The genus Enterococcus comprises a ubiquitous group of Gram-positive bacteria that are of great relevance to human health for their role as major causative agents of health care-associated infections. The enterococci are resilient and versatile species able to survive under harsh conditions, making them well adapted to the health care environment.
KEYWORDS: Enterococcus, antibiotic resistance, horizontal gene transfer
SUMMARY
The genus Enterococcus comprises a ubiquitous group of Gram-positive bacteria that are of great relevance to human health for their role as major causative agents of health care-associated infections. The enterococci are resilient and versatile species able to survive under harsh conditions, making them well adapted to the health care environment. Two species cause the majority of enterococcal infections: Enterococcus faecalis and Enterococcus faecium. Both species demonstrate intrinsic resistance to common antibiotics, such as virtually all cephalosporins, aminoglycosides, clindamycin, and trimethoprim-sulfamethoxazole. Additionally, a remarkably plastic genome allows these two species to readily acquire resistance to further antibiotics, such as high-level aminoglycoside resistance, high-level ampicillin resistance, and vancomycin resistance, either through mutation or by horizontal transfer of genetic elements conferring resistance determinants.
INTRODUCTION
Enterococci are leading causes of health care-associated infections (HAIs) globally, in particular urinary tract, soft tissue, and device-associated infections. Multidrug resistance is common, which prolongs hospitalization time, increases treatment cost, and increases the risk of treatment failure and death. In the past few decades, our knowledge of enterococcal biology, ecology, virulence, and genetics has steadily increased. However, there are still important questions about these pathogens that remain to be solved, in particular how to effectively treat multidrug-resistant strains. In this review, we present a general overview of the genus Enterococcus, the clinically relevant species, their mechanisms of infection and antibiotic resistance, the state of the art in treatment, and challenges and perspectives for the future.
BASIC MICROBIOLOGY
The enterococci are ubiquitous Gram-positive bacteria that have been isolated from soil, surface waters, and seawater; in association with plants; in fermented food products; as part of the gut microbiota of both vertebrates and invertebrates; and as causative agents of human disease (1–13). They have a low-GC genome content of about 34 to 45%, a genome size ranging from 2.3 to 5.4 Mb with 2,154 to 5,107 predicted genes, and a genus core genome with the number of genes ranging from 605 to 1,037 depending on the data set and criteria used for analysis (14, 15). The enterococcal pangenome is larger and reflects the highly plastic nature of their genomes and particular niche adaptations.
The term “entérocoque” was first coined by Thiercelin in 1899, when he described gut commensal bacteria with the ability to become pathogenic (16). Due to morphological and some biochemical similarities, the enterococci were considered part of the genus Streptococcus (17, 18) and classified as group D streptococci until the mid-1980s. Although four separated branches of streptococci were identified, the pyogenic streptococci, the viridans streptococci, the lactic streptococci, and the enterococcus (19), the term “enterococcus” was more considered a placeholder name for Gram-positive cocci isolated from the gut/feces rather than a monophyletic group. As Sherman stated in 1938, “‘The enterococcus’ as this term is commonly used among bacteriologists, has about as much biological meaning as the bear” (20). However, based on a detailed analysis of biochemical and culture characteristics, in 1970, Kalina (21) proposed that the so-called enteric streptococci should be placed in a genus of their own, the Enterococcus. It was only in 1984 that the formal proposal of the genus Enterococcus became more accepted (22), and it appeared as a properly recognized genus separated from the streptococci in an editorial addendum to the 1986 edition of Bergey’s Manual of Systematic Bacteriology (23). The genus Enterococcus has to date 58 described species with valid publications (according to compiled information from the List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.net/enterococcus.html#r]) (24). The family Enterococcaceae was first proposed by Ludwig and collaborators (25) in 2009 based on 16S rRNA gene similarity and originally comprised Enterococcus, Vagococcus, Tetragenococcus, and Melissococcus. Other presumptive genera within the Enterococcaceae are Catellicoccus (26) and Pilibacter (27); however, the precise phylogenetic position of Tetragenococcus, Melissococcus, Catellicoccus, and Pilibacter is not clear due to the limited number of species in each genus that have been described and sequenced and the observation that Melissococcus and at least one species of Tetragenococcus may branch within Enterococcus (15, 28–30). The Enterococcaceae are in the order Lactobacillales with other families of medical and economic importance, like the Lactobacillaceae and the Streptococcaceae, class Bacilli in the phylum Firmicutes. The number of predicted Lactobacillales-specific clusters of orthologous genes that appear to be essential for these bacteria is 567 (31).
SPECIES DIFFERENTIATION AND LABORATORY DIFFERENTIATION
Enterococci are non-spore-forming ovoid bacteria (22) that exist individually or as pairs, chains, or groups. They are chemo-organotrophic facultative anaerobes with homofermentative metabolism, with lactic acid as the predominant end product of carbohydrate fermentation (29).
Different selective media have been tested for the isolation and identification of enterococci; however, there are no definitive biochemical tests to differentiate Enterococcus from other Gram-positive catalase-negative cocci. Most enterococci are oxidase and catalase negative, salt tolerant (as high as 6.5%), resistant to 40% bile, esculin hydrolytic, and able to grow in the presence of sodium azide (up to 0.4%). In addition to the above-described characteristics, all described and tested species produce β-glucosidase; leucine arylamidase; acid from the sugars d-fructose, galactose, β-gentiobiose, glucose, lactose, maltose, d-mannose, ribose, trehalose, cellobiose, and N-acetylglucosamine; and the glycosides salicin, methyl β-d-glucoside, amygdalin, and arbutin. In general, enterococci are urease negative and do not produce acid from d-arabinose, erythritol, d- and l-fucose, methyl α-d-xyloside, and l-xylose; these metabolic characteristics have been used in the development of commercial testing kits. Growth occurs at between 10°C and 45°C, with optimal growth for most species at 35°C to 37°C (32). The enterococci are remarkably resistant to desiccation (32). Only two enterococcal species are reported to be mobile: Enterococcus gallinarum and E. casseliflavus/E. flavescens (33–36).
As early as 1919, Orla-Jensen (discussed in reference 37) proposed the separation of Streptococcus faecalis and Streptococcus faecium into two different species based on the ability of the former to tolerate potassium tellurite and produce black colonies. Additional biochemical tests, such as testing of the ability to reduce tetrazolium salts to the chromogenic formazan in the presence of glucose, were introduced along the way to improve species identification (38–42). A widely used system for classification and differentiation of enterococci was introduced by Lancefield in a seminal paper in 1933 based on serological groups (43). In this paper, the enteric streptococci were part of antigenic group D, and her classification system is still in use to differentiate Enterococcus from most Streptococcus species.
If grown on horse blood agar, enterococci can be alpha-, beta-, or nonhemolytic and form 1- to 2-mm colonies with a wet appearance (44). Based on their metabolic capabilities, different selective culture media have been developed for the isolation of enterococci; these selective media frequently contain bile salts, sodium azide, antibiotics, and esculin or tetrazolium salts. Not all enterococcal species are able to grow in these selective media, but the most clinically relevant species grow well. Most clinical testing for enterococcal identification includes catalase testing, pyrrolidonyl arylamidase/pyrrolidonyl-aminopeptidase (PYR) testing and a bile esculin hydrolysis test. Commercial kits have been developed to standardize and optimize the detection of enterococci in the clinical setting, all requiring previous isolation and culture of the organisms, potentially delaying diagnosis. Additionally, accurate differentiation between species in species groups is not always achieved based on phenotypic tests only (45).
The identification of enterococci to the species level has clinical relevance due to the antibiotic resistance profiles of the different pathogenic enterococci. Since the introduction of molecular techniques into clinical microbiology laboratories, improved species identification and expedited testing options have been developed; these techniques are also useful for epidemiology and surveillance and in the diagnosis of difficult cases. Molecular diagnosis techniques are gaining popularity; however, in resource-limited regions, they are still not widely in use in the clinical microbiology laboratory. Molecular-based methods have the potential advantages of increased diagnostic accuracy, providing information about antimicrobial resistance, and reduced time and cost compared to traditional cultivation and phenotypic testing.
Among the newer systems for classification and identification of enterococci are matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), nucleic acid amplification tests (NAATs), peptide nucleic acid fluorescent in situ hybridization (PNA-FISH), and multilocus sequence typing (MLST).
MALDI-TOF MS-based identification is a powerful, fast, and reliable method that is starting to gain traction more broadly for routine detection in clinical microbiology laboratories for species identification (46, 47). The clinical use of MALDI-TOF MS-based methods allows for rapid identification of enterococci directly from blood culture bottles, potentially reducing the time to antimicrobial treatment initiation (48). MALDI-TOF MS has a high sensitivity, being able to identify about 94% of isolates to the species level, including differentiating between closely related species (49, 50); additionally, it could potentially be useful for antibiotic resistance profiling, for instance, for detection of the presence of van genes, although it is not yet in use in clinical practice (51, 52).
NAAT methods are based on PCR amplification and subsequent sequencing or array/hybridization or real-time PCR amplification (53) of one or more genes that are useful for organism identification to the genus or species level and equally important to detect antimicrobial resistance genes. Different genes have been used for diagnostic and phylogenetic purposes. 16S rRNA gene sequencing is commonly used to identify bacterial species and allows discrimination of enterococci to the species level (45, 54–56); however, differentiating from species within a species group, such as the E. faecium group, can be less accurate (49, 57, 58). Several other genes have been proposed to help differentiate enterococcal species, such as ddl (d-alanine:d-alanine), atpA (ATP synthase), groES and groEL, sod (superoxide dismutase), and tuf (elongation factor Tu) (58–63), although, to our knowledge, no systematic comparison of the specificities and sensitivities of different genes has been done. Multiplexed real-time PCR permits testing for more than one gene, allowing the simultaneous determination of the species and potential antibiotic resistance genes (64), and genus- and species-specific assays have been developed, aimed at rapid detection (65). PNA-FISH targeting species-specific rRNA allows for rapid detection of the presence of enterococci from blood culture bottles. These tests allow differentiation of E. faecalis, E. faecium, and other less-common enterococcal species (66). Commercial clinically approved systems have been developed based on the different technologies described above, but a detailed description of commercial testing methods is beyond the scope of this review.
MLST provides strain identification and has been used to study molecular epidemiology and also to study outbreaks (67, 68), largely replacing pulsed-field gel electrophoresis (PFGE) analysis because of higher reproducibility and easier implementation. Recently, a new iteration to improve resolution has been implemented by performing core genome MLST (cgMLST), which expands the number of genes from 7 or so housekeeping genes to up to 1,423 (69) and is more cost-effective to implement than whole-genome sequencing (WGS) and average nucleotide difference analysis (70). In a study comparing MLST versus WGS for 495 clinical E. faecium isolates plus 11 reference genomes, the authors found high discrepancy between the two methods, and they mostly attributed these differences to a lack of robustness of MLST due to a high degree of recombination between isolates (71). Bayesian analysis of population structure (BASP) is a method that improves identification of deep-branching lineages and recombination and is more robust than MLST-based studies using DNA sequence or molecular marker data (72).
CLINICALLY SIGNIFICANT SPECIES AND LESS-COMMON SPECIES
Enterococci are considered commensal organisms of the human gastrointestinal tract; however, they can also be pathogenic, mostly linked to HAIs, commonly causing urinary tract infection (UTI), bacteremia, endocarditis, burn and surgical site wound infections, abdomen and biliary tract infections, and infection of catheters and other implanted medical devices. In most surveys, enterococci are the third most common cause of native valve endocarditis, after Staphylococcus aureus and viridans streptococci (73, 74). In humans, E. faecalis and E. faecium are the most abundant enterococcal species. All Lactobacillales comprise less than 1% of the gut microbiota in adults with westernized diets (75, 76). In Hadza hunter-gatherers and in a group of rural Papua New Guineans, there seems to be an enrichment for enterococci (77, 78).
MacCallum and Hastings (79) first reported a putative enterococcal infection in 1899, describing a case of endocarditis and offering a detailed description of the isolated bacteria, which they dubbed Micrococcus zymogenes. At around the same time, Thiercelin (16) described round commensal enteric bacteria (an entérocoque) capable of causing diarrheal disease and septicemia. Other early reports describe infections caused by Streptococcus/Enterococcus faecalis as the causative agent of endocarditis, puerperal fever, wound infections in First World War soldiers, bacteremia, and fever (80–84). Interestingly, the early literature also describes attempts at curing infections prior to the broad introduction of antibiotics by preparing a vaccine from the patient’s own fecal contents, which successfully cleared the symptoms (83). The incidence of enterococcal infections has been increasing steadily since the late 1970s (13, 85, 86). In both Europe and the United States, enterococci at the genus level are the 2nd most common pathogens associated with HAI, E. faecalis was the 5th most frequently isolated organism from catheter-associated urinary tract infections (CAUTIs) and third for central line-associated bloodstream infections (CLABSIs), and E. faecium was the 11th and 5th, respectively (87–89).
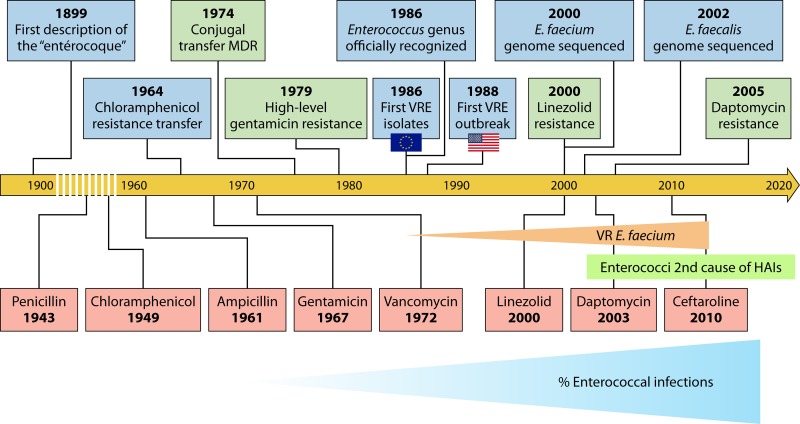
The success of enterococci in establishing themselves as HAI agents is partly due to their intrinsic resistance to many antimicrobials and their capacity to acquire new resistance traits. The most prevalent species in HAIs is E. faecalis, which is more virulent than E. faecium but with less-dramatic levels of intrinsic and acquired antimicrobial resistance. Historically, E. faecalis has been isolated in about 50.3% of all enterococcal HAIs; however, there is an increasing trend for E. faecium-caused infections, mostly associated with the rise of vancomycin- and β-lactam-resistant E. faecium strains (90). Roughly 10% of E. faecalis isolates are vancomycin resistant, compared to 80% of E. faecium isolates (89). Together, E. faecalis and E. faecium cause about 75% of all typed enterococcal infections (89). A timeline highlighting the major events in the establishment of enterococci as important HAI agents is shown in Fig. 1.
FIG 1.
Timeline of relevant events in the history of enterococci as human pathogens (blue rectangles), appearance of antibiotic resistance (green rectangles), and antibiotic clinical debut (red rectangles). The timeline begins in 1899 with the first formal description of putative enterococci, as round enteric bacteria. The timeline then jumps to 1964 to the first description of the transfer of chloramphenicol resistance, only 15 years after its clinical introduction. Similar stories occurred for aminoglycosides and glycopeptides. Since the late 1980s, the prevalence of vancomycin-resistant (VR) E. faecium has been increasing, as has the overall percentage of enterococcal HAIs. Resistance to the newest introduced antibiotics, linezolid and daptomycin, emerged very rapidly after their clinical introduction, but the majority of enterococci remain susceptible. MDR, multidrug resistance.
Nontyped enterococci, including nontyped E. faecium and E. faecalis and all other non-faecalis non-faecium enterococci (OE), comprise about 24.6% of all enterococcal infections (89); however, the percentage of OE is not reported separately from nontyped putative E. faecium and E. faecalis infections. The incidence of infections caused by OE has been on the increase; cases of OE bacteremia in U.S. hospitals ranked 10th among HAIs in the period comprising 2011 to 2014 (89), compared to 11th in the period comprising 2009 to 2010 (91). Species such as E. casseliflavus, E. gallinarum, E. durans, E. hirae, E. mundtii, E. avium, and E. raffinosus have been associated with human infection mostly in people with concurrent hematological malignancies, neutropenia, and previous corticosteroid treatment (92). E. durans, E. hirae, and E. mundtii belong to the E. faecium species group (29), suggesting that the capacity to become pathogenic was present in the shared common ancestor of this group. E. gallinarum and E. casseliflavus have intrinsically low-level resistance to vancomycin, which could be a potential treatment problem if rates of infections caused by these organisms continue to rise (93, 94). A recent study by Manfredo Vieira and colleagues (95) implicates E. gallinarum in the induction of autoantibodies linked to autoimmune disease after translocation from the gut to the liver in mice with autoimmune susceptibility and proposed that a similar mechanism could occur in people with autoimmune diseases such as lupus erythematosus, suggesting a new role for enterococci in human health. E. pallens, E. gilvus, and E. raffinosus belong to the same species groups (14, 15, 29). E. pallens has been associated with spontaneous peritonitis in patients with liver cirrhosis and has been isolated from ascites fluid, so far limited to 4 cases reported in Quebec Province, Canada (96, 97). The importance of this organism as a human pathogen is yet to be determined. E. gilvus was isolated as part of mixed infections with E. faecium and E. casseliflavus from the bile of a patient with cholecystitis (96). Because most infections caused by OE occur in severely ill patients with other comorbidities, it is difficult to establish the mortality rate of bacteremia caused by these organisms (98).
VIRULENCE
The enterococci are not highly virulent organisms, and the success of E. faecalis and E. faecium as pathogens in the hospital setting is primarily related to their survival capabilities in a hostile antimicrobial-rich environment. That said, several traits in both species have been linked with their pathogenic potential and ability to cause disease. These include the ability to evade the immune system; the capacity to attach to host cells, the extracellular matrix (EM), and inert materials, such as a variety of medical devices; and the ability to form biofilms that make them more resistant to antibiotic killing and phagocytic attack (99). Virulence factors are more evident in E. faecalis, perhaps explaining its still leading role in enterococcal infections.
Many proteins have been described as part of the virulence repertoire of pathogenic enterococci.
Microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) are surface elements that help enterococci to adhere to host tissues, helping in the initiation of infection (100).
In E. faecalis, MSCRAMM genes are found in most strains and are expressed in vivo during human infection (101). One of the best-characterized MSCRAMMs is Ace, a collagen-binding protein (102) that enhances early heart valve colonization, suggesting an important role in the early establishment of endocarditis (103).
MSCRAMM genes are enriched in clinical isolates of E. faecium, and genes of this family present in the genome were more abundant in endocarditis isolates (104). In E. faecium, Acm (a collagen-binding protein) is the best-characterized MSCRAMM. The acm gene is primarily present in health care-associated isolates (present in 99% of analyzed isolates), although one study found that it was disrupted by a transposon in commensal isolates, becoming nonfunctional (105).
Pilin gene clusters (PGCs) are present in both E. faecalis and E. faecium and encode LPxTG-like motif surface proteins that are responsible for the assembly of long filamentous structures extending from the surface, called pili. Like the MSCRAMMs, pili can function as adhesins (106). In E. faecalis, the ebp (endocarditis- and biofilm-associated pilus) PGC is associated with initial adherence and biofilm formation and has been implicated in the pathogenesis of endocarditis and UTI (107). In E. faecium, the role of the pilum is not entirely clear; however, there seem to be differential regulation of the PCG and differential assembly of pilus proteins between clinical isolates and commensal strains (108, 109).
Cytolysin (Cyl) (also called hemolysin), encoded by the cylLL and cylLS genes, contributes to virulence in E. faecalis infections. Cyl is a secreted two-peptide lytic protein that damages host cells and promotes infection. It also has bacteriocin activity, damaging other Gram-positive organisms (110–112). The cytolysin operon is normally located on mobile elements such as conjugative plasmids or within the pathogenicity island (PAI) and is often found in association with aggregation substance genes (113). Aggregation substance is a pheromone-induced surface protein that plays dual roles in mating pair formation during conjugation and virulence. It is involved in vegetation formation in infective endocarditis, extracellular matrix adherence, and phagocytosis protection, and it potentiates the pathogenic effect of Cyl (113–115).
Another virulence factor that increases the ability of E. faecalis to cause disease is gelatinase (GelE), a matrix metalloproteinase that hydrolyzes gelatin, collagen, and other proteins. Gelatinase plays a role in the development of endocarditis (116) and inhibits complement-mediated responses (117). gelE is cotranscribed with sprE, a gene encoding a serine protease; together, the two genes contribute to virulence (118). The expression of both genes is under the control of the fsr locus, a master regulator which also plays a role in biofilm formation, the expression of surface proteins, and metabolism (119). The cell wall-associated enterococcal surface protein (Esp) (120) contributes to cell adhesion in both E. faecalis and E. faecium, playing a role in urethral colonization (121) and endocarditis (122) and promoting biofilm formation (121, 123, 124). However, by itself, Esp is neither necessary nor sufficient to successfully establish infection and is not present in all clinical isolates. The phosphotransferase system (PTS) genes encode transmembrane proteins that participate in sugar intake. Diversification of the PTS allows enterococci to use a broad variety of sugars as carbohydrate sources and better adapt to changing environments. The PTS can act as part of the general stress response (125), as virulence factors helping the enterococci to colonize and survive within the host (126), and in biofilm and endocarditis development (127).
Genes encoding several of these virulence factors are often colocated in PAIs or mobile elements, facilitating their spread between isolates. PAIs are large elements that can be acquired by horizontal transfer and confer virulence to bacterial pathogens (128, 129). Several in-depth reviews of enterococcal virulence are recommended (130–133).
ANTIMICROBIAL SUSCEPTIBILITY AND INTRINSIC MECHANISMS OF RESISTANCE
E. faecalis and E. faecium are characterized by their reduced susceptibility to many agents that are quite active against streptococci and staphylococci. A list of antimicrobial agents to which enterococci are resistant is included in Table 1. Among the β-lactams, they are intrinsically resistant to virtually all cephalosporins (with possible exceptions being ceftaroline and ceftobiprole, which have in vitro activity against E. faecalis), antistaphylococcal penicillins, and aztreonam (134). E. faecalis strains can be susceptible in vitro to carbapenems, but there are few clinical data supporting the use of these agents for treatment of human infections. Enterococci are intrinsically susceptible to vancomycin but resistant to clindamycin, trimethoprim-sulfamethoxazole, and clinically achievable concentrations of aminoglycosides. They are intrinsically susceptible to tetracyclines and erythromycin, although acquired resistance to these agents is widespread (except for tigecycline) (135, 136). The newer agents linezolid, tedizolid, daptomycin, televancin, and oritavancin are active against enterococci, and the pristinamycin combination quinupristin-dalfopristin is active against E. faecium only. Fluoroquinolones have activity against enterococci, although ciprofloxacin’s MICs are borderline for non-urinary-tract infections, and fluoroquinolone resistance is common in clinical E. faecium strains (137). In the clinical setting, ampicillin remains the treatment of choice for susceptible strains in patients who can tolerate this agent.
TABLE 1.
Antimicrobial resistance in enterococci
| Antimicrobial class (agents) | Representative resistance gene(s)/operon(s) | Mechanism of resistance |
|---|---|---|
| Aminoglycosides (gentamicin, kanamycin) | aac-2′-aph-2″-le, aph-3′-IIIa | Modification of the aminoglycoside |
| β-Lactams | pbp4 (E. faecalis), pbp5 (E. faecium) | Reduced affinity for the antibiotic |
| Chloramphenicol | cat | Acetylation of chloramphenicol |
| Clindamycin | lsa(A) | Putative efflux |
| Daptomycin | liaFSR | Alteration in membrane charge and fluidity |
| Erythromycin | ermB | Ribosomal methylation |
| Fluoroquinolones | gyrA, parC | Modifications in quinolone resistance-determining region |
| Glycopeptides | vanA, vanB, vanD, vanM | Modified peptidoglycan precursors terminating in d-lactate |
| vanC, vanE, vanG, vanL, vanN | Modified peptidoglycan precursors terminating in d-serine | |
| Oxazolidinones | rRNA genes | Mutations reducing affinity |
| cfr | Methylation of 23S rRNA | |
| Rifampin | rpoB | Point mutations reducing affinity |
| Streptomycin | ant-6 | Modification of streptomycin |
| Tetracyclines | tet(L) | Efflux |
| tet(M) | Ribosomal protection | |
| Tigecycline | tet(L), tet(M) | Increased expression |
Enterococcal resistance to β-lactams is attributable to the expression of a low-affinity penicillin-binding protein (PBP) designated PBP4 in E. faecalis and PBP5 in E. faecium (138–140). Strains from which these pbp genes have been deleted exhibit reduced MICs for active β-lactams and reductions into the susceptible range for β-lactams that have poor activity against wild-type strains (141, 142). Many enterococcal strains also exhibit tolerance to the bactericidal activity of the active β-lactams, with minimal bactericidal concentrations greatly exceeding MICs (143). This tolerance has clinical significance in the treatment of endocarditis, with cure rates with β-lactam antibiotics alone being approximately 40% (144). The addition of streptomycin or gentamicin to an active β-lactam results in bactericidal synergism in vitro and yields clinical cure rates exceeding 70% (145). Experiments performed by Moellering and Weinberg (146) in the 1970s attributed this synergism to increased streptomycin penetration into the cell in the presence of penicillin or vancomycin, implying that the killing activity was provided by the aminoglycoside, once it achieved entry into the cell, facilitated by the cell wall-active agent. More recently, clinical data indicate that outcomes of E. faecalis endocarditis treatment are equivalent with combinations of ampicillin, which is active against E. faecalis, and ceftriaxone, which is not (147, 148). Although the mechanism for this apparent clinical synergism is not clear at present, it has been postulated that the combination of the two antibiotics inhibits all the E. faecalis PBPs more effectively than either antibiotic alone (149).
Resistance to clinically achievable concentrations of aminoglycosides has been attributed to the poor penetration of these agents through the enterococcal cell envelope (146). The reason for this poor penetration is not clear, but it has been postulated that enterococcal metabolism is essentially anaerobic, precluding aminoglycoside transport across the cytoplasmic membrane, which is an oxygen-dependent process. Clindamycin resistance in E. faecalis is attributable to the lsa(A) gene, which is believed to encode an ABC superfamily of proteins that confers resistance to lincosamides, pleuromutilins, and streptogramin A antibiotics from the cell (150). Resistance to trimethoprim-sulfamethoxazole in enterococci is an in vivo phenomenon. In vitro, wild-type enterococci appear to be susceptible to this combination, but trimethoprim-sulfamethoxazole is not effective in treating enterococcal infections in animal models. This appears to be due to the capacity of enterococci to absorb folate from the environment, thereby bypassing the steps toward folate synthesis blocked by the combination (151). There are no compelling clinical data on the effectiveness of trimethoprim-sulfamethoxazole in the treatment of human enterococcal infections.
ACQUIRED ANTIMICROBIAL RESISTANCE
Resistance to β-Lactams
As noted above, enterococci are intrinsically resistant to most β-lactams, being susceptible to only a limited number of penicillins (ampicillin, mezlocillin, penicillin, and piperacillin). Resistance to these penicillins is achievable through two mechanisms. The first, and least important, is the production of β-lactamase (152). A number of strains and some outbreak strains of E. faecalis that produce β-lactamase have been reported. Molecular analysis shows that in all cases, this β-lactamase is identical to that produced by S. aureus, in some cases within genetic regions identical to that of the S. aureus β-lactamase transposon Tn551 (153). The S. aureus β-lactamase is a narrow-spectrum enzyme that is active only against the penicillins that happen to have activity against E. faecalis. Expression of the β-lactamase in E. faecalis differs from that in S. aureus in that β-lactamase transcription is not inducible by exposure to β-lactam agents, and it appears that the enzyme remains membrane bound. The consequence of these two differences is that expression stays at a low level and does not confer significant resistance with a standard inoculum (154). With a high inoculum, however, animal studies suggest that enterococcal β-lactamase production compromises β-lactam therapy but can be counteracted by the addition of a β-lactamase inhibitor (155). Reports of β-lactamase production in strains of E. faecium are quite rare, and the strains expressing it have not been extensively analyzed.
High-level penicillin resistance in E. faecium is due to the expression of low-affinity PBP5 (142). Some resistant strains have been shown to express increased quantities of PBP5, although this has not been the most frequent mechanism of resistance. The most common mechanism is through a mutation in the pbp5 gene leading to amino acid substitutions in or near the active site of the enzyme (142, 156, 157). Molecular epidemiological data suggest that highly ampicillin-resistant strains fall into relatively few lineages that have spread widely, largely in hospitals, causing clinical infections and colonization of patients exposed to a variety of antibiotics (158). In many centers, rates of high-level ampicillin resistance in E. faecium exceed 70% (89).
Higher-level resistance to penicillins in E. faecalis is a much rarer event than in E. faecium. In one instance (138), increased expression of low-affinity PBP4 was implicated, but other cases have implicated amino acid changes within the enzyme itself. A recent report showed that reduced susceptibility in E. faecalis appeared due to the combination of increased expression of PBP4 (resulting from an adenine deletion upstream of the promoter sequence) and an alanine-to-tyrosine substitution adjacent to the active site (159). A second amino acid substitution was present in the N-terminal region of the protein but did not contribute to resistance. The mutated enzyme had a lower melting temperature, suggesting that it was less stable, offering a possible explanation as to why such mutant enzymes appear to be rare. In this case, the patient from whom the strain was isolated had been exposed to several years of treatment with aminopenicillins for a prosthetic knee infection.
The fact that deletion of PBP4 or PBP5 results in β-lactam susceptibility of E. faecalis and E. faecium, respectively, indicates that these proteins are required for resistance (141, 142). They are not, however, sufficient for resistance, since other proteins that are required for resistance expression have been found. In E. faecalis, the CroRS regulatory locus is required for cephalosporin resistance, as is a serine-threonine eukaryote-like kinase, IreK (also known as Stk) (160, 161). The presence of genes for two of the three E. faecalis class A PBPs (ponA and pbpZ) is also required for resistance to cephalosporins in E. faecalis (141). Deletion of the equivalent class A PBPs in E. faecium also results in increased cephalosporin susceptibility, but the susceptibility is restricted to certain cephalosporins (cefepime and ceftriaxone) that have a common side chain (141, 162). It is also an unstable phenotype that converts at a high frequency back to cephalosporin resistance, which in some cases is influenced by the E. faecium version of Stk (163). Cephalosporin resistance in this instance was also associated with the expression of a protein found associated with PBP5 by affinity chromatography, which has been designated P5AP (penicillin binding protein 5-associated protein) (163). In an E. faecium strain in which pbp5 was deleted, resistance to ampicillin emerged through the activity of an l,d-transpeptidase insensitive to inhibition by penicillins (but susceptible to carbapenems) (164). There is still much to be learned about how the low-affinity PBPs interact with their substrates and with β-lactam antibiotics and about the combination of cell wall synthesis proteins that leads to resistance in enterococci.
Resistance to Glycopeptides
The glycopeptide antibiotic vancomycin remained virtually universally active against E. faecalis and E. faecium for nearly three decades after its clinical introduction. In the early 1980s, strains began to emerge, first in Europe and then in the United States, that expressed inducible, high-level resistance to vancomycin and the more recently introduced antibiotic teicoplanin (165, 166). Resistance was attributable to the acquisition of operons that altered the nature of peptidoglycan precursors, substituting a d-lactate for the terminal d-alanine in the UDP-MurNac pentapeptide (167, 168). In the process of establishing the peptide cross-link essential for cell wall stability, the terminal d-alanine is removed from the chain to provide the energy for the transpeptidation reaction. Vancomycin binds to the terminal d-alanine of the cell wall precursor, preventing PBP access (vancomycin, because of its large size, also interferes somewhat with the adjacent transglycosylation reaction). Vancomycin binds to pentapeptide stems terminating in d-lactate with a roughly 1,000-fold-lower affinity than it does to those terminating in d-alanine and therefore is not an effective inhibitor of cell wall synthesis in these strains.
The first glycopeptide resistance operon that was described was the vanA operon (167), and this remains the most commonly encountered operon in the clinical setting. The operon consists of seven genes whose combined purpose is to replace the glycopeptide-susceptible pentapeptide terminating in d-Ala-d-Ala with a glycopeptide-resistant pentadepsipeptide precursor terminating in d-Ala-d-Lac. vanS encodes a transmembrane sensor kinase that is involved in detecting glycopeptides in the environment and phosphorylating VanR, whereby VanR is converted from a repressor of operon transcription to an activator (169). VanR regulates 3 downstream genes: vanH, vanA, and vanX. VanH is a dehydrogenase that reduces pyruvate to d-lactate, and VanA is a ligase that binds a d-alanine to the newly formed d-lactate to form a d-Ala-d-Lac depsipeptide (167), which is then ligated to the UDP-MurNAc tripeptide peptidoglycan precursor by the cellular adding enzyme. vanX encodes the VanX amidase, whose purpose is to cleave d-Ala-d-Ala, thereby reducing cellular quantities of d-Ala-d-Ala that can be used to create vancomycin-susceptible peptidoglycan precursors (170). Two additional genes that are not essential for glycopeptide resistance expression are included in the operon. vanY is a carboxypeptidase that cleaves the terminal d-alanine from cellular pentapeptide precursors, further reducing vancomycin-susceptible precursors (171). The final gene is vanZ, which encodes a protein of unknown function that contributes to resistance to the glycopeptide teicoplanin (172).
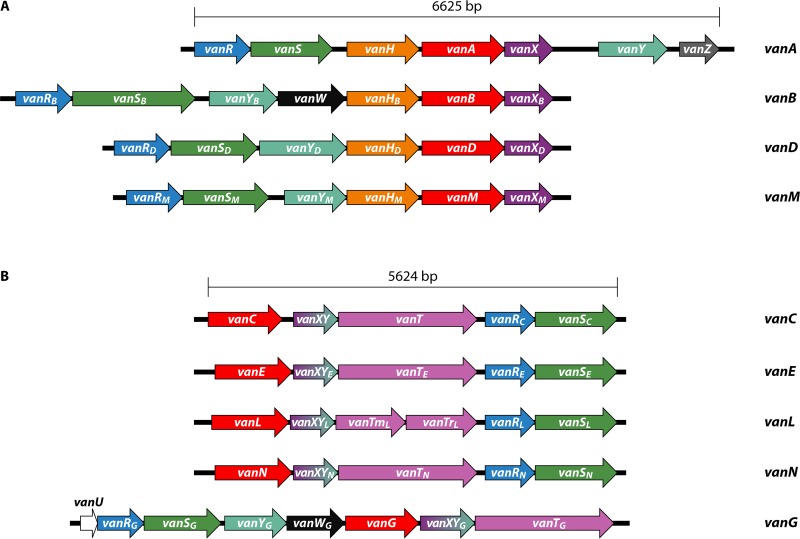
VanC-type vancomycin resistance operons, first described as intrinsic components of E. gallinarum and E. casseliflavus, produce peptidoglycan precursors terminating in d-Ala-d-Ser (93, 173). They encode a (serine) racemase (VanT), a d-Ala-d-Ser ligase (VanC), a combined dipeptidase-carboxypeptidase(VanXY), and the products of the regulatory genes vanR and vanS. The vanG operon has an additional carboxypeptidase, an analogue of VanW from the vanB operon, and an additional regulatory gene (vanU) (174).
There have been nine glycopeptide resistance operons described over the past few decades (Fig. 2). They fall into two general categories: those that replace the terminal d-Ala with a d-lactate (vanA, vanB, vanD, and vanM) (175–177) and those that replace the terminal d-Ala with a d-serine (vanC, vanE, vanG, vanL, and vanN) (94, 178–180). As opposed to the d-Lac-type operons, the operons encoding proteins that result in precursors terminating in d-Ser confer relatively lower levels of resistance to vancomycin but remain susceptible to teicoplanin. The mechanisms of the d-Lac operons all confer resistance to vancomycin and teicoplanin, although the vanB operon is not induced by the presence of teicoplanin, so strains in which the induction mechanism is intact will appear susceptible to teicoplanin (181). Clinical experience using this agent to treat VanB-type vancomycin-resistant enterococci (VRE) indicates that treatment failure is common, due to the emergence of strains with constitutive expression of the operon (182). The vanC operons confer resistance to vancomycin but not teicoplanin (173).
FIG 2.
Depictions of known glycopeptide resistance operons. (A) The four glycopeptide resistance operons that yield peptidoglycan precursors terminating in d-Ala-d-Lac. Arrows reflect the directions of transcription and relative sizes of the open reading frames. (B) The five glycopeptide resistance operons that yield peptidoglycan precursors terminating in d-Ala-d-Ser. See the text for descriptions of the open reading frame roles.
The vanA operon is carried by the Tn3 family transposon Tn1546 (167), which can be located on the chromosome or on transferable plasmids. The vanB operon is most commonly carried by Tn5382 (also referred to in some publications as Tn1549) (183), a Tn916 family element that also may be incorporated into the chromosome or a plasmid. The vanC operons are intrinsic to E. casseliflavus and E. gallinarum (94), which are rare causes of human infection (173). The remainder of the operons are found rarely, although local outbreaks of some of them have been reported. The vanN and vanG operons have been shown to be transferable, with vanG being found within different integrative and conjugative element (ICE)-type elements (184).
Resistance to Aminoglycosides
As noted above, enterococci are intrinsically resistant to clinically achievable concentrations of aminoglycosides. Aminoglycosides are useful for achieving bactericidal synergism in combination with cell wall-active agents, which is important in the treatment of enterococcal endocarditis (144, 146). Since the clinical utility of these combinations has been recognized, strains that have expressed high levels of resistance to aminoglycosides have emerged (MICs of >500 μg/ml for gentamicin and >2,000 μg/ml for streptomycin) (185). This level of resistance is due to the expression of aminoglycoside-modifying enzymes and negates the synergistic benefit of the combinations in the clinical setting. The gene encoding the most common enzyme conferring resistance to gentamicin (and other aminoglycosides except streptomycin) is aac-6′-Ie-aph-2″, classically found within Tn4001 in staphylococci and other variants in enterococci (186, 187). In some studies, this enzyme has been the exclusive cause of high-level gentamicin resistance in enterococci (188). Expression of a second phosphotransferase [APH(2″)-lc] has been associated with lower gentamicin MICs (ca. 256 μg/ml) but still negates ampicillin-aminoglycoside synergism. Such isolates may not be detected by clinical microbiology laboratories using concentrations of 500 or 1,000 μg/ml to screen for high-level resistance (189). Resistance to streptomycin in enterococci is most commonly encoded by the ant-6 gene (190). Very high levels of streptomycin resistance have also been attributed to ribosomal mutations (185). Finally, intrinsic resistance to kanamycin and tobramycin in E. faecium is attributable to chromosomally encoded AAC(6′)-li (191).
Resistance to Fluoroquinolones
Ciprofloxacin and levofloxacin have marginal activity against enterococci, and their use is restricted to the treatment of urinary tract infections due to susceptible strains. Moxifloxacin is more potent against Gram-positive bacteria than the other two but still exhibits only intermediate activity versus enterococci (192). High-level resistant strains have been shown to contain mutations in both gyrA and parC (193, 194). Some strains have mutations in only parC, suggesting that this topoisomerase may be the primary target of fluoroquinolones in enterococci. There has been suggestion in some studies that efflux pumps are also involved in enterococcal fluoroquinolone resistance, but specific efflux pumps have not been identified (195).
Resistance to Linezolid
Linezolid remains broadly active against both E. faecalis and E. faecium (196). Resistance frequently occurs through mutations in the rRNA genes. E. faecium has six such ribosomal genes, while E. faecalis has four, and the level of resistance expressed depends upon the number of these genes that contain the relevant mutations (197). Once a single such mutation occurs, continued selective pressure by linezolid has been associated with “gene conversion,” in which further genes acquire the same mutation through homologous recombination with the mutated gene. Conversely, if there remains a single such wild-type gene, then gene conversion can lead to restoration of susceptibility in the absence of antibiotics (198), suggesting that there is some selective disadvantage to these mutations in the absence of selective pressure. Resistance due to changes in ribosomal proteins L3, L4, and L22 appears to be extremely rare.
Enterococci can also develop resistance to linezolid through acquisition of the cfr or cfr(B) gene (199), which encodes a methyltransferase that modifies A2503 in bacterial 23S rRNA. This enzyme confers resistance to a variety of antimicrobial classes, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A, as well as decreased susceptibility to the 16-membered macrolides spiramycin and josamycin. Cfr is commonly plasmid encoded and transferable and has been associated with outbreaks of linezolid resistance in a variety of Gram-positive species (200). Finally, plasmid-mediated resistance has also been attributed to the acquisition of optRA, which encodes a putative ABC transporter (201).
Resistance to Daptomycin
Daptomycin is a cyclic lipopeptide that acts by interacting with the cytoplasmic membrane in the presence of physiological concentrations of calcium, resulting in a variety of alterations in cell membrane characteristics. It is a cationic peptide whose first attraction to the cell membrane is through its interaction with phosphatidylglycerol. In the presence of physiological concentrations of calcium, daptomycin aggregates and then inserts into the membrane. This membrane insertion is followed by the transition of phospholipids and daptomycin to the inner leaflet of the membrane. The result includes ion leakage, which may result from daptomycin “pores” in the membrane or through a “lipid extraction effect,” whereby lipids aggregate in areas on the membrane surface and are then extracted. Temporary water channels may be formed by this extraction, with associated ion leakage.
Resistance to daptomycin occurs through a variety of mutations that have different effects depending on the species. In E. faecalis, resistance is associated with a movement of membrane phospholipids away from the septum, which may divert daptomycin from the septum. In E. faecium, resistance is associated with repulsion of daptomycin from the cell membrane due to changes in membrane phospholipids, similar to that seen in resistant strains of S. aureus (202). Mutations resulting in daptomycin resistance are commonly identified in the liaFSR operon, which encodes a 3-component regulatory pathway involved in the response to cell membrane stress. Daptomycin-resistant E. faecium strains that have mutations in the liaFSR system also exhibit synergism between ampicillin and daptomycin (203). The clinical importance of this synergism remains to be established. Although overall rates of daptomycin resistance remain low, the risk of acquiring daptomycin resistance during therapy is substantial.
Resistance to Tetracyclines
Tetracycline resistance in enterococci is quite prevalent and frequently mediated by a ribosomal protection mechanism mediated by tet(M), which is most often carried by conjugative transposons (CTns) related to Tn916 (204, 205). Efflux-mediated resistance mechanisms encoded by genes such as tet(L) are also present in enterococci (205). Tigecycline is a minocycline derivative that is broadly active because it is not susceptible to most tetracycline resistance mechanisms, including Tet(M) and Tet(L). Despite this characteristic, E. faecium strains have been reported in which resistance to tigecycline has been tentatively attributed to the overexpression of plasmid-mediated tet(L) and tet(M) genes (206).
GENOME PLASTICITY
As a major cause of hospital-acquired infections, enterococci have become an important problem in clinical practice. Their abilities to survive in the environment and tolerate disinfectants, their intrinsic antimicrobial resistance, and their remarkable genome plasticity have helped to establish these organisms as frequent inhabitants of hospitals and other health care facilities. The majority of clinical isolates of both E. faecalis and E. faecium generally lack the adaptive immunity CRISPR-Cas loci (207), suggesting that the selective pressure faced by these organisms induced a trade-off of losing some protection from potentially harmful invading DNA versus gaining the ability to rapidly evolve new traits. Indeed, it has been shown that in strains with functional CRISPR-Cas loci and the innate immunity restriction-modification system, there was a 4-log reduction in the acquisition of pheromone-responsive plasmids (208). Horizontal gene transfer has played a key role in the evolution and success of clinical isolates of E. faecalis and E. faecium.
The most frequently reported mechanism for foreign DNA acquisition in enterococci is via conjugation, with less information about the role of phage-mediated transduction, although there is evidence that phages could be another important mechanism used by enterococci to share genes even between different species (209, 210). Contrary to streptococci, natural transformation has never been observed (211).
Conjugative Transposable Elements
Conjugation is the process of genetic material transfer from a donor to a recipient cell involving complex machineries encoded by mobilization (MOB) genes and mating pair formation (MPF) genes (212). Conjugation occurs via conjugative plasmids and conjugative transposons (or integrative conjugative elements). Conjugative plasmids encode the proteins required for their transfer from a donor to a recipient cell, during conjugation. The first report of conjugative plasmid transfer in the enterococci came from the observation of multiple-antibiotic-resistance transfer in E. faecalis by Jacob and Hobbs (213). Most clinical isolates carry plasmids and transposable elements that commonly encode antibiotic resistance factors, virulence factors, and bacteriocins (210, 211). Plasmids can harbor transposons that are capable of cotransfer and integration in the chromosome by site-specific recombination or by homologous recombination. Ten different plasmid families have been described in enterococci based on their replication initiation genes, but this might be only a small fraction of the actual diversity of enterococcal plasmids (214). Different types of plasmids are found in the two most relevant clinical species. In E. faecalis, pheromone-responsive plasmids (PRPs) are widely distributed and are a major source of antimicrobial resistance transfer, having high efficiency, with transfer rates of about 10−1 transconjugant cells per donor (215). PRPs have a narrow host range; the pheromones that induce plasmid transfer are heptapeptides or octapeptides chromosomally encoded by lipoprotein genes (derived from the signal peptide) and are released into the medium by the future recipient cell, which does not carry a PRP. PRPs encode specific receptors for a given pheromone; the formation of a mating pair is mediated by plasmid-encoded aggregation substance, which facilitates donor-recipient contact and is also a virulence factor, as discussed above (216–218). PRPs have a complex regulation (for excellent reviews, see references 219 and 220). Two of the best-studied families of PRPs in E. faecalis are pCF10 and pAD1, both of which have clinical relevance. pCF10 plasmids are mostly vehicles for antibiotic resistance genes (215, 219), whereas pAD1 plasmids carry cytolysin, bacteriocins, hemolysins, and UV light resistance (218, 220–222). Remarkably, PRPs are also able to mobilize large chromosomal regions (up to 857 kb) via the formation of a plasmid-chromosome cointegrate (223). Non-pheromone-responsive plasmids (NPRPs) conferring resistance to macrolides, aminoglycosides, and glycopeptides also occur in E. faecalis and can coexist with PRPs; these plasmids show a broader host range than PRPs (224, 225). Hybrid plasmids, derived from multiple plasmid recombination events, are a potential problem in the spread of multidrug resistance. pRE25 was identified from an E. faecalis food isolate, and it carries resistance to 12 antimicrobials and has a broad host range (226). It was subsequently determined that it was widespread in E. faecium isolates (227). Inc18-PRP hybrid plasmids are documented as disseminators of vanA resistance between E. faecalis strains (228).
In E. faecium, there is no evidence that a system such as the PRP of E. faecalis is largely used. There are a few reports about PRP-like systems (229, 230), but newer literature has not provided strong evidence for the widespread use of this mechanism in E. faecium strains. Transfer of large regions of the chromosome has also been observed in E. faecium, although the mechanism is different from what has been observed in E. faecalis because it is not mediated by PRPs (231–233). Interestingly, the pbp5 gene has been shown to be transferable as part of large chromosomal regions, and pbp5 horizontal gene transfer might be relevant in the acquisition of β-lactam resistance in clinical strains (233, 234). Two of the most prevalent plasmid types are the Inc18 group and the pRUM family (227). These plasmids commonly use as a maintenance mechanism a toxin-antitoxin system that ensures plasmid survival even in the absence of antibiotic selection (235). pRUM-like plasmids have a narrow host range, mostly confined to the E. faecium species complex (236); carry resistance to erythromycin, chloramphenicol, streptomycin, and streptothricin; and can carry vanA resistance (218, 228). Inc18 plasmids such as pAMβ1 and mosaic plasmids such as Inc18/pRUM show a high degree of shuffling, and they are a frequent finding in clinical isolates but are also present in sewage and animal isolates and are able to disseminate vanA glycopeptide resistance (228, 237).
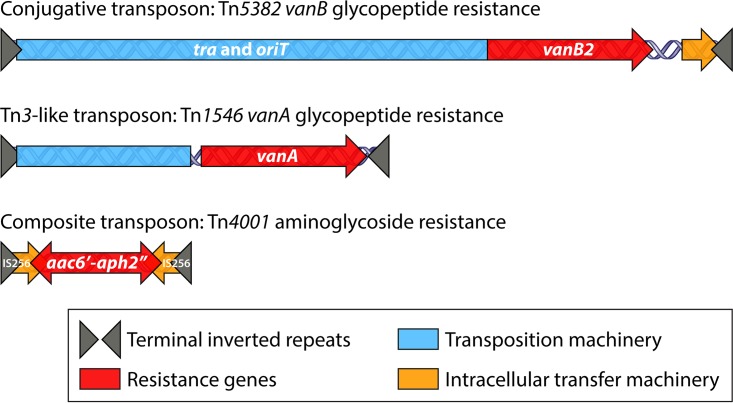
Transposable elements constitute the majority of mobile genetic elements (MGEs) present in enterococcal genomes (Fig. 3) (238–240). CTns have a broad host range and can cross between different species and genera and even transfer between Gram-positive and Gram-negative bacteria. CTns are mobile elements that possess the genetic information to mediate their own transfer within cells and between cells and are also able to comobilize other plasmids, transposons, and large chromosomal fragments and induce chromosomal deletions by excision (231, 241, 242). The first conjugative transposon known to carry antibiotic resistance was identified in E. faecalis in 1981 by Clewell and colleagues (224, 243). The authors described a chromosomally located element named Tn916 that carried tetracycline resistance and was able to mobilize via transposition to either the chromosome of a recipient cell or a conjugative plasmid. Incorporation of the transposon into a conjugative plasmid increases its frequency of transfer. Conjugative transposons can also facilitate the transfer of chromosomal genes even in the absence of the transfer of the transposon itself (204). After the description of Tn916, other conjugative transposons have been described in enterococci, mostly associated with resistance to macrolide-lincosamide-streptogramin B (MLSB) and, importantly, with glycopeptide resistance (vanB2 type) (235).
FIG 3.
Transposable elements in enterococci. The cartoon depicts the three kinds of transposable elements found in enterococci: conjugative transposons, such as Tn5382 carrying the vanB2 operon; insertion sequence elements (IS) in the Tn3 family, such as Tn1546 carrying vanA resistance; and composite transposons, such as Tn4001, for high-level aminoglycoside resistance. These three types of transposable elements can exist either in the chromosome or in plasmids as part of larger mobilizable elements.
Insertion Sequence Elements and Tn3-Like Transposons
Insertion sequences (ISs) are the most basic transposable elements, carrying only information necessary for their own transposition. However, ISs can form composite transposons by flanking resistance or pathogenesis genes, moving between replicons by a replicative transposition mechanism. This kind of transposable elements has been linked to high-level gentamicin resistance, such as Tn5281 or (in one instance) the vanB1 glycopeptide resistance element Tn1547 (244–246). Tn3-like transposons move intracellularly within or between replicons (i.e., chromosome to chromosome or chromosome to plasmid) via a replicative mechanism mediated by a transposase and a resolvase. Tn3-like transposons mostly reside within conjugative plasmids and are associated with MLSB resistance and with high-level glycopeptide resistance (vanA type) (167, 247).
Reports of antibiotic resistance transfer between strains of enterococci go back as early as 1964 for chloramphenicol resistance (248). Conjugal plasmid transfer of multiple antibiotic resistance determinants has been documented in enterococci since the 1970s (213, 249). In both E. faecalis and E. faecium, vancomycin resistance is mostly disseminated by non-pheromone-responsive plasmids; these plasmids can carry conjugative transposons but also nonconjugative transposons, such as the vancomycin resistance element Tn1546 hosting the vanA operon (250, 251). The first report of a vancomycin-resistant enterococcal outbreak appeared in 1988 and reported VRE recovered from patients in England since 1986 (both E. faecalis and E. faecium) (252). E. faecalis strain V583 was the first vancomycin-resistant strain reported from the United States (253).
POPULATION BIOLOGY
MLST schemes have been used extensively to understand the epidemiology and population structure of the two major enterococcal pathogens (67, 68) More recently, WGS methods have also been incorporated, and we have a fairly good picture about how these organisms emerged as important human pathogens and the challenges that they present.
E. faecalis strains have limited phylogenetic diversity in their core genome, with average nucleotide identity (ANI) values of 97.7% to 99.5%; in contrast, the shared gene content of these strains is more variable (70.95% to 96.5%) (254). This variability is due to genome size variation attributed to gains in genetic material via horizontal gene transfer; up to 25% of the genome in E. faecalis strains can comprise mobile elements acquired by horizontal gene transfer events, showing an even greater degree of recombination than E. faecium (223, 240). The genome sizes of different strains reflect this genome plasticity, with a probable minimal genome size of 2.74 Mb and genome sizes as large as 3.36 Mb (254). The phylogeny of E. faecalis does not show a significant association of strains based on their origin (hospital isolates, strains able to colonize hospitalized and nonhospitalized people, and animal isolates) and does not have a clear clade structure (72), although certain clonal complexes (CCs), such as CC2, CC16, and CC87, are more associated with multidrug-resistant strains and are enriched for mobile elements, and CC2 and CC87 are almost exclusively identified from HAIs (255). Clinical isolates tend to have larger genome sizes and carry more exogenously acquired DNA, such as transposons, ISs, plasmids, and phages, than commensal strains. E. faecalis clinical isolates are enriched for a PAI of approximately 150 kb, which harbors virulence factors and other traits that contribute to better adaptation to the host and contains different types of mobile elements. The E. faecalis PAI shows heterogeneity between isolates, including within the same genetic lineage, which indicates that different regions from the PAI can be mobilized independently (256–258). The differences observed in gene content and the prevalence of the PAI in clinical strains suggest the evolution of niche adaptation, suggesting that in the future, clearly separated subpopulations of E. faecalis could appear (259). Raven and colleagues (260) found limited international dissemination of E. faecalis, with local clonal expansion of dominant lineages.
In contrast to E. faecalis, the population structure of E. faecium is more complex and shows a clearer separation between clinical and commensal isolates (261, 262). E. faecium forms two distinct clades: clade A, which mostly comprises isolates from animal, environmental (clade A2), and clinical (clade A1) origins, and clade B, which mostly comprises isolates obtained from nonhospitalized people (commensal) (254, 261, 263). The ANI in the core genome between clades A and B ranges between 93.9 and 96% (239, 254). The accepted cutoff for bacterial species designation is an ANI of >94% (264). E. faecium clades A and B could be the result of a speciation process, which in the future will lead to two separate species, but there is still gene flow between the two groups, as evidenced by the finding of hybrid clade AB isolates (239, 263). The lack of competition between two populations is also a factor to determine if those two populations comprise two different species (265). Clade A1 isolates tend to have larger genomes that are enriched in mobile elements, importantly related to antibiotic resistance and carbohydrate utilization (263). Recombination plays a predominant role over mutation for the diversification of the species (266). Up to 38% of the genome can be of foreign origin (267). Clinical isolates have a larger genome than commensal strains and animal isolates (263). Interestingly, clade A1 strains have gained predominance in the clinical setting only since the 1980s. Earlier isolates do not cluster with clade A1 (158), perhaps due to their increased evolvability and capacity to gain new traits.
The split between clades A1 and A2 has been calculated to have occurred roughly 80 ± 30 years ago (263), probably several years before antibiotics started to be broadly used in human health and agriculture but well within the “antibiotic era.” A recent study with a large data set (495 isolates) challenges the subdivision of clade A into the A1 and A2 subgroups, instead suggesting a clonal expansion of clade A strains in the clinical setting (71); the original description of clades A1 and A2 was performed using a data set of 58 isolates, which perhaps explains the discrepancies. The same authors (71) also noticed that in their data set, about the same proportions of clade B isolates were recovered from the community setting as from the health care setting, in accordance with what was reported by Lebreton et al. (263), suggesting that clade B strains are also part of the HAI pool. Studies with larger data sets and broader geographic and ecological sampling would help better our understanding of the E. faecium population structure.
Using Escherichia coli and Bacillus anthracis mutation rates to calculate the divergence time of human commensal strains and clinical isolates, Galloway-Peña and collaborators determined that the divergence of the two clades occurred about 1 million to 300,000 years ago (261), Interestingly, the smaller time estimate coincides with new data pushing back the evolutionary history of our species, Homo sapiens, to about 300,000 years ago (268), perhaps showing the split of a human-associated clade as early as the dawn of our species. In another study based on the frequency of mutations to fosfomycin resistance, the divergence between the commensal clade (B) and the clinical and animal clade (A) was dated at 3,000 years ago, significantly later than the previous estimate (263). In this work, the authors proposed that the split between the animal clade and the human commensal clade coincided with an “increasing insulation between the flora of humans and animals, which likely stemmed from increased urbanization, increased domestication of animals providing restricted and specialized diets” (263). However, this interpretation is somewhat problematic because even if it is true that 3,000 years ago humans were establishing large urban centers, there was little insulation of animals and humans, and domestication of major animal groups occurred earlier and was not a one-time event; moreover, it occurred independently in different geographic areas and along a large time span and included phylogenetically unrelated species (269, 270). For many domestic species, bidirectional gene flow between the domestic species and their wild-type counterparts existed for a long time (271). Moreover, 3,000 years ago, humans and their domestic animals were already populating the entire globe, except for Antarctica, implying that the separation of clades A and B occurred in a very specific geographic place in the world and that later clade B isolates were introduced to other human populations worldwide and clade A isolates were introduced to their domestic animals. Although it is still not completely clear what drove E. faecium into its actual population structure, it is clear that there are two well-differentiated populations (clades A and B) occupying mostly nonoverlapping niches.
A wealth of information has been accumulated in the past two decades about the population biology of E. faecalis and E. faecium, but little is known about the population structure of the non-faecium non-faecalis enterococci.
TREATMENT OF ENTEROCOCCAL INFECTIONS
The cornerstones of antimicrobial therapy of enterococcal infections have been those β-lactams that demonstrate in vitro activity (predominantly ampicillin but also penicillin and piperacillin) and vancomycin. Therapy with a single such agent is generally adequate for routine infections for which bactericidal therapy is not required (skin and soft tissue infections, urinary tract infections, surgically drained intra-abdominal infections, and intravenous [i.v.] line-associated bloodstream infections). For those infections for which bactericidal therapy is optimal (endocarditis, osteomyelitis, and meningitis), traditional therapy has included an active β-lactam or vancomycin in combination with either streptomycin or gentamicin. Cure rates for enterococcal endocarditis predating the use of combination therapy were approximately 40% with penicillin alone (144). Combining penicillin with streptomycin elevated cure rates to 70% or higher (144, 145), and while there are no randomized controlled trials to reliably define cure rates, most studies using combinations report cure rates exceeding 70% (272).
The use of aminoglycosides to synergize with cell wall-active agents in the treatment of endocarditis was followed by the emergence of strains expressing aminoglycoside-modifying enzymes that resulted in high-level resistance to streptomycin or gentamicin and negated the in vitro synergism observed against strains that do not express these enzymes (185). Cure rates for monomicrobial therapy of endocarditis caused by strains expressing high-level aminoglycoside resistance appear to approximate the dismal results observed before synergistic therapy was used, necessitating alternative treatment strategies (272).
Mainardi and colleagues (149) first reported in vitro synergism between ampicillin and cefotaxime against E. faecalis, showing that in vitro susceptibility to either agent was enhanced in the presence of the other. They hypothesized that this synergism was due to the more complete inhibition of all the enterococcal PBPs by the combination of agents. These findings were later supported by Gavaldà and colleagues (273), whose animal studies showed similar synergisms between ampicillin and ceftriaxone. Subsequent human studies confirmed the effect of the combination of ampicillin and ceftriaxone against ampicillin-susceptible E. faecalis, whether or not the causative strains expressed high-level aminoglycoside resistance (147, 148, 274, 275). It is important to recognize that these studies are observational and not randomized, like all the data supporting the use of penicillin-aminoglycoside combinations. These results have led to a change in the consensus recommendations from a range of societies, including the American Heart Association and the Infectious Diseases Society of America, for the treatment of endocarditis (276), suggesting that ampicillin-ceftriaxone combination therapy is a reasonable treatment option for endocarditis due to high-level aminoglycoside-resistant strains and as a reasonable alternative for strains without high-level aminoglycoside resistance, for example, in patients with compromised renal function for whom the risks of renal damage from the aminoglycoside are significant.
The optimal duration of therapy for enterococcal endocarditis is 4 to 6 weeks, with no studies available to help distinguish between these two durations. Olaison and colleagues (277) reported a 78-patient study suggesting that the aminoglycoside component of the combination regimen could be discontinued after 15 days without a change in the cure rate but with beneficial effects for renal function. Retrospective data also suggest that one can achieve cure rates comparable to those for native valve disease in enterococcal prosthetic valve endocarditis caused by susceptible strains (272).
More recently, an impressive Danish study (278) showed that a regimen consisting of 17 days of intravenous treatment for endocarditis caused by one of four species (S. aureus, streptococci, E. faecalis, or coagulase-negative staphylococci) followed by a roughly equivalent number of days of active oral therapy was noninferior to a full course of i.v. therapy. A total of 97 patients with E. faecalis endocarditis (39 with prosthetic valve endocarditis) were included in that study, with results being essentially equivalent to the overall results for the combined endpoint and for each of the component endpoints. Caveats include that the patients had to be clinically stable at the time of randomization, that valve replacement surgery prior to a switch to oral therapy was permitted, and that the study was not blind. This study has the potential to have a major impact on costs for endocarditis patients whose condition has been stabilized after an initial 2 weeks of intravenous therapy or after surgical repair of an infected valve.
Of course, there are patients who are unable to tolerate β-lactam antibiotics or who are infected with E. faecium strains that are resistant to all β-lactams and vancomycin and produce aminoglycoside-modifying enzymes. In contrast to the 1990s, when there were no effective therapies for highly resistant strains, we now have several alternatives with in vitro activity against resistant strains. The first of these to become clinically available was the pristinamycin combination quinupristin-dalfopristin. This combination was synergistically active versus E. faecium strains lacking the erm macrolide resistance gene (but not E. faecalis). Although it was shown to be an effective therapy in the treatment of vancomycin-resistant enterococcal infections, its use was associated with vein inflammation and significant myalgias (279). Linezolid, an oxazolidinone antibiotic available in both i.v. and oral forms, was next licensed and has proven to be consistently active in vitro against resistant enterococci. Limitations on its use include bone marrow suppression when administered for more than 2 weeks (280) and an increased risk of serotonin syndrome occurring in patients being treated with selective serotonin reuptake inhibitors (SSRIs) (281). It is also strictly bacteriostatic, although it has been shown to be as effective in the treatment of bacteremias as more-bactericidal agents (282), and there have been instances of success in treating enterococcal endocarditis (283).
Daptomycin is a cyclic lipopeptide approved in 2003 (284). It is bactericidal against many enterococcal strains. Originally approved for a dose of 4 mg/kg of body weight/day, pharmacodynamic analyses indicated that higher doses would be needed for many strains (285). At present, physicians are using up to 8 to 10 mg/kg per day. One recent study (282) comparing daptomycin to linezolid for the treatment of enterococcal bacteremia showed linezolid to be superior to daptomycin at a dose of 6 mg/kg/day or lower but that the two regimens were equivalent when daptomycin was administered in a dose of >6 mg/kg/day.
Some E. faecium strains with higher MICs of daptomycin exhibit reduced MICs when exposed to a combination of daptomycin and a β-lactam antibiotic (to which the strains are resistant). The mechanism(s) underlying this apparent synergism remains unclear, although in some instances, greater binding of daptomycin to the cell membrane was seen in the presence of ampicillin (286). Other data suggest that such synergism occurs in strains with mutations of the liaFSR locus (203). Compelling clinical data to support the improved efficacy of these combinations against resistant strains are not available.
There remain more questions than answers for the treatment of serious multiresistant enterococcal infections. The Gram-Positive Committee of the Antimicrobial Resistance Leadership Group, an NIH-funded clinical trial consortium, recently outlined several unmet needs in the treatment of enterococcal infections, including the role of combination therapy with β-lactams for the treatment of enterococcal bloodstream infection and osteomyelitis, the role of combination β-lactam therapy against vancomycin-resistant enterococci, the optimal length of therapy for vancomycin-resistant enterococcal bloodstream infection, the optimal therapy for vancomycin-resistant enterococcal endocarditis, and the optimal therapy for vancomycin-resistant enterococcal infection caused by strains with elevated daptomycin MICs (287).
CONCLUSIONS
Enterococci, and particularly the clinically prevalent enterococcal species E. faecalis and E. faecium, continue to be important nosocomial pathogens. They are hearty species capable of surviving in important biological niches, such as the human gastrointestinal tract, and under stringent environmental conditions, facilitating their spread in institutions. Their broad spectrum of intrinsic resistance and tolerance to the bactericidal activity of many agents, combined with their prodigious ability to acquire resistance to available antibiotics, present ongoing therapeutic challenges to clinicians worldwide. They also express an increasing variety of virulence characteristics that promote colonization and infection. Moreover, their well-developed ability to acquire novel determinants for both resistance and virulence has kept them ahead of the many attempts to control the damage that they inflict on patients in our health care systems. Comprehensive strategies to contain their spread, limit their virulence, and eliminate them from infected sites will be required to prevent them from seriously limiting our ability to successfully treat a variety of serious diseases.
Biographies

Mónica García-Solache, M.D., Ph.D., is Assistant Professor of Medicine in the Department of Medicine in the Alpert Medical School of Brown University and Research Scientist at Rhode Island Hospital. She obtained her medical degree from the School of Medicine from the National Autonomous University of Mexico, followed by a Ph.D. in Developmental Genetics from the University of Cambridge. She received her microbiological training under the supervision of Dr. Arturo Casdevall and Dr. Louis B. Rice. Her research interests are the role of extracellular membrane-derived vesicles in antibiotic resistance, secretion and communication in Enterococcus faecium, and evolution of antibiotic resistance in Gram-positive bacteria, mostly focused on the evolutionary trajectories of penicillin-binding proteins in enterococci versus streptococci.

Louis B. Rice received his A.B. degree from Harvard College in 1977 and his M.D. from the Columbia University College of Physicians and Surgeons in 1983. After completing his residency in Internal Medicine at New York University and Bellevue Hospital Center, he trained in clinical infectious diseases at the New England Deaconess Hospital and Harvard Medical School, followed by 3 years working in the laboratories of Robert C. Moellering, Jr., M.D., at the Deaconess Hospital and George A. Jacoby, M.D., at Massachusetts General Hospital in Boston. In 1990, Dr. Rice moved to Cleveland, OH, where over the years he served as Chief of the Infectious Diseases Section and Chief of the Medical Service at the Cleveland VAMC and Vice Chairman of Medicine at University Hospitals of Cleveland. In 2010, Dr. Rice moved to his current position as Chair of the Department of Medicine at Warren Alpert Medical School of Brown University and Physician-in-Chief of Rhode Island Hospital and The Miriam Hospital in Providence, RI. Dr. Rice’s laboratory effort has been funded by the Department of Veterans Affairs and the National Institutes of Health, and he is an author of more than 200 original papers and invited reviews. He is a Fellow and former Director of the Infectious Diseases Society of America and is currently Editor-in-Chief of the journal Antimicrobial Agents and Chemotherapy. He is a member of the American Academy of Microbiology and a Fellow of the American College of Physicians. His research interests focus on the mechanisms of gene exchange and penicillin resistance in enterococci, the molecular epidemiology of resistant enterococcal infection, the molecular genetics of extended-spectrum β-lactamases in Gram-negative bacilli, and the influence of antibiotic administration on the emergence of resistance in the clinical setting.
REFERENCES
- 1.Mundt JO. 1963. Occurrence of enterococci in animals in a wild environment. Appl Microbiol 11:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mundt JO. 1963. Occurrence of enterococci on plants in a wild environment. Appl Microbiol 11:141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin JD, Mundt JO. 1972. Enterococci in insects. Appl Microbiol 24:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller T, Ulrich A, Ott EM, Muller M. 2001. Identification of plant-associated enterococci. J Appl Microbiol 91:268–278. doi: 10.1046/j.1365-2672.2001.01373.x. [DOI] [PubMed] [Google Scholar]
- 5.Svec P, Devriese LA, Sedlácek I, Baele M, Vancanneyt M, Haesebrouck F, Swings J, Doskar J. 2001. Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov., isolated from water. Int J Syst Evol Microbiol 51:1567–1574. doi: 10.1099/00207713-51-4-1567. [DOI] [PubMed] [Google Scholar]
- 6.Sedláček I, Holochová P, Mašlaňová I, Kosina M, Spröer C, Bryndová H, Vandamme P, Rudolf I, Hubálek Z, Švec P. 2013. Enterococcus ureilyticus sp. nov. and Enterococcus rotai sp. nov., two urease-producing enterococci from the environment. Int J Syst Evol Microbiol 63:502–510. doi: 10.1099/ijs.0.041152-0. [DOI] [PubMed] [Google Scholar]
- 7.Švec P, Vancanneyt M, Devriese LA, Naser SM, Snauwaert C, Lefebvre K, Hoste B, Swings J. 2005. Enterococcus aquimarinus sp. nov., isolated from sea water. Int J Syst Evol Microbiol 55:2183–2187. doi: 10.1099/ijs.0.63722-0. [DOI] [PubMed] [Google Scholar]
- 8.Švec P, Vancanneyt M, Sedláček I, Naser SM, Snauwaert C, Lefebvre K, Hoste B, Swings J. 2006. Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. Int J Syst Evol Microbiol 56:577–581. doi: 10.1099/ijs.0.63937-0. [DOI] [PubMed] [Google Scholar]
- 9.Naser SM, Vancanneyt M, De Graef E, Devriese LA, Snauwaert C, Lefebvre K, Hoste B, Švec P, Decostere A, Haesebrouck F, Swings J. 2005. Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. Int J Syst Evol Microbiol 55:2177–2182. doi: 10.1099/ijs.0.63752-0. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho MDGS, Shewmaker PL, Steigerwalt AG, Morey RE, Sampson AJ, Joyce K, Barrett TJ, Teixeira LM, Facklam RR. 2006. Enterococcus caccae sp. nov., isolated from human stools. Int J Syst Evol Microbiol 56:1505–1508. doi: 10.1099/ijs.0.64103-0. [DOI] [PubMed] [Google Scholar]
- 11.De Graef EM, Devriese LA, Vancanneyt M, Baele M, Collins MD, Lefebvre K, Swings J, Haesebrouck F. 2003. Description of Enterococcus canis sp. nov. from dogs and reclassification of Enterococcus porcinus Teixeira et al. 2001 as a junior synonym of Enterococcus villorum Vancanneyt et al. 2001. Int J Syst Evol Microbiol 53:1069–1074. doi: 10.1099/ijs.0.02549-0. [DOI] [PubMed] [Google Scholar]
- 12.Niemi RM, Ollinkangas T, Paulin L, Švec P, Vandamme P, Karkman A, Kosina M, Lindström K. 2012. Enterococcus rivorum sp. nov., from water of pristine brooks. Int J Syst Evol Microbiol 62:2169–2173. doi: 10.1099/ijs.0.038257-0. [DOI] [PubMed] [Google Scholar]
- 13.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong Z, Zhang W, Song Y, Liu W, Xu H, Xi X, Menghe B, Zhang H, Sun Z. 2017. Comparative genomic analysis of the genus Enterococcus. Microbiol Res 196:95–105. doi: 10.1016/j.micres.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. 2017. Tracing the enterococci from Paleozoic origins to the hospital. Cell 169:849.e13–861.e13. doi: 10.1016/j.cell.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiercelin ME. 1899. Sur un diplocoque saprophyte de l’intestin susceptible de devenir pathogène. C R Seances Soc Biol 50:269–271. [Google Scholar]
- 17.Frobisher M, Denny ER. 1928. A study of Micrococcus zymogenes. J Bacteriol 16:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrewes FW, Horder TJ. 1906. A study of the streptococci pathogenic for man. Lancet 168:708–713. doi: 10.1016/S0140-6736(01)31538-6. [DOI] [Google Scholar]
- 19.Sherman JM. 1937. The streptococci. Bacteriol Rev 1:3–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman JM. 1938. The enterococci and related streptococci. J Bacteriol 35:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalina AP. 1970. The taxonomy and nomenclature of enterococci. Int J Syst Evol Microbiol 20:185–189. doi: 10.1099/00207713-20-2-185. [DOI] [Google Scholar]
- 22.Schleifer KH, Kilpper-Bälz R. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Evol Microbiol 34:31–34. doi: 10.1099/00207713-34-1-31. [DOI] [Google Scholar]
- 23.Mundt JO. 1986. Enterococci, p 1065–1065. In Sneath PHA, Mair NS, Sharpe ME, Holt JG (ed), Bergey’s Manual of systematic bacteriology, vol 2 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 24.Parte AC. 2014. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res 42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig W, Schleifer K-H, Whitman WB. 2009. Family IV. Enterococcaceae fam. nov, p 594–623. In De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (ed), Bergey’s manual of systematic bacteriology, 2nd ed, vol 3 The Firmicutes. Springer, New York, NY. [Google Scholar]
- 26.Lawson PA, Collins MD, Falsen E, Foster G. 2006. Catellicoccus marimammalium gen. nov., sp. nov., a novel Gram-positive, catalase-negative, coccus-shaped bacterium from porpoise and grey seal. Int J Syst Evol Microbiol 56:429–432. doi: 10.1099/ijs.0.63874-0. [DOI] [PubMed] [Google Scholar]
- 27.Higashiguchi DT, Husseneder C, Grace JK, Berestecky JM. 2006. Pilibacter termitis gen. nov., sp. nov., a lactic acid bacterium from the hindgut of the Formosan subterranean termite (Coptotermes formosanus). Int J Syst Evol Microbiol 56:15–20. doi: 10.1099/ijs.0.63543-0. [DOI] [PubMed] [Google Scholar]
- 28.Okumura K, Arai R, Okura M, Kirikae T, Takamatsu D, Osaki M, Miyoshi-Akiyama T. 2011. Complete genome sequence of Melissococcus plutonius ATCC 35311. J Bacteriol 193:4029–4030. doi: 10.1128/JB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Švec P, Franz CMAP. 2014. The genus Enterococcus, p 175–211. In Holzapfel WH, Wood BJB (ed), Lactic acid bacteria: biodiversity and taxonomy. John Wiley & Sons, Ltd, Chichester, England. doi: 10.1002/9781118655252.ch15. [DOI] [Google Scholar]
- 30.Antunes LCS, Poppleton D, Klingl A, Criscuolo A, Dupuy B, Brochier-Armanet C, Beloin C, Gribaldo S. 2016. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. Elife 5:e14589. doi: 10.7554/eLife.14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova KS, Koonin EV. 2007. Evolutionary genomics of lactic acid bacteria. J Bacteriol 189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Švec P, Devriese LA. 2015. Enterococcus, p 1–25. In Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S (ed), Bergey’s manual of systematics of archaea and bacteria. John Wiley & Sons, Ltd, Chichester, England. doi: 10.1002/9781118960608.gbm00600. [DOI] [Google Scholar]
- 33.Hugh R. 1959. Motile streptococci isolated from the oropharyngeal region. Can J Microbiol 5:351–354. doi: 10.1139/m59-043. [DOI] [PubMed] [Google Scholar]
- 34.Mundt JO, Graham WF. 1968. Streptococcus faecium var. casseliflavus, nov. var. J Bacteriol 95:2005–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins MD, Jones D, Farrow JAE, Kilpper-Balz R, Schleifer KH. 1984. Enterococcus avium nom. rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int J Syst Evol Microbiol 34:220–223. doi: 10.1099/00207713-34-2-220. [DOI] [Google Scholar]
- 36.Naser SM, Vancanneyt M, Hoste B, Snauwaert C, Vandemeulebroecke K, Swings J. 2006. Reclassification of Enterococcus flavescens Pompei et al. 1992 as a later synonym of Enterococcus casseliflavus (ex Vaughan et al. 1979) Collins et al. 1984 and Enterococcus saccharominimus Vancanneyt et al. 2004 as a later synonym of Enterococcus italicus Fortina et al. 2004. Int J Syst Evol Microbiol 56:413–416. doi: 10.1099/ijs.0.63891-0. [DOI] [PubMed] [Google Scholar]
- 37.Heineman PG. 1920. Orla-Jensen’s classification of lactic acid bacteria. J Dairy Sci 3:143–155. doi: 10.3168/jds.S0022-0302(20)94257-1. [DOI] [Google Scholar]
- 38.Shattock PM. 1955. The identification and classification of Streptococcus faecalis and some associated streptococci. Ann Inst Pasteur Lille 7:95–100. [PubMed] [Google Scholar]
- 39.Barnes EM. 1956. Tetrazolium reduction as a means of differentiating Streptococcus faecalis from Streptococcus faecium. J Gen Microbiol 14:57–68. doi: 10.1099/00221287-14-1-57. [DOI] [PubMed] [Google Scholar]
- 40.Papavassiliou J. 1962. Species differentiation of group D streptococci. Appl Microbiol 10:65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittenbury R. 1965. The differentiation of Streptococcus faecalis and S. faecium. J Gen Microbiol 38:279–287. doi: 10.1099/00221287-38-2-279. [DOI] [PubMed] [Google Scholar]
- 42.Manero A, Blanch AR. 1999. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol 65:4425–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lancefield RC. 1933. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med 57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anonymous. 2014. Identification of Streptococcus species, Enterococcus species and morphologically similar organisms. UK standards for microbiology investigations. Public Health England, London, England: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality-and-consistency-in-clinical-laboratories. [Google Scholar]
- 45.Williams AM, Rodrigues UM, Collins MD. 1991. Intrageneric relationships of enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA. Res Microbiol 142:67–74. doi: 10.1016/0923-2508(91)90098-U. [DOI] [PubMed] [Google Scholar]
- 46.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 47.Singhal N, Kumar M, Kanaujia PK, Virdi JS. 2015. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagace-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ. 2012. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol 50:3324–3328. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintela-Baluja M, Böhme K, Fernández-No IC, Morandi S, Alnakip ME, Caamaño-Antelo S, Barros-Velázquez J, Calo-Mata P. 2013. Characterization of different food-isolated Enterococcus strains by MALDI-TOF mass fingerprinting. Electrophoresis 34:2240–2250. doi: 10.1002/elps.201200699. [DOI] [PubMed] [Google Scholar]
- 50.Stępień-Pyśniak D, Hauschild T, Różański P, Marek A. 2017. MALDI-TOF mass spectrometry as a useful tool for identification of Enterococcus spp. from wild birds and differentiation of closely related species. J Microbiol Biotechnol 27:1128–1137. doi: 10.4014/jmb.1612.12036. [DOI] [PubMed] [Google Scholar]
- 51.Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol 50:2918–2931. doi: 10.1128/JCM.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakano S, Matsumura Y, Kato K, Yunoki T, Hotta G, Noguchi T, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2014. Differentiation of vanA-positive Enterococcus faecium from vanA-negative E. faecium by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Int J Antimicrob Agents 44:256–259. doi: 10.1016/j.ijantimicag.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Ljungstrom L, Enroth H, Claesson BE, Ovemyr I, Karlsson J, Froberg B, Brodin AK, Pernestig AK, Jacobsson G, Andersson R, Karlsson D. 2015. Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis. BMC Infect Dis 15:199. doi: 10.1186/s12879-015-0938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bosshard PP, Abels S, Altwegg M, Böttger EC, Zbinden R. 2004. Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J Clin Microbiol 42:2065–2073. doi: 10.1128/JCM.42.5.2065-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel R, Piper KE, Rouse MS, Steckelberg JM, Uhl JR, Kohner P, Hopkins MK, Cockerill FR, Kline BC. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J Clin Microbiol 36:3399–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angeletti S, Lorino G, Gherardi G, Battistoni F, De Cesaris M, Dicuonzo G. 2001. Routine molecular identification of enterococci by gene-specific PCR and 16S ribosomal DNA sequencing. J Clin Microbiol 39:794–797. doi: 10.1128/JCM.39.2.794-797.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Devriese LA, Vancanneyt M, Descheemaeker P, Baele M, Van Landuyt HW, Gordts B, Butaye P, Swings J, Haesebrouck F. 2002. Differentiation and identification of Enterococcus durans, E. hirae and E. villorum. J Appl Microbiol 92:821–827. doi: 10.1046/j.1365-2672.2002.01586.x. [DOI] [PubMed] [Google Scholar]
- 58.Poyart C, Quesnes G, Trieu-Cuot P. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J Clin Microbiol 38:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naser S, Thompson FL, Hoste B, Gevers D, Vandemeulebroecke K, Cleenwerck I, Thompson CC, Vancanneyt M, Swings J. 2005. Phylogeny and identification of enterococci by atpA gene sequence analysis. J Clin Microbiol 43:2224–2230. doi: 10.1128/JCM.43.5.2224-2230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai J-C, Hsueh P-R, Lin H-M, Chang H-J, Ho S-W, Teng L-J. 2005. Identification of clinically relevant Enterococcus species by direct sequencing of groES and spacer region. J Clin Microbiol 43:235–241. doi: 10.1128/JCM.43.1.235-241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ke D, Picard FJ, Martineau F, Menard C, Roy PH, Ouellette M, Bergeron MG. 1999. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol 37:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan TY, Jiang B, Ng LSY. 2017. Faster and economical screening for vancomycin-resistant enterococci by sequential use of chromogenic agar and real-time polymerase chain reaction. J Microbiol Immunol Infect 50:448–453. doi: 10.1016/j.jmii.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Fang H, Ohlsson AK, Ullberg M, Ozenci V. 2012. Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur J Clin Microbiol Infect Dis 31:3073–3077. doi: 10.1007/s10096-012-1667-x. [DOI] [PubMed] [Google Scholar]
- 65.Jackson CR, Fedorka-Cray PJ, Barrett JB. 2004. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol 42:3558–3565. doi: 10.1128/JCM.42.8.3558-3565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deck MK, Anderson ES, Buckner RJ, Colasante G, Davis TE, Coull JM, Crystal B, Latta PD, Fuchs M, Fuller D, Harris W, Hazen K, Klimas LL, Lindao D, Meltzer MC, Morgan M, Shepard J, Stevens S, Wu F, Fiandaca MJ. 2014. Rapid detection of Enterococcus spp. direct from blood culture bottles using Enterococcus QuickFISH method: a multicenter investigation. Diagn Microbiol Infect Dis 78:338–342. doi: 10.1016/j.diagmicrobio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 67.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, van Embden JDA, Willems RJL. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol 40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruiz-Garbajosa P, Bonten MJM, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Cantón R, Baquero F, Murray BE, del Campo R, Willems RJL. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Been M, Pinholt M, Top J, Bletz S, Mellmann A, van Schaik W, Brouwer E, Rogers M, Kraat Y, Bonten M, Corander J, Westh H, Harmsen D, Willems RJL. 18 November 2015. A core genome MLST scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol doi: 10.1128/jcm.01946-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lytsy B, Engstrand L, Gustafsson Å, Kaden R. 2017. Time to review the gold standard for genotyping vancomycin-resistant enterococci in epidemiology: comparing whole-genome sequencing with PFGE and MLST in three suspected outbreaks in Sweden during 2013–2015. Infect Genet Evol 54:74–80. doi: 10.1016/j.meegid.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Raven KE, Reuter S, Reynolds R, Brodrick HJ, Russell JE, Torok ME, Parkhill J, Peacock SJ. 2016. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res 26:1388–1396. doi: 10.1101/gr.204024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willems RJL, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, Hanage WP, Corander J. 2012. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3:e00151-12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murdoch DR, Corey GR, Hoen B, Miró JM, Fowler VG, Bayer AS, Karchmer AW, Olaison L, Pappas PA, Moreillon P, Chambers ST, Chu VH, Falcó V, Holland DJ, Jones P, Klein JL, Raymond NJ, Read KM, Tripodi MF, Utili R, Wang A, Woods CW, Cabell CH, International Collaboration on Endocarditis-Prospective Cohort Study Investigators . 2009. Clinical presentation, etiology and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med 169:463–473. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slipczuk L, Codolosa JN, Davila CD, Romero-Corral A, Yun J, Pressman GS, Figueredo VM. 2013. Infective endocarditis epidemiology over five decades: a systematic review. PLoS One 8:e82665. doi: 10.1371/journal.pone.0082665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 66:2263–2266. doi: 10.1128/AEM.66.5.2263-2266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tendolkar PM, Baghdayan AS, Shankar N. 2003. Pathogenic enterococci: new developments in the 21st century. Cell Mol Life Sci 60:2622–2636. doi: 10.1007/s00018-003-3138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turroni S, Rampelli S, Centanni M, Schnorr SL, Consolandi C, Severgnini M, Peano C, Soverini M, Falconi M, Crittenden AN, Henry AG, Brigidi P, Candela M. 2016. Enterocyte-associated microbiome of the Hadza hunter-gatherers. Front Microbiol 7:865. doi: 10.3389/fmicb.2016.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 79.MacCallum WG, Hastings TW. 1899. A case of acute endocarditis caused by Micrococcus zymogenes (nov. spec.), with a description of the microorganism. J Exp Med 4:521–534. doi: 10.1084/jem.4.5-6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hicks JAB. 1912. An unusual organism (Micrococcus zymogenes) in a case of malignant endocarditis. Proc R Soc Med 5:126–130. [PMC free article] [PubMed] [Google Scholar]
- 81.Donaldson R. 1913. A case of puerperal fever associated with the enterococcus. J Pathol 18:469–477. doi: 10.1002/path.1700180146. [DOI] [Google Scholar]
- 82.Donaldson R. 1917. The characters of the enterococcus. Br Med J i:188–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subbarayudu G. 1931. Enterococcus infection. Ind Med Gaz 66:139–140. [PMC free article] [PubMed] [Google Scholar]
- 84.Geiger AJ, Greenman L. 1945. Streptococcus fecalis bacteremia and meningitis; report of a case cured with penicillin. Yale J Biol Med 18:77–80. [PMC free article] [PubMed] [Google Scholar]
- 85.Teixeira LM, Carvalho MDGS, Facklam RR, Shewmaker PL. 2015. Enterococcus, p 403–421. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW (ed), Manual of clinical microbiology, 11th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 86.Guzman Prieto AM, van Schaik W, Rogers MRC, Coque TM, Baquero F, Corander J, Willems RJL. 2016. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol 7:788. doi: 10.3389/fmicb.2016.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.European Centre for Disease Prevention and Control. 2013. Point prevalence survey of healthcare-associated infections and antimicrobial use in European acute care hospitals 2011–2012. European Centre for Disease Prevention and Control, Solna, Sweden. [Google Scholar]
- 88.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. 2008. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 89.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Top J, Willems R, Blok H, de Regt M, Jalink K, Troelstra A, Goorhuis B, Bonten M. 2007. Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin Microbiol Infect 13:316–319. doi: 10.1111/j.1469-0691.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 91.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 92.de Perio MA, Yarnold PR, Warren J, Noskin GA. 2006. Risk factors and outcomes associated with non-Enterococcus faecalis, non-Enterococcus faecium enterococcal bacteremia. Infect Control Hosp Epidemiol 27:28–33. doi: 10.1086/500000. [DOI] [PubMed] [Google Scholar]
- 93.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. 1992. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother 36:2005–2008. doi: 10.1128/AAC.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navarro F, Courvalin P. 1994. Analysis of genes encoding d-alanine-d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob Agents Chemother 38:1788–1793. doi: 10.1128/AAC.38.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, Barbieri A, Kriegel C, Mehta SS, Knight JR, Jain D, Goodman AL, Kriegel MA. 2018. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyrrell GJ, Turnbull L, Teixeira LM, Lefebvre J, Carvalho MDGS, Facklam RR, Lovgren M. 2002. Enterococcus gilvus sp. nov. and Enterococcus pallens sp. nov. isolated from human clinical specimens. J Clin Microbiol 40:1140–1145. doi: 10.1128/JCM.40.4.1140-1145.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levesque S, Longtin Y, Domingo MC, Masse C, Bernatchez H, Gaudreau C, Tremblay C. 2016. Enteroccocus [sic] pallens as a potential novel human pathogen: three cases of spontaneous bacterial peritonitis. JMM Case Rep 3:e005024. doi: 10.1099/jmmcr.0.005024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Monticelli J, Knezevich A, Luzzati R, Di Bella S. 2018. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J Infect Chemother 24:237–246. doi: 10.1016/j.jiac.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 99.Mohamed JA, Huang DB. 2007. Biofilm formation by enterococci. J Med Microbiol 56:1581–1588. doi: 10.1099/jmm.0.47331-0. [DOI] [PubMed] [Google Scholar]
- 100.Patti JM, Hook M. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol 6:752–758. doi: 10.1016/0955-0674(94)90104-X. [DOI] [PubMed] [Google Scholar]
- 101.Sillanpaa J, Xu Y, Nallapareddy SR, Murray BE, Hook M. 2004. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology 150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- 102.Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, Murray BE, Hook M. 1999. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem 274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 103.Singh KV, Nallapareddy SR, Sillanpaa J, Murray BE. 2010. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog 6:e1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. 2009. Distribution of genes encoding MSCRAMMs and pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol 47:896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. 2008. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun 76:4110–4119. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. 2006. Pili in gram-positive pathogens. Nat Rev Microbiol 4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 107.Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, Murray BE. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. 2008. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154:3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- 109.Hendrickx AP, Schapendonk CM, van Luit-Asbroek M, Bonten MJ, van Schaik W, Willems RJ. 2010. Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiology 156:2649–2659. doi: 10.1099/mic.0.041392-0. [DOI] [PubMed] [Google Scholar]
- 110.Ike Y, Hashimoto H, Clewell DB. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun 45:528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cox CR, Coburn PS, Gilmore MS. 2005. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci 6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 112.Haas W, Shepard BD, Gilmore MS. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 113.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother 37:2474–2477. doi: 10.1128/AAC.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ike Y, Clewell DB. 1992. Evidence that the hemolysin/bacteriocin phenotype of Enterococcus faecalis subsp. zymogenes can be determined by plasmids in different incompatibility groups as well as by the chromosome. J Bacteriol 174:8172–8177. doi: 10.1128/jb.174.24.8172-8177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coburn PS, Gilmore MS. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol 5:661–669. doi: 10.1046/j.1462-5822.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 116.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. 2010. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun 78:4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. 2007. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun 75:1861–1869. doi: 10.1128/IAI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586. doi: 10.1128/IAI.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol 188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun 67:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun 69:4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Heikens E, Singh KV, Jacques-Palaz KD, van Luit-Asbroek M, Oostdijk EA, Bonten MJ, Murray BE, Willems RJ. 2011. Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microbes Infect 13:1185–1190. doi: 10.1016/j.micinf.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Heikens E, Bonten MJ, Willems RJ. 2007. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol 189:8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penadés JR, Lasa I. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol 67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peng Z, Ehrmann MA, Waldhuber A, Niemeyer C, Miethke T, Frick JS, Xiong T, Vogel RF. 2017. Phosphotransferase systems in Enterococcus faecalis OG1RF enhance anti-stress capacity in vitro and in vivo. Res Microbiol 168:558–566. doi: 10.1016/j.resmic.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 126.Sauvageot N, Mokhtari A, Joyet P, Budin-Verneuil A, Blancato VS, Repizo GD, Henry C, Pikis A, Thompson J, Magni C, Hartke A, Deutscher J. 2017. Enterococcus faecalis uses a phosphotransferase system permease and a host colonization-related ABC transporter for maltodextrin uptake. J Bacteriol 199:e00878-16. doi: 10.1128/JB.00878-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Paganelli FL, Huebner J, Singh KV, Zhang X, van Schaik W, Wobser D, Braat JC, Murray BE, Bonten MJ, Willems RJ, Leavis HL. 2016. Genome-wide screening identifies phosphotransferase system permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J Infect Dis 214:189–195. doi: 10.1093/infdis/jiw108. [DOI] [PubMed] [Google Scholar]
- 128.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 129.Gal-Mor O, Finlay BB. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 130.Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology 155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 131.Gao W, Howden BP, Stinear TP. 2018. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol 41:76–82. doi: 10.1016/j.mib.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 132.Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. 2009. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol 27:301–305. doi: 10.4103/0255-0857.55437. [DOI] [PubMed] [Google Scholar]
- 133.Sava IG, Heikens E, Huebner J. 2010. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect 16:533–540. doi: 10.1111/j.1469-0691.2010.03213.x. [DOI] [PubMed] [Google Scholar]
- 134.Murray BE. 1990. The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65. doi: 10.1128/CMR.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Florescu I, Beuran M, Dimov R, Razbadauskas A, Bochan M, Fichev G, Dukart G, Babinchak T, Cooper CA, Ellis-Grosse EJ, Dartois N, Gandjini H, 307 Study Group . 2008. Efficacy and safety of tigecycline compared with vancomycin or linezolid for treatment of serious infections with methicillin-resistant Staphylococcus aureus or vancomycin-resistant enterococci: a phase 3, multicentre, double-blind, randomized study. J Antimicrob Chemother 62:i17–i28. doi: 10.1093/jac/dkn250. [DOI] [PubMed] [Google Scholar]
- 136.Roberts M. 1994. Epidemiology of tetracycline resistance determinants. Trends Microbiol 2:353–357. doi: 10.1016/0966-842X(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 137.de Lastours V, Maugy E, Mathy V, Chau F, Rossi B, Guerin F, Cattoir V, Fantin B, CIPHARES Study Group . 2017. Ecological impact of ciprofloxacin on commensal enterococci in healthy volunteers. J Antimicrob Chemother 72:1574–1580. doi: 10.1093/jac/dkx043. [DOI] [PubMed] [Google Scholar]
- 138.Duez C, Zorzi W, Sapunaric F, Amoroso A, Thamm I, Coyette J. 2001. The penicillin resistance of Enterococcus faecalis JH2-2R results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology 147:2561–2569. doi: 10.1099/00221287-147-9-2561. [DOI] [PubMed] [Google Scholar]
- 139.Infante VH, Conceicao N, de Oliveira AG, Darini AL. 2016. Evaluation of polymorphisms in pbp4 gene and genetic diversity in penicillin-resistant, ampicillin-susceptible Enterococcus faecalis from hospitals in different states in Brazil. FEMS Microbiol Lett 363:fnw044. doi: 10.1093/femsle/fnw044. [DOI] [PubMed] [Google Scholar]
- 140.Williamson R, Le Bouguénec C, Gutmann L, Horaud T. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol 131:1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- 141.Arbeloa A, Segal H, Hugonnet J-E, Josseaume N, Dubost L, Brouard J-P, Gutmann L, Mengin-Lecreulx D, Arthur M. 2004. Role of class A penicillin-binding proteins in PBP5-mediated β-lactam resistance in Enterococcus faecalis. J Bacteriol 186:1221–1228. doi: 10.1128/JB.186.5.1221-1228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rice LB, Bellais S, Carias LL, Hutton-Thomas R, Bonomo RA, Caspers P, Page MGP, Gutmann L. 2004. Impact of specific pbp5 mutations on expression of β-lactam resistance in Enterococcus faecium. Antimicrob Agents Chemother 48:3028–3032. doi: 10.1128/AAC.48.8.3028-3032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hodges TL, Zighelboim-Daum S, Eliopoulos GM, Wennersten C, Moellering RC. 1992. Antimicrobial susceptibility changes in Enterococcus faecalis following various penicillin exposure regimens. Antimicrob Agents Chemother 36:121–125. doi: 10.1128/AAC.36.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Geraci JE, Martin WJ. 1954. Subacute enterococcal endocarditis: clinical, pathologic and therapeutic considerations in 33 patients. Circulation 10:173–194. doi: 10.1161/01.CIR.10.2.173. [DOI] [PubMed] [Google Scholar]
- 145.Jawetz E, Sonne M. 1966. Penicillin-streptomycin treatment of enterococcal endocarditis—a reevaluation. N Engl J Med 274:710–715. doi: 10.1056/NEJM196603312741304. [DOI] [PubMed] [Google Scholar]
- 146.Moellering RC, Weinberg AN. 1971. Studies on antibiotic synergism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labelled streptomycin by enterococci. J Clin Invest 50:2580–2584. doi: 10.1172/JCI106758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fernández-Hidalgo N, Almirante B, Gavaldà J, Gurgui M, Peña C, de Alarcón A, Ruiz J, Vilacosta I, Montejo M, Vallejo N, López-Medrano F, Plata A, López J, Hidalgo-Tenorio C, Gálvez J, Sáez C, Lomas JM, Falcone M, de la Torre J, Martínez-Lacasa X, Pahissa A. 2013. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating Enterococcus faecalis infective endocarditis. Clin Infect Dis 56:1261–1268. doi: 10.1093/cid/cit052. [DOI] [PubMed] [Google Scholar]
- 148.Pericas JM, Cervera C, del Rio A, Moreno A, Garcia de la Maria C, Almela M, Falces C, Ninot S, Castaneda X, Armero Y, Soy D, Gatell JM, Marco F, Mestres CA, Miro JM, Hospital Clinic Endocarditis Study Group . 2014. Changes in the treatment of Enterococcus faecalis infective endocarditis in Spain in the last 15 years: from ampicillin plus gentamicin to ampicillin plus ceftriaxone. Clin Microbiol Infect 20:O1075–O1083. doi: 10.1111/1469-0691.12756. [DOI] [PubMed] [Google Scholar]
- 149.Mainardi JL, Gutmann L, Acar JF, Goldstein FW. 1995. Synergistic effect of amoxicillin and cefotaxime against Enterococcus faecalis. Antimicrob Agents Chemother 39:1984–1987. doi: 10.1128/AAC.39.9.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singh KV, Weinstock GM, Murray BE. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 46:1845–1850. doi: 10.1128/AAC.46.6.1845-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zervos MJ, Schaberg DR. 1985. Reversal of in vitro susceptibility of enterococci to trimethoprim-sulfamethoxazole by folinic acid. Antimicrob Agents Chemother 28:446–448. doi: 10.1128/AAC.28.3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rice LB, Murray BE. 1995. β-Lactamase-producing enterococci, p 107–114. In Brown F, Ferretti JJ (ed), Genetics of streptococci, enterococci and lactococci, vol 85 Developmental and biological standards. Karger, Basel, Switzerland. [Google Scholar]
- 153.Rice LB, Marshall SH. 1992. Evidence of incorporation of the chromosomal β-lactamase gene of Enterococcus faecalis CH19 into a transposon derived from staphylococci. Antimicrob Agents Chemother 36:1843–1846. doi: 10.1128/AAC.36.9.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zscheck KK, Murray BE. 1993. Genes involved in the regulation of β-lactamase production in enterococci and staphylococci. Antimicrob Agents Chemother 37:1966–1970. doi: 10.1128/AAC.37.9.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ingerman M, Pitsakis PG, Rosenberg A, Hessen MT, Abrutyn E, Murray BE, Levison ME. 1987. β-Lactamase production in experimental endocarditis due aminoglycoside-resistant Streptococcus faecalis. J Infect Dis 155:1226–1232. doi: 10.1093/infdis/155.6.1226. [DOI] [PubMed] [Google Scholar]
- 156.Rybkine T, Mainardi J-L, Sougakoff W, Collatz E, Gutmann L. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J Infect Dis 178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 157.Zorzi W, Zhou XY, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol 178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Galloway-Peña JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis 200:1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rice LB, Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Lonks J, Moon TM, D’Andréa ÉD, Page R, Peti W. 2018. Structural and regulatory changes in PBP4 trigger decreased β-lactam susceptibility in Enterococcus faecalis. mBio 9:e00361-18. doi: 10.1128/mBio.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kellogg SL, Little JL, Hoff JS, Kristich CJ. 2017. Requirement of the CroRS two-component system for resistance to cell wall-targeting antimicrobials in Enterococcus faecium. Antimicrob Agents Chemother 61:e02461-16. doi: 10.1128/AAC.02461-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kristich CJ, Wells CL, Dunny GM. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci U S A 104:3508–3513. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rice LB, Carias LL, Rudin S, Hutton R, Marshall S, Hassan M, Josseaume N, Dubost L, Marie A, Arthur M. 2009. Role of class A penicillin-binding proteins in the expression of β-lactam resistance in Enterococcus faecium. J Bacteriol 191:3649–3656. doi: 10.1128/JB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Dunman P, Coleman J, Arthur M, Rice LB. 2016. Involvement of the eukaryote-like kinase-phosphatase system and a protein that interacts with penicillin-binding protein 5 in emergence of cephalosporin resistance in cephalosporin-sensitive class A penicillin-binding protein mutants in Enterococcus faecium. mBio 7:e02188-15. doi: 10.1128/mBio.02188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C. 2006. Crystal structure of a novel β-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol 359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 165.Shlaes DM, Al-Obeid S, Shlaes JH, Boisivon A, Williamson R. 1989. Inducible, transferable resistance to vancomycin in Enterococcus faecium, D399. J Antimicrob Chemother 23:503–508. doi: 10.1093/jac/23.4.503. [DOI] [PubMed] [Google Scholar]
- 166.Shlaes DM, Bouvet A, Devine C, Shlaes JH, Al-Obeid S, Williamson R. 1989. Inducible, transferable resistance to vancomycin in Enterococcus faecalis A256. Antimicrob Agents Chemother 33:198–203. doi: 10.1128/AAC.33.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arthur M, Courvalin P. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother 37:1563–1571. doi: 10.1128/AAC.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol 179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reynolds PE, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol Microbiol 13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 171.Arthur M, Molinas C, Courvalin P. 1992. Sequence of the vanY gene required for production of a vancomycin-inducible D,D-carboxypeptidase in Enterococcus faecium BM4147. Gene 120:111–114. doi: 10.1016/0378-1119(92)90017-J. [DOI] [PubMed] [Google Scholar]
- 172.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87–92. doi: 10.1016/0378-1119(94)00851-I. [DOI] [PubMed] [Google Scholar]
- 173.Reid KC, Cockerill FR III, Patel R. 2001. Clinical and epidemiological features of Enterococcus casseliflavus/flavescens and Enterococcus gallinarum bacteremia: a report of 20 cases. Clin Infect Dis 32:1540–1546. doi: 10.1086/320542. [DOI] [PubMed] [Google Scholar]
- 174.Depardieu F, Mejean V, Courvalin P. 2015. Competition between VanU(G) repressor and VanR(G) activator leads to rheostatic control of vanG vancomycin resistance operon expression. PLoS Genet 11:e1005170. doi: 10.1371/journal.pgen.1005170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Evers S, Sahm DF, Courvalin P. 1993. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding D-ala:D-ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene 124:143–144. doi: 10.1016/0378-1119(93)90779-3. [DOI] [PubMed] [Google Scholar]
- 176.Perichon B, Reynolds P, Courvalin P. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother 41:2016–2018. doi: 10.1128/AAC.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F, Jacoby GA, Wang M. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 54:4643–4647. doi: 10.1128/AAC.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fines M, Perichon B, Reynolds P, Sahm DF, Courvalin P. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother 43:2161–2164. doi: 10.1128/AAC.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, Leclercq R, Courvalin P, Cattoir V. 2011. d-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother 55:4606–4612. doi: 10.1128/AAC.00714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sassi M, Guerin F, Lesec L, Isnard C, Fines-Guyon M, Cattoir V, Giard JC. 2018. Genetic characterization of a VanG-type vancomycin-resistant Enterococcus faecium clinical isolate. J Antimicrob Chemother 73:852–855. doi: 10.1093/jac/dkx510. [DOI] [PubMed] [Google Scholar]
- 181.Evers S, Courvalin P. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanS(B)-VanR(B) two-component regulatory system in Enterococcus faecalis V583. J Bacteriol 178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hayden MK, Trenholm GM, Schultz JE, Sahm DF. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis 167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 183.Carias LL, Rudin SD, Donskey CJ, Rice LB. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol 180:4426–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Srinivasan V, Metcalf BJ, Knipe KM, Ouattara M, McGee L, Shewmaker PL, Glennen A, Nichols M, Harris C, Brimmage M, Ostrowsky B, Park CJ, Schrag SJ, Frace MA, Sammons SA, Beall B. 2014. vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. mBio 5:e01386-14. doi: 10.1128/mBio.01386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hodel-Christian SL, Murray BE. 1991. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob Agents Chemother 35:1147–1152. doi: 10.1128/AAC.35.6.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lyon BR, May JW, Skurray RA. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol Gen Genet 193:554–556. doi: 10.1007/BF00382099. [DOI] [PubMed] [Google Scholar]
- 188.Padmasini E, Padmaraj R, Ramesh SS. 2014. High level aminoglycoside resistance and distribution of aminoglycoside resistant genes among clinical isolates of Enterococcus species in Chennai, India. ScientificWorldJournal 2014:329157. doi: 10.1155/2014/329157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chow JW, Zervos MJ, Lerner SA, Thal LA, Donabedian SM, Jaworski DD, Tsai S, Shaw KJ, Clewell DB. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob Agents Chemother 41:511–514. doi: 10.1128/AAC.41.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ounissi H, Derlot E, Carlier C, Courvalin P. 1990. Gene homogeneity for aminoglycoside-modifying enzymes in Gram-positive cocci. Antimicrob Agents Chemother 34:2164–2168. doi: 10.1128/AAC.34.11.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wright GD, Ladak P. 1997. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob Agents Chemother 41:956–960. doi: 10.1128/AAC.41.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tankovic J, Bachoual R, Ouabdesselam S, Boudjadja A, Soussy CJ. 1999. In-vitro activity of moxifloxacin against fluoroquinolone-resistant strains of aerobic gram-negative bacilli and Enterococcus faecalis. J Antimicrob Chemother 43:19–23. doi: 10.1093/jac/43.suppl_2.19. [DOI] [PubMed] [Google Scholar]
- 193.el Amin NA, Jalal S, Wretlind B. 1999. Alterations in GyrA and ParC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob Agents Chemother 43:947–949. doi: 10.1128/AAC.43.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kanematsu E, Deguchi T, Yasuda M, Kawamura T, Nishino Y, Kawada Y. 1998. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob Agents Chemother 42:433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Oyamada Y, Ito H, Fujimoto K, Asada R, Niga T, Okamoto R, Inoue M, Yamagishi J. 2006. Combination of known and unknown mechanisms confers high-level resistance to fluoroquinolones in Enterococcus faecium. J Med Microbiol 55:729–736. doi: 10.1099/jmm.0.46303-0. [DOI] [PubMed] [Google Scholar]
- 196.Vega S, Dowzicky MJ. 2017. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann Clin Microbiol Antimicrob 16:50. doi: 10.1186/s12941-017-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 46:3334–3336. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pillai SK, Sakoulas G, Wennersten C, Eliopoulos GM, Moellering RC Jr, Ferraro MJ, Gold HS. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J Infect Dis 186:1603–1607. doi: 10.1086/345368. [DOI] [PubMed] [Google Scholar]
- 199.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 200.Vester B. 2018. The cfr and cfr-like multiple resistance genes. Res Microbiol 169:61–66. doi: 10.1016/j.resmic.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 201.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 202.Miller WR, Bayer AS, Arias CA. 2016. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harb Perspect Med 6:a026997. doi: 10.1101/cshperspect.a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mishra NN, Bayer AS, Tran TT, Shamoo Y, Mileykovskaya E, Dowhan W, Guan Z, Arias CA. 2012. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7:e43958. doi: 10.1371/journal.pone.0043958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rice LB. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother 42:1871–1877. doi: 10.1128/AAC.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Speer BS, Shoemaker NB, Salyers AA. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev 5:387–399. doi: 10.1128/CMR.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, Werner G. 2016. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother 71:871–881. doi: 10.1093/jac/dkv420. [DOI] [PubMed] [Google Scholar]
- 207.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:e00064-16. doi: 10.1128/mSphere.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yasmin A, Kenny JG, Shankar J, Darby AC, Hall N, Edwards C, Horsburgh MJ. 2010. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J Bacteriol 192:1122–1130. doi: 10.1128/JB.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett Appl Microbiol 52:559–564. doi: 10.1111/j.1472-765X.2011.03043.x. [DOI] [PubMed] [Google Scholar]
- 211.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology, p 309–420. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 212.Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 213.Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol 117:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Jensen LB, Garcia-Migura L, Valenzuela AJS, Løhr M, Hasman H, Aarestrup FM. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods 80:25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 215.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dunny GM, Craig RA, Carron RL, Clewell DB. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454–465. doi: 10.1016/0147-619X(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 218.Palmer KL, Kos VN, Gilmore MS. 2010. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol 13:632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dunny GM. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc B Biol Sci 362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Clewell DB. 2007. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58:205–227. doi: 10.1016/j.plasmid.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 221.Gilmore MS, Segarra RA, Booth MC, Bogie CP, Hall LR, Clewell DB. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol 176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 223.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci U S A 107:12269–12274. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Clewell DB. 1981. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev 45:409–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lanza VF, Tedim AP, Martínez JL, Baquero F, Coque TM. 2015. The plasmidome of Firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr 3:PLAS-0039-2014. doi: 10.1128/microbiolspec.PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- 226.Schwarz FV, Perreten V, Teuber M. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 227.Rosvoll TCS, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, Jensen LB, Nielsen KM, Sundsfjord A. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol 58:254–268. doi: 10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 228.Freitas AR, Novais C, Tedim AP, Francia MV, Baquero F, Peixe L, Coque TM. 2013. Microevolutionary events involving narrow host plasmids influences local fixation of vancomycin-resistance in Enterococcus populations. PLoS One 8:e60589. doi: 10.1371/journal.pone.0060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Handwerger S, Pucci MJ, Kolokathis A. 1990. Vancomycin resistance is encoded on a pheromone response plasmid in Enterococcus faecium 228. Antimicrob Agents Chemother 34:358–360. doi: 10.1128/AAC.34.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Magi G, Capretti R, Paoletti C, Pietrella M, Ferrante L, Biavasco F, Varaldo PE, Facinelli B. 2003. Presence of a vanA-carrying pheromone response plasmid (pBRG1) in a clinical isolate of Enterococcus faecium. Antimicrob Agents Chemother 47:1571–1576. doi: 10.1128/AAC.47.5.1571-1576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rice LB, Carias LL, Marshall S, Rudin SD, Hutton-Thomas R. 2005. Tn5386, a novel Tn916-like mobile element in Enterococcus faecium D344R that interacts withTn916 to yield a large genomic deletion. J Bacteriol 187:6668–6677. doi: 10.1128/JB.187.19.6668-6677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rice LB, Carias LL, Rudin S, Laktičová V, Wood A, Hutton-Thomas R. 2005. Enterococcus faecium low-affinity pbp5 is a transferable determinant. Antimicrob Agents Chemother 49:5007–5012. doi: 10.1128/AAC.49.12.5007-5012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.García-Solache M, Lebreton F, McLaughlin RE, Whiteaker JD, Gilmore MS, Rice LB. 2016. Homologous recombination within large chromosomal regions facilitates acquisition of β-lactam and vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother 60:5777–5786. doi: 10.1128/AAC.00488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Novais C, Tedim AP, Lanza VF, Freitas AR, Silveira E, Escada R, Roberts AP, Al-Haroni M, Baquero F, Peixe L, Coque TM. 2016. Co-diversification of Enterococcus faecium core genomes and PBP5: evidences of pbp5 horizontal transfer. Front Microbiol 7:1581. doi: 10.3389/fmicb.2016.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 236.Werner G, Coque TM, Franz CMAP, Grohmann E, Hegstad K, Jensen L, van Schaik W, Weaver K. 2013. Antibiotic resistant enterococci—tales of a drug resistance gene trafficker. Int J Med Microbiol 303:360–379. doi: 10.1016/j.ijmm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 237.Tomita H, Pierson C, Lim SK, Clewell DB, Ike Y. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J Clin Microbiol 40:3326–3333. doi: 10.1128/JCM.40.9.3326-3333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PDR, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 194:2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Qin X, Galloway-Pena J, Sillanpaa J, Roh J, Nallapareddy S, Chowdhury S, Bourgogne A, Choudhury T, Muzny D, Buhay C, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino J, Worley K, Gibbs R, Weinstock G, Murray B. 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 241.Scott JR, Churchward GG. 1995. Conjugative transposition. Annu Rev Microbiol 49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 242.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Franke AE, Clewell DB. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol 145:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mahillon J, Kleckner N. 1992. New IS10 transposition vectors based on a Gram-positive replication origin. Gene 116:69–74. doi: 10.1016/0378-1119(92)90630-8. [DOI] [PubMed] [Google Scholar]
- 245.Quintiliani R Jr, Courvalin P. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 246.Siguier P, Filee J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 247.Shaw JH, Clewell DB. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol 164:782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Raycroft RE, Zimmerman LN. 1964. New mode of genetic transfer in Streptococcus faecalis var. liquefaciens. J Bacteriol 87:799–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tomich PK, An FY, Damle SP, Clewell DB. 1979. Plasmid-related transmissibility and multiple drug resistance in Streptococcus faecalis subsp. zymogenes strain DS 16. Antimicrob Agents Chemother 15:828–830. doi: 10.1128/AAC.15.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ike Y, Tanimoto K, Tomita H, Takeuchi K, Fujimoto S. 1998. Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J Bacteriol 180:4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tanimoto K, Ike Y. 2008. Complete nucleotide sequencing and analysis of the 65-kb highly conjugative Enterococcus faecium plasmid pMG1: identification of the transfer-related region and the minimum region required for replication. FEMS Microbiol Lett 288:186–195. doi: 10.1111/j.1574-6968.2008.01342.x. [DOI] [PubMed] [Google Scholar]
- 252.Uttley AHC, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet 331:57–58. doi: 10.1016/S0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 253.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 33:1588–1591. doi: 10.1128/AAC.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kuch A, Willems RJL, Werner G, Coque TM, Hammerum AM, Sundsfjord A, Klare I, Ruiz-Garbajosa P, Simonsen GS, van Luit-Asbroek M, Hryniewicz W, Sadowy E. 2012. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J Antimicrob Chemother 67:551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 256.Shankar N, Baghdayan AS, Gilmore MS. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 257.Coburn PS, Baghdayan AS, Dolan G, Shankar N. 2007. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Mol Microbiol 63:530–544. doi: 10.1111/j.1365-2958.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- 258.McBride SM, Coburn PS, Baghdayan AS, Willems RJ, Grande MJ, Shankar N, Gilmore MS. 2009. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J Bacteriol 191:3392–3402. doi: 10.1128/JB.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.McBride SM, Fischetti VA, LeBlanc DJ, Moellering RC, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Raven KE, Reuter S, Gouliouris T, Reynolds R, Russell JE, Brown NM, Török ME, Parkhill J, Peacock SJ. 2016. Genome-based characterization of hospital-adapted Enterococcus faecalis lineages. Nat Microbiol 1:15033. doi: 10.1038/nmicrobiol.2015.33. [DOI] [PubMed] [Google Scholar]
- 261.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kim EB, Marco ML. 2014. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl Environ Microbiol 80:154–165. doi: 10.1128/AEM.03108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cohan FM. 2002. What are bacterial species? Annu Rev Microbiol 56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 266.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11:821–828. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lam M, Seemann T, Tobias N, Chen H, Haring V, Moore R, Ballard S, Grayson L, Johnson P, Howden B, Stinear T. 2013. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 14:595. doi: 10.1186/1471-2164-14-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hublin J-J, Ben-Ncer A, Bailey SE, Freidline SE, Neubauer S, Skinner MM, Bergmann I, Le Cabec A, Benazzi S, Harvati K, Gunz P. 2017. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546:289–292. doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 269.Larson G, Fuller DQ. 2014. The evolution of animal domestication. Annu Rev Ecol Evol Syst 45:115–136. doi: 10.1146/annurev-ecolsys-110512-135813. [DOI] [Google Scholar]
- 270.Zeder MA. 2012. The domestication of animals. J Anthropol Res 68:161–190. doi: 10.3998/jar.0521004.0068.201. [DOI] [Google Scholar]
- 271.Marshall FB, Dobney K, Denham T, Capriles JM. 2014. Evaluating the roles of directed breeding and gene flow in animal domestication. Proc Natl Acad Sci U S A 111:6153–6158. doi: 10.1073/pnas.1312984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Rice LB, Calderwood SB, Eliopoulos GM, Farber BF, Karchmer AW. 1991. Enterococcal endocarditis: a comparison of native and prosthetic valve disease. Rev Infect Dis 13:1–7. doi: 10.1093/clinids/13.1.1. [DOI] [PubMed] [Google Scholar]
- 273.Gavaldà J, Cardona PJ, Almirante B, Capdevila JA, Laguarda M, Pou L, Crespo E, Pigrau C, Pahissa A. 1996. Treatment of experimental endocarditis due to Enterococcus faecalis using once-daily dosing regimen of gentamicin plus simulated profiles of ampicillin in human serum. Antimicrob Agents Chemother 40:173–178. doi: 10.1128/AAC.40.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gavaldà J, Len O, Miró JM, Muñoz P, Montejo M, Alarcón A, de la Torre-Cisneros J, Peña C, Martínez-Lacasa X, Sarria C, Bou G, Aguado JM, Navas E, Romeu J, Marco F, Torres C, Tornos P, Planes A, Falcó V, Almirante B, Pahissa A. 2007. Brief communication: treatment of Enterococcus faecalis endocarditis with ampicillin plus ceftriaxone. Ann Intern Med 146:574–579. doi: 10.7326/0003-4819-146-8-200704170-00008. [DOI] [PubMed] [Google Scholar]
- 275.Gavalda J, Onrubia PL, Gomez MT, Gomis X, Ramirez JL, Len O, Rodriguez D, Crespo M, Ruiz I, Pahissa A. 2003. Efficacy of ampicillin combined with ceftriaxone and gentamicin in the treatment of experimental endocarditis due to Enterococcus faecalis with no high-level resistance to aminoglycosides. J Antimicrob Chemother 52:514–517. doi: 10.1093/jac/dkg360. [DOI] [PubMed] [Google Scholar]
- 276.Baddour LM, Wilson WR, Bayer AS, Fowler VG Jr, Tleyjeh IM, Rybak MJ, Barsic B, Lockhart PB, Gewitz MH, Levison ME, Bolger AF, Steckelberg JM, Baltimore RS, Fink AM, O’Gara P, Taubert KA, American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council . 2015. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications. A scientific statement for healthcare professionals from the American Heart Association. Circulation 132:1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 277.Olaison L, Schadewitz K, Swedish Society of Infectious Diseases Quality Assurance Study Group for Endocarditis . 2002. Enterococcal endocarditis in Sweden, 1995-1999: can shorter therapy with aminoglycosides be used? Clin Infect Dis 34:159–166. doi: 10.1086/338233. [DOI] [PubMed] [Google Scholar]
- 278.Iversen K, Ihlemann N, Gill SU, Madsen T, Elming H, Jensen KT, Bruun NE, Hofsten DE, Fursted K, Christensen JJ, Schultz M, Klein CF, Fosboll EL, Rosenvinge F, Schonheyder HC, Kober L, Torp-Pedersen C, Helweg-Larsen J, Tonder N, Moser C, Bundgaard H. 28 August 2018. Partial oral versus intravenous antibiotic treatment of endocarditis. N Engl J Med doi: 10.1056/NEJMoa1808312. [DOI] [PubMed] [Google Scholar]
- 279.Delgado G Jr, Neuhauser MM, Bearden DT, Danziger LH. 2000. Quinupristin-dalfopristin: an overview. Pharmacotherapy 20:1469–1485. doi: 10.1592/phco.20.19.1469.34858. [DOI] [PubMed] [Google Scholar]
- 280.Gerson SL, Kaplan SL, Bruss JB, Le V, Arellano FM, Hafkin B, Kuter DJ. 2002. Hematologic effects of linezolid: summary of clinical experience. Antimicrob Agents Chemother 46:2723–2726. doi: 10.1128/AAC.46.8.2723-2726.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lawrence KR, Adra M, Gillman PK. 2006. Serotonin toxicity associated with the use of linezolid: a review of postmarketing data. Clin Infect Dis 42:1578–1583. doi: 10.1086/503839. [DOI] [PubMed] [Google Scholar]
- 282.Chuang YC, Lin HY, Chen PY, Lin CY, Wang JT, Chang SC. 2016. Daptomycin versus linezolid for the treatment of vancomycin-resistant enterococcal bacteraemia: implications of daptomycin dose. Clin Microbiol Infect 22:890.e1–890.e7. doi: 10.1016/j.cmi.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 283.Lauridsen TK, Bruun LE, Rasmussen RV, Arpi M, Risum N, Moser C, Johansen HK, Bundgaard H, Hassager C, Bruun NE. 2012. Linezolid as rescue treatment for left-sided infective endocarditis: an observational, retrospective, multicenter study. Eur J Clin Microbiol Infect Dis 31:2567–2574. doi: 10.1007/s10096-012-1597-7. [DOI] [PubMed] [Google Scholar]
- 284.Eisenstein BI, Oleson FB Jr, Baltz RH. 2010. Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin Infect Dis 50:S10–S15. doi: 10.1086/647938. [DOI] [PubMed] [Google Scholar]
- 285.Foolad F, Taylor BD, Shelburne SA, Arias CA, Aitken SL. 13 March 2018. Association of daptomycin dosing regimen and mortality in patients with VRE bacteraemia: a review. J Antimicrob Chemother doi: 10.1093/jac/dky072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sakoulas G, Bayer AS, Pogliano J, Tsuji BT, Yang SJ, Mishra NN, Nizet V, Yeaman MR, Moise PA. 2012. Ampicillin enhances daptomycin- and cationic host defense peptide-mediated killing of ampicillin- and vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother 56:838–844. doi: 10.1128/AAC.05551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Doernberg SB, Lodise TP, Thaden JT, Munita JM, Cosgrove SE, Arias CA, Boucher HW, Corey GR, Lowy FD, Murray B, Miller LG, Holland TL, Gram-Positive Committee of the Antibacterial Resistance Leadership Group . 2017. Gram-positive bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 64:S24–S29. doi: 10.1093/cid/ciw828. [DOI] [PMC free article] [PubMed] [Google Scholar]