics (ECA6: 80 499 826 to 81 809 066) was further analyzed for positional candidate genes. Using NCBI and the Ensembl genome browser with EquCab2 as the reference genome, a total of 16 positional candidate genes were identified, comprising 3 RNA genes, 2 pseudogenes, and 11 protein coding genes (Figure 3C). A search of the PubMed literature database for known biological function and relevance in other species resulted in the prioritization of HMGA2 and IRAK3 as biological positional candidate genes. HMGA2 was the only protein‐coding gene within the smaller 81 161 980 to 81 583 507 region fine mapped by haplotype analysis.
The HMGA2 c.83G>A variant in exon 1 reported by Frischnecht et al.44 was identified in our 51 horse multi‐breed cohort (6 Morgan horses, 6 Arabian horses, 6 Tennessee Walking horses, 12 Quarter horses, 3 Miniature horses, and 18 Welsh ponies); however, no additional HMGA2 or IRAK3 exonic variants were detected. All individuals (n = 823) were then genotyped for the HMGA2 c83G>A variant. In the Welsh pony (n = 294) cohort, the A allele frequency was 0.76 and the G allele frequency was 0.24 (Table 2). The HMGA2 A allele frequencies across the 5 sections of the Welsh ponies present in our population were 1.0 for section A, 0.74 for section B, 0.83 for section C, 0.03 for section D, and 0.64 for section H (Table 2, Supporting Information Table 1). In the large breed horses (n = 529), there were only 5 horses heterozygous for the HMGA2 A allele (2 Tennessee Walking horses, 1 Morgan horse, 1 Mustang, and 1 Kentucky Mountain horse), resulting in an overall A allele frequency of 0.005.
Table 2.
Genotyping results for the HMGA2 c.83G > A variant in Welsh ponies and large breed horses
| Breed | n | G/G (WT) | G/A (HET) | A/A (MUT) | A allele frequency | G allele frequency |
|---|---|---|---|---|---|---|
| Welsh ponies | 294 | 30 | 80 | 184 | 0.76 | 0.24 |
| Section A | 78 | 78 | 1.0 | 0.00 | ||
| Section B | 150 | 8 | 62 | 80 | 0.74 | 0.26 |
| Section C | 3 | 1 | 2 | 0.83 | 0.17 | |
| Section D | 15 | 14 | 1 | 0.03 | 0.97 | |
| Section H | 37 | 8 | 11 | 18 | 0.64 | 0.37 |
| Unregistered | 11 | 5 | 6 | 0.77 | 0.23 | |
| All large breed horses | 530 | 525 | 5 | 0.005 | 0.995 | |
| Morgan horses | 293 | 292 | 1 | 0.002 | 0.998 | |
| Quarter horses | 59 | 59 | 1.0 | |||
| Tennessee Walking horses | 48 | 46 | 2 | 0.021 | 0.98 | |
| Arabians | 64 | 64 | 1.0 | |||
| Other large breed horses | 66 | 64 | 2 | 0.015 | 0.985 |
Results are also shown for specific breeds including: Sections of Welsh ponies, Morgan horses, Quarter horses, Tennessee Walking horse, and Arabians. Allele frequencies are provided for the G (wild‐type) and A (mutant) allele.
Abbreviations: HET, heterozygote; MUT, homozygous for the mutant allele; WT, Homozygous for the wild‐type allele.
3.7. Correlations among HMGA2 genotype, EMS traits, and ACTH
Correlation analyses between HMGA2 genotype and the measured traits were performed in Welsh ponies. A negative (−0.75; 95% CI: −0.80 to −0.70; P‐value <.001) correlation was identified between the A allele and height. Pairwise comparisons of the least square means of height and HMGA2 genotype revealed statistically significant differences between all 3 genotypes (Figure 4A). Although the ANOVA F‐statistic did not differentiate among the 3 possible modes of inheritance, an additive model was favored over recessive and dominant models based on AIC (Supporting Information Table 5).
Figure 4.
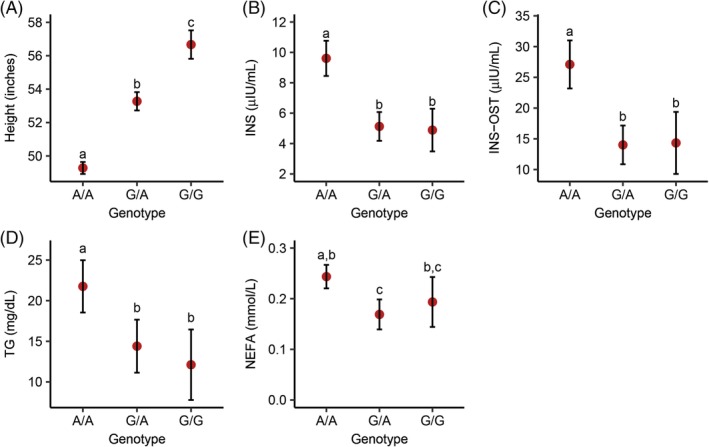
Least‐square mean estimates and 95% confidence intervals for the HMGA2 c.83G>A variant and various phenotypes in a population of 294 Welsh ponies. Height (A), insulin (B), insulin after an oral sugar test (INS‐OST) (C), triglycerides (D), and non‐esterified fatty acids (NEFA) (E)
Positive correlations with P‐values <.005 were also identified between the HMGA2 A allele and 4 of the 9 measured EMS traits in the ponies, including insulin (0.32; 95% CI: 0.21‐0.42), INS‐OST (0.25; 95% CI: 0.14‐0.35), NEFA (0.19; 95% CI: 0.075‐0.30), and triglycerides (0.22; 95% CI: 0.10‐0.32). Correlations for traits that were not statistically significant included: glucose, GLU‐OST, leptin, adiponectin, and ACTH. Pairwise comparisons for insulin, INS‐OST, and triglycerides revealed that the predicted marginal means for the A/A genotype were statistically different (P‐value <.001) from both the G/G and G/A genotypes, but that the predicted marginal means for the G/G and G/A genotypes were not statistically different from each other, suggesting recessives model of inheritance for these measurements (Figure 4B‐D). Although the P‐values for the F‐statistic linear regression modeling slightly favored recessive models for insulin, INS‐OST, and NEFA, the AIC values showed minimal separation between additive and recessive models for all 4 biochemical measurements (Supporting Information Table 5). Pairwise comparisons between the marginal means and genotype for NEFA also revealed statistically significant differences between the A/A and G/A genotypes (Figure 4E).
4. DISCUSSION
It is well recognized that ponies are at high risk for developing EMS; however, the mechanisms underlying this increased susceptibility, and the roles that genetic factors might play, are not understood. In our study, we demonstrated that baseline insulin values, a major component of the EMS phenotype, were correlated to height in Welsh ponies. With complementary genome‐wide analysis methods with high‐density SNP genotype data, we identified and fine‐mapped a locus on ECA6 associated with both of these traits in Welsh ponies, which we estimated to be contributing ~40% and ~20% of the total h2 SNP for height and insulin, respectively. The positional candidate genes HMGA2 and IRAK3 were prioritized based on known biological function and evidence in other species. Sequencing of the promoters, coding exons, and flanking intronic regions revealed only a c.83G>A variant (p.G28E) in HMGA2, previously described in other small stature horse breeds.44 Correlations between HMGA2 genotype and critical metabolic measures of EMS in the Welsh ponies suggested a previously unrecognized pleiotropic effect of this locus and its candidate HMGA2 functional variant.
Similar to what has been found in humans, an inverse correlation between height and 5 EMS measurements (insulin, glucose, triglycerides, leptin, and adiponectin) as well as ACTH were found in the large cohort of horses and Welsh ponies. However, we determined that the ponies were predominately driving the correlations in this cohort for baseline insulin, as statistically significant correlations were not identified for any of the 4 other individual breeds. This led us to investigate whether genetic loci for height, EMS measures, and ACTH in Welsh ponies could be one and the same.
High‐density SNP genotype data enabled us to use an FST‐based approach (di) to detect regions of low heterogeneity that exist because of selection for a phenotype, as well as identify genomic regions containing variants associated with both height and insulin on ECA6. We identified several breed‐specific loci undergoing selection in the Welsh pony; however, the region with the highest number of significant di windows, as well as those at the top of the empirical distribution, was a ~782 kb segment on ECA6 that was within the boundaries of the 1.3 Mb ROI identified by association analysis. Although the di statistic is blinded to phenotype, given the extensive breeding selection for short stature in ponies and the overlapping results with the association analysis, we surmised that selection for height was responsible for this genomic signature. Based on our cohort and the high heritability of height and baseline insulin, our association analysis had adequate power to identify alleles with moderate to high effect size51 and readily detected the ECA6 locus in Welsh ponies for both traits.
With genomic partitioning, we estimated that the ROI (ECA6: 80 499 826 to 81 809 066) contributed to 39% of the genetic variation for height and 24% for baseline insulin. However, this approach leads to inclusion of SNPs that were top predictors from association analysis, violating the effect size assumption when using a restricted estimated maximum likelihood analysis. Thus, we also performed a top predictors approach after pruning for highly correlated SNPs that resulted in an estimate of genomic contribution of 47% for height and 16% for baseline insulin. Although these estimates were not performed in an independent population, which can lead to over fitting of the data, it does suggest that the ECA6 locus is contributing ~40% of the genetic variation of height and ~20% for baseline insulin in our population. Unaccounted for population stratification or cryptic relatedness can lead to overestimation of h2 SNP. However, the mean h2 SNP estimates and SDs after randomly subsetting the data did not significantly differ from the original estimates, indicating that population substructure or cryptic relatedness was not significantly biasing our estimates (Supporting Information Supplemental Methods).
We identified a haplotype block that spanned the entire height ROI on ECA6 found by association analysis, while haplotype blocks in the same region for baseline insulin contained distinct major and minor peaks. This likely reflects differences in variant effect size, nonshared factors affecting the traits, and selection for height. We showed that 39%‐47% of the genetic variance in height could be explained by our ROI on ECA6; thus, the locus has a large effect on height in ponies. In contrast, the effect on insulin is smaller at 21%‐25% of the genetic variation. This is consistent with the results from the Pearson's correlation between height and insulin, which was −0.26, indicating that not all the variation in insulin could be explained by its relationship to height with nonshared factors present between the traits. Finally, short stature has been strongly selected in ponies through extensive breeding; however, hyperinsulinemia is not a desirable trait. The long haplotype on height likely reflects extensive hitchhiking secondary to selective breeding for that trait. Thus, haplotype analysis allowed us to fine map our ROI for height and insulin to bp positions: 81161980 to 81 583 507, where HMGA2 was the only annotated coding gene.
The HMGA2 protein interacts with AT‐rich regions of DNA through 3 DNA binding domains (AT hooks). This interaction alters the chromatin structure and promotes protein‐protein interactions necessary for assembly and stabilization of the enhanceosome during initiation of transcription.52 HMGA2's main functional role is thought to be in cellular proliferation and differentiation, which has been supported by the numerous studies in humans linking HMGA2 with height.53, 54, 55, 56, 57, 58, 59 The HMGA2 locus was also identified as being 1 of 4 loci explaining 83% of the genetic variation of height in horses and 1 of 6 loci explaining 46%‐52.5% of the genetic variation of height in dogs.60, 61 Furthermore, knockout mouse models for HMGA2 result in a lean, pygmy phenotype62, whereas, gain‐of‐function mutations of this gene led to gigantism, excessive fat formation, and lipomatosis in both mice and humans.63, 64 In addition to the alterations in fat metabolism noted above, HMGA2 has been associated with other causes of metabolic derangements, particularly type II diabetes in humans.65 Voight et al. hypothesized that an HMGA2 variant was likely affecting insulin levels independent of an obesity‐driven mechanism.65 Since then, both genome‐wide association and meta‐analyses have replicated this result.66, 67, 68 The only HMGA2 variant found in our panel of 48 horses was a missense mutation (c.83G>A) in exon 1, which was previously described as associated with decreased height in Shetland and other pony breeds.44 The variant, with its glycine to glutamate substitution at residue 28, is predicted to affect the first AT hook, and the authors demonstrated that the mutant nucleotide sequence had decreased binding affinity for DNA. This is additional evidence supporting the likely functional impact of this mutation.
In our pony cohort, the HMGA2 variant had an allele frequency of 0.76, was distributed across the sections of the Welsh pony breed consistent with their height distribution, was negatively correlated (−0.75) with height, and its effect was explained by an additive model of inheritance in our population of ponies. We also identified a negative correlation for the A allele with 4 EMS traits, including insulin, NEFA, INS‐OST, and triglycerides. This provides evidence that HMGA2 is having an effect on EMS traits beyond modulating height.
Notably, pairwise comparisons of NEFA between genotypes revealed that, although there was a statistical difference between the A/A and G/A genotypes, there was not a difference between either of the homozygous genotypes. This is most likely due to the large 95% confidence intervals identified when assessing the least square means for genotype and NEFA concentrations in the ponies, particularly those with the G/G genotype (Figure 4E). Pairwise comparisons between the least squared means for genotype and insulin, INS‐OST, and triglycerides suggested a recessive model of inheritance; however, model analyses were unable to differentiate between an additive or recessive model. The lack of distinction is likely because of the large variation within EMS traits, as well as bias owing to unequal sampling among our ponies, as our cohort only included 3 section Cs and 15 section Ds. Therefore, inclusion of more samples from these sections would likely improve our power to differentiate between an additive and recessive model.
IRAK3 was included as a biological candidate gene due to evidence in other species and its close proximity to the fine mapped ROI. IRAK3 is downregulated in individuals with obesity and metabolic syndrome and is believed to be a key inhibitor of inflammation during metabolic derangements.69 Furthermore, IRAK3 mutant mouse phenotypes include reduced body size, decreased femur diameter, and abnormal bone morphology,70 as well as impaired glucose tolerance.71 We sequenced the IRAK3 gene in our sample panel of horses but did not find any variants. Although a predicted miRNA (MIR763) was within our refined ROI, its function is unknown and does not have any associated orthologues.
In conclusion, through genome‐wide analyses, we identified an allele for a known height gene, HMGA2, as contributing to both height and several EMS traits in a cohort of Welsh ponies. Additional functional analysis would determine if the HMGA2 mutation has a pleiotropic effect on these traits or if another unidentified variant within our ROI independently contributes to the EMS traits and has been inadvertently selected for due to genomic hitchhiking. Although our study focused on Welsh ponies, the HMGA2 variant has been correlated with height in other pony breeds; thus, it is likely that this variant is also having an effect on metabolic traits in these individuals, as supported by the correlation analysis with the addition of 3 Shetland, 2 Hackney, and 3 British Riding ponies to our cohort (Supporting Information Table 7). Moreover, although height was not correlated with EMS traits in the large breed horses in our study, this does not rule out stature as contributing to these traits in that population. In humans, leg‐length‐to‐torso ratios are consistently correlated with metabolic traits over total height.11 Therefore, length‐to‐torso ratios in large breed horses might reveal a correlation not identified in this analysis. These data are a major step forward towards understanding genetic influences on EMS that could also have implications for improving equine health and understanding contributors to MetS.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
This work was approved by the University of Minnesota's IACUC, approval number 1501‐32254A.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Appendix S1: Supporting information
Supplemental Figure 1 Local di values for the equine chromosome 6 (ECA6) segment analyzed further in this study. Each d i value is plotted on the y axis and the ECA6 position in bp is shown on the x axis.
Supplemental Figure 2: Least‐square mean estimates and 95% confidence intervals for height or EMS phenotypes and section in a population of 283 registered Welsh ponies adjusting for both age and sex. Height (A), insulin (B), INS‐OST (C), triglycerides (D), and NEFA (E). Abbreviations: INS‐OST = insulin after an oral sugar test, NEFA = non‐esterified fatty acids.
Supplemental Table 1 Breed requirements for Welsh pony sections based on pedigree and height requirements.
Supplemental Table 2: Reference population used for the calculation of di.
Supplemental Table 3: PCR primers for Sanger sequencing and annotation for HMGA2 exon sequencing. Based on poor annotation of the HMGA2 gene in EquCab2 reference genome, we did a full reconstruction of the gene. Notably, Ensembl has this gene positioned for the horse at equine chromosome 6 (ECA6): 81197462‐81 402 841 in contrast to NCBI position at ECA6: 81389151‐81 518 054. Neither assembly included the ~1.4 Kb annotated by Frishchknecht et al, including exon 1 and the 5’ UTR (GenBank: LN8490000.1). Based on our annotation of exons 2‐5, the NCBI position appears more accurate and corresponds with the most predominant peak identified in the haplotype analysis for baseline insulin (ECA6: 81381221‐81 583 507). Base pair locations for EquCab2 and EquCab3 are also provided.
Supplemental Table 4 (cont): PCR primers and annotation for IRAK3 exon sequencing.
Supplemental Table 5: anova results and Akaike information criterion (AIC) values for models of inheritance between the HMGA2 c.83G > A variant and height and the four EMS traits significantly correlated with genotype. anova results and AIC values for models of inheritance between the HMGA2 c.83G > A variant and height and the four EMS traits significantly correlated with genotype. Deciding values are highlighted in red. For height, an additive model was the best fit model (lowest AIC). For the EMS traits, P‐value for the F‐statistic slightly favored the recessive model but the AIC could not differentiate between a recessive and additive model. For example, the AIC for the recessive insulin model was 249.2 and 251.2 for the additive model, which can be interpreted as the additive model being 0.36 [exp^([249.2‐251.2]/2)] times as likely as the recessive model, concluding that there is insufficient information to support picking either model. Abbreviations: INS‐OST = insulin post oral sugar test, NEFA = non‐esterified fatty acids.
Supplemental Table 6: EquCab2 and EquCab3 base pair (bp) position for SNPs on the Axiom MCEc2M within the region of interest on equine chromosome 6 (ECA6) bp positions 80 499 826‐ 81 809 066. SNPs (presented by their Axiom MCEc2M SNP ID) within the entire region of interest were remapped to EquCab3 (manuscript in preparation: Beeson S., Schaefer R., Mason V., McCue M.. “Robust remapping of equine SNP array coordinates to EquCab3.”). EquCab3 coordinates were not provided for three SNPs as they did not have probes that mapped uniquely to EquCab3. SNPs which exceeded the threshold for genome wide significance on association analysis (Assoc) for height and baseline insulin are indicated by an X. Significant di windows are based on the average base pair position within a 10Kb window of SNPs. SNPs marked with an X represent 5Kb upstream and 5Kb downstream of the base pair location.
Supplemental Table 7: Correlations between height and biochemical traits with the addition of seven ponies. Pearson's correlation coefficients were repeated with the inclusion of seven ponies representing three Shetland ponies, two Hackney ponies, and three British Riding ponies. Presented in the table are: Pearson's correlation coefficients, 95% confidence intervals and P‐values for height, eight EMS biochemical traits, and ACTH for the entire cohort as well as just the ponies. All traits were corrected for age and sex prior to analysis. Significant P‐values (<0.005) are in bolded text. Abbreviations: INS = insulin, INS‐OST = insulin post oral sugar test, GLU = glucose, GLU‐OST = glucose post oral sugar test, NEFA = non‐esterified fatty acids, TG = triglycerides, ADIPON = adiponectin.
ACKNOWLEDGMENTS
This work was presented at the 2016 American College of Veterinary Internal Medicine Forum, Denver, CO; Havemeyer Endocrinology Summit, January 2017, Miami, FL; and Equine Science Society Annual Meeting, May 2017, Minneapolis, MN.
Norton EM, Avila F, Schultz NE, Mickelson JR, Geor RJ, McCue ME. Evaluation of an HMGA2 variant for pleiotropic effects on height and metabolic traits in ponies. J Vet Intern Med. 2019;33:942–952. 10.1111/jvim.15403
Funding information Foundation for the National Institutes of Health, Grant/Award Number: T32 OD010993; Morris Animal Foundation, Grant/Award Number: D14EQ‐033; National Institute of Food and Agriculture, Grant/Award Number: 2009‐55205‐052542012‐67015‐19432
REFERENCES
- 1. Frank N, Geor RJ, Bailey SR, Durham AE, Johnson PJ, American College of Veterinary Internal Medicine . Equine metabolic syndrome. J Vet Intern Med. 2010;24:467‐475. [DOI] [PubMed] [Google Scholar]
- 2. Karikoski NP, Horn I, McGowan TW, et al. The prevalence of endocrinopathic laminitis among horses presented for laminitis at a first‐opinion/referral equine hospital. Domest Anim Endocrinol. 2011;41:111‐117. [DOI] [PubMed] [Google Scholar]
- 3. Jeffcott LB, Field JR. Current concepts of hyperlipaemia in horses and ponies. Vet Rec. 1985;116:461‐466. [DOI] [PubMed] [Google Scholar]
- 4. Bamford NJ, Potter SJ, Baskerville CL, Harris PA, Bailey SR. Effect of increased adiposity on insulin sensitivity and adipokine concentrations in different equine breeds adapted to cereal‐rich or fat‐rich meals. Vet J. 2016;214:14‐20. [DOI] [PubMed] [Google Scholar]
- 5. Bamford NJ, Potter SJ, Harris PA, Bailey SR. Breed differences in insulin sensitivity and insulinemic responses to oral glucose in horses and ponies of moderate body condition score. Domest Anim Endocrinol. 2014;47:101‐107. [DOI] [PubMed] [Google Scholar]
- 6. Dugdale AH, Curtis GC, Cripps PJ, et al. Effects of season and body condition on appetite, body mass and body composition in ad libitum fed pony mares. Vet J. 2011;190:329‐337. [DOI] [PubMed] [Google Scholar]
- 7. Batty GD, Barzi F, Woodward M, et al. Adult height and cancer mortality in Asia: the Asia Pacific Cohort Studies Collaboration. Ann Oncol. 2010;21:646‐654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. La Vecchia C, Decarli A, Negri E, et al. Height and the prevalence of chronic disease. Rev Epidemiol Sante Publique. 1992;40:6‐14. [PubMed] [Google Scholar]
- 9. Perelman J. Are chronic diseases related to height? Results from the Portuguese National Health Interview Survey. Econ Hum Biol. 2014;15:56‐66. [DOI] [PubMed] [Google Scholar]
- 10. Lawlor DA, Ebrahim S, Davey SG. The association between components of adult height and type II diabetes and insulin resistance: British Women's Heart and Health Study. Diabetologia. 2002;45:1097‐1106. [DOI] [PubMed] [Google Scholar]
- 11. Smith GD, Greenwood R, Gunnell D, Sweetnam P, Yarnell J, Elwood P. Leg length, insulin resistance, and coronary heart disease risk: the Caerphilly study. J Epidemiol Community Health. 2001;55:867‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guerrero‐Igea FJ, Lepe‐Jimenez JA, Garrido‐Serrano A, Palomo‐Gil S. Association among hyperinsulinemia, family history of diabetes, and diminutive stature in normoglycemic premenopausal women. Diabetes Care. 2001;24:602‐603. [DOI] [PubMed] [Google Scholar]
- 13. Silva EC, Martins IS, de Araujo EA. Metabolic syndrome and short stature in adults from the metropolitan area of Sao Paulo city (SP, Brazil). Cien Saude Colet. 2011;16:663‐668. [DOI] [PubMed] [Google Scholar]
- 14. Bozorgmanesh M, Hadaegh F, Zabetian A, Azizi F. Impact of hip circumference and height on incident diabetes: results from 6‐year follow‐up in the Tehran lipid and glucose study. Diabet Med. 2011;28:1330‐1336. [DOI] [PubMed] [Google Scholar]
- 15. Janghorbani M, Amini M. Associations of hip circumference and height with incidence of type 2 diabetes: the Isfahan diabetes prevention study. Acta Diabetol. 2012;49(Suppl 1):S107‐S114. [DOI] [PubMed] [Google Scholar]
- 16. Asao K, Kao WH, Baptiste‐Roberts K, et al. Short stature and the risk of adiposity, insulin resistance, and type 2 diabetes in middle age: the third National Health and Nutrition Examination Survey (NHANES III), 1988‐1994. Diabetes Care. 2006;29:1632‐1637. [DOI] [PubMed] [Google Scholar]
- 17. Brown DC, Byrne CD, Clark PM, et al. Height and glucose tolerance in adult subjects. Diabetologia. 1991;34:531‐533. [DOI] [PubMed] [Google Scholar]
- 18. Nuesch E, Dale C, Palmer TM, et al. Adult height, coronary heart disease and stroke: a multi‐locus Mendelian randomization meta‐analysis. Int J Epidemiol. 2016;45:1927‐1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bosy‐Westphal A, Plachta‐Danielzik S, Dorhofer RP, et al. Short stature and obesity: positive association in adults but inverse association in children and adolescents. Br J Nutr. 2009;102:453‐461. [DOI] [PubMed] [Google Scholar]
- 20. La Batide‐Alanore A, Tregouet DA, Sass C, et al. Family study of the relationship between height and cardiovascular risk factors in the STANISLAS cohort. Int J Epidemiol. 2003;32:607‐614. [DOI] [PubMed] [Google Scholar]
- 21. Olatunbosun ST, Bella AF. Relationship between height, glucose intolerance, and hypertension in an urban African black adult population: a case for the "thrifty phenotype" hypothesis? J Natl Med Assoc. 2000;92:265‐268. [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson CP, Hamby SE, Saleheen D, et al. Genetically determined height and coronary artery disease. N Engl J Med. 2015;372:1608‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barker DJ. The intrauterine origins of cardiovascular and obstructive lung disease in adult life. The Marc Daniels lecture 1990. J R Coll Physicians Lond. 1991;25:129‐133. [PMC free article] [PubMed] [Google Scholar]
- 24. Palmer JR, Rosenberg L, Shapiro S. Stature and the risk of myocardial infarction in women. Am J Epidemiol. 1990;132:27‐32. [DOI] [PubMed] [Google Scholar]
- 25. Liu G, Liu J, Li N, et al. Association between leg length‐to‐height ratio and metabolic syndrome in Chinese children aged 3 to 6 years. Prev Med Rep. 2014;1:62‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meyre D, Boutin P, Tounian A, et al. Is glutamate decarboxylase 2 (GAD2) a genetic link between low birth weight and subsequent development of obesity in children? J Clin Endocrinol Metab. 2005;90:2384‐2390. [DOI] [PubMed] [Google Scholar]
- 27. Wegner L, Andersen G, Sparso T, et al. Common variation in LMNA increases susceptibility to type 2 diabetes and associates with elevated fasting glycemia and estimates of body fat and height in the general population: studies of 7,495 Danish whites. Diabetes. 2007;56:694‐698. [DOI] [PubMed] [Google Scholar]
- 28. Schaefer RJ, Schubert M, Bailey E, et al. Developing a 670k genotyping array to tag ~2M SNPs across 24 horse breeds. BMC Genomics. 2017;18:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing‐data inference for whole‐genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCoy AM, McCue ME. Validation of imputation between equine genotyping arrays. Anim Genet. 2014;45:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Purcell S, Neale B, Todd‐Brown K, et al. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet. 2007;81:559‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Akey JM, Ruhe AL, Akey DT, et al. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci U S A. 2010;107:1160‐1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petersen JL, Mickelson JR, Rendahl AK, et al. Genome‐wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet. 2013;9:e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schultz N. Characterization of equine metabolic syndrome and mapping of candidate genetic loci In:. Veterinary Population Medicine. Saint Paul, MN: University of Minnesota; 2016. [Google Scholar]
- 35. Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou X, Stephens M. Genome‐wide efficient mixed‐model analysis for association studies. Nat Genet. 2012;44:821‐824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome‐wide association studies. Nat Methods. 2011;8:833‐835. [DOI] [PubMed] [Google Scholar]
- 38. Li MX, Yeung JM, Cherny SS, et al. Evaluating the effective numbers of independent tests and significant p‐value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Speed D, Cai N, Consortium U, et al. Reevaluation of SNP heritability in complex human traits. Nat Genet. 2017;49:986‐992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome‐wide SNPs. Am J Hum Genet. 2012;91:1011‐1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gusev A, Lee SH, Trynka G, et al. Partitioning heritability of regulatory and cell‐type‐specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu H, Guan Y. Detecting local haplotype sharing and haplotype association. Genetics. 2014;197:823‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frischknecht M, Jagannathan V, Plattet P, et al. A non‐synonymous HMGA2 variant decreases height in Shetland Ponies and other small horses. PLoS One. 2015;10:e0140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woodward J. Bi‐allelic SNP genotyping using the TaqMan(R) assay. Methods Mol Biol. 2014;1145:67‐74. [DOI] [PubMed] [Google Scholar]
- 46. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 47. Pinheiro J, Bates D, DebRoy S, et al. nlme: Linear and nonlinear mixed effects models. In: R package version 3.1‐131; 2017.
- 48. Lenth RV. Least‐squares means: the R package lsmeans. J Stat Softw. 2016;69:33. [Google Scholar]
- 49. Gonzalez JR, Armengol L, Sole X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644‐645. [DOI] [PubMed] [Google Scholar]
- 50. Norton EM, Schultz NE, Rendahl AK, et al. Heritability of metabolic traits associated with equine metabolic syndrome in Welsh ponies and Morgan horses. Equine Vet J. 10.1111/evj.13053 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 51. Feng S, Wang S, Chen CC, Lan L. GWAPower: a statistical power calculation software for genome‐wide association studies with quantitative traits. BMC Genet. 2011;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (review). Int J Oncol. 2008;32:289‐305. [PubMed] [Google Scholar]
- 53. Lettre G, Jackson AU, Gieger C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weedon MN, Lango H, Lindgren CM, et al. Genome‐wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weedon MN, Lettre G, Freathy RM, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245‐1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang TL, Guo Y, Zhang LS, et al. HMGA2 is confirmed to be associated with human adult height. Ann Hum Genet. 2010;74:11‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609‐615. [DOI] [PubMed] [Google Scholar]
- 58. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lanktree MB, Guo Y, Murtaza M, et al. Meta‐analysis of dense Genecentric association studies reveals common and uncommon variants associated with height. Am J Hum Genet. 2011;88:6‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Makvandi‐Nejad S, Hoffman GE, Allen JJ, et al. Four loci explain 83% of size variation in the horse. PLoS One. 2012;7:e39929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rimbault M, Beale HC, Schoenebeck JJ, et al. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 2013;23:1985‐1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou X, Benson KF, Ashar HR, et al. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI‐C. Nature. 1995;376:771‐774. [DOI] [PubMed] [Google Scholar]
- 63. Battista S, Fidanza V, Fedele M, et al. The expression of a truncated HMGI‐C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793‐4797. [PubMed] [Google Scholar]
- 64. Ligon AH, Moore SD, Parisi MA, et al. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large‐scale association analysis. Nat Genet. 2010;42:579‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bai H, Liu H, Suyalatu S, et al. Association analysis of genetic variants with type 2 diabetes in a Mongolian population in China. J Diabetes Res. 2015;2015:613236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ng MC, Shriner D, Chen BH, et al. Meta‐analysis of genome‐wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saxena R, Elbers CC, Guo Y, et al. Large‐scale gene‐centric meta‐analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hulsmans M, Geeraert B, De Keyzer D, et al. Interleukin‐1 receptor‐associated kinase‐3 is a key inhibitor of inflammation in obesity and metabolic syndrome. PLoS One. 2012;7:e30414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li H, Cuartas E, Cui W, et al. IL‐1 receptor‐associated kinase M is a central regulator of osteoclast differentiation and activation. J Exp Med. 2005;201:1169‐1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tan Q, Majewska‐Szczepanik M, Zhang X, et al. Irak‐M deficiency promotes the development of type 1 diabetes in NOD mice. Diabetes. 2014;63:2761‐2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Supplemental Figure 1 Local di values for the equine chromosome 6 (ECA6) segment analyzed further in this study. Each d i value is plotted on the y axis and the ECA6 position in bp is shown on the x axis.
Supplemental Figure 2: Least‐square mean estimates and 95% confidence intervals for height or EMS phenotypes and section in a population of 283 registered Welsh ponies adjusting for both age and sex. Height (A), insulin (B), INS‐OST (C), triglycerides (D), and NEFA (E). Abbreviations: INS‐OST = insulin after an oral sugar test, NEFA = non‐esterified fatty acids.
Supplemental Table 1 Breed requirements for Welsh pony sections based on pedigree and height requirements.
Supplemental Table 2: Reference population used for the calculation of di.
Supplemental Table 3: PCR primers for Sanger sequencing and annotation for HMGA2 exon sequencing. Based on poor annotation of the HMGA2 gene in EquCab2 reference genome, we did a full reconstruction of the gene. Notably, Ensembl has this gene positioned for the horse at equine chromosome 6 (ECA6): 81197462‐81 402 841 in contrast to NCBI position at ECA6: 81389151‐81 518 054. Neither assembly included the ~1.4 Kb annotated by Frishchknecht et al, including exon 1 and the 5’ UTR (GenBank: LN8490000.1). Based on our annotation of exons 2‐5, the NCBI position appears more accurate and corresponds with the most predominant peak identified in the haplotype analysis for baseline insulin (ECA6: 81381221‐81 583 507). Base pair locations for EquCab2 and EquCab3 are also provided.
Supplemental Table 4 (cont): PCR primers and annotation for IRAK3 exon sequencing.
Supplemental Table 5: anova results and Akaike information criterion (AIC) values for models of inheritance between the HMGA2 c.83G > A variant and height and the four EMS traits significantly correlated with genotype. anova results and AIC values for models of inheritance between the HMGA2 c.83G > A variant and height and the four EMS traits significantly correlated with genotype. Deciding values are highlighted in red. For height, an additive model was the best fit model (lowest AIC). For the EMS traits, P‐value for the F‐statistic slightly favored the recessive model but the AIC could not differentiate between a recessive and additive model. For example, the AIC for the recessive insulin model was 249.2 and 251.2 for the additive model, which can be interpreted as the additive model being 0.36 [exp^([249.2‐251.2]/2)] times as likely as the recessive model, concluding that there is insufficient information to support picking either model. Abbreviations: INS‐OST = insulin post oral sugar test, NEFA = non‐esterified fatty acids.
Supplemental Table 6: EquCab2 and EquCab3 base pair (bp) position for SNPs on the Axiom MCEc2M within the region of interest on equine chromosome 6 (ECA6) bp positions 80 499 826‐ 81 809 066. SNPs (presented by their Axiom MCEc2M SNP ID) within the entire region of interest were remapped to EquCab3 (manuscript in preparation: Beeson S., Schaefer R., Mason V., McCue M.. “Robust remapping of equine SNP array coordinates to EquCab3.”). EquCab3 coordinates were not provided for three SNPs as they did not have probes that mapped uniquely to EquCab3. SNPs which exceeded the threshold for genome wide significance on association analysis (Assoc) for height and baseline insulin are indicated by an X. Significant di windows are based on the average base pair position within a 10Kb window of SNPs. SNPs marked with an X represent 5Kb upstream and 5Kb downstream of the base pair location.
Supplemental Table 7: Correlations between height and biochemical traits with the addition of seven ponies. Pearson's correlation coefficients were repeated with the inclusion of seven ponies representing three Shetland ponies, two Hackney ponies, and three British Riding ponies. Presented in the table are: Pearson's correlation coefficients, 95% confidence intervals and P‐values for height, eight EMS biochemical traits, and ACTH for the entire cohort as well as just the ponies. All traits were corrected for age and sex prior to analysis. Significant P‐values (<0.005) are in bolded text. Abbreviations: INS = insulin, INS‐OST = insulin post oral sugar test, GLU = glucose, GLU‐OST = glucose post oral sugar test, NEFA = non‐esterified fatty acids, TG = triglycerides, ADIPON = adiponectin.


