Abstract
Animal model is an essential tool in the life sciences research, notably in understanding the pathogenesis of the diseases and for further therapeutic intervention success. Rodents have been the most frequently used animals to model human disease since the establishment of gene manipulation technique. However, they remain inadequate to fully mimic the pathophysiology of human brain disease, partially due to huge differences between rodents and humans in terms of anatomy, brain function, and social behaviors. Nonhuman primates are more suitable in translational perspective. Thus, genetically modified animals have been generated to investigate neurologic and psychiatric disorders. The classical transgenesis technique is not efficient in that model; so, viral vector‐mediated transgene delivery and the new genome‐editing technologies have been promoted. In this review, we summarize some of the technical progress in the generation of an ad hoc animal model of brain diseases by gene delivery and real transgenic nonhuman primate.
Keywords: animal models, neuroscience, nonhuman primates
1. INTRODUCTION
Worldwide, brain disorders are among the principal cause of disease burden. However, the development of new and better clinical treatment progresses slowly, in part, due to the lack of good animal models.1 Animal models of brain diseases are essential for understanding the pathophysiology and investigating the brain‐behavior relationship that cannot be studied in humans.2
Studies on nonhuman primates (NHP) are still limited. The number of monkeys used for research was anecdotal compared to others species such as the rodents (less than 0.1% vs 80%, respectively).3 Of all animal models used in neurosciences, the brain of monkey is most similar to the human brain.4 Hence, the NHP brings a physiological, genetic, and morphological environment close to human, which makes it more suitable than rodent models.
Various methodologies have been explored to obtain the most similar models to human disease. Surgical and drug‐inducible methods have been used for a long time but nowadays transgenic models are under development. Viral vector delivery has been largely used to obtain and ad hoc animal model. For that purpose, the method of vector delivery is as important as the vector itself to achieve transgenesis. In rodent, transgenic animals have also been obtained by introducing genetic modified embryonic stem cells (ESC) into host blastocysts. Until recently, ESCs of nonhuman primate do not have the competency to generate chimeric animals. That is why the new genome‐editing tools, such as zinc finger nuclease (ZFNs), transcription activator‐like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat/CRISPR‐associated protein 9 (CRISPR/Cas9), were used to disrupt a target gene (knockout [KO]) or to replace (knockin [KI]) a gene of interest.5
To date, there are few validated primate models of central nervous system (CNS) disorders. However, advances in the knowledge of genetic risk factors for brain disorders, including autism, schizophrenia, bipolar disorder, Alzheimer's disease, and Parkinson's disease (PD), open new perspectives. These progressive acquaintances in human disease genetics associated with the recent genetic targeting promise to revolutionize the generation of transgenic animal models in a translational perspective.
2. WHY USING NONHUMAN PRIMATE MODELS?
Within neuroscience, the use of NHPs has led to a greater understanding of the brain physiology and pathology. In rodents, some important aspects of human brain function are difficult to address. Some higher cognitive functions are too complex and evolutionarily recent to be meaningfully studied in rodents.3 Notably, psychiatric diseases are difficult or impossible to model in rodents since they are diagnosed purely through behavioral symptoms.1
Primates are the closest species to humans phylogenetically, sharing a common ancestor with macaques (Old World monkeys) and marmoset (New World monkeys).6 NHPs are the closest to humans in aspects of anatomy, immunology, endocrinology, genome constitution, social behavior, cognitive, metabolic, reproductive, and brain functions.7, 8, 9, 10, 11 They share a similar brain network organization.4, 12, 13, 14 In contrast to most mammalian models, the primate brain has kept characteristics of human brain such as a stable neuronal density with the increased number of neurons,15 complex motor skills, and a highly developed prefrontal cortex. The prefrontal cortex has no clear structural or functional homolog in rodents but is responsible for higher cognitive function in primates and it contains vulnerable domains involved in some psychiatric disorders.8, 16, 17 In the rodent, the striatum is a single entity, whereas cortical fibers in humans and NHPs are grouped into the internal capsule, separating the striatum dorsomedially into the caudate nucleus and ventrolaterally into the putamen, an anatomical characteristic with particular relevance to PD.18 Another interesting feature is the presence of neuromelanin pigment in dopaminergic neurons in the substantia nigra (SN) which can contribute to the vulnerability of this brain region in PD.19
Nonhuman primates are more sophisticated behaviorally.10, 11, 12, 20 The motor, cognitive abilities, and social behavior of NHPs closely mimic humans.1, 21 In humans and NHP, the brain is extremely modified via experience‐dependent plasticity and learning during the juvenile phase.1 Many biological processes implicated or disrupted in brain diseases differ between primates and rodents, such as the detailed organization of the primate oxytocine system, the neurotransmitter and neuromodulatory system, the specific receptor system, and the neuronal connectivity.1
Several genes that are expressed only in primate neuronal system have been described.22 Even if the precise function of these genes is still unknown, it has been hypothesized that they play important roles in higher cognitive function.23 It enables a more accurate assessment of the impact of the pathology on motor outcomes, neuroimaging procedures, and non‐motor symptoms. There are also differences in cell types and noncoding sequences of the genome between primate and rodent brains.1 The mapping of the rhesus (Macaca mulatta) and common marmosets (Callithrix jacchus) genomes accomplished, respectively, in 2007 and 2014 improved NHP models of disease.24
The high similarity in brain anatomy allows noninvasive, high‐resolution imaging of the brain structures and functions by functional magnetic resonance imaging (fMRI) and positron emission topography (PET). Furthermore, for studying neurodegenerative diseases and the physiologic decline of aging in monkeys, various cognitive and behavioral tests are available.7 Moreover, in progressive pathology such as PD where age is the main risk factor, the use of long‐lived animals is crucial. It will allow studying developmental phenotypes and prodromal disease stage, which is difficult to assess in human patients.1
All that characteristics allow researchers to study not only physiological processing (such as visual information processing, working memory, decision‐making, reward processing, social relationship, facial expression and recognition, etc.) but also pathological behaviors (anxiety and depressive‐like phenotypes, addiction, autism, or schizophrenia).2, 3 Primates are already used in pharmaceutical research for pharmacokinetic and toxicology studies that precede human clinical trials but the few number of validated primate models prevent their use for studies on drug efficacy.1 The new precise gene‐editing techniques promise to change this research field, notably after the demonstration of genetic changes in embryos from many species, including NHPs and nonviable human embryos.23, 25, 26, 27, 28, 29, 30, 31
3. THE DIFFERENT TYPES OF NHP TRANSGENIC MODELS
Genetically manipulated NHPs were used to investigate disease pathophysiology. However, researchers have to face to political and technically challenge, such as the long monofetal gestation, the low chances of success due to the possible random insertion of the foreign DNA. The low efficiency of assisted reproductive technologies in producing oocytes and embryos suitable for genetic engineering and the technical challenges of embryonic stem cells derivation and cloning prevent the large‐scale production of transgenic monkeys. Moreover, the high cost of production and maintenance associated with ethical concerns explain the low use of transgenic NHP models.32 For instance, the first transgenic NHP was created more than 25 years after the first transgenic mouse.33 To date, few studies have been carried out using viral‐mediated delivery of gene or gene‐editing methods, notably TALEN or CRISPR‐Cas9.
3.1. The viral vector‐mediated gene transfer models
Before the development of gene‐based methodologies to model diseases, a number of experimental models had been developed, including in NHPs.34, 35 They come essentially in two flavors: pharmacological (eg, reserpine) and toxic (eg, 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine). These models can also be recast by some authors as pathogenic and symptomatic/pathophysiologic, as each may contribute to our understanding of the cause, the mechanisms, and the treatment of PD.34 The MPTP NHP models come into many different regimens of intoxication.36 Although the construct validity could be put into question regarding the mechanisms of cell death, the symptomatic face validity is without equivalent as these models are perfect phenocopies of PD, featuring moor and non‐motor symptoms, as well as the subserving anatomopathology.36, 37, 38, 39, 40, 41 Further understanding the etiology and the mechanisms of cell death required the development of gene‐based models. Among the initial proposals was the local transgenesis that brings the advantages to induce a prolonged effect and to mimic the ongoing disease pathophysiology by promoting the production of the pathological protein.
3.1.1. Postnatal stereotaxic delivery
Among the various viral vectors available for transgenesis, adenoassociated virus (AAV) and lentivirus are the main virus used. As a prelude to translational studies, as well as clinical trials in patients with PD for example, the transduction efficiency of AAV2/1‐GFP, AAV2/5‐GFP. and AAV2/8‐GFP was examined in NHPs. Viral particles were injected into the either striatum or SN via stereotaxic surgery. AAV2/5‐GFP and AAV2/1‐GFP transduce significantly higher number of cells than AAV2/8‐GFP. The transduction was predominantly neuronal with a GFP expression around the injection site: in the cell bodies and fibers in the striatal region or throughout the SN.42 This study confirms that transgene expression in some brain areas can be achieved by postnatal stereotaxic viral delivery.
In PD, new models have emerged in the last few years based on native or mutated ∝‐synuclein (∝‐syn) overexpression. Viral vector‐mediated gene transfer models appear to be promising by mimicking the time course of neuropathology. For the first time, Kirik and colleagues created a NHP ∝‐syn overexpression model43 based on the protocol tested initially in rats.44 They injected high‐titer recombinant AAV2 unilaterally in the SN in adult marmosets to express the wild‐type (WT) or A53T human ∝‐syn. 90%‐95% of all nigral dopamine neurons expressed the ∝‐syn protein which was distributed by anterograde transport throughout axonal and dendritic projections. The overexpression of either WT or mutant ∝‐syn is sufficient to induce a PD‐like neuropathology in the primate including dopaminergic neuron loss (30%‐60%) and motor impairments 16 weeks post‐transduction. Another study was performed by the same group with 24 adult marmosets injected unilaterally into the SN with rAAV2/5‐WT ∝‐syn or rAAV2/5‐A53T∝‐syn. The follow‐up was performed during 1 year. A decline in motor coordination, associated with a decrease in TH+ fibers density with neurodegeneration (especially in the A53T‐∝‐syn injected animals), the presence of pathological Ser129‐phosphorylated ∝‐syn, PK‐resistant ∝‐syn aggregates, and ubiquitin containing aggregates were described.45 In 2015, Yang et al used lentivirus to deliver A53T ∝‐syn unilaterally in the SN of Old World Rhesus monkeys. Aging increased the accumulation of A53T in neurites and its associated neuropathology.46 In the same year, our team reported the lack of an additive role of aging in nigrostriatal neurodegeneration after the injection of rAAV2/9‐A53T ∝‐syn unilaterally in the SN pars compacta in two age groups marmosets (1‐2 years old : n = 8, >6 years old: n = 5).47 Another team described different experiments of injection in order to obtain the best NHP model to test developmental therapeutics. They designed a rAAV1/2‐A53T ∝‐syn injected bilaterally in six cynomolgus macaques. ∝‐Synuclein was expressed in >85% of nigral neurons, the number of TH+ neurons decreased by 50%, and dopamine neurochemistry showed a 21%‐72% reduction according to the groups.48 Based on α‐syn delivery, these studies demonstrated the efficacy of this strategy to model PD. Recently, recombinant AAV1/2 vector was also used to deliver human α‐synuclein under the myelin basic protein promoter to obtain oligodendroglial α‐syn expression following striatal delivery. The aim was to obtain a model for multiple system atrophy with >60% expression in oligodendrocytes.49
In the Huntington's disease (HD), NHP models were first generated by stereotaxic delivery of lentivirus expressed mutant HTT. The lentiviral vector encoding for the first 171 amino acids of the Huntingtin protein with 19 (wild‐type) or 82 (mutated: Htt171‐82Q) polyglutamine repeats was injected into the dorsolateral sensorimotor putamen of macaques. The monkeys injected with the Htt171‐82Q presented the classic symptoms of HD in humans, associating to neuritic and nuclear hungtintin aggregates, astrocytosis and neuronal loss.50
These studies show that stereotaxic delivery allows the generation of different brain disease models but that the transgene expression is relatively concentrated around the injection site. To induce a wider and global central nervous transduction, postnatal systemic delivery was performed.
3.1.2. Postnatal systemic delivery
In a systemic injection approach, using viral vector‐mediated transgene delivery in mice, the cell types targeted shifted from neurons to astrocytes when the viral vector was injected in adults compared to neonate injection.51, 52 In monkeys, injections have also been performed either in newborn or adults. A male cynomolgus macaque was injected intravenously with 1‐3 × 1014 particles of scAAV9‐GFP on postnatal day 1 (P1). Robust GFP expression was described within the dorsal root ganglia and motor neurons along the entire neuraxis.53 Dehay et al also reported intravenously injection with four different AAV9 viral titers (from 9.5 × 109 to 1.33 × 1012 particles/g of body weight) in newborn rhesus macaques (P1). All rAAV2/9‐GFP injections resulted in efficient neuronal transduction of brain areas such as the cortex, midbrain, hippocampus and cerebellum.54 Different time points for the intravenous injection were tested by another group: P1, P30, P90 animals injected with 1‐3 × 1014 vg/kg of sc‐AAV2/9‐GFP and 3‐year‐old cynomolgus macaques injected with 2.7 × 1013 vg/kg of sc‐AAV2/9‐GFP. An efficient motor neurons targeting was described but injection in adults was less powerful compared to injection in young animals (partially explained by the lower dose injected). However, all GFP‐injected animals had extensive transgene expression throughout the entire brain. The cortical region, lateral geniculate, midbrain, pons, and medulla had the higher GFP+ cell density compared to subcortical structures. Interestingly, the glial transduction with microglia and astrocytes were the most prominent cell types targeted whatever the age of the animals.55 In 3‐ to 4‐year‐old rhesus macaques, a reduction in brain transduction was observed compared to mice, along with a clear shift toward mostly glial transduction after intravenous or intra‐arterial injection of sc‐AAV9‐GFP.56 The results obtained with AAV‐GFP in primates confirmed the preliminary results observed in mice with a global neuronal and glial brain transduction but with a preferentially glial transduction, and that particularly in older animals. In another study, different routes of viral vector administration were compared: infusion of AAV‐9 GFP into the internal carotid artery (ICA) or into the cisterna magna (CM). A mainly astrocytic transduction was described in both routes of administration but CM infusion led to a stronger expression throughout the cortex and cerebellum.57 This last study confirms that the transgene expression was mostly observed in superficial postnatally developed structures.
Since the neonatal injection allows a better neuronal transduction compared to adult injection, earlier injections have been performed via in utero delivery. The goal was to obtain an exclusive and efficient neuronal expression.
3.1.3. In utero delivery
In utero viral vector delivery has already shown widespread brain transduction in rodents using retrovirus,58, 59 adenovirus60 or AAV.61, 62, 63 In 2003, Garrett et al performed gene transfer with rAAV2‐GFP via an in utero approach in a rhesus macaque. The recombinant virus was delivered by transabdominal infection into the amniotic fluid under ultrasound control. The tissues analyzed at 15 months of age revealed GFP expression in lung cells and intestines villi.64 Another report focused on the transduction of the central and peripheral nervous systems. scAAV2/9‐CMV‐eGFP vectors genomes (1.1013) were infused slowly into the intrahepatic vein at E140 of a 155‐day gestation. The authors showed that GFP expression throughout the CNS was variable following in utero gene transfer for up to 14 weeks. The transduction was clearly observed in the cerebral cortex. Moreover, both the injected offspring showed transduction of the majority of Purkinje cells in the cerebellum and neurons, including motor neurons in the spinal cord, compared with the greater variation in neuronal transduction found within the cerebrum.65 Further efforts have been done to obtain a homogeneous transgene expression in the cortex and deep brain structures. A widespread neuronal transduction was obtained after in utero delivery of scAAV2/2‐GFP to monkey embryos via intracerebroventricular injection.66
All these promising studies are probably the prelude to the production of animal models after in utero transgene injection. Moreover, researchers have focused on the development of the classical transgenesis in NHP.
3.2. The classical transgenesis
The criteria for a successful transgenic animal are (a) stable integration of the transgene into the host cell genome, (b) expression of the transgene and bioactivity, and (c) germline transmission.67 The advancement in transgenic technology in the end of the 1990 has led to the generation of the first transgenic monkeys in 2001.68, 69 The first transgenic monkey model of human disease, associating genetic modification and disease phenotype, was generated in 2008.70 The germline transmission of the transgene was described in 2009 in common marmoset.71 The efficiency of transgenesis was improved in this study by injecting a higher volume of lentiviral solution into dehydrated embryos with a large perivitelline space and by selecting the transgenic embryos before transfer. Transgenic F1 and F2 animals have been obtained.5 A new era in modeling human genetic disorders has been opened by this success.
3.2.1. What are the available methodologies?
Pluripotent stem cells
Different methodologies were published with the goal to generate chimeric offspring. In mice, genetic modified pluripotent stem cells, such as embryonic stem cells (ESCs) or induced pluripotent cells (iPS), are powerful tool to obtain KO mice.23 The first attempts to translate the methodology used for transgenesis in mouse, such as linear DNA injection into the fertilized eggs and blastocyst injection of marmoset pluripotent stem cells carrying transgene, failed to produce transgenic NHP.72, 73 Established ESCs and freshly isolated inner cell mass (ICMs) failed to produce chimeras when injected into host blastocysts. However, ICMs developed into separate fetuses with placental support from the host embryos. Aggregating of several four cells embryos efficiently produced live chimeric offspring.74 Few years later, Chen et al published the production of chimeric monkey fetuses by injection of ESCs grown in adjusted culture conditions into host morulae.75 Nuclear transfer embryo derived from embryonic blastomeres or fetal fibroblasts was also achieved for the cloning of rhesus monkeys. The transfer of embryonic cell nuclear transfer embryos resulted in a term pregnancy.76 However, for a transgenic model, a foreign gene needs to be introduced into the somatic cell donor of nucleus. This method fails to produce any live monkey.77, 78, 79 Until now, these techniques are not optimal in NHPs. Others strategies have also been explored.
Viral vector delivery
Viral vector delivery, such as lentiviral gene transfer, has shown to be a powerful tool and is one of the most efficient methods in creating transgenic animals.7 A simian immunodeficiency virus (SIV)‐based vector that encodes EGFP was used to infect one‐ to two‐cell stage embryos or four‐ to eight‐cell stage rhesus monkey embryos. Two offspring exhibited whole‐body expression of the EGFP reporter.80 Then, lentivirus was used to create transgenic marmoset and cynomolgus monkey models.81, 82, 83, 84
This methodology was then used to model diseases. To study HD, transgenic rhesus monkeys were generated by microinjection of lentivirus carrying exon‐1 of the htt gene containing 147 CAG repeats into MII oocytes before in vitro fertilization. Animals died following a premature birth at 4 months of gestation and showed abundant neuropils aggregates in swelling neuronal processes.85 The same methodology was performed by Yang et al. where high‐titer lentivirus expressing exon 1 of the human htt gene with 84 CAG repeats was injected. Five monkeys were born, all carrying the transgenic mutant HTT. No clear sign of neurodegeneration was detected but animals develop chorea, dystonia, and other involuntary motor deficiencies similar to HD.70 The germline transmission of the pathogenic mutant HTT in HD monkey was demonstrated by the production of embryos and subsequent derivation of HD monkey embryonic stem cells.86 Longitudinal studies until the 2 or 5 years old of transgenic animals described progressive motor dysfunction and a lack of behavioral inhibitory control, suggesting a functional decline of the frontostriatal pathway in HD monkeys. It is associated to a decreased brain volume and neuronal loss. Thus, these transgenic models closely display the progressive clinical features of HD.87, 88 The emerging symptoms of HD were explored in two male transgenic HD rhesus monkeys. They expressed anxiety and irritability/aggression to an acute stressor as compared to control. Moreover, HD monkey exhibited increased proinflammatory cytokines and higher induction of immune pathway genes compared to controls.89 Another seven transgenic monkeys were produced for longitudinal studies to study the transcriptomic pattern (n = 4)90 or the whole brain white matter integrity (n = 3).91 Interestingly, germline transmission was confirmed by the detection of the transgene directly not only in the male germ cells, but also in embryonic stem cells derived from blastocyst obtained after microinjection of spermatozoa from transgenic male. Moreover, transgenic F1 offspring was produced after artificial insemination.92 These different transgenic animals recapitulate the characteristics of the disease and offer the opportunities to create a transgenic line.
For a Rett syndrome monkey model, Liu et al reported that the transgenic monkeys obtained after the microinjection of lentivirus expressing human MECP2 into the perivitelline space of oocytes exhibited autism‐like behaviors. Male germline transmission of the transgene was also obtained. Five F1 monkeys with defective social behaviors carrying the human MECP2 transgene were born.93 In another report, transgenic animals were generated after ICSI with spermatozoa collected in immunodeficient mice after xenografting of testicular tissue from one transgenic monkey.94 This technique shortens the time necessary to obtain mature spermatozoa and enables the researchers to accelerate the reproduction of transgenic monkeys.16
For PD, Niu et al injected a lentivirus expressing A53T‐∝‐syn into the perivitelline space of 133 MII oocytes before fertilization. Six of the seven live newborns and five of the eight aborted fetuses were positive for transgenic A53T.95 They described in the stillborn monkey brain a number of neurons expressing A53T in the SN and some punctate staining that may reflect the enrichment A53T in synaptic terminals but without S129‐phosphorylated synuclein labeling. Moreover, no neurodegeneration was obtained in the SN, cortex, and striatum. The oldest A53T transgenic animal began to display cognitive defects and an anxiety phenotype at the age of 2.5 years old which can be consistent with the non‐motor symptoms of PD patients at the early disease stage. However, MRI analysis revealed no obvious degeneration in the transgenic monkey brain.95
More recently, a transgenic marmoset model of the polyglutamine disease, a neurodegenerative disease, was reported which recapitulates progressive neurological symptoms 3‐4 months after birth, accumulation of misfolded protein, and neurodegeneration.83 In this study, a self‐inactivating lentiviral vector carrying full‐length human ataxin 3 cDNA with 120 CAGs was introduced in four‐cell to morula‐stage embryos. Six of the seven offspring expressed the transgene and three of them showed age‐related neurological symptoms. The same group successfully generated transgenic marmosets using a tetracyclin‐inducible transgene expression (tet‐on) system. The mutant human ataxin 3 gene controlled by the tet‐on system was injected into marmoset's embryos via lentiviral transduction. Four of the seven live offspring were transgenic and carried the transgene controlled by the tet‐on system.84 With the objective to monitor in vivo neural activity, Park et al reported the generation of a transgenic marmoset expressing GCaMP, a genetically encoded calcium indicator, under ubiquitous and neuronal promoters.81 Probably, others transgenic monkey models will be published in few years since the generation of transgenic models of various neurodegenerative diseases (ie, amyotrophic lateral sclerosis, Alzheimer's disease, and PD) are being planned.16, 96
Viral vector‐mediated gene transfer has shown its efficacy but has limitations, such as size limitation of the transgene, random insertion in the genome, and uncontrollable expression levels.16 The recent breakthroughs in techniques for site‐specific gene editing and transfection is paving a new way forward for studying disease progression in NHPs.
Site‐specific gene editing
The most popular techniques are the ZFN, the TALENs, and the CRISPRs (Figure 1). Once the breakpoint done, the cell's endogenous homologous recombination machinery allows the insertion of exogenous DNA sequence. The ZFNs combine the N‐terminal zinc finger DNA‐binding domain, a variable peptide linker, and a C‐terminal Fn domain with a nuclease activity for digesting target genes.97 The TALENs are chimeric proteins comprising the transcription activator‐like effector for the DNA binding and FokI nuclease to induce a specific DNA double‐strand break.98 The CRISPR‐Cas9 system associates a specific RNA guide to the Cas9 nuclease to digest double‐strand target DNA.99
Figure 1.
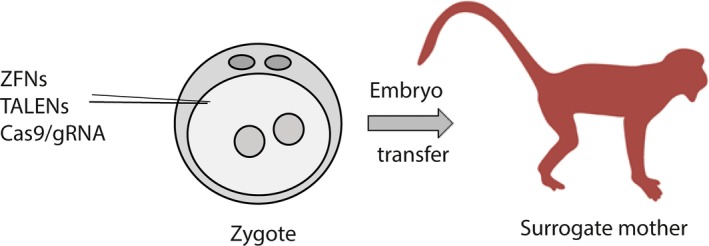
Illustration of the method for transgenesis in nonhuman primates (NHP) with the new genome‐editing technologies. Metaphase II oocytes were subjected to intracytoplasmic sperm injection (ICSI). At 8‐10 h after ICSI, the fertilization was checked and two pronuclei zygotes were injected with ZFN, TALEN or CRISPR/Cas9. The zygotes were then transferred into surrogate females and pregnancy was confirmed by ultrasonography
Using these recent technologies, the first models were generated in 2014. The CRISPR/Cas 9 system was used in one‐cell embryos to modify the monkey genome and obtain founder animals harboring two gene modifications (disruption of Ppar‐γ and Rag1 in founder animals).29 In another studies, CRISPR/Cas9 was used via zygote injection to KO the p53 gene or to realize the dystrophin gene KO to generate Duchenne muscular dystrophy monkey models.26, 100 This technology was also used in order to model the X‐linked adrenal hypoplasia congenital and hypogonadotropic hypogonadism obtained from DAX1 KO. The DAX‐1–deficient monkey displayed defect in adrenal gland development with abnormal architecture of the adrenal cortex and of the testis.101 A recent paper also demonstrated the generation of a SIRT6‐null cynomolgus monkey. SIRT6‐specific sgRNAs and Cas9 mRNA were injected into 98 monkey zygotes. Three female infants were born with efficient KO of the SIRT6 gene and no detectable off‐target. All three animals showed a global development delay in utero compared to wild‐type newborns with prematurity and developmental retardation at the tissue level. Notably, a smaller brain and thinner cortical layers in the cerebral and cerebellar hemispheres were described in the transgenic newborn monkeys, evoking neuronal maturation delay. It was consistent to the loss‐of‐function mutation of SIRT6 described in humans that causes late fetal loss with intrauterine growth restriction.102 So, the use of CRISPR/Cas9 appears to be a promising way to obtain new transgenic animal model. However, technological limitations induce difficulties to obtain homozygous mutations at a high rate in NHPs, compared to mice. Nowadays, the challenge consists in developing more precise gene‐editing technologies, by increasing the efficiency, producing more homozygotes, and reducing the mosaicism and the off‐targets,33 even if cases of off‐targeting were still limited in previous studies.29, 101
Direct injection of transcription activator‐like effector nuclease (TALEN) plasmids in rhesus and cynomolgus monkey zygotes was also performed for genetic modeling of human disease. This technology has been employed for effective gene editing of MECP2 gene in modeling Rett syndrome but without any behavioral deficits.28 Further detailed analyses using eye tracking and magnetic resonance imaging showed homologies between Rett syndrome in humans and the phenotype observed in monkeys.103 Another five fetuses were developed, all but one were stillborn. The remaining one failed to survive after birth but was the only one who showed a specific deletion in the MECP2 gene.104 Others report detailed the feasibility to generate KO monkey using ZFN and TALEN.9, 105, 106 Notably, TALEN was used to produce microcephaly‐associated MCPH1 KO cynomolgus monkeys. One monkey recapitulated most of the important clinical features observed in microcephalic patients, including reduction in head circumference, premature chromosome condensation, hypoplasia of the corpus callosum, and upper limb spasticity.105
4. CONCLUSIONS
The new genome‐editing tools associated to the development of NHP‐assisted reproductive technologies have allowed for the development of NHP disease models, notably by transgene overexpression and mutagenesis of endogenous genome DNA. Many studies have been conducted, especially in the neuroscience field and in particular in neurodegenerative diseases, psychiatric disorders, and cognitive functions. Further experiments are needed for evaluating the phenotype of the transgenic animals and for demonstrating the reproducibility while keeping in mind the three “R” principles (replacement, reduction, and refinement) for NHP experimental technique.
The research community expects that new findings will be obtained, since these new animal models have been developed in order to better understand the pathophysiology and to identify novel therapies. It will therefore be desirable to improve the emerging reproductive technologies which appear to be a promising and efficient way to establish animal models. Various strategies used in mice and rats could be translated to NHP, for example, genome editing of spermatogonial stem cells, gene modification of androgenetic (male) and parthogenetic (female) haploid ES cells used to fertilized an oocyte, testis xenografting, nuclear transfer from cultured cell lines, differentiation of ES cells into primordial germ cells followed by ovary or testis transplantation.8, 94, 107, 108, 109, 110, 111
All these strategies developed to model human disease can also be diverted for gene therapy. In humans, the use of viral vector, notably the AAVs, offers a promising way in clinical trials such as muscular dystrophy, hemophilia, PD, Leber's congenital amaurosis, and macular degeneration.112 In human preimplantation embryos, CRISPR‐Cas9 was used to correct the heterozygous MYPBC3 mutation which causes hypertrophic cardiomyopathy. To minimize the mosaicism, the coinjection of sperm and CRISPR‐Cas9 components was performed into metaphase II oocytes. The yield of MYBCP3 WT/WT embryo (72.4%) in the injected group was significantly higher than in untreated controls (47.4%).113 Another team demonstrated efficient correction of point mutation in HBB and G6PD gene.114 The clinical application is not for today but these studies open the field of efficient gene therapy in human embryos.
To conclude, the development of all these novel models is paving a new way forward for investigating disease pathogenesis and for preclinical drug studies, notably for intractable neurodegenerative diseases such as PD and amyotrophic lateral sclerosis, as well as mental disorder such as schizophrenia and autism spectrum disorders.
CONFLICT OF INTEREST
None.
ACKNOWLEDGEMENTS
E.B. received the Grand Prix from Foundation de France.
Chansel‐Debordeaux L, Bezard E. Local transgene expression and whole‐body transgenesis to model brain diseases in nonhuman primate. Animal Model Exp Med. 2019;2:9‐17. 10.1002/ame2.12055
REFERENCES
- 1. Jennings CG, Landman R, Zhou Y, et al. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci. 2016;19:1123‐1130. [DOI] [PubMed] [Google Scholar]
- 2. Camus S, Ko WK, Pioli E, Bezard E. Why bother using non‐human primate models of cognitive disorders in translational research? Neurobiol Learn Mem. 2015;124:123‐129. [DOI] [PubMed] [Google Scholar]
- 3. Roelfsema PR, Treue S. Basic neuroscience research with nonhuman primates: a small but indispensable component of biomedical research. Neuron. 2014;82:1200‐1204. [DOI] [PubMed] [Google Scholar]
- 4. Mantini D, Corbetta M, Romani GL, Orban GA, Vanduffel W. Evolutionarily novel functional networks in the human brain? J Neurosci. 2013;33:3259‐3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sato K, Sasaki E. Genetic engineering in nonhuman primates for human disease modeling. J Hum Genet. 2018;63(2):125‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marmion DJ, Kordower JH. alpha‐Synuclein nonhuman primate models of Parkinson's disease. J Neural Transm. 2018;125:385‐400. [DOI] [PubMed] [Google Scholar]
- 7. Chan AW, Yang SH. Generation of transgenic monkeys with human inherited genetic disease. Methods. 2009;49:78‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Izpisua Belmonte JC, Callaway EM, Caddick SJ, et al. Brains, genes, and primates. Neuron. 2015;86:617‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kishi N, Sato K, Sasaki E, Okano H. Common marmoset as a new model animal for neuroscience research and genome editing technology. Dev Growth Differ. 2014;56:53‐62. [DOI] [PubMed] [Google Scholar]
- 10. Rhesus Macaque Genome Sequencing and Analysis Consortium , Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222‐234. [DOI] [PubMed] [Google Scholar]
- 11. Sereno MI, Tootell RB. From monkeys to humans: what do we now know about brain homologies? Curr Opin Neurobiol. 2005;15:135‐144. [DOI] [PubMed] [Google Scholar]
- 12. Dehay B, Fernagut PO. Alpha‐synuclein‐based models of Parkinson's disease. Rev Neurol. 2016;172:371‐378. [DOI] [PubMed] [Google Scholar]
- 13. Herculano‐Houzel S. The human brain in numbers: a linearly scaled‐up primate brain. Front Hum Neurosci. 2009;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wallis JD. Cross‐species studies of orbitofrontal cortex and value‐based decision‐making. Nat Neurosci. 2011;15:13‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herculano‐Houzel S, Manger PR, Kaas JH. Brain scaling in mammalian evolution as a consequence of concerted and mosaic changes in numbers of neurons and average neuronal cell size. Front Neuroanat. 2014;8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okano H, Kishi N. Investigation of brain science and neurological/psychiatric disorders using genetically modified non‐human primates. Curr Opin Neurobiol. 2018;50:1‐6. [DOI] [PubMed] [Google Scholar]
- 17. Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends Neurosci. 2008;31:599‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parent A. Comparative Neurobiology of the Basal Ganglia. New York, NY: Wiley; 1986. [Google Scholar]
- 19. Double KL, Halliday GM. New face of neuromelanin. J Neural Transm Suppl. 2006;70:119‐123. [DOI] [PubMed] [Google Scholar]
- 20. Prins NW, Pohlmeyer EA, Debnath S, et al. Common marmoset (Callithrix jacchus) as a primate model for behavioral neuroscience studies. J Neurosci Methods. 2017;284:35‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Capitanio JP, Emborg ME. Contributions of non‐human primates to neuroscience research. Lancet. 2008;371:1126‐1135. [DOI] [PubMed] [Google Scholar]
- 22. Yamamori T. Selective gene expression in regions of primate neocortex: implications for cortical specialization. Prog Neurobiol. 2011;94:201‐222. [DOI] [PubMed] [Google Scholar]
- 23. Sasaki E. Prospects for genetically modified non‐human primate models, including the common marmoset. Neurosci Res. 2015;93:110‐115. [DOI] [PubMed] [Google Scholar]
- 24. Emborg ME. Nonhuman primate models of neurodegenerative disorders. ILAR J. 2017;58:190‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carlson DF, Tan W, Lillico SG, et al. Efficient TALEN‐mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382‐17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Zheng Y, Kang Y, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet. 2015;24:3764‐3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9‐mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu H, Chen Y, Niu Y, et al. TALEN‐mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell. 2014;14:323‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niu Y, Shen B, Cui Y, et al. Generation of gene‐modified cynomolgus monkey via Cas9/RNA‐mediated gene targeting in one‐cell embryos. Cell. 2014;156:836‐843. [DOI] [PubMed] [Google Scholar]
- 30. Wang H, Yang H, Shivalila CS, et al. One‐step generation of mice carrying mutations in multiple genes by CRISPR/Cas‐mediated genome engineering. Cell. 2013;153:910‐918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitelaw CB, Sheets TP, Lillico SG, Telugu BP. Engineering large animal models of human disease. J Pathol. 2016;238:247‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gama Sosa MA, De Gasperi R, Elder GA. Modeling human neurodegenerative diseases in transgenic systems. Hum Genet. 2012;131:535‐563. [DOI] [PubMed] [Google Scholar]
- 33. Chen Y, Niu Y, Ji W. Genome editing in nonhuman primates: approach to generating human disease models. J Intern Med. 2016;280:246‐251. [DOI] [PubMed] [Google Scholar]
- 34. Bezard E, Przedborski S. A tale on animal models of Parkinson's disease. Mov Disord. 2011;26:993‐1002. [DOI] [PubMed] [Google Scholar]
- 35. Bezard E, Imbert C, Gross CE. Experimental models of Parkinson's disease : from the static to the dynamic. Rev Neurosci. 1998;9:71‐90. [DOI] [PubMed] [Google Scholar]
- 36. Porras G, Li Q, Bezard E. Modeling Parkinson's disease in primates: the MPTP model. Cold Spring Harb Perspect Med. 2012;2:a009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bezard E, Dovero S, Prunier C, et al. Relationship between the appearance of symptoms and the level of nigrostriatal degeneration in a progressive MPTP‐lesioned macaque model of Parkinson's disease. J Neurosci. 2001;21:6853‐6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prunier C, Bezard E, Mantzarides M, et al. Presymptomatic diagnosis of experimental parkinsonism with 123I‐PE2I SPECT. NeuroImage. 2003;19:810‐816. [DOI] [PubMed] [Google Scholar]
- 39. Meissner W, Prunier C, Guilloteau D, Chalon S, Gross CE, Bezard E. Time course of nigrostriatal degeneration in a progressive MPTP‐lesioned macaque model of Parkinson's disease. Mol Neurobiol. 2003;28:87‐96. [DOI] [PubMed] [Google Scholar]
- 40. Engeln M, De Deurwaerdere P, Li Q, Bezard E, Fernagut PO. Widespread monoaminergic dysregulation of both motor and non‐motor circuits in Parkinsonism and Dyskinesia. Cereb Cortex. 2015;25:2783‐2792. [DOI] [PubMed] [Google Scholar]
- 41. Fernagut PO, Li Q, Dovero S, et al. Dopamine transporter binding is unaffected by L‐DOPA administration in normal and MPTP‐treated monkeys. PLoS One. 2010;5:e14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dodiya HB, Bjorklund T, Stansell J III, Mandel RJ, Kirik D, Kordower JH. Differential transduction following basal ganglia administration of distinct pseudotyped AAV capsid serotypes in nonhuman primates. Mol Ther. 2010;18:579‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kirik D, Annett LE, Burger C, Muzyczka N, Mandel RJ, Bjorklund A. Nigrostriatal alpha‐synucleinopathy induced by viral vector‐mediated overexpression of human alpha‐synuclein: a new primate model of Parkinson's disease. Proc Natl Acad Sci USA. 2003;100:2884‐2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kirik D, Rosenblad C, Burger C, et al. Parkinson‐like neurodegeneration induced by targeted overexpression of alpha‐synuclein in the nigrostriatal system. J Neurosci. 2002;22:2780‐2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eslamboli A, Romero‐Ramos M, Burger C, et al. Long‐term consequences of human alpha‐synuclein overexpression in the primate ventral midbrain. Brain. 2007;130:799‐815. [DOI] [PubMed] [Google Scholar]
- 46. Yang W, Wang G, Wang CE, et al. Mutant alpha‐synuclein causes age‐dependent neuropathology in monkey brain. J Neurosci. 2015;35:8345‐8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bourdenx M, Dovero S, Engeln M, et al. Lack of additive role of ageing in nigrostriatal neurodegeneration triggered by alpha‐synuclein overexpression. Acta Neuropathol Commun. 2015;3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koprich JB, Johnston TH, Reyes G, Omana V, Brotchie JM. Towards a non‐human primate model of alpha‐synucleinopathy for development of therapeutics for Parkinson's Disease: optimization of AAV1/2 delivery parameters to drive sustained expression of alpha synuclein and dopaminergic degeneration in Macaque. PLoS One. 2016;11:e0167235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bassil F, Guerin PA, Dutheil N, et al. Viral‐mediated oligodendroglial alpha‐synuclein expression models multiple system atrophy. Mov Disord. 2017;32:1230‐1239. [DOI] [PubMed] [Google Scholar]
- 50. Palfi S, Brouillet E, Jarraya B, et al. Expression of mutated Huntingtin fragment in the putamen is sufficient to produce abnormal movement in non‐human primates. Mol Ther. 2007;15:1444‐1451. [DOI] [PubMed] [Google Scholar]
- 51. Duque S, Joussemet B, Riviere C, et al. Intravenous administration of self‐complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187‐1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Foust KD, Wang X, McGovern VL, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271‐274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54. Dehay B, Dalkara D, Dovero S, Li Q, Bezard E. Systemic scAAV9 variant mediates brain transduction in newborn rhesus macaques. Sci Rep. 2012;2:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bevan AK, Duque S, Foust KD, et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol Ther. 2011;19:1971‐1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gray SJ, Matagne V, Bachaboina L, Yadav S, Ojeda SR, Samulski RJ. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol Ther. 2011;19:1058‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Samaranch L, Salegio EA, San Sebastian W, et al. Adeno‐associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum Gene Ther. 2012;23:382‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rahim AA, Wong AM, Howe SJ, et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non‐integrating lentiviral vectors. Gene Ther. 2009;16:509‐520. [DOI] [PubMed] [Google Scholar]
- 59. Stott SR, Kirik D. Targeted in utero delivery of a retroviral vector for gene transfer in the rodent brain. Eur J Neurosci. 2006;24:1897‐1906. [DOI] [PubMed] [Google Scholar]
- 60. Shen JS, Meng XL, Maeda H, Ohashi T, Eto Y. Widespread gene transduction to the central nervous system by adenovirus in utero: implication for prenatal gene therapy to brain involvement of lysosomal storage disease. J Gene Med. 2004;6:1206‐1215. [DOI] [PubMed] [Google Scholar]
- 61. Chansel‐Debordeaux L, Bourdenx M, Dovero S, et al. In utero delivery of rAAV2/9 induces neuronal expression of the transgene in the brain: towards new models of Parkinson's disease. Gene Ther. 2017;24:801‐809. [DOI] [PubMed] [Google Scholar]
- 62. Haddad MR, Donsante A, Zerfas P, Kaler SG. Fetal brain‐directed AAV gene therapy results in rapid, robust, and persistent transduction of mouse choroid plexus epithelia. Mol Ther Nucleic Acids. 2013;2:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rahim AA, Wong AM, Ahmadi S, et al. In utero administration of Ad5 and AAV pseudotypes to the fetal brain leads to efficient, widespread and long‐term gene expression. Gene Ther. 2012;19:936‐946. [DOI] [PubMed] [Google Scholar]
- 64. Garrett DJ, Larson JE, Dunn D, Marrero L, Cohen JC. In utero recombinant adeno‐associated virus gene transfer in mice, rats, and primates. BMC Biotechnol. 2003;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mattar CN, Waddington SN, Biswas A, et al. Systemic delivery of scAAV9 in fetal macaques facilitates neuronal transduction of the central and peripheral nervous systems. Gene Ther. 2013;20:69‐83. [DOI] [PubMed] [Google Scholar]
- 66. Bourdenx M, Chansel‐Debordeaux L, Dovero S, et al. In utero delivery of rAAV mediates widespread brain transduction in rats and monkeys: towards new models of Parkinson's disease. Gene Ther; Society of Neuroscience Conference. 2015; 24:801‐809. [DOI] [PubMed] [Google Scholar]
- 67. Yang SH, Chan AW. Transgenic animal models of Huntington's disease. Curr Top Behav Neurosci. 2011;7:61‐85. [DOI] [PubMed] [Google Scholar]
- 68. Chan AW, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309‐312. [DOI] [PubMed] [Google Scholar]
- 69. Wolfgang MJ, Eisele SG, Browne MA, et al. Rhesus monkey placental transgene expression after lentiviral gene transfer into preimplantation embryos. Proc Natl Acad Sci USA. 2001;98:10728‐10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang SH, Cheng PH, Banta H, et al. Towards a transgenic model of Huntington's disease in a non‐human primate. Nature. 2008;453:921‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sasaki E, Suemizu H, Shimada A, et al. Generation of transgenic non‐human primates with germline transmission. Nature. 2009;459:523‐527. [DOI] [PubMed] [Google Scholar]
- 72. Sasaki E, Hanazawa K, Kurita R, et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus). Stem Cells. 2005;23:1304‐1313. [DOI] [PubMed] [Google Scholar]
- 73. Shiozawa S, Kawai K, Okada Y, et al. Gene targeting and subsequent site‐specific transgenesis at the beta‐actin (ACTB) locus in common marmoset embryonic stem cells. Stem Cells Dev. 2011;20:1587‐1599. [DOI] [PubMed] [Google Scholar]
- 74. Tachibana M, Sparman M, Ramsey C, et al. Generation of chimeric rhesus monkeys. Cell. 2012;148:285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen Y, Niu Y, Li Y, et al. Generation of cynomolgus monkey chimeric fetuses using embryonic stem cells. Cell Stem Cell. 2015;17:116‐124. [DOI] [PubMed] [Google Scholar]
- 76. Mitalipov SM, Yeoman RR, Nusser KD, Wolf DP. Rhesus monkey embryos produced by nuclear transfer from embryonic blastomeres or somatic cells. Biol Reprod. 2002;66:1367‐1373. [DOI] [PubMed] [Google Scholar]
- 77. Ng SC, Chen N, Yip WY, et al. The first cell cycle after transfer of somatic cell nuclei in a non‐human primate. Development. 2004;131:2475‐2484. [DOI] [PubMed] [Google Scholar]
- 78. Niu Y, Yang S, Yu Y, et al. Impairments in embryonic genome activation in rhesus monkey somatic cell nuclear transfer embryos. Cloning Stem Cells. 2008;10:25‐36. [DOI] [PubMed] [Google Scholar]
- 79. Zhou Q, Yang SH, Ding CH, et al. A comparative approach to somatic cell nuclear transfer in the rhesus monkey. Hum Reprod. 2006;21:2564‐2571. [DOI] [PubMed] [Google Scholar]
- 80. Niu Y, Yu Y, Bernat A, et al. Transgenic rhesus monkeys produced by gene transfer into early‐cleavage‐stage embryos using a simian immunodeficiency virus‐based vector. Proc Natl Acad Sci USA. 2010;107:17663‐17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Park JE, Zhang XF, Choi SH, Okahara J, Sasaki E, Silva AC. Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci Rep. 2016;6:34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seita Y, Tsukiyama T, Iwatani C, et al. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci Rep. 2016;6:24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tomioka I, Ishibashi H, Minakawa EN, et al. Transgenic monkey model of the polyglutamine diseases recapitulating progressive neurological symptoms. eNeuro. 2017;4 10.1523/ENEURO.0250-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tomioka I, Nogami N, Nakatani T, et al. Generation of transgenic marmosets using a tetracyclin‐inducible transgene expression system as a neurodegenerative disease model. Biol Reprod. 2017;97:772‐780. [DOI] [PubMed] [Google Scholar]
- 85. Wang CE, Tydlacka S, Orr AL, et al. Accumulation of N‐terminal mutant Huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum Mol Genet. 2008;17:2738‐2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Putkhao K, Kocerha J, Cho IK, Yang J, Parnpai R, Chan AW. Pathogenic cellular phenotypes are germline transmissible in a transgenic primate model of Huntington's disease. Stem Cells Dev. 2013;22:1198‐1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chan AW, Jiang J, Chen Y, et al. Progressive cognitive deficit, motor impairment and striatal pathology in a transgenic Huntington disease monkey model from infancy to adulthood. PLoS One. 2015;10:e0122335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chan AW, Xu Y, Jiang J, et al. A two years longitudinal study of a transgenic Huntington disease monkey. BMC Neurosci. 2014;15:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Raper J, Bosinger S, Johnson Z, Tharp G, Moran SP, Chan AWS. Increased irritability, anxiety, and immune reactivity in transgenic Huntington's disease monkeys. Brain Behav Immun. 2016;58:181‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kocerha J, Liu Y, Willoughby D, et al. Longitudinal transcriptomic dysregulation in the peripheral blood of transgenic Huntington's disease monkeys. BMC Neurosci. 2013;14:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Meng Y, Jiang J, Bachevalier J, Zhang X, Chan AW. Developmental whole brain white matter alterations in transgenic Huntington's disease monkey. Sci Rep. 2017;7:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moran S, Chi T, Prucha MS, et al. Germline transmission in transgenic Huntington's disease monkeys. Theriogenology. 2015;84:277‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu Z, Li X, Zhang JT, et al. Autism‐like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. 2016;530:98‐102. [DOI] [PubMed] [Google Scholar]
- 94. Liu Z, Nie YH, Zhang CC, et al. Generation of macaques with sperm derived from juvenile monkey testicular xenografts. Cell Res. 2016;26:139‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Niu Y, Guo X, Chen Y, et al. Early Parkinson's disease symptoms in alpha‐synuclein transgenic monkeys. Hum Mol Genet. 2015;24:2308‐2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Okano H, Sasaki E, Yamamori T, et al. Brain/MINDS: a Japanese national brain project for marmoset neuroscience. Neuron. 2016;92:582‐590. [DOI] [PubMed] [Google Scholar]
- 97. Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967‐973. [DOI] [PubMed] [Google Scholar]
- 98. Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143‐148. [DOI] [PubMed] [Google Scholar]
- 99. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wan H, Feng C, Teng F, et al. One‐step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2015;25:258‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kang Y, Zheng B, Shen B, et al. CRISPR/Cas9‐mediated Dax1 knockout in the monkey recapitulates human AHC‐HH. Hum Mol Genet. 2015;24:7255‐7264. [DOI] [PubMed] [Google Scholar]
- 102. Zhang W, Wan H, Feng G, et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature. 2018;560:661‐665. [DOI] [PubMed] [Google Scholar]
- 103. Chen Y, Yu J, Niu Y, et al. Modeling rett syndrome using TALEN‐Edited MECP2 mutant cynomolgus monkeys. Cell. 2017;169(945‐955):e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liu Z, Zhou X, Zhu Y, et al. Generation of a monkey with MECP2 mutations by TALEN‐based gene targeting. Neurosci Bull. 2014;30(3):381‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ke Q, Li W, Lai X, et al. TALEN‐based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res. 2016;26:1048‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sato K, Oiwa R, Kumita W, et al. Generation of a nonhuman primate model of severe combined immunodeficiency using highly efficient genome editing. Cell Stem Cell. 2016;19:127‐138. [DOI] [PubMed] [Google Scholar]
- 107. Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64‐66. [DOI] [PubMed] [Google Scholar]
- 108. Kanatsu‐Shinohara M, Ogonuki N, Inoue K, et al. Long‐term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612‐616. [DOI] [PubMed] [Google Scholar]
- 109. Sato T, Sakuma T, Yokonishi T, et al. Genome editing in mouse spermatogonial stem cell lines using TALEN and double‐nicking CRISPR/Cas9. Stem Cell Reports. 2015;5:75‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang H, Liu Z, Ma Y, et al. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013;23:1187‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhou Q, Wang M, Yuan Y, et al. Complete meiosis from embryonic stem cell‐derived germ cells in vitro. Cell Stem Cell. 2016;18:330‐340. [DOI] [PubMed] [Google Scholar]
- 112. Bartel MA, Weinstein JR, Schaffer DV. Directed evolution of novel adeno‐associated viruses for therapeutic gene delivery. Gene Ther. 2012;19:694‐700. [DOI] [PubMed] [Google Scholar]
- 113. Ma H, Marti‐Gutierrez N, Park SW, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413‐419. [DOI] [PubMed] [Google Scholar]
- 114. Tang L, Zeng Y, Du H, et al. CRISPR/Cas9‐mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics. 2017;292:525‐533. [DOI] [PubMed] [Google Scholar]


