Abstract
Fungal pathogens pose an increasing threat to public health. Limited clinical drug regimens and emerging drug-resistant isolates challenge infection control. The global burden of human fungal pathogens is estimated at 1 billion infections and 1.5 million deaths annually. In addition, plant fungal pathogens increasingly threaten global food resources. Novel strategies are needed to combat emerging fungal diseases and pan-resistant fungi. An untapped mechanistically novel approach is to pharmacologically activate the intrinsic cell death pathways encoded by pathogenic fungi. This strategy is analogous to new anti-cancer therapeutics now entering the clinic. Here we summarize the best understood examples of cell death mechanisms encoded by pathogenic fungi, contrast these to mammalian cell death pathways, and highlight the gaps in knowledge towards identifying potential death effectors as druggable targets.
Keywords: fungi, cell death, pathogen, yeast, necrosis, apoptosis
Graphical Abstract

Fungal species appear to undergo forms of programmed cell death though the detailed mechanisms are not yet known.
1. Introduction
An estimated 3.8 million fungal species make up a significant portion of global biodiversity [1]. Of these, the small fraction that is pathogenic has significant impact on global health and mortality. Annual global incidence of acute invasive fungal diseases was recently estimated at 1.9 million, primarily attributed to cryptococcal meningitis, invasive aspergillosis, pneumocystis pneumonia, and invasive candidiasis [2]. Another three million live with chronic pulmonary aspergillosis, while over 10% of the world population has non-invasive fungal infections [2, 3]. Despite the importance to public health, fungal infections are more neglected than other major killers. Diagnostics for fungal infections have greatly improved but remain limited or unavailable. Treatment strategies for invasive species are challenged by the small arsenal of anti-fungal agents, the susceptibility of immunocompromised populations, the rise of drug resistance across major fungal pathogens including Candida, Cryptococcus, Aspergillus and new emerging strains [2, 4–6]. In addition, fungal infections of unknown proportions affect terrestrial, avian and marine wildlife and cause significant agricultural losses [7–11].
The rise in outbreaks of fungal pathogens has been attributed to a number of factors including contaminated medical devices, organ transplants, and patient immune status [7, 12, 13]. Rising global temperatures are predicted to select for fungal thermal tolerance, which may facilitate breaching mammalian defenses, though direct evidence is limited to date [8, 9, 14]. Beyond human pathogens, plant fungal pathogens such as Magnaporthe oryzae (rice blast) threaten global food security by infecting economically significant cereal crops, typically claiming 10–30% of rice harvests in parts of the Americas, Asia and Africa [15–17]. Epidemics of rice blast can devastate entire fields, potentially impacting approximately half the world’s population dependent on rice as a primary staple, compounded by the high costs of anti-fungals for treating crops [15].
In light of these challenges, new out-of-the box strategies are needed to combat fungal pathogens. One possibility on the horizon is pharmacologic manipulation of intrinsic cell death mechanisms encoded by fungi. Precedence for this concept is provided by the cancer field. A new class of drugs emerged from the discovery of a deep binding cleft on human anti-apoptotic proteins BCL2 and BCLxL where their natural inhibitors bind, and where small molecule mimics of these inhibitors also bind [18]. In 2016, three decades after the discovery of BCL2 [19–23], a BCL2 antagonist (Venetoclax/ABT-199) was approved for clinical use in a subset of cancer patients [24–27], and many related compounds are currently in clinical trails [28] – an exciting new era. While similar approaches are being explored for the BCL2 homologs in viruses [29–31], fungi lack BCL2 homologs and therefore are not amenable to this approach. Nevertheless, there is interest in this general direction [32], and feasibility is suggested by growing evidence indicating that molecular death mechanisms exist in multicellular and filamentous fungal pathogens (e.g. Magnaporthe and Neurospora crassa) and likely also exist in single-cell yeast pathogens (e.g. Cryptococcus and Candida).
2. Distinctions between cell death programs in animals versus fungi
The systematic disappearance/death of cells observed in developing animals was once assumed to result from depletion of essential resources for those cells. In this case, cell death could simply be the absence of life without any active participation by the cell destined to die. However this philosophy changed with delineation of the first molecularly defined cell death pathway, a cell autonomous caspase-dependent process genetically mapped in C. elegans and mammals [33–36]. This apoptotic death pathway is inhibited by the CED9/BCL2 proteins and is required to eliminate many cells during embryonic development [23]. Apoptosis can be induced in mammalian cells by a variety of stimuli from within the cell (e.g. DNA damage) and by extracellular ligand-induced signaling pathways that converge to activate caspase 3, the primary effector molecule of apoptosis (Fig 1). The morphological features of apoptotic mammalian cells are attributed to actions of caspase 3 that prepare apoptotic cell corpses for engulfment and degradation by neighboring cells. Caspases are also widely studied for their roles in non-death related cellular processes including differentiation, proliferation, and neuronal function [37–41]. However, biochemical mechanisms analogous to mammalian caspase-dependent apoptosis have not been identified in fungi (see nomenclature conflict, section 4).
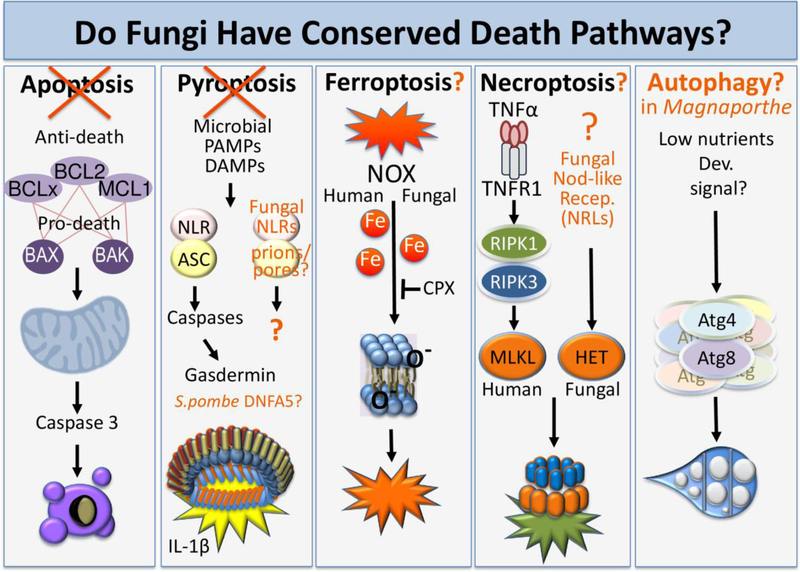
Figure 1. Are there conserved molecular death pathways in mammals and fungi?
Features of the best characterized mammalian cell death pathways and potentially analogous mechanisms present in fungal species. Fungi lack the mammalian apoptosis pathway in which caspase 3 activation is regulated by BCL2 family proteins, and also lack the caspases 1, 4, 5 and 11, and pore-forming gasdermins (unlike related fungal proteins) that mediate programmed necrosis by pyroptosis, although fungal NLR-like receptors can trigger cell death upon cell-cell fusion of highly related but incompatible fungal cells. Iron-dependent cell death via ferroptosis due to lipid peroxidation may be generalizable across a wide range of species. The fungal pore-forming domain of HET-S thought to mediate incompatibility cell death has predicted structural similarity to the mammalian pore-forming domain of MLKL, mediator of necroptosis.
Most mammalian caspases do not promote apoptosis but several caspases can activate pyroptosis (programmed necrosis) by cleaving and activating the pore-forming protein gasdermin D that lyses mammalian cells from the inside of the plasma membrane [42, 43] (Fig 1). Cell death pathways can become intertwined, for example when apoptotic caspase 3 promotes pyroptosis by cleaving and activating a different gasdermin protein, gasdermin E [44]. The human gasdermin E/DNFA5 C-terminal domain is conserved in some species of yeast (Cdc23/Mcm10), but these yeast proteins lack the relevant death-inducing pore-forming domain and are not known to promote yeast cell death [45]. Other widely studied non-apoptotic (necrotic) mammalian death pathways are necroptosis and more recently ferroptosis, which play important roles in disease pathogenesis including neurodegeneration and infection [46–51]. Fungi may have semblances of mammalian necrosis pathways (see section 5).
Unlike mammalian cells, fungi have cell walls, live in distinctly different environments, and are genetically diverse. Thus, it seems likely that fungal mechanisms of cell death would have been selected under equally diverse driving forces. Thus, fungi can be expected to have diverse biochemical death processes to build morphologically distinct structures or suppress the spread of fungal viruses, and therefore may share more dissimilarities than similarities with mammals. By extension, applying mammalian apoptosis assays and reporters (e.g. TUNEL assays, zVAD caspase inhibitors/reporters, and Annexin V to detect exposed phosphatidylserine) to study fungi may not answer the important mechanistic details of fungal cell death [52–54]. Mammalian apoptosis assays have been applied in studies of Saccharomyces cerevisiae, Cryptococcus neoformans, Candida species, and Aspergillus fumigatus among other pathogenic fungi but without identifying the precise causes of reporter activity or the direct effectors of cell death [54–58].
3. Rationale for the existence of cell death in multi- and unicellular fungi
Definitive molecular evidence for a deliberate program of self-inflicted injury in fungi has not yet been achieved, at least not in a manner analogous to the salient discoveries that finally squelched remaining skepticism about the existence of programmed cell death in animals [20, 59, 60]. The existence of cell death mechanisms in multi-cellular filamentous fungi (e.g. Aspergillus) is more easily rationalized because it was originally assumed that programmed cell death machinery arose during evolution with multicellular species. Among multi-cellular fungal species, there are clear examples where fungal cell death appears to be deliberately orchestrated and even required for the fungal life cycle, for example in the plant pathogen Magnaporthe oryzae [61] and in Podospora anserina and related filamentous fungi [62]. Some of the molecular mechanisms have been partially delineated, providing potential pharmacologic targets (see section 5).
Even the communities of single cell yeast species such as the human pathogens Candida albicans and Cryptococcus neoformans as well as the laboratory workhorse Saccharomyces cerevisiae exhibit characteristics of multi-cellularity [63]. Like multicellular organisms that undergo distinct pathways of differentiation to fulfill specific roles required for organismal viability, unicellular fungi also encode differentiation processes to form multicellular communities. Cell morphological changes between smooth, wrinkled and pseudohyphae have long been observed to correlate with phenotypic/functional changes including virulence in Cryptococcus neoformans and Candida albicans [64, 65]. Different gene expression profiles in Saccharomyces cerevisiae cells are associated with different cell behaviors in aggregates, hyphae and biofilms [63, 66–68]. The transition from colonies to biofilms by Saccharomyces cerevisiae is orchestrated in part by the opposing actions of two transcription factors Tup1, which promotes biofilm formation, and Cyc8/Ssn6, which inhibits Flo11 expression and represses biofilm formation [69]. Long non-coding RNAs (lncRNAs) are differentially expressed in Saccharomyces cerevisiae colonies versus biofilms [70]. Although a role for cell death in prokaryote biofilm formation is not understood, dying bacterial cells have been observed within biofilms and proposed to be beneficial by preventing extensive disruption of colony architecture [71, 72].
Even within simple colonies of Saccharomyces cerevisiae, transcriptional profiling has revealed some of the inner workings of yeast colony life and the changes that occur during aging [73, 74]. Simple yeast colonies of S. cerevisiae are stratified in cell layers with distinguishable morphological and metabolic characteristics [75]. In general, yeast cells in upper colony layers are larger and undergo autophagy, while cells in lower layers are smaller, rely more on respiration and are more sensitive to stress [75]. By occupying specific niches, cells in yeast communities can perform unique roles such as adhesion, invasion, and nutrient recycling that can reasonably be expected to facilitate community function and survival [67, 69]. Yeast colony subdomains that are prone to cell death also could potentially benefit the surviving population [76].
Beneficial yeast cell death has been suggested to occur during failed mating attempts, stress responses to adverse environmental conditions, and cell death could conceivably have a role in self-recognition that also limits the spread of viral pathogens [77–79]. Altruistic cell death of aged yeast cells has also been reported [80, 81]. In search of gene-dependent yeast cell death programs, we and others have identified candidate genes in genome-wide screens of the yeast Saccharomyces knockout collection in response to heat stress, heat shock, ER stress or acetate treatment [82–86], but the precise death mechanisms and death effectors are not yet known. Thus, the possibility of unicellular programmed cell death remains a philosophical question until compelling molecular pathways are defined [87].
4. Evidence for and against Apoptosis-Like Cell Death in Fungi
A conspicuous nomenclature problem stems from the application of the term “apoptosis” to yeast and other fungi. The original definition and rationale for coining the word apoptosis in 1972 was to infer a deliberate cell death process occurring in mammalian tissues, long before any molecular mechanisms were known [88]. The original evidence for such was limited to morphological/pathological changes, some of which resemble dying yeast cells [55]. Thus, apoptosis began to be defined morphologically. The nomenclature conflict arises because yeast biologists/geneticists continue to apply the term apoptosis as originally defined or by morphological features and mammalian apoptosis reporters. Meanwhile, the definition of apoptosis in the larger human/animal cell death field further evolved over the decades beginning with the identification of pro-apoptotic caspases in the 1990’s (see section 2). Thus, those who were taught that apoptosis is caspase-dependent might assume incorrectly that the prevalent use of the term ‘yeast apoptosis’ in the literature is supported by a body of evidence demonstrating caspase-dependent death. However, this appears unlikely, as fungi do not encode orthologs of mammalian apoptosis mediators (e.g. true caspases or BCL2 family proteins). Thus, it is more likely that fungi die by alternative necrotic processes [52, 89, 90].
Fungi encode metacaspases, which are related to mammalian caspases but differ in substrate specificity and mechanisms of activation [91]. Saccharomyces cerevisiae has a single metacaspase Mca1 (renamed Yca1), while Aspergillus fumigatus has two, CasA and CasB [92]. While Saccharomyces Yca1 has been implicated in cell death under some circumstances [93], Aspergillus CasA and CasB appear not to promote death and are even protective [92]. Given that caspases have both death-jobs and day-jobs in mammalian cells (see section 2), Saccharomyces metacaspase Mca1/Yca1 may also have a day-job, such as clearing protein aggregates [94]. A few metacaspase substrates have been reported, for example GAPDH, which is also cleaved in mammalian cells by caspases [95, 96]. How these substrates contribute to yeast cell death is unclear. An alternative interpretation is that the enhanced cell survival by MCA1/YCA1 deletion strains are an indirect or compensatory effect of MCA1/YCA1 deficiency [52]. Several other yeast homologs of mammalian cell death regulators have been reported to have some involvement in yeast cell death processes, including orthologs of EndoG, AIF, cytochrome c and others, but the detailed death mechanisms are not clarified [97–101] (note that some of these references use apoptosis to mean programmed/regulated cell death).
5. Programmed Cell Death in Growth and Development of Fungi
In our view, the best examples of fungal cell death occurring in natural settings, and for which some of the molecular death machinery is known, are the economically critical plant pathogen Magnaporthe oryzae and the model organism Podospora anserina.
5.1. Autophagy-dependent cell death in fungal development
Plant pathogen Magnaporthe oryzae (also known as Pyricularia oryzae) is a filamentous ascomycete fungus that infects the world’s most valuable cereal crops, including rice, barley, wheat, millet, oat, and ryegrass [15, 102]. A three-celled spore (conidium) produces an infectious structure, the appressorium, which has sufficient turgor to facilitate entry through the outer cuticle of the plant cell wall to reach the inner plasma membrane and invade [102, 103]. The fungus then develops invasive hyphae that spread within the plant and release new spores through lesions formed in the host [102, 104]. The fungus induces necrotic plant cell death and can spread across fields within days. Estimated annual losses are sufficient to feed millions of people, underscoring the need for quick acting fungicidal compounds [61, 105, 106].
Growing evidence indicates that programmed cell death of Magnaporthe oryzae conidia is required for successful invasion of plants, and the mechanism of cell death involves autophagy (Fig 1). Upon adhesion of a conidial spore to a leaf surface, a germ tube emerges from the conidium and its distal end subsequently differentiates into the infective appressorium. To generate the appressorium, the conidium undergoes one round of mitosis allowing one daughter nucleus to migrate through the germ tube to the maturing appressorium. Within hours, the conidium undergoes nuclear collapse, which is accompanied by the formation of autophagosomes and vacuoles [61, 105]. Interestingly, conidia lacking the autophagy factor Mgatg8 (homolog of mammalian LC3 and hallmark of autophagy), which exhibit phenotypes indicative of defective autophagy as expected, fail to die and also fail to form the appressorium required for infectivity [61]. Thus, conidial cell death appears to be essential for the life cycle of Magnaporthe oryzae. Based on sequence homology, other components of the autophagy pathway were identified in Magnaporthe oryzae. Remarkably, individual deletion of 16 non-selective macroautophagy genes resulted in continued survival of conidia and failure to form the appressorium or to infect plants. [107]. Thus, autophagy is required for the fungus to develop its appressorium structure needed to traverse the plant cell wall [108, 109]. In contrast, deletion of genes predicted to be involved in pexophagy or mitophagy (forms of selective autophagy) did not affect development of the appressorium or subsequent infection [170].
How is autophagy related to conidial cell death? The differentiation process is stimulated by nutrient deprivation, as conidia incubated in high nutrients do not proceed to form an appressorium, but instead form undifferentiated hyphae and continue to undergo mitosis [108, 109]. Nutrient depletion in fungi and mammals is known to downregulate cell growth by inhibiting the Tor1 kinase complex (TORC1) to derepress autophagy [110]. We recently identified Saccharomyces cerevisiae Whi2 as a novel inhibitor of TORC1 activity specifically in low amino acid conditions [111, 112]. Because TORC1 suppresses autophagy, sustained TORC1 activity in yeast lacking Whi2 results in failure to induce autophagy [113]. These functions of yeast Whi2 appear to be conserved in the human KCTD proteins, and children with bi-allelic mutations in KCTD7 have a severe neurodevelopmental disorder, potentially due to defective autophagy required to recycle nutrients and sustain neuronal functional and viability [111, 113]. Conversely in the case of Magnaporthe conidia, autophagy genes are critical for conidial cell death, likely because the nutrients derived from conidia are donated to build the appended appressorium, resulting in conidial cell sacrifice. However, it is not known if the Whi2 homolog in Magnaporthe oryzae is required for developmental cell death and appressorium formation. If so, Whi2 inhibition is a potential strategy to control Magnaporthe oryzae. Saccharomyces cerevisiae lacking Whi2 have reduced fitness and are super sensitive to many death stimuli, further supporting this approach [83, 114]. Other pro-survival proteins, such as the conserved BIR1 (baculovirus inhibitor of apoptosis repeat) proteins of human pathogen Aspergillus fumigatus, may also be viable therapeutic targets based on a recent study [58]. Mammalian IAP family proteins are also being targeting in pre-clinical trials as therapeutics [115–117], potentially paving the way for fungal inhibitors.
Definitive demonstration that autophagy is a direct versus indirect mediator of Magnaporthe oryzae conidial cell death is challenging. There is a distinction between autophagy-dependent and autophagy-mediated cell death. In the latter case, cells die as a direct consequence of overzealous autophagy, while in the former case, autophagy could be indirectly involved, for example by degrading a key pro-survival factor required to sustain conidial cell viability. An analogous situation occurs during Drosophila development. The essential Drosophila inhibitor of apoptosis (IAP) protein DIAP1 constitutively inhibits the Drosophila caspase Dronc [118–121]. Degradation of DIAP1 by the proteasome and possibly by autophagy leads to rapid activation of Dronc and cell death [118, 121, 122]. Therefore, the caspase Dronc, and not the proteasome/autophagosome, is the direct mediator of death. Furthermore, evidence for the existence of autophagic cell death in mammalian cells is limited and has been reclassified as autophagy-dependent cell death [123, 124]. Despite efforts, no alternative death effectors have been identified thus far in Magnaporthe, and the death of conidia concomitant with building the appressorium under nutrient deprivation conditions, makes a case for autophagic cell death. In either case, it can surely be agreed that conidial cell death is dependent on autophagy and that conidial cell death constitutes a deliberate developmental form of programmed cell death in the life cycle of Magnaporthe.
Magnaporthe oryzae also causes the death of its host plant cell upon egress. Inhibition of a single fungal kinase Pmk1 results in trapping of the fungus in the host cell and blocking its migration to infect a second cell, thereby protecting the plant and rendering the fungus non-infective [104]. Interestingly, pharmacologic inhibition of Pmk1 also blocked appressorium formation. This raises the exciting possibility for targeted inhibition of Pmk1 and potentially other inhibitors of appressorium formation as potential control strategies for agriculture.
5.2. Heterokaryon Incompatibility may resemble mammalian necroptosis
The second example of apparent fungal programmed cell death that occurs in some filamentous fungi is known as vegetative or heterokaryon incompatibility (HI). Hyphal cells can fuse to form multinucleated cells termed heterokaryons, a process allowing cytoplasmic exchange and the formation of a syncytial network that facilitates the flow of resources and biochemical signals [125, 126]. Cell fusion and formation of the heterokaryon involves mechanisms to avoid fusion between incompatible non-self cells. The ability to distinguish between self and non-self is determined by genetic differences at het loci [125, 127, 128]. Fusion between cells with identical/compatible het loci will form stable heterokaryons. In contrast, fusion between cells with distinct het loci are incompatible and will trigger a type of programmed cell death characterized by rapid compartmentalization, septal plugging and eventual death of the original two cells [125, 127, 129, 130]. The compartmentalization and septal plugging during HI has been reported to restrict horizontal transfer of mycoviruses, suggesting one possible benefit for the evolution of programmed cell death mechanisms [131–133].
Work done particularly in Podospora anserine, Neurospora crassa and Cryphonectria parasitica (chestnut blight) [125, 126, 130, 134–139] has uncovered some of the mechanisms of cell death due to incompatibility [140]. These fungi encode multiple HI systems that have been identified molecularly or genetically [125]. A prominent morphological feature of HI cell death is vacuolization, raising the possibility that vacuolar leakage or rupture contributes to cell death. This would be akin to cell death by lysosomal membrane permeability (LMP) in mammals, such as occurs during mammary gland involution [141], and has been implicated in fungal cell death in different scenarios and species [84, 97, 137]. However, biochemical explanations for the morphological changes during HI death are uncertain, but appear not to require metacaspases [134]. Transcriptional profiling implicates increased reactive oxygen species production along with calcium and phosphatidylinositol signaling pathways [134]. Autophagy is also induced during HI [139], but is not required for cell death [138]. In N. crassa, transcription factors CZT-1 [135] and VIB1 [136] are required for activation of the HI cell death program, and the P. anserina transcription factor idi-4, has been implicated in cell death induction [140]. However, the link between these transcription factors and the cell death effector proteins directly responsible for cytological-morphological features of HI are only partially delineated, predominantly in P. anserina, but are potential drug targets in pathogenic strains.
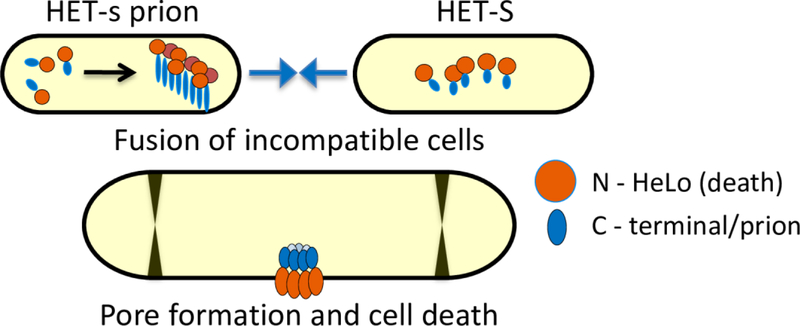
One candidate mechanism of cell death during HI in Podospora anserina may share features reminiscent of mammalian necroptosis (Fig 1) (see section 2). Different subpopulations of P. anserina can be distinguished by two allelic variants at the het-s locus, encoding two highly similar proteins HET-S and HET-s. Cell-cell fusion of populations differing at this locus results in cell death due to HI [142, 143]. The het-s allelic variant encodes a soluble cytoplasmic protein that can spontaneously fold into an amyloid-like prion state via a C-terminal domain [142, 144, 145]. Despite being 96% identical to het-s, the het-S allelic variant product does not spontaneously adopt the prion-like state [143, 146]. However, HET-S proteins can be induced to form prion-like structures upon physical interaction with the prion-like state of HET-s as a consequence of heterokaryon formation. This leads to adaptation of HET-S to a prion-like fold and subsequent partial unfolding of the HET-S N-terminal HeLo protein domain [143, 145, 147]. This HeLo domain unfolding leads to conversion of HET-S into an integral membrane protein that appears to oligomerize into a pore-forming complex mediating cell death during HI [147] (Fig 2). Interestingly, the HeLo domain has predicted structural similarity to the N-terminal domain of the mammalian pseudokinase MLKL (e.g. Phyre2). Mammalian MLKL is thought to oligomerize and form pores from the cytosolic side of the plasma membrane resulting in cell lysis, and thus acts as the effector protein of necroptosis through its pore-forming properties [62, 148–150] (Fig 1). HeLo-like domains termed HELLP present in other fungal species are suggested to have some sequence similarity to the pore-forming domain of MLKL, even though true homology may not be present [62]. Pore-forming proteins in fungal programmed cell death represents a potentially conserved mechanism across many fungal species and regulation of such may serve as a platform for the development of antifungal drugs.
Figure 2. Fungal cell death regulated by HET proteins during heterokaryon incompatibility.
The C-terminal prion-like domain is thought to trigger conformational changes and membrane association by the N-terminal HeLo domain to induce pore formation following cell-cell fusion between incompatible fungal cells with non-self het loci.
In addition to pore formation, the amyloid-prion assembly phenomenon to induce fungal cell death may have additional conceptual counterparts in the fungal kingdom (Fig 1). Like other types of intracellular signaling platforms, cell death pathways in mammals, including apoptosis, have an amplification step that is mediated by large dedicated protein complexes (apoptosome, inflammasome, necrosome) that recruit and activate death effectors such as caspases or other. To build these large death-osomes, their protein components undergo oligomerization. The mammalian necrosome turns out to be an amyloid with a function. Unstructured regions of necrosome components RIPK1 and RIPK3 preferentially assemble into a stable hetero-amyloid that is required for necroptotic cell death [150, 151]. The prion protein HET-s of filamentous fungi also folds into an amyloid fibril to promote fungal cell death by heterokaryon incompatibility [152, 153]. Recently, an new HI mechanism in Neurospora crassa was reported that involves a compatibility locus encoding two interacting proteins, the SNARE protein SEC-9 and a NOD-like receptor (NLR) protein PLP-1 with features similar to NLRs encoded by plants, mammals and other fungi [154]. Fusion of germinated asexual spores with incompatible SEC-9– PLP-1 proteins induces PLP-1 oligomerization and cell death. In this way, fungal incompatibility mechanisms resemble mammalian innate responses where NLRs recognize foreign molecular signatures and pathogens leading to NLR oligomerization to signal cell death by pyroptosis [155, 156].
The mammalian pyroptosis pathway adaptor protein ASC can assemble into filaments through the cooperative interactions of its PYD domain, which binds to the PYD domain of NOD-like inflammasome receptor protein (NLRP3) to facilitate caspase 1 recruitment and activation [153]. One study found that swapping the PYD domains of both NLRP3 and its adaptor ASC with the corresponding region of a NOD-like fungal protein and the prion domain of fungal HET-s, respectively, could induce inflammasome assembly [157]. The yeast prion protein SUP35 found in Saccharomyces cerevisiae and other fungi could also functionally replace a region of mammalian inflammatory response protein MAVS [157]. Although many details remain unaddressed, it is possible that prion-like amplifications are a cell death theme across species. This concept has been capitalized by a number of investigators using yeast Saccharomyces cerevisiae to study mechanisms of neuronal toxicity induced by human disease-associated amyloids, made popular by Susan Lindquist [158].
Ferroptosis is an emerging and exciting new research area in part because of its relevance to disease states involving neuronal or renal damage [50]. Ferroptosis is thought to result from loss of cell integrity due to lipid peroxidation that damages cell membranes, but the exact molecular effectors and mediators of mammalian cell ferroptosis are just beginning to be delineated. The working definition of ferroptosis is cell death inhibited by iron chelators [48], a phenomenon observed to occur in diverse cell types including fungi [159] (Fig 1).
6. Conclusions
Fungi represent a significant public health burden and require a reinforcement of current therapeutics. One attractive therapeutics approach is pharmacological manipulation of intrinsic cell death pathways to induce the death of fungi. Targeting intrinsic fungal cell death programs may allow for the development of fast acting and pathogen-specific anti-fungal agents. The existence for programmed cell death mechanisms in multi- and unicellular fungal species is currently challenged for the lack of molecularly defined death mechanisms equivalent to what is available for animals. However, death programs can be reasonably assumed to exist based on observations of fungal cell death that occurs in normal development, for example during host cell invasion by the economically important plant pathogen Magnaporthe oryzae, as well as in germlings and mycelial growth of several filamentous fungi that undergo incompatibility loci-induced cell death. Furthermore, new evidence reveals the complex order within colonies of unicellular fungi that potentially require cell death as a mechanism to maintain long-term population dynamics and pathogen evasion. However, with these exceptions, little is known about fungal cell death effectors in either multi- or unicellular species and further work is needed to address the gaps in knowledge about cell death modalities across the diverse kingdom of fungi. New nomenclature may assist in addressing the nuances of fungal cell death that differ from mammalian pathways and to avoid confusion when applying mammalian assays to fungi [53]. With stringency in the assays applied to fungi, the field will move to uncover the effectors of cell death in fungi. Delineating these pathways can lead to a better understanding of novel targets for controlling pathogenic fungi.
New therapeutic approaches are justified as there are only four major groups of antifungal drugs currently available, azoles (e.g. fluconazole affecting ergosterol biosynthesis), polyenes (e.g. amphotericin B and nystatin affecting membrane ergosterol), flucytosine (anti-metabolite) and echinocandins (interfere with cell wall biosynthesis), which have been used for decades, have off target effects and have contributed to resistant infections [160]. Pan-resistant fungi such as the transmissible human pathogen Candida auris and Lomentospora prolificans are emerging currently [161–163]. Fungal infections can be difficult to clear. A small portion of fungal cell populations are not killed by anti-fungals either because they are drug-resistant (e.g. enhanced efflux mechanisms) or because they are persisters in a metabolic state that renders them drug insensitive. Persisters are not drug-resistant if they reenter the growing state, but can be highly drug insensitive in their more dormant persister state [164]. Thus, persisters are insensitive to drugs directed only to active metabolic pathways. Approaches that target viability per se may be more effective against persisters, which likely contribute to clinically reticent infections. Counter intuitively, slow growing fungal persister cells with decreased ergosterol or cell wall synthesis can be more tolerant to metabolic drug pressures [164]. Persistence of Candida albicans and Candida glabrata has been attributed to defects in ribosome synthesis and mutations in the target of rapamycin (TOR) pathway that promotes cell growth and protein translation [160]. Development of combination anti-fungal therapies are needed to effectively kill both growing and persistent fungal pathogens, analogous to anti-bacterials for tuberculosis [163]. The 9–12 month treatment for Mycobacteria tuberculosis was reduced to 6 months by including pyrazinamide, which kills bacterial persisters, in combination with rifampin and isoniazid, which kill growing mycobacteria [165].
An important next step is to identify and characterize the key molecular players in microbial cell death. Similar to the current situation for fungi and other microbes, the early mammalian/animal cell death field was viewed with skepticism until the molecular effectors of animal cell death were identified, making the existence of programmed cell death irrefutable. Thus, the current fungal cell death field is in a similar position as the mammalian and animal cell death field about 30 years ago, but faster progress can be expected since the paths to the major concepts and potentially to some of the molecular similarities have been paved by the animal cell death field.
Acknowledgements
Supported by the National Institutes of Health USA grants R01NS083373 and R01GM077875 to J.M.H., T32AI007417 to M.K. and the Wendy Klag Center to M.K. and J.M.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest. The authors have no conflicts of interest.
References
- [1].Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 2018; https://www.ncbi.nlm.nih.gov/pubmed/30442909. [DOI] [PubMed]
- [2].Bongomin F, Gago S, Oladele RO, Denning DW. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J Fungi (Basel) 2017; 3: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bulpa P, Dive A, Sibille Y. Invasive pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Eur Respir J 2007; 30:782–800. [DOI] [PubMed] [Google Scholar]
- [4].Bongomin F, Harris C, Foden P, Kosmidis C, Denning DW. Innate and Adaptive Immune Defects in Chronic Pulmonary Aspergillosis. J Fungi (Basel) 2017; 3: 10.3390/jof3020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zheng YH, Ma YY, Ding Y, Chen XQ, Gao GX. An insight into new strategies to combat antifungal drug resistance. Drug Des Devel Ther 2018; 12:3807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Alcazar-Fuoli L, Mellado E. Current status of antifungal resistance and its impact on clinical practice. Br J Haematol 2014; 166:471–84. [DOI] [PubMed] [Google Scholar]
- [7].Benedict K, Richardson M, Vallabhaneni S, Jackson BR, Chiller T. Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect Dis 2017; 17:e403–e11 10.1016/S473-3099(17)30443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Casadevall A Fungal Diseases in the 21st Century: The Near and Far Horizons. Pathog Immun 2018; 3:183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de SAGR, Souza W, Frases S. The hidden pathogenic potential of environmental fungi. Future Microbiol 2017; 12:1533–40. [DOI] [PubMed] [Google Scholar]
- [10].Altizer S, Ostfeld RS, Johnson PT, Kutz S, Harvell CD. Climate change and infectious diseases: from evidence to a predictive framework. Science 2013; 341:514–9. [DOI] [PubMed] [Google Scholar]
- [11].Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJ, et al. Ecological responses to recent climate change. Nature 2002; 416:389–95. [DOI] [PubMed] [Google Scholar]
- [12].Talpaert MJ, Balfour A, Stevens S, Baker M, Muhlschlegel FA, Gourlay CW. Candida biofilm formation on voice prostheses. J Med Microbiol 2015; 64:199–208. [DOI] [PubMed] [Google Scholar]
- [13].Vallabhaneni S, Haselow D, Lloyd S, Lockhart S, Moulton-Meissner H, Lester L, et al. Cluster of Cryptococcus neoformans Infections in Intensive Care Unit, Arkansas, USA, 2013. Emerg Infect Dis 2015; 21:1719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnston SA, Voelz K, May RC. Cryptococcus neoformans Thermotolerance to Avian Body Temperature Is Sufficient For Extracellular Growth But Not Intracellular Survival In Macrophages. Sci Rep 2016; 6:20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 2012; 13:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Inoue Y, Vy TTP, Yoshida K, Asano H, Mitsuoka C, Asuke S, et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 2017; 357:80–3. [DOI] [PubMed] [Google Scholar]
- [17].Wang GL, Valent B. Durable resistance to rice blast. Science 2017; 355:906–7. [DOI] [PubMed] [Google Scholar]
- [18].Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435:677–81. [DOI] [PubMed] [Google Scholar]
- [19].Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985; 228:1440–3. [DOI] [PubMed] [Google Scholar]
- [20].Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell 1986; 44:817–29. [DOI] [PubMed] [Google Scholar]
- [21].Cleary ML, Smith SD, Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 1986; 47:19–28. [DOI] [PubMed] [Google Scholar]
- [22].Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988; 335:440–2. [DOI] [PubMed] [Google Scholar]
- [23].Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 1992; 356:494–9. [DOI] [PubMed] [Google Scholar]
- [24].Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med 2016; 374:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rossi D Venetoclax: a new weapon to treat high-risk CLL. Lancet Oncol 2016; 17:690–1. [DOI] [PubMed] [Google Scholar]
- [26].Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013; 19:202–8. [DOI] [PubMed] [Google Scholar]
- [27].Roberts AW, Huang D. Targeting BCL2 With BH3 Mimetics: Basic Science and Clinical Application of Venetoclax in Chronic Lymphocytic Leukemia and Related B Cell Malignancies. Clin Pharmacol Ther 2017; 101:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016; 538:477–82. [DOI] [PubMed] [Google Scholar]
- [29].Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci U S A 1993; 90:8479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nava VE, Cheng EH, Veliuona M, Zou S, Clem RJ, Mayer ML, et al. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J Virol 1997; 71:4118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, et al. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci U S A 1997; 94:690–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Leiter E, Csernoch L, Pocsi I. Programmed cell death in human pathogenic fungi - a possible therapeutic target. Expert Opin Ther Targets 2018; 22:1039–48. [DOI] [PubMed] [Google Scholar]
- [33].Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 1993; 75:641–52. [DOI] [PubMed] [Google Scholar]
- [34].Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, et al. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 1996; 384:368–72. [DOI] [PubMed] [Google Scholar]
- [35].Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, et al. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 1998; 94:325–37. [DOI] [PubMed] [Google Scholar]
- [36].Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, et al. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell 1998; 94:339–52. [DOI] [PubMed] [Google Scholar]
- [37].Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 2003; 4:687–97. [DOI] [PubMed] [Google Scholar]
- [38].Tang HL, Tang HM, Fung MC, Hardwick JM. In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci Rep 2015; 5:9015 10.1038/srep09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Erturk A, Wang Y, Sheng M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J Neurosci 2014; 34:1672–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gyorffy BA, Kun J, Torok G, Bulyaki E, Borhegyi Z, Gulyassy P, et al. Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc Natl Acad Sci U S A 2018; 115:6303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mollereau B, Perez-Garijo A, Bergmann A, Miura M, Gerlitz O, Ryoo HD, et al. Compensatory proliferation and apoptosis-induced proliferation: a need for clarification. Cell Death Differ 2013; 20:181 10.1038/cdd.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016; 535:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 2018; 557:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017; 547:99–103. [DOI] [PubMed] [Google Scholar]
- [45].Gregan J, Van Laer L, Lieto LD, Van Camp G, Kearsey SE. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim Biophys Acta 2003; 1638:179–86. [DOI] [PubMed] [Google Scholar]
- [46].Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol 2003; 4:95–104. [DOI] [PubMed] [Google Scholar]
- [47].Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1:112–9. [DOI] [PubMed] [Google Scholar]
- [48].Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012; 149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shan B, Pan H, Najafov A, Yuan J. Necroptosis in development and diseases. Genes Dev 2018; 32:327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fearnhead HO, Vandenabeele P, Vanden Berghe T. How do we fit ferroptosis in the family of regulated cell death? Cell Death Differ 2017; 24:1991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137:1112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hardwick JM. Do Fungi Undergo Apoptosis-Like Programmed Cell Death? MBio 2018; 9: 10.1128/mBio.00948-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vachova L, Palkova Z. Caspases in yeast apoptosis-like death: facts and artefacts. FEMS Yeast Res 2007; 7:12–21. [DOI] [PubMed] [Google Scholar]
- [54].Aouacheria A, Cunningham KW, Hardwick JM, Palkova Z, Powers T, Severin FF, et al. Comment on “Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death”. Science 2018; 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 1997; 139:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Semighini CP, Averette AF, Perfect JR, Heitman J. Deletion of Cryptococcus neoformans AIF ortholog promotes chromosome aneuploidy and fluconazole-resistance in a metacaspase-independent manner. PLoS Pathog 2011; 7:e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Leger T, Garcia C, Ounissi M, Lelandais G, Camadro JM. The metacaspase (Mca1p) has a dual role in farnesol-induced apoptosis in Candida albicans. Mol Cell Proteomics 2015; 14:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shlezinger N, Irmer H, Dhingra S, Beattie SR, Cramer RA, Braus GH, et al. Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science 2017; 357:1037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 1998; 93:519–29. [DOI] [PubMed] [Google Scholar]
- [60].Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002; 111:331–42. [DOI] [PubMed] [Google Scholar]
- [61].Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006; 312:580–3. [DOI] [PubMed] [Google Scholar]
- [62].Daskalov A, Habenstein B, Sabate R, Berbon M, Martinez D, Chaignepain S, et al. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci U S A 2016; 113:2720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Palkova Z, Vachova L. Yeast cell differentiation: Lessons from pathogenic and non-pathogenic yeasts. Semin Cell Dev Biol 2016; 57:110–9. [DOI] [PubMed] [Google Scholar]
- [64].Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun 1999; 67:6076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Radford DR, Challacombe SJ, Walter JD. A scanning electronmicroscopy investigation of the structure of colonies of different morphologies produced by phenotypic switching of Candida albicans. J Med Microbiol 1994; 40:416–23. [DOI] [PubMed] [Google Scholar]
- [66].Hope EA, Amorosi CJ, Miller AW, Dang K, Heil CS, Dunham MJ. Experimental Evolution Reveals Favored Adaptive Routes to Cell Aggregation in Yeast. Genetics 2017; 206:1153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Marsikova J, Wilkinson D, Hlavacek O, Gilfillan GD, Mizeranschi A, Hughes T, et al. Metabolic differentiation of surface and invasive cells of yeast colony biofilms revealed by gene expression profiling. BMC Genomics 2017; 18:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Stovicek V, Vachova L, Palkova Z. Yeast biofilm colony as an orchestrated multicellular organism. Commun Integr Biol 2012; 5:203–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nguyen PV, Hlavacek O, Marsikova J, Vachova L, Palkova Z. Cyc8p and Tup1p transcription regulators antagonistically regulate Flo11p expression and complexity of yeast colony biofilms. PLoS Genet 2018; 14:e1007495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wilkinson D, Vachova L, Hlavacek O, Marsikova J, Gilfillan GD, Palkova Z. Long Noncoding RNAs in Yeast Cells and Differentiated Subpopulations of Yeast Colonies and Biofilms. Oxid Med Cell Longev 2018; 2018:4950591 10.1155/2018/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Miguelez EM, Hardisson C, Manzanal MB. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J Cell Biol 1999; 145:515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 2012; 109:18891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wilkinson D, Marsikova J, Hlavacek O, Gilfillan GD, Jezkova E, Aalokken R, et al. Transcriptome Remodeling of Differentiated Cells during Chronological Ageing of Yeast Colonies: New Insights into Metabolic Differentiation. Oxid Med Cell Longev 2018; 2018:4932905 10.1155/2018/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vachova L, Palkova Z. Diverse roles of Tup1p and Cyc8p transcription regulators in the development of distinct types of yeast populations. Curr Genet 2018; [DOI] [PubMed]
- [75].Cap M, Stepanek L, Harant K, Vachova L, Palkova Z. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 2012; 46:436–48. [DOI] [PubMed] [Google Scholar]
- [76].Vachova L, Palkova Z. How structured yeast multicellular communities live, age and die? FEMS Yeast Res 2018; 18: 10.1093/femsyr/foy033. [DOI] [PubMed] [Google Scholar]
- [77].Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol 2005; 168:257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Barriuso J, Hogan DA, Keshavarz T, Martinez MJ. Role of quorum sensing and chemical communication in fungal biotechnology and pathogenesis. FEMS Microbiol Rev 2018; 42:627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ivanovska I, Hardwick JM. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J Cell Biol 2005; 170:391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol 2004; 166:1055–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wloch-Salamon DM, Fisher RM, Regenberg B. Division of labour in the yeast: Saccharomyces cerevisiae. Yeast 2017; 34:399–406. [DOI] [PubMed] [Google Scholar]
- [82].Teng X, Cheng WC, Qi B, Yu TX, Ramachandran K, Boersma MD, et al. Gene-dependent cell death in yeast. Cell Death Dis 2011; 2:e188 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Teng X, Dayhoff-Brannigan M, Cheng WC, Gilbert CE, Sing CN, Diny NL, et al. Genome-wide consequences of deleting any single gene. Mol Cell 2013; 52:485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim H, Kim A, Cunningham KW. Vacuolar H+-ATPase (V-ATPase) promotes vacuolar membrane permeabilization and nonapoptotic death in stressed yeast. J Biol Chem 2012; 287:19029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sousa M, Duarte AM, Fernandes TR, Chaves SR, Pacheco A, Leao C, et al. Genome-wide identification of genes involved in the positive and negative regulation of acetic acid-induced programmed cell death in Saccharomyces cerevisiae. BMC Genomics 2013; 14:838 10.1186/471-2164-14-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jarolim S, Ayer A, Pillay B, Gee AC, Phrakaysone A, Perrone GG, et al. Saccharomyces cerevisiae genes involved in survival of heat shock. G3 (Bethesda) 2013; 3:2321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ 2002; 9:367–93. [DOI] [PubMed] [Google Scholar]
- [88].Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 1972; 26:239–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Skulachev VP. Phenoptosis: programmed death of an organism. Biochemistry (Mosc) 1999; 64:1418–26. [PubMed] [Google Scholar]
- [90].Pandey SS, Singh S, Pathak C, Tiwari BS. “Programmed Cell Death: A Process of Death for Survival” - How Far Terminology Pertinent for Cell Death in Unicellular Organisms. J Cell Death 2018; 11:1179066018790259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bell RAV, Megeney LA. Evolution of caspase-mediated cell death and differentiation: twins separated at birth. Cell Death Differ 2017; 24:1359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Richie DL, Miley MD, Bhabhra R, Robson GD, Rhodes JC, Askew DS. The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress. Mol Microbiol 2007; 63:591–604. [DOI] [PubMed] [Google Scholar]
- [93].Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell 2002; 9:911–7. [DOI] [PubMed] [Google Scholar]
- [94].Lee RE, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci U S A 2010; 107:13348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Silva A, Almeida B, Sampaio-Marques B, Reis MI, Ohlmeier S, Rodrigues F, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochim Biophys Acta 2011; 1813:2044–9. [DOI] [PubMed] [Google Scholar]
- [96].Sundstrom JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, et al. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 2009; 11:1347–54. [DOI] [PubMed] [Google Scholar]
- [97].Eastwood MD, Cheung SW, Meneghini MD. Programmed nuclear destruction in yeast: self-eating by vacuolar lysis. Autophagy 2013; 9:263–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, et al. Endonuclease G regulates budding yeast life and death. Mol Cell 2007; 25:233–46. [DOI] [PubMed] [Google Scholar]
- [99].Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, et al. An AIF orthologue regulates apoptosis in yeast. J Cell Biol 2004; 166:969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell 2002; 13:2598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Klim J, Gladki A, Kucharczyk R, Zielenkiewicz U, Kaczanowski S. Ancestral State Reconstruction of the Apoptosis Machinery in the Common Ancestor of Eukaryotes. G3 (Bethesda) 2018; 8:2121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Martin-Urdiroz M, Oses-Ruiz M, Ryder LS, Talbot NJ. Investigating the biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol 2016; 90:61–8. [DOI] [PubMed] [Google Scholar]
- [103].Ryder LS, Talbot NJ. Regulation of appressorium development in pathogenic fungi. Curr Opin Plant Biol 2015; 26:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sakulkoo W, Oses-Ruiz M, Oliveira Garcia E, Soanes DM, Littlejohn GR, Hacker C, et al. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 2018; 359:1399–403. [DOI] [PubMed] [Google Scholar]
- [105].Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 2003; 57:177–202. [DOI] [PubMed] [Google Scholar]
- [106].Armed Pennisi E. and dangerous. Science 2010; 327:804–5. [DOI] [PubMed] [Google Scholar]
- [107].Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A 2009; 106:15967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Marroquin-Guzman M, Sun G, Wilson RA. Glucose-ABL1-TOR Signaling Modulates Cell Cycle Tuning to Control Terminal Appressorial Cell Differentiation. PLoS Genet 2017; 13:e1006557 10.1371/journal.pgen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sun G, Qi X, Wilson RA. A feed-forward subnetwork emerging from integrated TOR- and cAMP/PKA-signaling architecture reinforces Magnaporthe oryzae appressorium morphogenesis. Mol Plant Microbe Interact 2018; https://www.ncbi.nlm.nih.gov/pubmed/30431400. [DOI] [PubMed]
- [110].Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-sensing mechanisms across evolution. Cell 2015; 161:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chen X, Wang G, Zhang Y, Dayhoff-Brannigan M, Diny NL, Zhao M, et al. Whi2 is a conserved negative regulator of TORC1 in response to low amino acids. PLoS Genet 2018; 14:e1007592 10.1371/journal.pgen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Teng X, Yau E, Sing C, Hardwick JM. Whi2 signals low leucine availability to halt yeast growth and cell death. FEMS Yeast Res 2018; 18: 10.1093/femsyr/foy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Metz KA, Teng X, Coppens I, Lamb HM, Wagner BE, Rosenfeld JA, et al. KCTD7 deficiency defines a distinct neurodegenerative disorder with a conserved autophagy-lysosome defect. Ann Neurol 2018; 84:766–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ 2008; 15:1838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Goncharov T, Hedayati S, Mulvihill MM, Izrael-Tomasevic A, Zobel K, Jeet S, et al. Disruption of XIAP-RIP2 Association Blocks NOD2-Mediated Inflammatory Signaling. Mol Cell 2018; 69:551–65 e7. [DOI] [PubMed] [Google Scholar]
- [116].Qi J, Dong Z, Liu J, Peery RC, Zhang S, Liu JY, et al. Effective Targeting of the Survivin Dimerization Interface with Small-Molecule Inhibitors. Cancer Res 2016; 76:453–62. [DOI] [PubMed] [Google Scholar]
- [117].Berezov A, Cai Z, Freudenberg JA, Zhang H, Cheng X, Thompson T, et al. Disabling the mitotic spindle and tumor growth by targeting a cavity-induced allosteric site of survivin. Oncogene 2012; 31:1938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol 2008; 10:1440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Steller H Regulation of apoptosis in Drosophila. Cell Death Differ 2008; 15:1132–8. [DOI] [PubMed] [Google Scholar]
- [120].Li X, Wang J, Shi Y. Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE. Nat Commun 2011; 2:408. [DOI] [PubMed] [Google Scholar]
- [121].Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P, et al. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet 2011; 7:e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lee TV, Kamber Kaya HE, Simin R, Baehrecke EH, Bergmann A. The initiator caspase Dronc is subject of enhanced autophagy upon proteasome impairment in Drosophila. Cell Death Differ 2016; 23:1555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018; 25:486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2012; 19:107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Daskalov A, Heller J, Herzog S, Fleissner A, Glass NL. Molecular Mechanisms Regulating Cell Fusion and Heterokaryon Formation in Filamentous Fungi. Microbiol Spectr 2017; 5: 10.1128/microbiolspec.FUNK-0015-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hickey PC, Jacobson D, Read ND, Glass NL. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet Biol 2002; 37:109–19. [DOI] [PubMed] [Google Scholar]
- [127].Glass NL, Dementhon K. Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol 2006; 9:553–8. [DOI] [PubMed] [Google Scholar]
- [128].Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell 2003; 2:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Goncalves AP, Heller J, Daskalov A, Videira A, Glass NL. Regulated Forms of Cell Death in Fungi. Front Microbiol 2017; 8:1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Jacobson DJ, Beurkens K, Klomparens KL. Microscopic and Ultrastructural Examination of Vegetative Incompatibility in Partial Diploids Heterozygous at het Loci in Neurospora crassa. Fungal Genet Biol 1998; 23:45–56. [DOI] [PubMed] [Google Scholar]
- [131].van Diepeningen AD, Debets AJ, Hoekstra RF. Heterokaryon incompatibility blocks virus transfer among natural isolates of black Aspergilli. Curr Genet 1997; 32:209–17. [DOI] [PubMed] [Google Scholar]
- [132].Zhang DX, Spiering MJ, Dawe AL, Nuss DL. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 2014; 197:701–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Debets F, Yang X, Griffiths AJ. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet 1994; 26:113–9. [DOI] [PubMed] [Google Scholar]
- [134].Hutchison E, Brown S, Tian C, Glass NL. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology 2009; 155:3957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Goncalves AP, Hall C, Kowbel DJ, Glass NL, Videira A. CZT-1 is a novel transcription factor controlling cell death and natural drug resistance in Neurospora crassa. G3 (Bethesda) 2014; 4:1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dementhon K, Iyer G, Glass NL. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot Cell 2006; 5:2161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Pinan-Lucarre B, Paoletti M, Clave C. Cell death by incompatibility in the fungus Podospora. Semin Cancer Biol 2007; 17:101–11. [DOI] [PubMed] [Google Scholar]
- [138].Pinan-Lucarre B, Balguerie A, Clave C. Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell 2005; 4:1765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Pinan-Lucarre B, Paoletti M, Dementhon K, Coulary-Salin B, Clave C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol 2003; 47:321–33. [DOI] [PubMed] [Google Scholar]
- [140].Dementhon K, Saupe SJ, Clave C. Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Mol Microbiol 2004; 53:1625–40. [DOI] [PubMed] [Google Scholar]
- [141].Kreuzaler P, Watson CJ. Killing a cancer: what are the alternatives? Nat Rev Cancer 2012; 12:411–24. [DOI] [PubMed] [Google Scholar]
- [142].Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A 1997; 94:9773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Riek R, Saupe SJ. The HET-S/s Prion Motif in the Control of Programmed Cell Death. Cold Spring Harb Perspect Biol 2016; 8: 10.1101/cshperspect.a023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol 2011; 22:460–8. [DOI] [PubMed] [Google Scholar]
- [145].Saupe SJ, Daskalov A. The [Het-s] prion, an amyloid fold as a cell death activation trigger. PLoS Pathog 2012; 8:e1002687 10.1371/journal.ppat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Turcq B, Deleu C, Denayrolles M, Begueret J. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet 1991; 228:265–9. [DOI] [PubMed] [Google Scholar]
- [147].Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, et al. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 2012; 10:e1001451 10.1371/journal.pbio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kajava AV, Klopffleisch K, Chen S, Hofmann K. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep 2014; 4:7436 10.1038/srep07436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Daskalov A, Paoletti M, Ness F, Saupe SJ. Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes. PLoS One 2012; 7:e34854 10.1371/journal.pone.0034854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mompean M, Li W, Li J, Laage S, Siemer AB, Bozkurt G, et al. The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell 2018; 173:1244–53 10.016/j.cell.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Meng H, Liu Z, Li X, Wang H, Jin T, Wu G, et al. Death-domain dimerization-mediated activation of RIPK1 controls necroptosis and RIPK1-dependent apoptosis. Proc Natl Acad Sci U S A 2018; 115:E2001–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein ML, et al. The mechanism of prion inhibition by HET-S. Mol Cell 2010; 38:889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Wu H, Fuxreiter M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell 2016; 165:1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Heller J, Clave C, Gladieux P, Saupe SJ, Glass NL. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc Natl Acad Sci U S A 2018; 115:E2292–E301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 2016; 16:407–20. [DOI] [PubMed] [Google Scholar]
- [156].Lamkanfi M, Dixit VM. In Retrospect: The inflammasome turns 15. Nature 2017; 548:534–5. [DOI] [PubMed] [Google Scholar]
- [157].Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014; 156:1207–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci 2010; 11:436–49. [DOI] [PubMed] [Google Scholar]
- [159].Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc 2014; 136:4551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Bojsen R, Regenberg B, Folkesson A. Persistence and drug tolerance in pathogenic yeast. Curr Genet 2017; 63:19–22. [DOI] [PubMed] [Google Scholar]
- [161].Sears D, Schwartz BS. Candida auris: An emerging multidrug-resistant pathogen. Int J Infect Dis 2017; 63:95–8. [DOI] [PubMed] [Google Scholar]
- [162].CDC. Tracking Candida auris https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html 2018.
- [163].Perfect JR. The antifungal pipeline: a reality check. Nat Rev Drug Discov 2017; 16:603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Wuyts J, Van Dijck P, Holtappels M. Fungal persister cells: The basis for recalcitrant infections? PLoS Pathog 2018; 14:e1007301 10.1371/journal.ppat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang Y, Yew WW, Barer MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother 2012; 56:2223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]