Abstract
Clownfishes are an iconic group of coral reef fishes, especially known for their mutualism with sea anemones. This mutualism is particularly interesting as it likely acted as the key innovation that triggered clownfish adaptive radiation. Indeed, after the acquisition of the mutualism, clownfishes diversified into multiple ecological niches linked with host and habitat use. However, despite the importance of this mutualism, the genetic mechanisms allowing clownfishes to interact with sea anemones are still unclear. Here, we used a comparative genomics and molecular evolutionary analyses to investigate the genetic basis of clownfish mutualism with sea anemones. We assembled and annotated the genome of nine clownfish species and one closely related outgroup. Orthologous genes inferred between these species and additional publicly available teleost genomes resulted in almost 16,000 genes that were tested for positively selected substitutions potentially involved in the adaptation of clownfishes to live in sea anemones. We identified 17 genes with a signal of positive selection at the origin of clownfish radiation. Two of them (Versican core protein and Protein O-GlcNAse) show particularly interesting functions associated with N-acetylated sugars, which are known to be involved in sea anemone discharge of toxins. This study provides the first insights into the genetic mechanisms of clownfish mutualism with sea anemones. Indeed, we identified the first candidate genes likely to be associated with clownfish protection form sea anemones, and thus the evolution of their mutualism. Additionally, the genomic resources acquired represent a valuable resource for further investigation of the genomic basis of clownfish adaptive radiation.
Keywords: anemonefish, Amphiprion, coral reef fish, positive selection, key-innovation
Introduction
The spectacular diversity of life on Earth that Darwin sought to explain in On the origin of Species (Darwin 1859) emerged through a variety of complex biological processes. One of these is adaptive radiation, during which a single ancestral species diversifies into many descendants adapted to a wide range of ecological conditions. It is considered of crucial importance and potentially responsible for much of the diversity of life (Simpson 1953; Schluter 2000). However, the process of adaptive radiation is an extremely complex process influenced by a variety of ecological, genetic, and developmental factors, and since decades researchers have been trying to understand the causes, consequences, and mechanisms of this process (Simpson 1953; Givnish and Sytsma 1997; Schluter 2000; Givnish 2015; Soulebeau et al. 2015).
Current theories postulate that adaptive radiations start with ecological opportunity, in which an ancestral species occupies an environment with abundant and underused resources (Yoder et al. 2010; Stroud and Losos 2016). Divergent natural selection among these different resources should subsequently drive the adaptive diversification of the ancestral species through ecological speciation (Rundell and Price 2009). This starting ecological opportunity is seen in empirical studies, with clades diversifying after the colonization of isolated areas (e.g. Galapagos finches: Grant and Grant 2008; African Rift Lake cichlids: Seehausen 2006; Caribbean Anolis lizards: Losos 2009), following the appearance of new habitat and resources (e.g. grasses and grazing horses in MacFadden 2005), after an extinction event (e.g. Erwin 2007), or following the evolution of traits (i.e., key innovation) allowing the interaction with the environment in a novel way (e.g. the evolution of flight in bats in Simmons et al. 2008; the evolution of the pharyngeal jaw apparatus in cichlids and labrid fishes in Mabuchi et al. 2007; the evolution of antifreeze glycoproteins in Antarctic notothenioid fishes in Near et al. 2012).
The importance of ecological opportunity was also emphasized by modeling approaches aiming at identifying the general patterns that should be observed during adaptive radiations (Gavrilets and Vose 2005; Gavrilets and Losos 2009). Other general patterns predicted by these studies include patterns of evolutionary rates, geographical components of speciation, selection intensity, and genomic architecture (Gavrilets and Vose 2005; Gavrilets and Losos 2009). Until recently, however, empirical studies describing adaptive radiations were not able to fully assess the predictions made by those models, as the necessary deep genomic data were missing. This data start to be available for iconic clades such as cichlids (Brawand et al. 2014), sticklebacks (Jones et al. 2012), Heliconius butterflies (Dasmahapatra et al. 2012; Supple et al. 2013), and Darwin’s finches (Lamichhaney et al. 2015). These studies revealed the first insight on the genomic mechanisms of adaptive radiations, with for example, the reuse of standing variation having an important role in the evolution of sticklebacks and cichlids (Jones et al. 2012; Brawand et al. 2014), and introgressive hybridization playing a role in Heliconius and Darwin’s finches diversification (Dasmahapatra et al. 2012; Lamichhaney et al. 2015).
Despite these empirical studies, modeling approaches and acquired genomic data, much remains to be understood about the general mechanisms of adaptive radiations. This is particularly true for marine ecosystems, where described cases of adaptive radiations remain scarce (i.e., the nothotenioids fish in Antarctica, Near et al. 2012) as barriers to dispersal are uncommon, making ecological speciation less likely than in more isolated landscapes (Puebla 2009). Therefore, to obtain a wider overview of the processes underlying adaptive radiations, it is essential to step back from classical textbook examples of adaptive radiations and gather data from less studied clades occurring in different ecosystems. One interesting case of recently described adaptive radiation in marine environments is represented by clownfishes (family Pomacentridae, genera Amphiprion and Premnas, Litsios et al. 2012).
Clownfishes are an iconic group of coral reef fishes distributed in the tropical belt of the Indo-Pacific Ocean, and it includes 26 currently recognized species and 2 natural hybrids (Fautin and Allen 1997; Ollerton et al. 2007; Gainsford et al. 2015). A distinctive characteristic of this group is the mutualistic interaction they maintain with sea anemones (Fautin and Allen 1997; fig. 1). This mutualism is particularly important as it was proposed to act as the key innovation that triggered clownfish adaptive radiation (Litsios et al. 2012). Indeed, after the acquisition of the mutualism, clownfishes diversified into multiple ecological niches linked with both host and habitat use (Litsios et al. 2012).
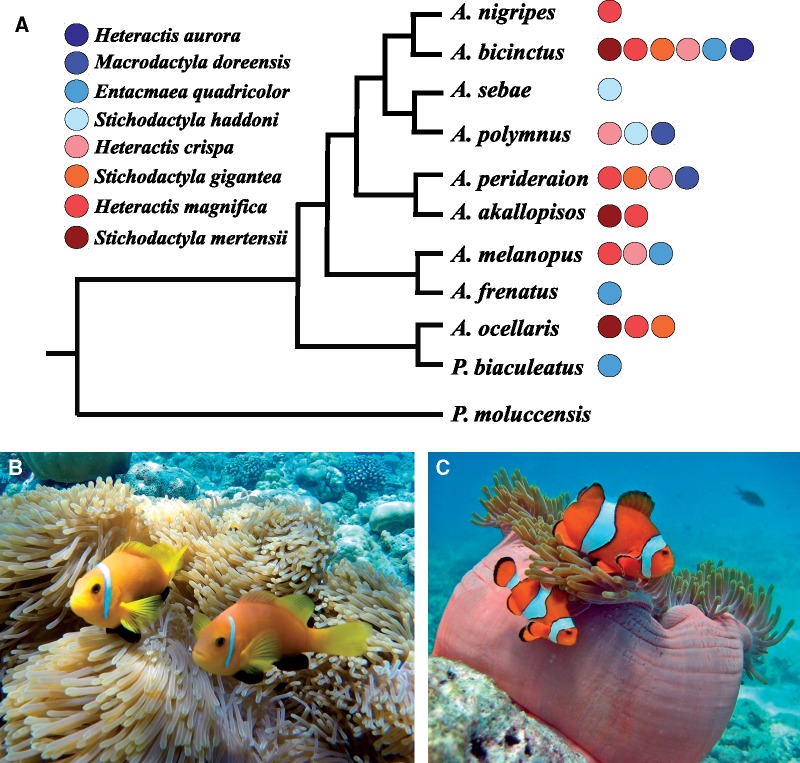
Fig. 1.
—(A) Phylogenetic relationship of the nine selected clownfish species, Amphiprion frenatus (available from Marcionetti et al. 2018), and the outgroup species Pomacentrus moluccensis. Circles represent the sea anemones species with whom each clownfish can interact (Fautin and Allen 1997). Closely related species with divergent host usages were selected. (B) and (C) show, respectively, A. nigripes and A. ocellaris in their host sea anemone Heteractis magnifica.
Although this mutualism is seen as the key innovation driving the adaptive radiation of clownfishes, the underlying mechanisms that are at the basis of the evolution of the mutualism are still unclear. Sea anemones are sessile organisms that have evolved a variety of toxins used for protection and hunting, which can be extremely harmful to the fishes (Nedosyko et al. 2014). These toxins are released from specialized cells (i.e., cnidocytes) after the combination of chemical and mechanical stimuli (Anderson and Bouchard 2009), or they are secreted in the mucus of sea anemones (Mebs 2009). Clownfishes must have evolved specific characteristics to counteract these toxins and it was suggested that the mucus coating of clownfishes played a central role in this protection (Schlichter 1976; Lubbock 1980, 1981; Miyagawa and Hidaka 1980; Miyagawa 2010; Balamurugan et al. 2014). For instance, some evidence (Abdullah and Saad 2015) suggests that the mucus of A. ocellaris has a significantly low level of N-acetylneuraminic acid, which was shown to stimulate cnidocytes discharge (Ozacmak et al. 2001; Anderson and Bouchard 2009). Additionally, a resistance against sea anemones cytolytic toxins was observed in several clownfish species (Mebs 1994), suggesting a potential role of specific immune response mechanisms (Mebs 2009).
We can today take advantage of next-generation sequencing technologies to obtain genomes of different clownfish species to better understand the mechanisms of clownfish adaptation to sea anemones. By considering the mutualism as a new and advantageous phenotype that evolved in clownfishes, we can investigate the role of selection on the genetic basis of the adaptation. Indeed, phenotypic evolution may occur through alterations of the structure of protein-coding genes, which can be fixed by positive selection if they confer an advantage (as, e.g., in Spady et al. 2005; Hoekstra et al. 2006; Protas et al. 2006; Lynch 2007). In this study, we obtained genomic data for several clownfish species and test the genetic mechanisms underlying clownfish protection from sea anemone toxins using comparative genomic and molecular evolution analyses. We hypothesized that this protection could be granted by positively selected substitutions modifying the original function of protein-coding genes in a way that ultimately prevent the release of sea anemone toxins or provide immunity to these toxins. These mechanisms resulted in the mutualism with sea anemones, which acted as the probable key innovation that triggered clownfish adaptive radiation. Thus, this study will not only improve our understanding of the genetic mechanisms involved at the beginning of an adaptive radiation but it will also provide data for further investigation of the diversification process in marine environments.
Materials and Methods
Species Selection, DNA Extraction, Library Preparation
We selected nine clownfish species (Premnas biaculeatus, Amphiprion ocellaris, A. perideraion, A. akallopisos, A. polymnus, A. sebae, A. melanopus, A. bicinctus, A. nigripes) spanning the whole clownfish divergence and the whole distribution range of the group. Genomic data from one additional species (A. frenatus) were already available (Marcionetti et al. 2018). This total of ten species forms five pairs of closely related but ecologically divergent species in their host and habitat usage (fig. 1). The lemon damselfish (Pomacentrus moluccensis) was selected as a closely related outgroup species whose estimated divergence with clownfishes ranged from 21.5 to 38.5 Ma depending on the study (Litsios et al. 2012; Sanciangco et al. 2016).
One individual of each clownfish species and P. moluccensis was obtained from a local aquarium shop. Because all individuals were acquired from an aquarium shop, their exact origin is not available. All individuals passed away beforehand at the aquarium shop, and samples from deceased fish were received. Thus, all the individuals sampled did not undergo any manipulation or experimentation in the laboratory. All remaining samples are stored at the Department of Computational Biology, University of Lausanne (Switzerland).
For each species, genomic DNA (gDNA) was extracted from 50 mg of fin tissue using DNeasy Blood & Tissue Kit (Quiagen, Hilden, Germany) and following manufacturer’s instructions. Short-insert (350 bp) paired-end (PE) libraries were prepared from 100 ng of gDNA at the Lausanne Genomic Technologies Facility (LGTF, Switzerland), using TruSeq Nano DNA LT Library Preparation Kit (Illumina). PE libraries of A. ocellaris and P. moluccensis were sequenced on two lanes of Illumina HiSeq2000 at the LGTF, while PE libraries for the other species were each sequenced on one lane. For A. ocellaris, a long-insert (3 kb) mate pairs (MP) library was prepared from 4 μg of gDNA at Fasteris SA (Geneva, Switzerland) using the Nextera Mate Pair Library Preparation Kit from Illumina. This MP library was sequenced on a half lane of Illumina HiSeq2500 at Fasteris.
Whole-Genome Assemblies
Because we needed to acquire genomic data for ten different species, we investigated an alternative strategy for genome assembly that allowed for reduced coverage and library types, as well as decreased computational time and memory usage during the assembly process. This strategy consisted of using an available reference genome of a species as the substrate to reconstruct the genome of a second species. Such approach is conceivable only if the divergence between the considered species is low, and if large genomic rearrangements did not occur since the split of those species. Because clownfishes are a fast diversifying group with most of the diversification occurring 5 Ma (Litsios et al. 2012), we did not expect to observe high divergence and large genomic rearrangements within the group. Thus, we investigated the feasibility of such reference-based approach in clownfishes by assembling A. ocellaris genome with both de novo and reference-based strategies, and by comparing then the results. Similar methods taking advantage of reference genomes from closely related species for the assembly of new species are also reported in the literature (Buza et al. 2015; Lischer and Shimizu 2017), with for instance the genomes of Arabidopsis thaliana (Schneeberger et al. 2011) or Tetrao tetrix (Wang et al. 2014) being obtained successfully by using a reference to guide their assemblies.
The processing of sequenced reads for all species and the de novo genome assembly of A. ocellaris were performed as reported in Marcionetti et al. (2018; more details in supplementary material and methods, Supplementary Material online). Reference-based assembly of A. ocellaris was performed using half of the original coverage (1 Illumina lane, ∼50×) and employing A. frenatus genome as the reference. For this, we mapped processed PE reads of A. ocellaris against the assembly of A. frenatus using Stampy (v1.0.28; Lunter and Goodson 2011), setting the expected substitution rate parameter to 0.05 to allow the mapping of reads including substitutions. We retrieved the consensus sequences with SAMtools (v1.3; Li et al. 2009) and we closed gaps with GapCloser (from SOAPdenovo2, v2.04.240; Luo et al. 2012). The remaining species were also assembled following this reference-guided assembly strategy, and using the entire set of processed reads (total of 1 Illumina lane per species).
Validation of the Reference-Based Assembly Strategy
To validate the reference-based approach, we compared assembly statistics and mapping rates of the de novo and reference-guided assemblies of A. ocellaris. Because it is difficult to perform synteny analysis with fragmented assemblies, we used SynMap2 (Haug-Baltzell et al. 2017) to investigate the synteny and collinearity between the recently available A. percula genome (Lehmann et al. 2018) and the two A. ocellaris assemblies. We reordered A. ocellaris scaffolds according to the alignments regions of A. percula genome and we plotted the synteny in R (R Core Team 2013).
To confirm that the reference-guided assembly method resulted in the correct reconstruction of species sequences, we reconstructed a phylogeny containing additional publicly available clownfish samples. Only eight nuclear gene sequences were available for these additional samples (BMP-4, Glyt, Hox6, RAG1, RH, S7, SVEP1, Zic1; GenBank ID in supplementary table S1, Supplementary Material online). We extracted these genes from the obtained assemblies based on the functional annotation of the genomes. We aligned the genes using Mafft (v7.305 Katoh and Standley 2013) and we concatenated the alignments within Geneious (v10.0.5; Kearse et al. 2012.). We constructed the gene trees for each separate alignment and for the concatenate one with PhyML (v.3.3, GTR + Γ model, bootstrap 100, Guindon et al. 2010). The trees were plotted with Dendroscope (v1.4, Huson et al. 2007) and they were visually examined for inconsistency in topology.
Genome Quality Investigation and Genome Annotation
We assessed the quality of all the obtained assemblies (the de novo A. ocellaris assembly and all the reference-guided assemblies) and we structurally and functionally annotated them as performed in Marcionetti et al. (2018, more details in supplementary material and methods, Supplementary Material online). The completeness of the genome annotation was investigated with BUSCO (v1.0, data set: vertebrates; Simão et al. 2015). For each species, we calculated the sequence coverage (proportion of the sequence covered by mapped reads) and average depth (average number of reads mapping to the gene) with bedtools coverage (v2.22.1, Quinlan and Hall 2010).
Orthology Inference, HOG Filtering, and Classification
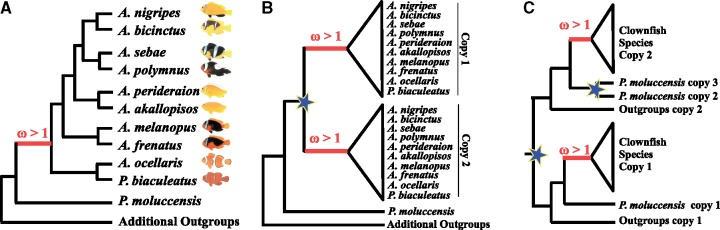
We inferred orthologous genes between the ten clownfish species, P. moluccensis and 12 publicly available Actinopterygii species (Astyanax mexicanus, Danio rerio, Gadus morhua, Gasterosteus aculeatus, Lepisosteus oculatus, Oreochromis niloticus, Oryzias latipes, Poecilia formosa, Takifugu rubripes, Tetraodon nigroviridis, Xiphophorus maculeatus, and Stegastes partitus, supplementary table S2 and fig. S1, Supplementary Material online). The use of additional Actinopterygii species was necessary for the positive selection analysis. Indeed, the power in detecting patterns of positive selection is increasing with increasing taxa (Anisimova et al. 2002). Orthology inference was performed with OMA standalone (Altenhoff et al. 2013) on the proteomes of the 23 species, using the species tree represented in supplementary figure S1, Supplementary Material online, to guide the clustering of orthologous pairs. For each species and gene, the longest protein isoform was used for orthology inference. The resulting Hierarchical Orthologous Groups (HOGs) were filtered to keep only HOGs containing both clownfish and outgroup species, with a minimum number of species required set to six species. Additionally, only HOGs containing sequences for P. moluccensis were kept, as this species corresponds to the most closely related species to clownfish, and it is necessary for specifically aiming at the ancestral branch of clownfish group (fig. 2).
Fig. 2.
—Examples of gene trees for 1-to-1 OG (A), clownfish-specific duplicated genes (B), and overall multicopy HOGs (C). Mutualism with sea anemones appeared on the ancestral basis of clownfishes (in red in A). Gene were tested for positive selection (ω > 1) on branches specific to all clownfishes (in red in A, B, and C). Gene duplication events are visualized with blue stars.
HOGs were classified as single-copy orthologs (1-to-1 OG), clownfish-specific duplicated genes (i.e., genes with potential duplications event on the branch leading to clownfish), and overall multicopy orthologs. Single-copy orthologs were obtained by selecting HOGs with one sequence per species at different taxonomic levels. We defined “level 1” as all species being kept, “level 2” where L. oculatus was removed, “level 3” where L. oculatus, D. rerio, and A. mexicanus were removed and “level 4” where L. oculatus, D. rerio, A. mexicanus, and G. morhua were removed (supplementary fig. S1, Supplementary Material online). HOGs were classified as clownfish-specific duplicated genes when the minimal number of gene copies in clownfishes was higher than the maximum number of gene copies in the outgroup species. This strategy allows for possible incomplete annotation of both clownfish and outgroup genomes to be accounted for. A minimum number of two outgroups was required for all analyses, and the four different taxonomic level (supplementary fig. S1, Supplementary Material online) were considered. To identify potential false positives, we investigated the coverage (proportion of sequence covered by mapped reads, and the number of mapped reads) and length of clownfish-specific duplicated genes. The remaining HOGs were classified as overall multicopy orthologs.
Positive Selection Analysis
All HOGs resulting from orthology inference were composed by the longest protein isoforms of each gene and species. For each HOG, we performed protein alignments with MAFFT (v7.305, G-INS-i strategy; Katoh and Standley 2013), with the option “–allowshift.” Codon alignments were inferred from protein alignments with PAL2NAL (Suyama et al. 2006). Because positive selection analyses are sensitive to alignment errors (Fletcher and Yang 2010), we filtered the alignments to keep only highly confident homologous regions. For this, we followed a stringent filtering approach proposed in the Selectome database (Moretti et al. 2014). Details are available in supplementary material and methods, Supplementary Material online. The strict filtering strategy also allows reducing false positives potentially arising from the use of different isoforms for different species in each HOG, as mentioned in Villanueva-Canas et al. (2013). Gene trees were obtained with PhyML (v3.3; Guindon et al. 2010) from the unfiltered codon alignments. For each HOG, the gene tree was reconstructed with both HKY85 and GTR substitution models (100 bootstrap). The best model was selected with a likelihood ratio test (df= 4).
For 1-to-1 OGs, positive selection was tested with CodeML implemented in the PAML package (v4.9; Yang 2007), using the filtered codon alignments and obtained gene trees. We tested for positive selection at the onset of the clownfish radiation with the “branch-site model,” by setting the branch leading to the clownfish as foreground branch and all other branches as the background (fig. 2A). The null model (with foreground ω constrained to be smaller or equal to 1) was compared with the alternative model (with estimation of foreground ω) with a likelihood ratio test (df = 1). We corrected for multiple-testing with the Benjamin–Hochberg method implemented in the q value package in R (FDR threshold of 0.1; Dabney et al. 2010). Additional information is reported in supplementary material and methods, Supplementary Material online.
For clownfish-specific duplicated genes and overall multicopy HOGs, positive selection was tested with the method aBSREL implemented in HyPhy (v2.3.7; Smith et al. 2015). The analysis was run in an exploratory way, testing for positive selection at each branch (fig. 2B and C). Although this approach reduces the power due to multiple testing, it was preferred as we do not know a priori which copy of the genes may be positively selected. We corrected for multiple-testing with the Benjamin–Hochberg method implemented in the q value package in R (FDR threshold of 0.1; Dabney et al. 2010).
Positively selected HOGs were annotated by retrieving the SwissProt ID annotation of genes forming the HOGs. We ensured that all genes of different species forming the HOGs were annotated with the same function. Gene trees were plotted with FigTree (v.1.4.2; Rambaut 2014).
Comparison of Gene Trees versus Species Tree Approaches
The tree topology has an effect on the inference of positive selection (Diekmann and Pereira-Leal 2015), and the use of either gene trees or the species tree may lead to different results if topology incongruence is present. We investigated the effect of using gene trees or species tree in the positive selection analysis by randomly selecting 5,000 1-to-1 OGs and inferring positive selection using the species trees as input tree. We investigated the level of topology incongruence in the randomly selected data set by calculating the unweighted Robinson–Foulds (uRF) distance between the species tree and the gene tree using the python library DendroPy (Sukumaran and Holder 2010) and compared it with the results of positive-selection measured as the number of significant results, both before (P values <0.05) and after (q values < 0.05) multiple-testing correction. More information is available in supplementary material and methods, Supplementary Material online.
Power and Type I Error in Positive Selection Analyses
We investigated the power to detect positive selection on the branch leading to clownfishes by simulating data using the software evolver in the PAML package (v4.9; Yang 2007), and by testing positive selection on the simulated data with CodeML. We simulated codon alignments (alignment length: 5,000, 1,000, and 550 codons) under the branch-site model, with ω varying both among sites and branches, to match the model used in the positive selection analyses. We generated trees following the species tree topology, and with branch lengths randomly drawn from the branch lengths distributions obtained from all gene trees of analyzed HOGs. Different selection strengths were simulated, with ω values ranging from 2 to 900. To assess the level of Type I errors in the analysis, we also simulated codon alignments without positive selection (ω = 0.5 and ω = 1 on the foreground branch). For each alignment length, randomly generated tree, and ω value, we simulated four set of sequences (supplementary table S3, Supplementary Material online).
Simulated codon alignments were tested for positive selection with CodeML (PAML v4.9; Yang 2007), applying the same pipeline developed for the test of positive selection on 1-to-1 OG. We investigated the power to detect positive selection and the number of false positive (Type I errors) by recording the number of significant LRT (P value <0.05) between the null model and the alternative model. More information on this analysis is available in supplementary material and methods, Supplementary Material online.
Results
Genome Assemblies, Quality Assessment, and Annotations
For all species, paired-end (PE) sequencing and reads processing with ALLPATH-LG module (Gnerre et al. 2011) performed well. This resulted in an average coverage of 125.8× for A. ocellaris (sequenced on two Illumina lanes), while an average coverage between 36.5× (A. sebae) and 54.7× (A. polymnus) was obtained for the other species (sequenced on a single Illumina lane; supplementary table S4, Supplementary Material online). The sequencing of long-insert mate-pairs (MP) for A. ocellaris resulted in a low level of unique reads (31.8%), which corresponds to a final genomic coverage of 3.5× (supplementary table S4, Supplementary Material online).
The higher coverage and different library types for A. ocellaris were necessary because a classical de novo approach was also used to assemble the genome of this species. The best de novo assembly for A. ocellaris was obtained with ALLPATH-LG processed reads assembled with PLATANUS (total assembly size of 744 Mb, 27,951 scaffolds, N50 of 136 kb; table 1 and supplementary table S5A, Supplementary Material online). The fragmentation of the assembly is mainly due to the low number of unique MP, which prevented an optimal scaffolding. Reference-guided assemblies for A. ocellaris (obtained with only half of the original PE coverage and without the use of MP) and for the additional species were less fragmented. This is because they were constructed based on the genome of A. frenatus, and therefore statistics for these assemblies mainly reflect the ones of A. frenatus genome (Marcionetti et al. 2018; table 1 and supplementary table S5B, Supplementary Material online).
Table 1.
Genome Assembly and Annotation Statistics for the Nine Assembled Clownfish Species and Pomacentrus moluccensis
| De Novo Assembly |
Reference-Guided Assembly |
|||||
|---|---|---|---|---|---|---|
| Amphiprion ocellaris | A. ocellaris | A. bicinctus | A. nigripes | A. polymnus | A. sebae | |
| Total assembly size (Mb) | 744 | 798 | 799 | 800 | 800 | 799 |
| Number of scaffolds | 27,951 | 16,543 | 16,953 | 16,995 | 17,050 | 16,941 |
| N50 (bp) | 136,417 | 246,482 | 246,127 | 246,124 | 246,119 | 245,870 |
| non-ATGC characters (%) | 4.6 | 3.6 | 2.8 | 2.7 | 2.7 | 2.9 |
| Paired-ends mapping rate (%) | 95.3 | 98.2 | 98.9 | 98.9 | 99.0 | 99.0 |
| Number of genes | 24,383 | 29,913 | 28,891 | 28,558 | 28,640 | 28,727 |
| Number of proteins | 27,606 | 33,845 | 33,219 | 32,905 | 33,128 | 33,271 |
| Functional annotated proteins (%) | 94.0 | 92.7 | 93.2 | 93.1 | 92.9 | 92.9 |
| CEGMA genes in assembly (%) | 97.2 | 99.6 | 99.6 | 99.6 | 99.6 | 100 |
| BUSCOs genes in annotation (%) | 87 | 93 | 94 | 95 | 95 | 95 |
|
Reference-guided assembly |
||||||
| A. akallopisos | A. perideraion | A. melanopus | P. biaculeatus | P. moluccensis | ||
| Total Assembly Size (Mb) | 801 | 801 | 803 | 797 | 794 | |
| Number of scaffolds | 17,172 | 17,212 | 17,399 | 16,164 | 15,505 | |
| N50 (bp) | 246,052 | 246,037 | 245,703 | 247,121 | 246,470 | |
| non-ATGC characters (%) | 1.9 | 2.0 | 1.4 | 2.9 | 7.9 | |
| Paired-ends mapping rate (%) | 99.0 | 99.0 | 97.4 | 96.9 | 81.2 | |
| Number of genes | 28,730 | 29,014 | 29,408 | 28,170 | 28,885 | |
| Number of proteins | 33,120 | 33,320 | 33,768 | 32,385 | 32,027 | |
| Functional annotated proteins (%) | 93.1 | 92.9 | 92.7 | 93.8 | 94.0 | |
| CEGMA genes in assembly (%) | 99.6 | 99.6 | 99.6 | 99 | 99.6 | |
| BUSCOs genes in annotation (%) | 95 | 94 | 94 | 95 | 89 | |
Note.—For A. ocellaris, statistics of both de novo and reference-guided assemblies are reported. Reference-guided assemblies were obtained using A. frenatus (Marcionetti et al. 2018) as reference genome. N50 index indicates the shortest scaffold length above which 50% of the genome is assembled. CEGMA and BUSCOs genes represent the completeness of the genome assemblies and annotations, respectively.
The completeness of the obtained assemblies was assessed with CEGMA. As for A. frenatus genome, reference-guided assemblies resulted in 99% to 100% of the core genes being either completely or partially represented in the assembly of the different species. Because of the larger fragmentation, this number is slightly decreased in A. ocellaris de novo assembly, with only 97.2% of the genes being retrieved (table 1 and supplementary table S6, Supplementary Material online).
To assess the correct reconstruction of the genomic sequence for each species, we investigated the mapping statistics of PE against the assembled genomes. Here as well, slightly better results were obtained for reference-guided assemblies compared with the A. ocellaris de novo assembly. Indeed, depending on the species, between 97% and 99% of reads mapped against the corresponding reference-guided assembly, while only 95% of PE reads of A. ocellaris mapped against its de novo assembly (table 1 and supplementary table S7, Supplementary Material online). Additionally, to validate the reference-guided assembly strategy, we performed synteny analysis of the de novo and reference-guided assemblies of A. ocellaris and the recently available A. percula genome (Lehmann et al. 2018). As expected, we found that overall the synteny and collinearity pattern is consistent between the two assembly strategies and A. percula genome (supplementary figs. S2 and S3, Supplementary Material online).
Structural annotation of A. ocellaris de novo assembly resulted in 24,383 predicted genes. This number is increased in reference-based assemblies, for which the number of predicted genes ranged from 28,170 to 29,913 depending on the species (table 1 and supplementary table S8, Supplementary Material online). The number of annotated genes in two recent assemblies of A. percula (Lehmann et al. 2018) and A. ocellaris (Tan et al. 2018) genomes were 26,597 and 27,420, respectively. This suggests that several genes predictions are missing in our de novo assembly of A. ocellaris, but not in our reference-based assemblies. Evidence for this is also provided by BUSCO analyses, which showed that 13% of BUSCO genes were missing in the A. ocellaris de novo assembly, while only 5% to 6% of genes were missing in the reference-guided assemblies of clownfishes (table 1 and supplementary table S9, Supplementary Material online). The missing gene predictions in the de novo A. ocellaris assembly are due to the increased fragmentation of this assembly compared with the reference-based assemblies (table 1).
For all assemblies, most of the predicted proteins (92% to 94%) were functionally annotated (table 1 and supplementary table S10, Supplementary Material online), with proteins in the reference-based assemblies showing an overall good coverage with proteins from the SwissProt database (supplementary fig. S4, Supplementary Material online, in red). This coverage was reduced for proteins predicted in the A. ocellaris de novo assembly (supplementary fig. S4, Supplementary Material online, in blue), suggesting a lower quality of gene structure prediction for the de novo assembly.
The phylogeny reconstructed based on all the publicly available clownfish sequences and sequences extracted from the assembled genomes resulted in the expected topology (supplementary fig. S5, Supplementary Material online). Most of the assembled individuals branched with individuals of the same species. Three exceptions were observed for A. ocellaris, A. akallopisos, and A. melanopus. However, these inconsistencies are mainly due to a lack of resolution, as suggested by the low support of these nodes.
Taken together, these results indicate that the genome of A. ocellaris obtained by reference-guided assembly is at least as good as the one obtained with the de novo assembly strategy. Thus, through a reference-based approach, we managed to obtain overall good quality assemblies for all the species while reducing the sequencing and computational costs. Amphiprionocellaris de novo assembly was not considered for further analysis.
Orthology Inference, HOG Filtering, and Classification
Orthology inference performed with OMA on Actinopterygii proteomes (10 clownfish species and 13 outgroup species, supplementary fig. S1, Supplementary Material online) resulted in a total of 35,976 Hierarchical Orthologous Groups (HOGs). To investigate the level of selective pressure on genes at the origin of clownfishes, HOGs composed by both clownfish species and outgroup Actinopterygii species are necessary (fig. 2). For this reason, we discarded 14,903 HOGs that were formed by either only clownfish sequences (i.e., clownfish-specific HOGs) or by only outgroup sequences (i.e., outgroup-specific HOGs). These discarded HOGs were mainly composed by inaccurately predicted proteins, as suggested by them being composed by only few species with overall shorter sequences compared with the remaining HOGs (supplementary fig. S6, Supplementary Material online). In addition, 5,133 HOGs were discarded because they were formed by fewer than six species or because they did not contain any sequence from P. moluccensis, which is necessary to specifically target our estimation of positive selection on the ancestral branch of clownfishes. This filtering resulted in a total of 15,940 HOGs being retained for positive selection analysis.
Out of the 15,940 HOGs, 13,215 were single-copy when considering the four taxonomic levels (supplementary fig. S1, Supplementary Material online). As HOGs may be formed by several 1-to-1 OG (i.e., single-copy OG at a given taxonomic level) when considering the different taxonomic level, these 13,215 HOGs corresponded to a total of 13,500 1-to-1 OG. Only 19 HOGs were found specifically duplicated in clownfishes when considering the four taxonomic levels (i.e., clownfish-specific duplicated genes), while the remaining 2,706 HOGs were classified as overall multicopy genes. Most of the genes in the 23 Actinopterygii genomes were part of these 15,940 HOGs tested for signature of positive selection at the basis of the clownfishes (supplementary table S11, Supplementary Material online).
Positive Selection on Single-Copy Genes
We tested for positive selection at the basis of the clownfishes clade on the 13,500 1-to-1 OG. After correction for multiple testing, we found a total of 13 genes that evolved under positive selection in the branch leading to clownfishes (table 2). The functions of the positively selected genes are diverse and they are reported in table 3. Examples of positively selected genes include genes involved in cell adhesion, such as protocadherin-15 (HOG4335_1a), vezatin (HOG16495), and Cadherin-related family member 2 (HOG4262). Other examples include the Versican Core Protein (HOG1437), which is involved in hyaluronic acid binding, and the Protein O-GlcNAcase (HOG16500), which plays a role in the N-acetylglucosamine metabolic process.
Table 2.
Results for the Positive Selection Analysis on 1-to-1 OG
| HOG Name | logL (Null Model) | logL (Alternative Model) | LRT P Values | q Values | Positively Selected Sites (%) | ω |
|---|---|---|---|---|---|---|
| HOG11195 | −41,547.05 | −41,526.90 | 2.19E-010 | 2.28E-007 | 0.8 | 233.5 |
| HOG16495 | −13,655.66 | −13,642.13 | 1.96E-007 | 1.53E-004 | 0.5 | 999.0 |
| HOG1437 | −16,835.23 | −16,825.39 | 9.19E-006 | 4.79E-003 | 0.3 | 248.0 |
| HOG9295 | −4,960.03 | −4,950.14 | 8.71E-006 | 4.79E-003 | 1.0 | 102.8 |
| HOG5827_3b | −2,138.88 | −2,129.50 | 1.48E-005 | 6.61E-003 | 1.1 | 999.0 |
| HOG11468 | −14,064.81 | −14,055.92 | 2.47E-005 | 7.85E-003 | 0.4 | 760.6 |
| HOG4335_1a | −23,361.23 | −23,352.35 | 2.51E-005 | 7.85E-003 | 0.5 | 340.4 |
| HOG11290 | −10,498.06 | −10,489.15 | 2.42E-005 | 7.85E-003 | 0.4 | 999.0 |
| HOG14257 | −23,503.25 | −23,495.89 | 1.24E-004 | 3.53E-002 | 0.1 | 999.0 |
| HOG16500 | −11,287.91 | −11,280.90 | 1.79E-004 | 3.87E-002 | 0.2 | 999.0 |
| HOG21171 | −69,291.90 | −69,284.86 | 1.75E-004 | 3.87E-002 | 1.3 | 25.3 |
| HOG4262 | −31,942.98 | −31,935.94 | 1.76E-004 | 3.87E-002 | 2.0 | 27.8 |
| HOG16343 | −6,212.65 | −6,205.67 | 1.86E-004 | 3.87E-002 | 0.5 | 359.5 |
Note.—The 13 positively selected genes are reported here, with information on the log-likelihood of the null model (no positive selection) and alternative model (positive selection on the branch leading to clownfishes, fig. 2A). Likelihood-ratio test (LRT) P values, multiple-testing corrected q values, the proportion of sites under positive selection on the tested branch (ω classes 2a and 2b) and the corresponding ω values are reported for each gene.
Table 3.
Annotation of the Positively Selected 1-to-1 OG
| HOG Name | SwissProt ID | SwissProt Name |
|---|---|---|
| HOG11195 | P0C5E4 | Phosphatidylinositol phosphatase PTPRQ |
| HOG16495 | Q5RFL7 | Vezatin |
| HOG1437 | Q90953 | Versican core protein |
| HOG9295 | Q3UHZ5 | Leiomodin-2 |
| HOG5827_3b | Q803L0 | Protein lin-28 homolog A |
| HOG11468 | Q9D805 | Calpain-9 |
| HOG4335_1a | Q0ZM14 | Protocadherin-15 |
| HOG11290 | Q92581 | Sodium/hydrogen exchanger 6 |
| HOG14257 | Q8WXG6 | MAP kinase-activating death domain protein |
| HOG16500 | Q9EQQ9 | Protein O-GlcNAcase |
| HOG21171 | Q9TU53 | Cubilin |
| HOG4262 | Q9BYE9 | Cadherin-related family member 2 |
| HOG16343 | P37892 | Carboxypeptidase E |
The use of either genes trees or species tree for the positive selection analysis on a subset of the data produced similar results. Before multiple testing correction, 86 genes were found consistently positively selected (i.e., significant in both species tree and gene trees analysis). Twelve additional genes were found positively selected when using the gene trees, and 16 when using the species tree (supplementary table S12, Supplementary Material online). However, these differences are no longer present after multiple-testing correction, which resulted in seven genes consistently being detected as positively selected with both species and gene trees (supplementary table S12, Supplementary Material online). Thus, the use of either gene or species trees does not affect the results of the analysis after correcting for multiple testing.
The simulations showed that the positive selection analysis performed on data simulated under neutral or purifying selection scenarios resulted in no false positive detected, and this independently of the simulated sequence length (supplementary fig. S7, Supplementary Material online). The power to detect positive selection is increased when the strength of selection is larger (i.e., increasing ω; supplementary fig. S8, Supplementary Material online) until it reaches a maximum of 75% for large ω (ω > 200, supplementary fig. S8, Supplementary Material online). This pattern is observed also for shorter simulated sequences, although the maximum power for large ω is reduced.
Transcriptomic analysis (see Supplementary Material and Methods online) provided evidence of expression of at least seven positively selected 1-to-1 OG in A. ocellaris epidermis (TPM > 2, supplementary table S14, Supplementary Material online), which is the layer of interaction with sea anemones tentacles. Taken together, all these results provide a set of candidate genes that may be linked with the acquisition of the particular life-history traits of clownfishes, such as the mutualism with sea anemones.
Positive Selection on Duplicated Genes
For the overall multicopy genes (i.e., genes with duplications not specific to clownfishes), no evidence of positive selection on gene copies specific to clownfish was found. Out of the 19 clownfish-specific duplicated HOGs, we found four genes with a signature of positive selection in at least one gene copy specific to clownfishes (table 4 and supplementary fig. S9, Supplementary Material online). All these positively selected clownfish-specific duplicated genes were annotated with SwissProt IDs (table 5). One of these positively selected gene is the T-cell receptor alpha (HOG5488), which plays a role in immunity responses. Two other genes, the Glutathione S-transferase (HOG5344) and Cytochrome P450 (HOG4655), are involved in the detoxification of various endogenous and exogenous substances. Transcriptomic analysis (see Supplementary Material and Methods online) showed evidence of expression of Glutathione S-transferase (HOG5344) in A. ocellaris epidermis (TPM > 2, supplementary table S14, Supplementary Material online), supporting a potential role of this gene in the interaction with sea anemones.
Table 4.
Results for the Positive Selection on Clownfish-Specific Duplicated Genes
| Node | LRT | Corrected P Value | ω1 | ω2 | |
|---|---|---|---|---|---|
| HOG4655 | |||||
| Node172 | 26.5139 | 0.0001 | 0.0681 (97%) | 46.2 (2.9%) | |
| Node119 | 24.3395 | 0.0004 | 1.00 (98%) | 10,000 (2.2%) | |
| Node70 | 19.7309 | 0.0041 | 0.00 (100%) | 10,000 (0.21%) | |
| HOG5344 | |||||
| Node89 | 23.3766 | 0.0006 | 0.00 (85%) | 15.9 (15%) | |
| Node204 | 20.9201 | 0.002 | 0.0401 (87%) | 11.1 (13%) | |
| Node142 | 17.5192 | 0.0109 | 0.00 (95%) | 10,000 (5.1%) | |
| AMPSE31855 | 15.4114 | 0.0314 | 0.00 (92%) | 92.5 (7.6%) | |
| Node120 | 14.6422 | 0.046 | 0.00 (98%) | 10,000 (2.4%) | |
| HOG5488 | |||||
| ENSDARG00000098394 | 20.6963 | 0.001 | 0.184 (81%) | 111 (19%) | |
| Node63 | 20.8642 | 0.001 | 0.00 (67%) | 9,410 (33%) | |
| Node32 | 15.7701 | 0.0121 | 0.00 (92%) | 10,000 (8.4%) | |
| HOG19886 | |||||
| Node7 | 21.4722 | 0.001 | 0.484 (93%) | 47.7 (7.1%) | |
| Node26 | 17.837 | 0.0061 | 0.439 (95%) | 21.3 (4.9%) |
Note.—We report the nodes with inferred positive selection, the Likelihood Ratio Test (LRT) statistic for selection, the corrected P value and the value of the inferred ω classes, with the proportion of sites in each class. The reported nodes correspond to nodes from the inferred gene trees (supplementary fig. S9, Supplementary Material online).
Table 5.
Annotation of the Clownfish-Specific Duplicated Genes
Discussion
The knowledge on the genomic mechanisms underlying adaptive radiations is still scarce, and this is particularly true when the radiations occurred in a marine ecosystem. In this study, we acquired genomic data for nine clownfishes species and one closely related outgroup, in addition to the previously available genome of A. frenatus (Marcionetti et al. 2018). These are valuable resources that may be further exploited for advancing our understanding of the genomic patterns observed in adaptive radiations.
In this study, these genomic data sets were exploited to obtain the first insights on the genetic mechanisms underlying the clownfish protection from sea anemone toxins, which resulted in the mutualism that acted as the probable key innovation that triggered clownfish adaptive radiation. Out of the almost 16,000 genes tested, we only found a total of 17 genes showing a signal of positive selection at the origin of clownfishes. Even if a causal link cannot be confirmed without further experimental validation, some of these positively selected genes show functions that are likely to be associated with the protection from sea anemone toxins.
Genomic Resources for Clownfishes and P. moluccensis
To reduce sequencing and computational effort, genomes assemblies for the clownfish species and P. moluccensis were obtained using a reference-based approach. Similar approaches were successfully used in the literature (Buza et al. 2015; Lischer and Shimizu 2017), with for instance the genomes of Arabidopsis thaliana (Schneeberger et al. 2011) or Tetrao tetrix (Wang et al. 2014) being obtained by using a reference to guide their assemblies. These methods may nevertheless raise concerns about the validity of the final genomic sequences obtained, especially in the case of nonconserved synteny and collinearity between the reference and the newly assembled species.
Teleost genomes have been found to be evolutionary stable, with genetic content of chromosomes being conserved over nearly 200 Myr of evolution (Schartl et al. 2013). Almost complete synteny and large blocks of collinearity were also observed between the sea bass (Dicentrarchus labrax) and three teleost genomes: Oreochromis niloticus, Gasterosteus aculeatus, and Tetraodon nigroviridis (Tine et al. 2014). The divergence time between D. labrax and these three species is >100 Ma (126.8 Ma for O. niloticus, 104.8 for T. nigroviridis and G. aculeatus;Sanciangco et al. 2016). Nonconserved synteny and noncollinearity were therefore not expected to be a concern here, especially considering that clownfishes started to diversify between 12.1 (Santini et al. 2009) and 18.9 Ma (Litsios et al. 2012).
The observed synteny and collinearity between the two A. ocellaris assemblies (i.e., de novo and reference-guided) and the available genome of A. percula (Lehmann et al. 2018, supplementary figs. S2 and S3, Supplementary Material online) confirmed this expectation. This clearly indicates that the use of A. frenatus as reference did not introduce a striking bias in the reconstructed genomic sequences of clownfishes. Evidence for this is also given by the good mapping statistics of paired-end reads (and mate-reads for A. ocellaris) for all reference-based assemblies (supplementary table S7, Supplementary Material online), which imply that most reads mapped with the expected insertion size and orientation on the assembled genomes. Therefore, the use of A. frenatus assembly as reference resulted in all the assemblies having an overall quality that is comparable to the reference used (Marcionetti et al. 2018) but achieved with only half of the original coverage and only one library type.
Although we verified the validity of the obtained genomes, we should keep in mind that the reference-guided assemblies may still miss characteristics that are specific to newly assembled species but not found in the used reference. For instance, species-specific gene duplications or losses may be omitted when looking exclusively at the resulting assembled genomes. However, these features may be identified by taking advantage of the gene coverage, in a similar way of what it is done for copy-number variation detection (e.g. Yoon et al. 2009; Trost et al. 2018). Here, the distribution of the gene coverage was overall normally distributed, with the mean centered on the expected average coverage (supplementary fig. S10, Supplementary Material online), suggesting the absence of high levels of species-specific duplication or losses. Species-specific features are in any case out of the scope of this study, as we investigated here what is common to all clownfish species.
Candidate Genes Involved in Clownfish Protection from Sea Anemones Toxins
Evolutionary mechanisms that may result in the appearance of new advantageous traits (such as the protection from toxins) include positive selection on protein-coding genes, where mutations altering the function of genes are fixed in the population because they are favorable. Examples of this process have already been reported (e.g. Spady et al. 2005; Hoekstra et al. 2006; Protas et al. 2006; Lynch 2007). By contrast, purifying selection is the mechanism preventing the fixation of deleterious mutations, as those mutations are detrimental for the organism. Therefore, the appearance of an advantageous trait by positive selection in an ancestral species may be followed by a switch in the selective pressure, with this trait undergoing purifying selection in the descendant species (i.e., if this trait is still advantageous for them). Examples of this scenario with pattern of positive selection in internal branches of a phylogeny, followed by a switch to purifying selection are found in primates (Perry et al. 2012; Daub et al. 2017), grasses (Schwerdt et al. 2015), seagrasses (Wissler et al. 2011), and rust fungi (Silva et al. 2015).
This scenario of a switch in selective pressure in the internal branches was tested in this study as it fits well with the appearance of clownfish-specific life-history traits, such as their mutualism with sea anemones. For this, the presence of the outgroup P. moluccensis was necessary, as it allowed to specifically aim for the ancestral branch of clownfishes. Thus, after the acquisition of the advantageous traits such as the ability to live unharmed in sea anemones on this specific branch, these traits must have been conserved (i.e., underwent purifying selection) across the whole clownfish group.
A total of 17 genes (either single copy or duplicated genes) were found to have evolved under positive selection at the origin of clownfishes, and showed a later switch to purifying selection in the other branches of the clade. Simulations showed that the level of false positive results that we can expect in our data sets is very low, which suggests that we can have a high confidence in these results. In addition to the mutualism with sea anemones, these positively selected genes that are specific to the evolution of clownfishes may be associated with other clownfish-specific traits, such as their outstanding estimated lifespan (Buston and García 2007) or their hierarchical social structure (Buston 2003). Similarly, although none of the positively selected genes are documented as involved in the evolution of coloration in teleosts (Lorin et al. 2018), we cannot exclude their potential role in the evolution of clownfishes particular coloration.
One of the detected positively selected genes is the HOG1437, which is annotated as coding for the Versican Core Protein. This protein plays a role in intercellular signaling, in connecting cells with the extracellular matrix, and it may also take part in the regulation of cell motility, growth, and differentiation. Additionally, it is binding hyaluronic acid (Bignami et al. 1989; Perides et al. 1989), a glycosaminoglycan distributed widely throughout connective, epithelial, and neural tissues. Glycosaminoglycans are polysaccharides consisting of repeating amino-sugar units, such as N-acetylglucosamine (GlcNAc). Another gene found positive selected is the HOG16500, annotated as coding for Protein O-GlcNAse, which function is to cleave GlcNAc from O-glycosylated proteins (Toleman et al. 2006).
These observations are interesting since N-acetylated sugars (such as GlcNAc) have been shown to trigger the discharge of sea anemones cnidocytes, leading to the release of toxins (Anderson and Bouchard 2009). Chemoreceptors of N-acetylated sugars are located in cells surrounding cnidocytes, which change in morphology in response to stimulation of these receptors by N-acetylated sugars. These structural modifications alter the mechanical properties of the hair bundles, and tune them to the frequencies of vibrations emitted by swimming prey, resulting in an increase in the baseline discharge of cnidocytes when the anemone touches the prey (Thorington and Hessinger 1988a; Mire‐Thibodeaux and Watson 1994). One N-acetylated sugar shown to trigger cnidocytes discharges is the N-acetylneuraminic acid (NANA; Ozacmak et al. 2001). This compound was found to be significantly lacking in A. ocellaris mucus (Abdullah and Saad 2015). GlcNAc is another N-acetylated sugar that may be recognized by N-acetylated chemoreceptors, and thus trigger the discharge of sea anemone toxins. This is supported by studies showing that hyaluronic acid, which is composed by GlcNAc, was the only polysaccharide able to strongly excite cnidocytes and trigger their discharge (Lubbock 1979; Thorington and Hessinger 1988b).
The two positively selected genes HOG1437 (Versican Core Protein) and HOG16500 (Protein O-GlcNAse) display therefore interesting functions associated with N-acetylated sugars. The Versican Core Protein is observed to be expressed in A ocellaris epidermis (supplementary table S14, Supplementary Material online), that is, the layer of interaction with sea anemones tentacles. A low signal of Protein O-GlcNAse expression was also detected in A. ocellaris epidermis (supplementary table S14, Supplementary Material online). With these evidence, we hypothesize that these genes might play a role in the masking (GlcNAc binding by versican core protein) or removal (cleavage by Protein O-GlcNAse) of N-acetylated sugars. This would therefore help decrease or prevent the stimulation of the chemoreceptors for N-acetylated sugars, thus preventing or decreasing cnidocytes discharge and the release of toxins. Clownfishes might thus not necessarily be fully resistant to toxins released by cnidocytes, but they could have evolved a system that prevents these toxins to be discharged (as previously suggested in Lubbock 1980, 1981).
Sea anemones toxicity is not only due to the discharge of cnidocytes but also by the presence of secreted toxins in sea anemone mucus such as cytolytic toxins. A resistance against sea anemone cytolytic toxins was effectively observed in some clownfish species (Mebs 1994), suggesting that this resistance may be mediated through specific mechanisms such as immune response (Mebs 2009). Clownfish-specific duplicated genes involved in immunity response as the T-cell receptor alpha (HOG5488), or involved in detoxification such as Cytochrome P450 (HOG4655; Manikandan and Nagini 2018) and Glutathione S-transferases (HOG5344; Sheehan et al. 2001) are found positively selected at the origin of clownfishes. These genes are part of gene families having a large number of different roles (Sheehan et al. 2001; Manikandan and Nagini 2018), thus making it difficult to define their precise function. In addition, genes involved in immune responses are often seen as subject to positive selection (Schlenke and Begun 2003; Jiggins and Kim 2007), and have been seen to evolve faster than nonimmune genes (McTaggart et al. 2012). For these reasons, direct links between these positively selected genes and a potential role in the protection from sea anemones secreted toxins cannot be drawn without further experimental evidence.
Furthermore, as only some clownfishes species showed resistance to cytolytic toxins (Mebs 1994), this resistance could have appeared later in the evolution of clownfishes, and be specific to only some clownfish species.
In addition to positive selection on protein-coding genes (i.e., coding changes), the acquisition of new phenotypes may also occur through regulatory changes that alter gene expression profiles (e.g. Wittkopp et al. 2003; Shapiro et al. 2004). However, the identification and analysis of noncoding elements such as transcription factor binding sites in nonmodel organisms remain challenging. Therefore, although not analyzed here, we may expect that regulatory sequences evolution has acted in concert with the coding changes (i.e., positive selection on coding genes) identified in this study in the built up of clownfish mutualism with sea anemones.
Conclusions
In this study, we acquired genomic data for nine clownfishes species and one closely related outgroup. These data are a valuable resource that may be further exploited for advancing our understanding of the genomic patterns observed in adaptive radiations.
Using these newly assembled genomes, we investigated here the mechanisms underlying clownfish protection from sea anemone toxins, which resulted in the acquisition of the mutualism that likely acted as the key innovation triggering clownfish adaptive radiation. We identified 17 genes with a signal of positive selection at the origin of clownfishes. Some of these genes showed interesting function associated with N-acetylated sugars, which are known to be involved in sea anemones discharge of toxins. Although further experimental validations are necessary to find a causal link between these genes and the ability to interact with sea anemones, this study provides the first genomic approach to try to disentangle the mechanisms behind the mutualism between sea anemones and clownfishes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank the Vital-IT infrastructure from the Swiss Institute of Bioinformatics for the computational resources and the Lausanne Genomic Technology Facility for the sequencing. Funding: University of Lausanne funds, Swiss National Science Foundation, Grant Number: 31003A-163428.
Data deposition: Raw Illumina reads are available in the Sequence Read Archive in the NCBI database (BioProject ID is enough for SRA reference BioProject ID: PRJNA515163). The assembled genomes and their annotation are available in Zenodo repository (doi: 10.5281/zenodo.2540241).
Literature cited
- Abdullah NS, Saad S.. 2015. Rapid detection of N-acetylneuraminic acid from false clownfish using HPLC-FLD for symbiosis to host sea anemone. Asian J Appl Sci. 03(5):858–864. [Google Scholar]
- Altenhoff AM, Gil M, Gonnet GH, Dessimoz C.. 2013. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One 8(1):e53786.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PA, Bouchard C.. 2009. The regulation of cnidocyte discharge. Toxicon 54(8):1046–1053. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z.. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19(6):950–958. [DOI] [PubMed] [Google Scholar]
- Balamurugan J, Kumar TA, Kannan R, Pradeep HD.. 2014. Acclimation behaviour and bio-chemical changes during anemonefish (Amphiprion sebae) and sea anemone (Stichodactyla haddoni) symbiosis. Symbiosis 64(3):127–138. [Google Scholar]
- Bignami A, Lane WS, Andrews D, Dahl D.. 1989. Structural similarity of hyaluronate binding proteins in brain and cartilage. Brain Res Bull. 22(1):67–70. [DOI] [PubMed] [Google Scholar]
- Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buston P. 2003. Social hierarchies: size and growth modification in clownfish. Nature 424(6945):145.. [DOI] [PubMed] [Google Scholar]
- Buston PM, García MB.. 2007. An extraordinary life span estimate for the clown anemonefish Amphiprion percula. J Fish Biol. 70(6):1710–1719. [Google Scholar]
- Buza K, Wilczynski B, Dojer N.. 2015. RECORD: reference-assisted genome assembly for closely related genomes. Int J Genomics. 2015:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney A, Storey JD, Warnes GR.. 2010. qvalue: q-value estimation for false discovery rate control. R Package Version 1.38.0, http://github.com/jdstorey/qvalue, last accessed November 2018. [Google Scholar]
- Darwin C. 1859. The origin of species by means of natural election, or the preservation of favored races in the struggle for life. London: Murray. [Google Scholar]
- Dasmahapatra KK, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487(7405):94.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub JT, Moretti S, Davydov II, Excoffier L, Robinson-Rechavi M.. 2017. Detection of pathways affected by positive selection in primate lineages ancestral to humans. Mol Biol Evol. 34(6):1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann Y, Pereira-Leal JB.. 2015. Gene tree affects inference of sites under selection by the branch-site test of positive selection. Evol Bioinformatics. 11:EBO-S30902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH. 2007. Increasing returns, ecological feedback and the Early Triassic recovery. Palaeoworld 16(1–3):9–15. [Google Scholar]
- Fautin DG, Allen GR.. 1997. Anemonefishes and their host sea anemones. Perth: Western Australian Museum. [Google Scholar]
- Fletcher W, Yang Z.. 2010. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 27(10):2257–2267. [DOI] [PubMed] [Google Scholar]
- Gainsford A, Van Herwerden L, Jones GP.. 2015. Hierarchical behaviour, habitat use and species size differences shape evolutionary outcomes of hybridization in a coral reef fish. J Evol Biol. 28(1):205–222. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Losos JB.. 2009. Adaptive radiation: contrasting theory with data. Science 323(5915):732–737. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Vose A.. 2005. Dynamic patterns of adaptive radiation. Proc Natl Acad Sci U S A. 102(50):18040–18045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ. 2015. Adaptive radiation versus ‘radiation’ and ‘explosive diversification’: why conceptual distinctions are fundamental to understanding evolution. New Phytol. 207(2):297–303. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Sytsma KJ.. 1997. Molecular evolution and adaptive radiation. New York: Cambridge University Press. [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR.. 2008. How and why species multiply: the radiation of Darwin’s finches. Princeton (NJ: ): Princeton University Press. [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E.. 2017. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics 33(14):2197–2198. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP.. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313(5783):101–104. [DOI] [PubMed] [Google Scholar]
- Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8(1):460.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kim KW.. 2007. A screen for immunity genes evolving under positive selection in Drosophila. J Evol Biol. 20(3):965–970. [DOI] [PubMed] [Google Scholar]
- Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484(7392):55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518(7539):371.. [DOI] [PubMed] [Google Scholar]
- Lehmann R, et al. 2018. Finding Nemo’s Genes: a chromosome‐scale reference assembly of the genome of the orange clownfish Amphiprion percula. Mol Ecol Resour. Advance online publication. doi:10.1111/1755-0998.12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischer HE, Shimizu KK.. 2017. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinformatics 18(1):474.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios G, et al. 2012. Mutualism with sea anemones triggered the adaptive radiation of clownfishes. BMC Evol Biol. 12(1):212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorin T, Brunet FG, Laudet V, Volff JN.. 2018. Teleost fish-specific preferential retention of pigmentation gene-containing families after whole genome duplications in vertebrates. G3 (Bethesda). 8(5):1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of Anoles .Berkeley (CA: ): University of California Press. [Google Scholar]
- Lubbock R. 1979. Chemical recognition and nematocyte excitation in a sea anemone. J Exp Biol. 83(1):283–292. [Google Scholar]
- Lubbock R. 1980. Why are clownfishes not stung by sea anemones? Proc Biol Sci. 207(1166):35–61. [Google Scholar]
- Lubbock R. 1981. The clownfish/anemone symbiosis: a problem of cellular recognition. Parasitology 82(01):159–173. [Google Scholar]
- Lunter G, Goodson M.. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21(6):936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1(1):18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ. 2007. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A 2 genes. BMC Evol Biol. 7(1):2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi K, Miya M, Azuma Y, Nishida M.. 2007. Independent evolution of the specialized pharyngeal jaw apparatus in cichlid and labrid fishes. BMC Evol Biol. 7(1):10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadden BJ. 2005. Fossil horses–evidence for evolution. Science 307(5716):1728–1730. [DOI] [PubMed] [Google Scholar]
- Manikandan P, Nagini S.. 2018. Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets. 19(1):38–54. [DOI] [PubMed] [Google Scholar]
- Marcionetti A, Rossier V, Bertrand JA, Litsios G, Salamin N.. 2018. First draft genome of an iconic clownfish species (Amphiprion frenatus). Mol Ecol Resour. 18(5):1092–1101. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ, Obbard DJ, Conlon C, Little TJ.. 2012. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol Biol. 12(1):63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebs D. 1994. Anemonefish symbiosis: vulnerability and resistance of fish to the toxin of the sea anemone. Toxicon 32(9):1059–1068. [DOI] [PubMed] [Google Scholar]
- Mebs D. 2009. Chemical biology of the mutualistic relationships of sea anemones with fish and crustaceans. Toxicon 54(8):1071–1074. [DOI] [PubMed] [Google Scholar]
- Mire‐Thibodeaux P, Watson GM.. 1994. Morphodynamic hair bundles arising from sensory cell/supporting cell complexes frequency‐tune nematocyst discharge in sea anemones. J Exp Zool. 268(4):282–292. [DOI] [PubMed] [Google Scholar]
- Miyagawa K. 2010. Experimental analysis of the symbiosis between anemonefish and sea anemones. Ethology 80(1–4):19–46. [Google Scholar]
- Miyagawa K, Hidaka T.. 1980. Amphiprion clarkii juvenile: innate protection against and chemical attraction by symbiotic sea anemones. Proc Jpn Acad B. 56(6):356–361. [Google Scholar]
- Moretti S, et al. 2014. Selectome update: quality control and computational improvements to a database of positive selection. Nucleic Acids Res. 42(D1):D917–D921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, et al. 2012. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci U S A. 109(9):3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedosyko AM, Young JE, Edwards JW, da Silva KB.. 2014. Searching for a toxic key to unlock the mystery of anemonefish and anemone symbiosis. PLoS One 9(5):e98449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerton J, McCollin D, Fautin DG, Allen GR.. 2007. Finding NEMO: nestedness engendered by mutualistic organization in anemonefish and their hosts. Proc Biol Sci. 274(1609):591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozacmak VH, Thorington GU, Fletcher WH, Hessinger DA.. 2001. N-acetylneuraminic acid (NANA) stimulates in situ cyclic AMP production in tentacles of sea anemone (Aiptasia pallida): possible role in chemosensitization of nematocyst discharge. J Exp Biol. 204(11):2011–2020. [DOI] [PubMed] [Google Scholar]
- Perides G, Lane WS, Andrews D, Dahl D, Bignami A.. 1989. Isolation and partial characterization of a glial hyaluronate-binding protein. J Biol Chem. 264(10):5981–5987. [PubMed] [Google Scholar]
- Perry GH, et al. 2012. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 22(4):602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, et al. 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 38(1):107.. [DOI] [PubMed] [Google Scholar]
- Puebla O. 2009. Ecological speciation in marine v. freshwater fishes. J Fish Biol. 75(5):960–996. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2014. FigTree version 1.4.2. Computer program distributed by the author. Available from: http://tree.bio.ed.ac.uk/software/figtree/ [internet]; last accessed November 2018.
- R Core Team. 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: http://www.R-project.org/ [Internet], last accessed January 2019.
- Rundell RJ, Price TD.. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol Evol. 24(7):394–399. [DOI] [PubMed] [Google Scholar]
- Sanciangco MD, Carpenter KE, Betancur-R R.. 2016. Phylogenetic placement of enigmatic percomorph families (Teleostei: percomorphaceae). Mol Phylogenet Evol. 94:565–576. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME.. 2009. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 9(1):194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 45(5):567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ.. 2003. Natural selection drives Drosophila immune system evolution. Genetics 164(4):1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichter D. 1976. Macromolecular mimicry: substances released by sea anemones and their role in the protection of anemone fishes In: Coelenterate ecology and behavior. Boston (MA: ): Springer; p. 433–441. [Google Scholar]
- Schluter D. 2000. The ecology of adaptive radiation. Oxford: Oxford University Press. [Google Scholar]
- Schneeberger K, et al. 2011. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci U S A. 108(25):10249–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdt JG, MacKenzie K, Oehme D, et al. 2015. Evolutionary dynamics of the cellulose synthase gene superfamily in grasses. Plant Physiol. 168(3):968–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc Biol Sci. 273(1597):1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, et al. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428(6984):717.. [DOI] [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA.. 2001. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 360(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DN, et al. 2015. Genomic patterns of positive selection at the origin of rust fungi. PLoS One 10(12):e0143959.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simmons NB, Seymour KL, Habersetzer J, Gunnell GF.. 2008. Primitive early eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451(7180):818.. [DOI] [PubMed] [Google Scholar]
- Simpson GG. 1953. The major features of evolution. New York: Columbia University Press. [Google Scholar]
- Smith MD, et al. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32(5):1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulebeau A, et al. 2015. The hypothesis of adaptive radiation in evolutionary biology: hard facts about a hazy concept. Organ Divers Evol. 15(4):747–761. [Google Scholar]
- Spady TC, et al. 2005. Adaptive molecular evolution in the opsin genes of rapidly speciating cichlid species. Mol Biol Evol. 22(6):1412–1422. [DOI] [PubMed] [Google Scholar]
- Stroud JT, Losos JB.. 2016. Ecological opportunity and adaptive radiation. Annu Rev Ecol Evol Syst. 47(1):507–532. [Google Scholar]
- Sukumaran J, Holder MT.. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26(12):1569–1571. [DOI] [PubMed] [Google Scholar]
- Supple MA, et al. 2013. Genomic architecture of adaptive color pattern divergence and convergence in Heliconius butterflies. Genome Res. 23(8):1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, et al. 2018. Finding Nemo: hybrid assembly with Oxford Nanopore and Illumina reads greatly improves the clownfish (Amphiprion ocellaris) genome assembly. GigaScience 7(3):gix137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorington GU, Hessinger DA.. 1988a. Control of discharge: factors affecting discharge of cnidae. In: The biology of nematocysts. pp. 233–253, San Diego: Academic Press. [Google Scholar]
- Thorington GU, Hessinger DA.. 1988b. Control of cnida discharge: i. Evidence for two classes of chemoreceptor. Biol Bull. 174(2):163–171. [DOI] [PubMed] [Google Scholar]
- Tine M, et al. 2014. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun. 5:5770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman C, Paterson AJ, Shin R, Kudlow JE.. 2006. Streptozotocin inhibits O-GlcNAcase via the production of a transition state analog. Biochem Biophys Res Commun. 340(2):526–534. [DOI] [PubMed] [Google Scholar]
- Trost B, et al. 2018. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet. 102(1):142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Canas JL, Laurie S, Alba MM.. 2013. Improving genome-wide scans of positive selection by using protein isoforms of similar length. Genome Biol Evol. 5(2):457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ekblom R, Bunikis I, Siitari H, Höglund J.. 2014. Whole genome sequencing of the black grouse (Tetrao tetrix): reference guided assembly suggests faster-Z and MHC evolution. BMC Genomics 15(1):180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissler L, et al. 2011. Back to the sea twice: identifying candidate plant genes for molecular evolution to marine life. BMC Evol Biol. 11(1):8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Williams BL, Selegue JE, Carroll SB.. 2003. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc Natl Acad Sci U S A. 100(4):1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yoder JB, et al. 2010. Ecological opportunity and the origin of adaptive radiations. J Evol Biol. 23(8):1581–1596. [DOI] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J.. 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 19(9):1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.