This review provides a comprehensive summary of issues associated with treating polyclonal bacterial biofilms in chronic diabetic wounds. We use this as a foundation and discuss the alternatives to conventional antibiotics and the emerging need for suitable drug delivery systems.
KEYWORDS: chronic diabetic wounds, biofilms, drug delivery systems, engineering, nanoparticles, quorum sensing
SUMMARY
This review provides a comprehensive summary of issues associated with treating polyclonal bacterial biofilms in chronic diabetic wounds. We use this as a foundation and discuss the alternatives to conventional antibiotics and the emerging need for suitable drug delivery systems. In recent years, extraordinary advances have been made in the field of nanoparticle synthesis and packaging. However, these systems have not been incorporated into the clinic for treatments other than for cancer or severe genetic diseases. We present a unifying perspective on how the field is evolving and the need for an early amalgamation of engineering principles and a biological understanding of underlying phenomena in order to develop a therapy that is translatable to the clinic in a shorter time.
INTRODUCTION
Diabetes mellitus affects about 34.6 million adults in the United States, which is approximately 12.6% of the population (1). The global prevalence of diabetes among adults is 8.5% (2), thereby making it a pandemic. Neuropathy or nerve damage due to hyperglycemia is a common cause of chronic foot ulcers and wounds in diabetic patients. As a result, 5% of diabetic patients undergo lower-extremity amputation (3). The direct annual expenditure toward managing these foot ulcers was recorded to be $9 billion to $13 billion in the United States alone (4). These management costs, along with limb amputations, create a significant economic burden on the health care system. Limb amputation also deteriorates the quality of life of the affected individuals.
The phenomenon of wound healing has been described in detail by Demidova-Rice et al. and by Reinke and Sorg (5, 6). Briefly, wound healing occurs in three stages: (i) the coagulation and inflammation phase, (ii) the proliferative phase, and (iii) the remodeling phase. Upon injury, the platelets migrate to the damaged blood vessels to initiate blood clotting. They release chemokines that attract inflammatory cells such as leukocytes, neutrophils, and macrophages to the site of injury. These cells release reactive oxygen species (ROS) and different proteases that eliminate any cell debris or bacteria at the open wound site. Simultaneously, they induce and maintain the proliferation of dermal and epidermal cells to replace the damaged tissue. Vascularization of these cells needs to happen to maintain cell growth and the nutrient supply. Therefore, proangiogenic cytokines, such as vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF), are released by platelets and inflammatory cells. Subsequently, fibroblasts differentiate to form an extracellular matrix (ECM) with the aid of a class of enzymes called matrix metalloproteinases (MMPs). Eventually, the fibroblasts are cleared by apoptosis, and acellular scar tissue finally seals off the wound.
When a person is suffering from underlying pathologies such as diabetes, it gives rise to chronic wounds. The hallmark of chronic wounds is prolonged and excessive inflammation, recurrent infections, the inability of dermal and epidermal cells to respond to regenerative stimuli, and, most importantly, vascular impairment due to microvascular pathologies related to hyperglycemia (7). Since these wounds do not heal, they are a favorable breeding ground for bacteria, which, in turn, further delay wound healing.
Several strategies for wound management have been described previously, including the delivery of growth factors to accelerate healing (8–10). Magana et al. also described in detail the laboratory techniques used to monitor, characterize, and quantify bacterial biofilms (11). The focus of this review article is on infection management strategies for chronic wounds. First, we describe the complications associated with bacterial infections in chronic wounds. A large portion of this review, however, focuses on drug delivery systems from an engineering perspective that are in an exploratory phase for infection prevention and treatment.
BACTERIAL INFECTIONS IN CHRONIC WOUNDS
The Gram-positive bacterium Staphylococcus aureus is the most commonly found bacterial species in diabetic ulcers. Other microorganisms such as beta-hemolytic streptococci and a mixture of Gram-negative species such as Escherichia coli, Klebsiella, and Pseudomonas aeruginosa are also present in wounds (12). Staphylococcus epidermidis, a Gram-positive bacterium native to human skin, may also turn pathogenic when exposed to systemic circulation in the wound bed (13). Conventionally, bacterial infections have been treated with oral or intravenous antibiotics depending upon the severity of infection and sometimes bioabsorption of the antibiotics. However, infections of chronic wounds are canny. Wounds can become infected by bacteria that encapsulate themselves in biofilms over time or when the body’s natural defense mechanisms are impaired. Biofilm consists of bacteria encapsulated in a protective layer of bacterially derived extracellular polymeric substances (EPSs) that provides them with a favorable environment for proliferation and survival (14). These biofilms are generally surface associated, and bacterial cells in these biofilms communicate with each other via quorum sensing (QS). QS signals regulate the expression of genes, production of proteases, and other cues that enable the high-density bacterial cluster to thrive (15). Biofilms exhibit enhanced tolerance to antibiotics compared to free-living bacteria, which makes treatment of wound infections challenging.
The ineffectiveness of traditional antibiotics in treating biofilms has been attributed to a combination of different factors. The multilayered defense against antibiotics includes poor penetration into biofilms, adaptive stress responses, and metabolic inactivation due to nutrient and gas limitation (16). Charged pockets on biofilm surfaces have been identified by Kurniawan and Fukuda (17). A negatively charged biofilm membrane may limit the penetration of positively charged antibiotics through the biofilm (18), referred to as charge- and size-based limitations in engineering jargon. Even if the antibiotic molecule enters the biofilm, it has to diffuse through the aqueous matrix in order to reach the bacterial cells. Aminoglycosides and beta-lactams may be inactivated or sequestered by binding to any solutes present in the matrix, making it impossible for them to diffuse to the depths of the biofilm (19, 20), also referred to as mass transport limitation. The same principle limits the diffusion of nutrients and gas transfer to the bacteria that grow at the bottom core of the biofilms. As a result, they cannot divide and grow as actively as bacteria that are at the top surface of the biofilm. Since most antibiotics work only on growing bacterial cells (21), these drugs would not have an effect on bacteria that are metabolically inactive and hence in a stationary phase. Bacteria in the nutrient-limited zone upregulate their stress responses and switch their metabolic pathways from growth to persistence (16). Some bacterial cells change their phenotype such that they can survive for prolonged periods in the presence of antibiotics. These cells are known as persisters (22). Interestingly, biofilms containing S. aureus have a higher number of persister cells than free-growing bacteria (23). The nutrient and oxygen limitations in the biofilms provide the environmental cues necessary for transforming regular cells into persisters.
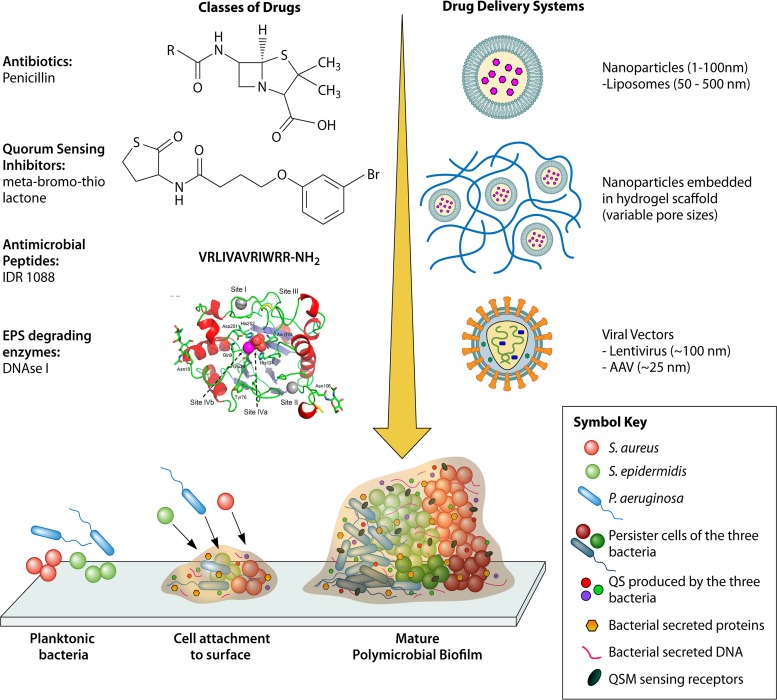
Additionally, cells sense their environmental changes and the presence of other bacterial cells and modify their physiological processes through QS. QS enables bacterial cells to coordinate gene expression and nucleotide signaling to help them survive collectively as a community within the biofilm (24). Signaling through QS suppresses the expression of virulence factors until bacterial cells reach a high cell density, which helps ensure that virulence is not suppressed by the host immune system. Additionally, QS also changes the phenotype of bacterial cells in polymicrobial biofilms, thereby making it more difficult to treat the infection (25). In spite of the complex biological landscape described above, tremendous progress has been made in engineering treatment options for chronic wound infections. A schematic of biofilm formation with different drug molecules and drug delivery systems used in treating chronic wound infections is presented in Fig. 1.
FIG 1.
Biofilm formation and treatment options for chronic wounds. Planktonic bacteria secrete extracellular proteins and DNA and form a glycocalyx containing polysaccharide film around them, which marks the beginning of the formation of a biofilm. As the number of bacterial cells in the polysaccharide matrix increases due to cell division and from the environment, the matrix thickens and forms a mature biofilm. Each bacterial species proliferates in its own “territory” until nutrient and gas supplies are not limiting and secretes quorum-sensing molecules. Several classes of drug molecules exist for treating bacterial infections, but their efficacy is limited since they either cannot penetrate the matrix or are degraded by matrix components. Drug delivery systems have evolved to attenuate the problem.
ALTERNATIVES TO ANTIBIOTICS
Four classes of compounds have emerged in response to the rapid spread of antibiotic resistance among bacterial species. These include antimicrobial peptides (AMPs), biofilm-degrading agents, QS inhibitors, and miscellaneous compounds. Each class of molecules was initially identified from natural sources, followed by the creation of synthetic analogs to increase their potency. Other mechanisms for treating biofilm infections, such as debridement, energy transfer, and augmentation of innate and/or adaptive mechanisms, etc. (26–28), differ in their modes of action from the approaches described here and are therefore not included in this review.
Antimicrobial Peptides
AMPs are produced by both eukaryotic and prokaryotic organisms, and they are particularly attractive as antimicrobials due to their small size (15 to 50 amino acids) and positive charge, which attracts them toward the negatively charged biofilm surface (29). Although the mechanism of action of AMPs depends on their structure and sequence, many AMPs are believed to act by perturbing the cell membrane (30). Bionda et al. took cyclic lipopeptides belonging to the fusaricidin/LI-F class and structurally modified the amino acid sequence, thereby creating 12 synthetic analogs. They showed that cyclic lipopeptides 1 and 3 were effective at both eradication and inhibition of biofilm formation by methicillin-resistant S. aureus (MRSA) and P. aeruginosa PA14 due to a higher hydrophobicity and net positive charge (31).
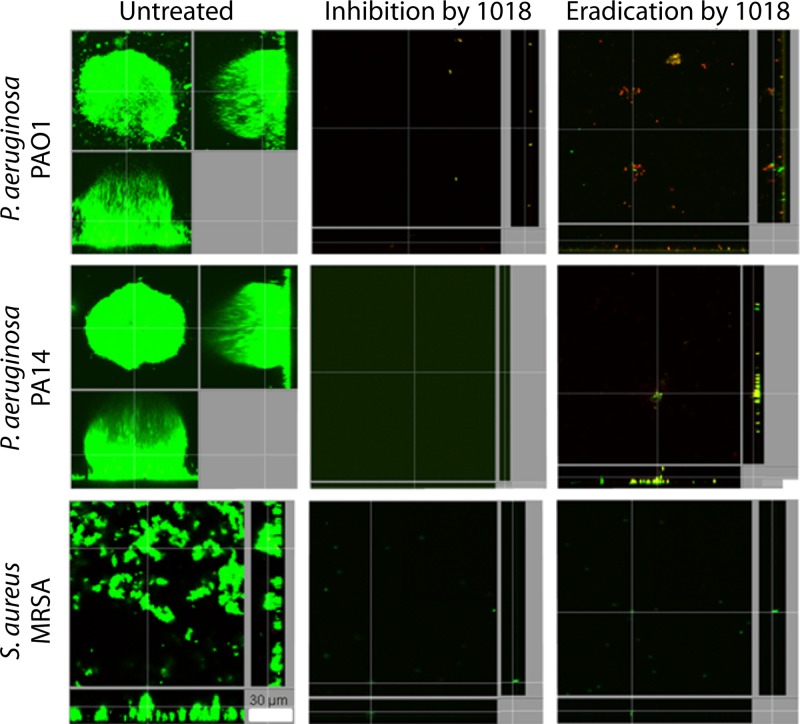
One mechanism by which bacterial cells respond to environmental stress is by using the secondary messenger metabolite (p)ppGpp. (p)ppGpp sets off a cascade of effects at the molecular level called the “stringent response.” This stress response enables the cells to develop into a persister phenotype, which confers antibiotic resistance to these cells (32). Therefore, the development of (p)ppGpp inhibitors is an active area of research. The effectiveness of AMPs such as IDR-1088, DJK-5, and DJK-6 against ppGpp in both Gram-positive and -negative organisms makes them clinically viable potential broad-spectrum antibiofilm therapeutics (33) (Fig. 2).
FIG 2.
IDR-1018 inhibits bacterial biofilm formation and eradicates preformed biofilms in Gram-negative and Gram-positive bacteria. Gram-positive and -negative bacterial biofilm formation was monitored for up to 3 days after treating the surface with IDR-1018 at sub-MICs. Biofilm eradication was evaluated 2 days after the flow cell surface came into contact with the bacteria. Bacterial presence was tested using live/dead staining using confocal microscopy. (Adapted from reference 33 [published under a Creative Commons license].)
Interestingly, bacteria themselves also produce AMPs when in the vicinity of other competing species of bacteria. Bacterial cross talk between S. epidermidis and S. aureus has been reported. This is particularly interesting for the field of chronic wound infections since S. aureus is the dominant species in diabetic ulcers. It is important to note that (i) not all AMPs reported in the literature are effective against the biofilm-forming bacteria, although they may be effective against all antibiotic-resistant planktonic bacteria, and (ii) bacterial cells may develop resistance to AMPs just as they develop tolerance to antibiotics by forming phenotypically mutant cells. In fact, S. aureus develops resistance to AMPs such as lysozyme via the biofilm formation-regulatory system GraRS, which produces virulence factors in S. aureus (34). These AMPs are also susceptible to proteolytic degradation within biofilms since they are proteinaceous in nature.
Biofilm-Degrading Agents
When bacterial cells adhere to a solid surface, they secrete polymeric substances such as proteins, nucleic acids, and polysaccharides. These EPSs form the biofilm architecture. Both synthetically and naturally derived enzymes such as a DNase I, amylase, dispersin B, lysostaphin, and alginate lyase have been shown to degrade the EPSs (29). Precoating of vascular catheters with dispersin B and triclosan resulted in significantly lower rates of colonization by S. aureus, S. epidermidis, and E. coli (35). Pretreatment of biofilms formed by Aggregatibacter actinomycetemcomitans with dispersin B made the biofilms more sensitive to detachment by the surfactant SDS (36). Similarly, recombinant human DNase I (rhDNase I) alone and in combination with dispersin B inhibited biofilm formation by S. aureus and S. epidermidis in microtiter plates and increased their sensitivity to biocide by other antibiotics (37). DNase I, when used in combination with different antibiotics, has also been shown to decrease the biomass of established Gram-positive and -negative bacteria biofilms but was not observed to be as effective when used alone (38). These studies indicate that biofilm-degrading enzymes can be used as supplementary agents in the treatment of biofilm-forming bacteria. Additionally, the higher cost of enzyme production may make their widespread use prohibitive. Other strategies that are employed in the clinic for degrading biofilms are changing the pH of the wound to slightly acidic, high osmolality, and the use of surfactants to break down the polymeric biofilm matrix (39, 40).
QS Inhibitors
QS in Gram-positive bacteria is controlled by autoinducing peptides (AIPs). These molecules are secreted constitutively or in response to environmental cues such as high cell density. AIPs bind to transmembrane receptors to communicate with other cells and also regulate the transcription of target genes. QS in Gram-negative bacteria is mainly controlled by the LuxI and LuxR proteins and their analogs. LuxI homologs produce autoinducers called acyl homoserine lactones (AHLs) that bind to LuxR homologs to regulate the transcription of target genes within the bacterial cell (41). Just like AMPs, QS inhibition is an actively expanding field of investigation, particularly because many QS pathways are generic for both Gram-positive and -negative bacteria. QS inhibitors may work by either blocking the detection of an environmental cue by a bacterial cell, inhibiting signal exchange between different bacterial cells, or inhibiting signal propagation to downstream targets within a cell. Balaban et al. demonstrated that an RNAIII-inhibiting peptide suppressed TRAP (target of RNAIII-activating protein)-agr systems in MRSA, resulting in reduced MRSA graft infections in rats (42). A compound called meta-bromo-thiolactone has been shown to reduce biofilm formation in P. aeruginosa by inhibiting the production of pyocyanin, a QS molecule (QSM) (43).
Bacteria produce molecules to inhibit the growth of other bacterial species while existing in a polymicrobial community. A serine protease, Esp, produced by S. epidermidis inhibits biofilm formation by S. aureus in vitro (44). Similarly, a synthetic derivative of the agr pheromone produced by S. epidermidis has been shown to potently inhibit the S. aureus agr system that is responsible for the production of virulence factors associated with biofilm formation (45). Another method for inhibiting quorum sensing is signal degradation or quorum quenching. Two types of AHL-degrading enzymes have been described. The enzyme AHL-lactonase hydrolyzes the lactone ring in AHL, whereas the enzyme acylase breaks down the amide bond in AHL (46).
A detailed study of naturally derived QS inhibition molecules, focusing on plants, prokaryotes, and marine life, has been described elsewhere (47). Plant metabolites such as coumaric acid, catechin, and salicylic acid demonstrate QS inhibition properties. Ajoene and iberin, isolated from garlic and horseradish, respectively, attenuate virulence factors such as rhamnolipid produced by P. aeruginosa (48, 49).
Miscellaneous Compounds
Several antibiofilm compounds that cannot be classified into one of the three categories described above have been reported in the literature. For instance, gallium-containing compounds interfere with bacterial iron metabolism, which is believed to be vital for bacterial growth and virulence (50). Amyloid blockers are another such category of compounds. Bacteria such as E. coli produce amyloids, such as pili and curli, that adhere to surfaces to form biofilms. Peptidomimetics such as FN075, which hinder protein assembly and curli and pilus biogenesis, alleviate the initial biofilm attachment of E. coli to surfaces (51). Small molecules such as LP 3134 and LP 3145 can be used to inhibit diguanylate cyclase (DGC) enzyme production, which synthesizes cyclic di-GMP, the secondary messenger responsible for the stress response in bacteria. While the chemical structures of these molecules were not discussed by Sambanthamoorthy et. al, these molecules have demonstrated antibiofilm activity against P. aeruginosa (52).
DRUG DELIVERY SYSTEMS
The need to engineer a drug delivery system for treating chronic infections in wounds has emerged from the underlying biochemical properties of a biofilm. Extracellular components of the biofilm matrix either sequester, inactivate, or inhibit the drug, thus preventing the required amount of drug from reaching the target cells. A delivery system protects the drug from these inhibitory components to some extent, thereby augmenting both its pharmacodynamic and pharmacokinetic effects. Two types of drug delivery systems have been widely explored for treating wound infections: nanoparticles (NPs) and scaffolds embedding nanoparticles.
Nanoparticles
NPs are particles that have dimensions of between 1 nm and 100 nm (53). Based on the material used for their synthesis, they are generally classified into six groups: metallic NPs, nonmetal NPs, polymeric NPs, lipid NPs, quantum dots, and ceramic NPs. While quantum dots and ceramic NPs are biocompatible and soluble in water and have demonstrated the potential for use in the treatment of chronic wound infections (54, 55), they have been excluded from this review since research is still in the early stages.
Metal NPs.
Metal ions and their oxides found their place as topical antimicrobial agents several decades ago. Since metals are also liable to inactivation by components of the wound bed (56), metallic NPs were explored as a solution. Oxides such as CuO, Fe2O3, Al2O3, Au2O3, ZnO, and Ag2O have been used to synthesize NPs. Of these, silver and zinc NPs by themselves have been reported to be effective antimicrobials. Both these metals act by disrupting the cell membrane of bacteria and interfering with their metabolism, thereby preventing cell growth. Kalishwaralal et al. incubated cultures of P. aeruginosa and S. epidermidis with various concentrations of silver NPs and quantified biofilm formation by measuring the binding of crystal violet dye to adherent cells. Compared to the untreated group, the plates incubated with silver NPs inhibited biofilm formation by ∼95% (57). The antimicrobial activity of ZnO, CuO, and Fe2O3 NPs has also been tested against both Gram-positive (S. aureus and Bacillus subtilis) and Gram-negative (E. coli and P. aeruginosa) bacteria in vitro using the well diffusion method (58). In the well diffusion method, as described by Holder and Boyce and Balouiri et al., the microbial inoculum is spread on an agar plate, and a hole is aseptically created in the center of the plate by using a micropipette tip. Antimicrobial solutions (NPs in this case) of the desired concentrations are introduced into the hole, and they diffuse into the agar medium to exert an antimicrobial effect. The output of this test is measurement of the zone of inhibition. The zone-of-inhibition test is a qualitative method of determining whether the bacterium is sensitive to the test antimicrobial (NPs). If the bacterium is susceptible to the test antimicrobial, a zone of inhibition emerges around the punched hole described above, where the bacterium does not grow after sufficient incubation of the cells with the NPs (59, 60). It was observed that ZnO NPs showed a greater bactericidal activity than the other tested metallic NPs (58). The antibiofilm effectiveness of ZnO NPs against S. aureus and P. aeruginosa has also been evaluated. Lee et al. tested 36 metal ions for antibiofilm activity against P. aeruginosa. ZnO NPs were found to be the most effective among the tested metal NPs (61). S. aureus biofilms were cultured with different concentrations of ZnO NPs on polyvinyl chloride (PVC) microscope slide surfaces. The reduction in biofilm formation was quantified using crystal violet staining. A 55% reduction in biofilm growth on the composite surface was observed (62). In fact, Zn NPs synthesized from natural polysaccharides demonstrate broad-spectrum antimicrobial activity against several pathogenic bacterial species when tested using the agar well diffusion method (63). The antibacterial properties of gold NPs (Au-NPs) have also been evaluated. In one study, Au-NPs with a size of 20 to 30 nm showed no antibacterial effect on S. aureus ATCC 6538 and E. coli K-12 NCTC 10538 (64). The MIC of Au-NPs against S. aureus was also shown to be significantly higher than that of Ag-NPs (65). However, Au-NPs can be used as effective antimicrobials in a modified state or as drug carriers (discussed below).
Although generally effective, the use of these metallic NPs as antimicrobials has two major drawbacks. First, a very high dose is required for them to be effective against bacterial cells. Doses as high as 100 μg/ml (66) and 1 mM (61) have been reported for zinc NPs. These high doses are difficult to scale and deliver to the wound bed from an engineering perspective. Moreover, at high doses, metals may also become toxic to human cells (67, 68), which could inhibit the recruitment of immune cells, the regrowth of epidermal cells, and, ultimately, wound healing. Similar to the case with antibiotics, prolonged treatment with metal ions may result in the emergence of resistant bacterial strains. A silver resistance gene (silE) was reported in MRSA isolates cultured from wound and nasal passages of dogs and household pets that are capable of transmitting infections to humans (69). Finally, metallic NPs exhibit size-dependent cytotoxicity. Gold NPs with a size of <2 nm have been shown to have toxic effects on epithelial cells, macrophages, fibroblasts, and other human cell lines (70, 71). Interestingly, this toxicity has not been reported for Au-NPs with a diameter of >2 nm (71–73).
Metal NPs can also be used as drug carriers. Au-NPs in particular have attracted considerable attention in recent years due to their negative surface charge and ability to be synthesized in multiple shapes and sizes. Conjugation of the Au-NP surface with carboxyl groups in methotrexate has been shown to decrease the growth rate of cancer cells (74). Attachment of doxorubicin to Au-NPs via a pH-sensitive linker allows for the site-directed release of the drug inside tumor cells that have an acidic environment (75). Wadhwani et al. functionalized gold NPs with cationic AMPs and showed that the conjugated peptides are biologically active and more resistant to degradation by proteases than free peptides (76).
Nonmetal NPs.
Since the discovery of fullerene, a spherical 60-carbon atom molecule, in 1991 (77), carbon-based NPs have been garnering interest from the bioengineering community. Graphene and diamonds are two other allotropes of carbon that have generated interest recently in the bioengineering discipline because of their unique chemical properties. Graphene- and graphene oxide (GO)-based nanocomposites can be functionalized with biocompatible polymers such as polyethylene glycol (PEG) and exhibit electrical and thermal conductivity (78). Graphene oxide can be dually functionalized with PEG and polyethylenimine (PEI). Since PEI forms a complex with DNA in order to enter a cell (referred to as transfection), the concept behind the GO-PEG-PEI-DNA complex is to increase the transfection efficiency of the DNA. Compared to the free PEI-DNA complex, and the GO-PEI-DNA complex without PEGylation, nano-GO (NGO)–PEG–PEI shows superior DNA transfection efficiency. The NGO-PEG-PEI-DNA complex is also effective in the presence of fetal bovine serum (FBS), a component required for cell growth, and is less toxic to cells than the PEI-DNA complex (79).
Nanoscale diamonds are roughly 4 to 5 nm in size and are widely applied in imaging, magnetic sensors, and conjugating biomolecules for delivery (80) because of their high aqueous solubility and biocompatibility. Producing nanodiamonds (NDs) is less expensive than producing viral vehicles or liposomes for gene delivery. Since thousands of surface modifications can be made on the ND surface, biocompatibility data are difficult to assess. NDs are also known to aggregate (81). Like graphene NPs, NDs can be modified by immobilizing 800-molecular-weight PEI (PEI800) on their surface. NDs modified with PEI800 exhibit a transfection efficiency comparable to that of 25,000-molecular-weight PEI (PEI25K), without its high cytotoxicity (associated with the higher molecular weight of PEI). The enhanced delivery properties are due to hybrid ND-PEI800 (82). These studies with graphene and NDs as vehicles for DNA delivery open up opportunities to explore their use as peptide and antibiotic delivery vehicles. Since PEI has been shown to exhibit antimicrobial activity against S. aureus and E. coli (83), ND-PEG in combination with PEI is worthy of investigation for its potentially enhanced antimicrobial properties.
Other nonmetals, such as silica and selenium, have also been used to synthesize NPs. Selenium NPs created by the colloidal synthesis method have been shown to be effective against S. aureus (84). Incubation of these bacterial cells with selenium NPs for up to 5 h reduces their growth by 60-fold compared to untreated bacterial cells, as determined by optical density (OD) measurements. Mesoporous silica NPs (MSNs) have attracted considerable attention in cancer theranostics due to their large surface area and adjustable pore sizes but have not yet been studied extensively for antimicrobial applications. Recent advances in the field of MSN drug delivery systems have been described by Wang et al. (85).
Polymer NPs.
Poly(lactic-co-glycolic acid) (PLGA) NPs are one of the most widely studied categories of NPs for antimicrobial applications. PLGA is a biocompatible and biodegradable polymer. The antibacterial activity of PLGA NPs loaded with rifampin was compared to that of the free drug rifampin in a 24-h zone-of-inhibition study conducted using agar plates. The PLGA-rifampin NPs showed higher bactericidal activity than the free drug against all three Gram-positive bacteria (S. aureus, MRSA, and B. subtilis) due to better penetration into bacterial cells and targeted delivery of rifampin to the site of action. Since Gram-negative bacteria are resistant to rifampin, the NPs were not effective against P. aeruginosa and E. coli (86).
Several other polymers, such as chitosan and poly(β-amino esters) (PBAEs), are also used for gene and cytokine delivery into the wound beds (10). Fewer studies have been conducted with polymer-based NPs as drug delivery vehicles for antimicrobial compounds in wound beds than with metal and lipid NPs. Of particular note among polymer particles are the dendrimeric NPs and molecularly imprinted particles (MIPs). Dendrimers are characterized by a highly branched structure with small empty pockets. This structure makes it possible to package drugs and use them as drug delivery systems. An arginine-grafted cationic dendrimer, PAM-RG4, was combined with a plasmid encoding vascular endothelial growth factor (VEGF), and the PAM-RG4–plasmid complex was transfected by subcutaneous injection into the wounds of diabetic mice. The wounds treated with the PAM-RG4–plasmid complex healed significantly faster than the wounds treated with the naked VEGF plasmid and demonstrated sustained release (87). This study indicates that it might be feasible to potentially test dendrimers for packaging small peptides and small molecules such as antibiotics. MIPs, on the other hand, are synthesized using acrylic or methacrylic monomers in the presence of an epitope to which the particle is intended to bind. The target molecules are removed, and the MIPs are then divided into particles that can selectively rebind to the target when they are exposed to it again (lock-and-key analogy). Although MIPs are low-cost alternatives to a monoclonal antibody to be used in bioassays or sensor applications, their use has been limited by the presence of residual target templates and unstandardized synthesis methods. Recently, a solid-phase synthesis method with an affinity purification step has been developed for synthesizing template-free MIPs that can bind selectively to molecules of ≤500 Da (88). These advances in the field lay the groundwork for potentially producing MIPs selective for bacterial cell membrane epitopes.
Lipid NPs.
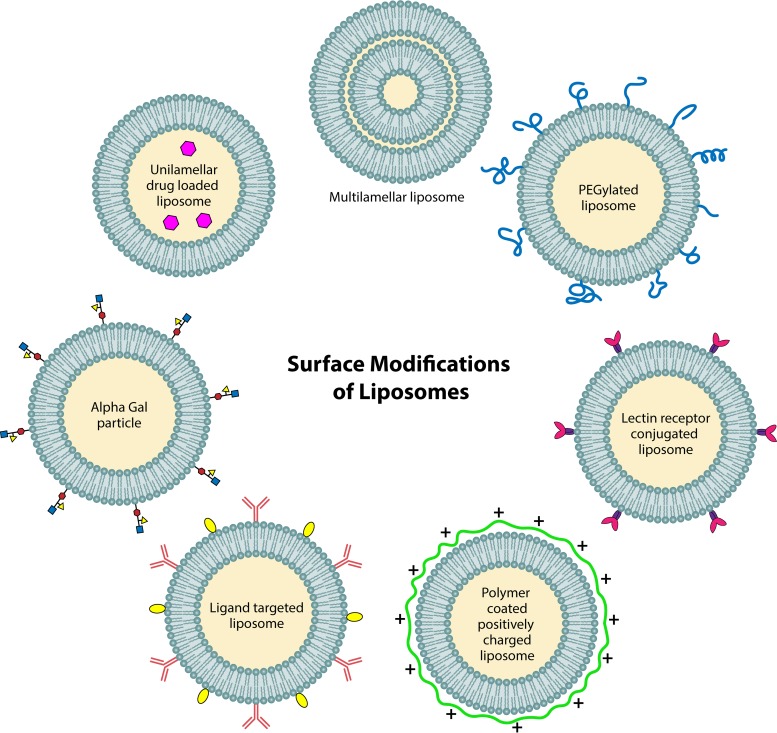
Liposomes have been extensively studied as delivery vehicles for conventional antibiotics. Studies with antibiotics packaged in liposomes to treat bacterial biofilms have been reviewed extensively elsewhere (89, 90). Here, we aim to provide a fundamental understanding of why liposomes have gained impetus as drug carriers in recent years and the versatility of these vehicles for treating biofilm infections in wounds. Figure 3 illustrates various surface modifications on liposome NPs that could potentially find their use in treating chronic wound infections.
FIG 3.
Various surface modifications of liposomes. Structure, composition, and surface modifications increase the utility of liposomes as drug carriers. A multilamellar (multiple lipid bilayer) liposome is more useful for controlled drug release. Surface modifications such as positive charge, PEGylation, and ligand conjugation are governed for the liposome to penetrate the biofilm better, protect it from degradation by proteases, and selectively bind to the target molecule, respectively.
Liposomes are spherical NPs that have a hydrophobic shell and a hydrophilic inner core, as opposed to micelles that are self-assembling colloidal NPs with a hydrophobic core and a hydrophilic shell. The hydrophobic shell of a liposome consists of a lipid bilayer that has a hydrophilic head and a hydrophobic tail. In the presence of water, the polar heads orient toward the water, while the nonpolar sites are oriented inward toward each other. This lipid arrangement mimics that of a living cell membrane. Since the lipid bilayer composition can be changed according to the target, these NPs are highly biocompatible and have relatively low immunogenicity. Liposomes fuse with cell membranes and, in this process, empty their payload inside cells (91). Physicochemical properties of liposomes may be modified as necessary and have been deemed critical for the stability of the NPs and for the sustained release of the packaged drug molecule.
Smaller lipid NPs (≤500 nm) are associated with more-effective penetration (89). Similarly, it has been observed that positively charged liposomes are able to bind to bacterial cells better than negatively charged or neutral liposomes in P. aeruginosa and S. aureus biofilms (92), since the bacterial cell membrane is negatively charged.
Hydrophilic cargo can be packaged in a liposome in the inner aqueous core, whereas hydrophobic cargo may be packaged between the two layers of phospholipids within the lipid bilayer, which makes liposomes universal carriers. Changing the composition of the lipid bilayer and making it rigid by the addition of long-chain fatty acids and cholesterol, for example, reduce drug leakage, a property that can be harnessed for the controlled release of the drug.
Surface modifications can be made almost effortlessly on liposomes and micelles. Coating the liposomes with PEG stabilizes them against components of the complement system and prolongs their half-life in blood. Although contradictory results have been reported regarding the impact of PEGylation on the affinity of liposomes for the biofilm surface (93, 94), PEGylation can be beneficial for liposomes traveling through the extracellular polymeric matrix of a biofilm. Similarly, coating a liposome with lectins would enable the particle to bind with glycans in the glycocalyx secreted by the bacteria. The glycocalyx is an integral part of the biofilm matrix and surrounds bacterial cells by forming a protective barrier (95).
A promising approach facilitating wound healing with α-Gal NPs was recently reported (96). α-Gal NPs are essentially liposomes made with glycolipids whose surface is modified to express several epitopes of a glycan named α-Gal. These epitopes bind to α-Gal antibodies that recruit macrophages to the wound bed and accelerate the inflammatory phase of wound healing. α-Gal antibodies are naturally produced in humans. A diabetic mouse model producing α-Gal antibodies has been developed for the purpose of testing these NPs in vivo. α-Gal NPs injected into diabetic wounds in these mice resulted in a significant regeneration of epidermal cells in 12 days, compared to the control group that was treated with saline (96).
While promising, the engineering of nanoparticle delivery systems is in its infancy. The current understanding of how these NPs are cleared from the body is insufficient. More importantly, immunogenicity effects of different classes of NPs have not yet been fully delineated. The immune response to NPs can render them inefficacious or lead to an autoimmune response if the surface modification on NPs is also a molecule native to the human body. A major obstacle limiting particle research at the cellular level is the lack of high-resolution techniques to visualize these NPs inside cells. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM), while high-resolution tools, work only for electron-dense materials like metal NPs. Fluorescent labeling of a NP requires knowledge of the stability of these tags in various cellular compartments, which have different pHs (97). Another potential safety concern is the presence of residual organic solvents that are used in the synthesis of lipid NPs (97).
Scaffolds Embedding NPs
Biomaterials are a Pandora’s box for treating diabetic wounds. While they inherently do not exhibit antibacterial properties, the value of biomaterials lies in the ability to use them as scaffolds to embed drug particles or drug carriers. Among various types of biomaterials, hydrogels have emerged as the material of choice for use in diabetic wounds. Hydrogels can be formulated as particles, sponges, films, and other three-dimensional (3D) structures, and their porosity can be controlled as desired to embed particles of various sizes (98). Hydrogels swell upon absorption of water and retain the water, thereby maintaining a moist microenvironment in the wound bed, which is essential for healing some wounds (99).
Both alginate and chitosan hydrogels embedded with silver NPs exhibit antibacterial activity against S. aureus and E. coli in diabetic rats (100). Conventional antibiotics such as mupirocin can also be packaged in liposomes that are embedded in hydrogel and delivered to the wound bed. These delivery systems are nontoxic to keratinocytes, a type of skin cell (101). A detailed discussion of applications of hydrogels as a scaffold for the delivery of antibiotics or antibiotic substitutes has been reported elsewhere (102).
Viral Vector-Based Gene Delivery Systems
Viruses are yet another mode of delivering drugs to the wound bed. Genetically modified viruses act as vehicles and have been shown to be effective in delivering functional copies of genes to target cells (103, 104). The field of viral vector gene therapy has matured immensely in the last decade. Lentiviral vector (LVV)-based products manufactured by Kite Pharma and Novartis were approved by the U.S. FDA for immuno-oncology applications last year. Similarly, adeno-associated virus (AAV)-based products, such as Luxturna, were approved by the U.S. FDA for RPE65 mutation-associated retinal dystrophy. LVVs integrate into the host cell genome randomly but have a large packaging capacity, as opposed to AAVs, which have a limited packaging capacity and do not integrate into the host cell genome. The use of viral vectors for wound healing applications has also been evaluated.
Human β-defensin 4 (hBD4), an antimicrobial peptide that is naturally produced by human cells, was tested for its efficacy and persistence against P. aeruginosa infection in mice when delivered using a viral vehicle. Deep burns were induced in mice by burning their dorsal skin. hBD4-encoding mRNA was packaged in Newcastle disease virus (NDV) and transduced into Madin-Darby bovine kidney (MDBK) cells. These MDBK cells transduced with NDV were introduced into the wound beds of mice, and P. aeruginosa colony counts and wound size measurements were performed at 7, 14, and 21 days posttreatment. Compared to the untreated group, the treated group showed significantly lower colony counts and smaller wound areas (105).
Since viral vectors exhibit risks of random integration and mutagenesis, precise genome editing has also been reported to deliver growth factors such as platelet-derived growth factor B (PDGF-B) in mouse fibroblasts in the wound bed. Transcription activator-like effector nucleases (TALENs) were used to create a double-strand break at the desired locus in the genome, followed by the insertion of the gene for PDGF-B expression. The double-strand break was repaired by the mammalian cells’ native DNA repair mechanism, viz., homologous recombination. These edited fibroblasts persisted in the wound bed for up to 5 months (106).
Viral vectors and precise genome editing offer great value, owing to the persistent expression of the transduced gene in the genome. They also prevent the proteolytic degradation of peptide-based molecules as observed otherwise. However, the cost of manufacturing good-manufacturing-practice (GMP)-grade viral vectors is currently prohibitive. Transduction of patient cells in an ex vivo setting remains a major challenge from a scale-up/scale-out perspective for treating hundreds of thousands of patients. Most importantly, since the field has matured only in the last decade, there are not enough long-term data on the off-target effects, immunogenicity, and persistence of viral vector-based therapeutics for use in non-life-threatening conditions for large populations.
PERSPECTIVE
Biofilm infections are distinctly harder to treat than those caused by planktonic bacteria due to the underlying biology associated with complex adaptive microbial communities coexisting within the protective barrier of EPSs. Therefore, the field of discovery is ripe for broad-spectrum drugs that can annihilate both Gram-positive and Gram-negative bacteria present in biofilms. Our fundamental understanding of how bacteria behave within a biofilm is burgeoning. It is clear that quorum sensing is a vital pathway for bacteria to communicate and express genes necessary to survive in nutrient-limited environments. AMPs and QS inhibitors are attractive alternatives to conventional antibiotics. However, while overcoming some challenges, such as the potential development of resistance, they are subject to some of the same limitations as conventional antibiotics. These molecules are peptides and small organic compounds and therefore are susceptible to enzymatic degradation and inactivity due to pH changes. In fact, potential drug candidates are often discarded during discovery screening as inefficacious, since they cannot penetrate biofilms. The need arises to design thermodynamically stable drug delivery systems that can overcome these obstacles. Cross talk between engineering and biology is essential for this field to progress and to resolve several critical issues on all three fronts, viz., the clinic, microbiology, and engineering.
A principal challenge in clinical care is the rapid and accurate determination of the composition of the biofilms necessary prior to the initiation of treatment, thereby curbing antibiotic abuse. The development of biosensors that can identify electrochemically active quorum sensing molecules (QSMs) produced by bacteria present in various bodily fluids and with minimal sample preparation at the point of care, as demonstrated by Sismaet et al. (112), is an interesting approach. The development of a panel of biomarkers that could determine the susceptibility of a chronic wound to a biofilm infection, or monitor wounds for the transition from colonization to infection by bacteria, would be an efficient preventive strategy.
Many questions regarding the price of implementing these technologies remain, but given the progress that has already been made, the cost to commercialize these promising approaches is expected to be orders of magnitude lower than for new drug development. Per-unit costs of wound dressings would increase, but the real costs must be framed in terms of preventative care and improved patient outcomes. All of the approaches that we have described would require only a few dollars worth of materials per dressing when manufactured in reasonable quantities. Insurance companies are beginning to realize that spending a few extra dollars on an antimicrobial material to significantly lower the chances of being admitted to a hospital for complicated infection is a worthwhile investment. There is also the larger societal cost savings associated with reducing infection rates and slowing the emergence of antimicrobial resistance.
Fundamental principles of mass transfer govern sustained drug delivery. The penetration and transport of NPs from the surface to the core depths of a biofilm are governed and can be predicted by the balance of physical and chemical forces. The release of drugs from NPs can be regulated by controlling the inherent properties of a material, which can be altered by the composition of the coating or lipid bilayer of the NP. The packaging capacity of a NP may be modified using size-, charge-, and polarity-based compartmentalization. Once there is biological knowledge about the target to which a drug molecule binds, concepts of molecular chemisorption can be employed to determine the binding affinity. One potentially ideal drug delivery system for chronic wound infections is a bilamellar liposome, a NP with two concentric compartments, where the outer compartment releases a payload to degrade the extracellular matrix, whereas the inner bilayer is coated with a bacterial cell-specific epitope and releases drug from the inner compartment at the cell surface. These NPs are less expensive to manufacture and exhibit higher stability than biologics. Properties such as drug loading capacity, leakiness, and sustained release of payload need to be reproducibly controlled to make them a clinical reality.
From a drug delivery perspective, the main question that is yet to be answered adequately is whether NPs generate an immune response when delivered to chronic wounds. This is difficult to determine, since a prolonged presence of immune cells is observed in infected diabetic wounds. While it is well accepted that biofilms in vivo and in vitro differ both phenotypically and genotypically (107, 108), it would be helpful to understand what these differences are to develop an acceptable in vitro model for testing the effectiveness of antimicrobials on a biofilm. It would also be interesting to develop “smart” drug delivery mechanisms that adapt to the changing conditions in biofilms.
Another research avenue that needs further exploration is bacterial cross talk (45, 109). While it is known that mobile genetic elements (MGEs) are unilaterally transferred from S. epidermidis to S. aureus (110), it will be interesting to see if MGEs can be exploited for antimicrobial applications. Advancements in the development of restriction endonucleases and ligases have made it possible to engineer custom bacterial species. Precise genome-editing tools such as megaTALs, zinc finger nucleases, and CRISPR/Cas9 may also be used to engineer a commensal bacterial species, such as S. epidermidis (111). An engineered commensal, ideally, would not become an opportunistic pathogen upon exposure to the wound bed and would produce cues to inhibit the transcription of genes required for survival in pathogenic species, such as S. aureus.
Finally, the research area of treating biofilm infections in wounds is expanding rapidly. As the field matures, more testing of the drug delivery systems is required to understand their safety and efficacy profiles. Integrating biological understanding with engineering principles is central to translating these nanocomposites into the clinic in a shorter time frame, given the rate at which resistant strains of bacteria are emerging.
ACKNOWLEDGMENTS
We thank Northeastern University for providing financial support, and we have no conflict of interest to declare.
Biographies

Pranali J. Buch is currently a second-year Ph.D. student in the department of Chemical Engineering at Northeastern University, where she has been a recipient of a Kanojia Engineering Research Fellowship. She received her M.S. degree in Biotechnology from Northeastern University in 2013 and worked as a lentiviral vector process development scientist at bluebird bio before joining the Ph.D. program. Her research interest lies in developing scalable and sustainable solutions in the health care sector that can be easily made available to the masses. For her Ph.D. project, she is designing a drug delivery system to treat biofilm infections in chronic wounds.

Yunrong Chai is an associate professor in the Department of Biology at Northeastern University (Boston, MA, USA). He earned his B.S. and M.S. in Microbiology from Fudan University (Shanghai, China) and Ph.D. in Microbiology from Cornell University (Ithaca, NY). From 2006 to 2012, Dr. Chai was a Jane Coffin Childs postdoctoral fellow and then a BASF research fellow in the Department of Molecular and Cellular Biology at Harvard University. The research in the Chai laboratory focuses on understanding fundamental mechanisms controlling bacterial biofilm formation and the role of biofilms in bacterium-host interactions. The lab is also interested in inhibitory mechanisms targeting key processes in bacterial biofilm development.

Edgar D. Goluch is an associate professor in the Department of Chemical Engineering at Northeastern University (Boston, MA, USA), with courtesy appointments in Bioengineering, Biology, and Civil and Environmental Engineering. He earned his B.S. in Chemical Engineering from the University of Illinois at Urbana-Champaign in 2003. He remained at Illinois for graduate school, receiving a M.S. in Mechanical Engineering and a Ph.D. in Bioengineering. From 2008 to 2010, he was a NSF Postdoctoral Fellow at Delft University of Technology in the Netherlands. He has over 50 journal publications and conference proceedings covering the fields of microfluidics and sensors. Since 2010, his research group has focused on developing engineering solutions in the areas of clinical and environmental microbiology. In 2014, he founded QSM Diagnostics, Inc., to commercialize bacterial identification technology developed in his laboratory.
REFERENCES
- 1.National Center for Health Statistics. 2017. Health, United States, 2016: with chartbook on long-term trends in health. National Center for Health Statistics, Hyattsville, MD. [PubMed] [Google Scholar]
- 2.Mathers CD, Loncar D. 2006. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. 2017. National diabetes statistics report, 2017. CDC, Atlanta, GA. [Google Scholar]
- 4.Raghav A, Khan ZA, Labala RK, Ahmad J, Noor S, Mishra BK. 2018. Financial burden of diabetic foot ulcers to world: a progressive topic to discuss always. Ther Adv Endocrinol Metab 9:29–31. doi: 10.1177/2042018817744513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demidova-Rice TN, Hamblin MR, Herman IM. 2012. Acute and impaired wound healing: pathophysiology and current methods for drug delivery, part 1. Normal and chronic wounds: biology, causes, and approaches to care. Adv Skin Wound Care 25:304–314. doi: 10.1097/01.ASW.0000416006.55218.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinke JM, Sorg H. 2012. Wound repair and regeneration. Eur Surg Res 49:35–43. doi: 10.1159/000339613. [DOI] [PubMed] [Google Scholar]
- 7.Falanga V. 2005. Wound healing and its impairment in the diabetic foot. Lancet 366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- 8.Schultz GS, Sibbald RG, Falanga V, Ayello EA, Dowsett C, Harding K, Romanelli M, Stacey MC, Teot L, Vanscheidt W. 2003. Wound bed preparation: a systematic approach to wound management. Wound Repair Regen 11:S1–S28. [DOI] [PubMed] [Google Scholar]
- 9.Demidova-Rice TN, Hamblin MR, Herman IM. 2012. Acute and impaired wound healing: pathophysiology and current methods for drug delivery, part 2. Role of growth factors in normal and pathological wound healing: therapeutic potential and methods of delivery. Adv Skin Wound Care 25:349–370. doi: 10.1097/01.ASW.0000418541.31366.a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalashnikova I, Das S, Seal S. 2015. Nanomaterials for wound healing: scope and advancement. Nanomedicine (Lond) 10:2593–2612. doi: 10.2217/NNM.15.82. [DOI] [PubMed] [Google Scholar]
- 11.Magana M, Sereti C, Ioannidis A, Mitchell CA, Ball AR, Magiorkinis E, Chatzipanagiotou S, Hamblin MR, Hadjifrangiskou M, Tegos GP. 2018. Options and limitations in clinical investigation of bacterial biofilms. Clin Microbiol Rev 31:e00084-16. doi: 10.1128/CMR.00084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner SE, Frantz RA. 2008. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs 10:44–53. doi: 10.1177/1099800408319056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen GJM, Brüggemann H. 2014. Bacterial skin commensals and their role as host guardians. Benef Microbes 5:201–215. doi: 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 14.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 15.Whiteley M, Diggle SP, Greenberg EP. 2017. Progress in and promise of bacterial quorum sensing research. Nature 551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart PS. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 17.Kurniawan A, Fukuda Y. 2016. Electric charge characteristics of biofilms formed on various surfaces. J Pure Appl Chem Res 5:95–100. doi: 10.21776/ub.jpacr.2016.005.02.267. [DOI] [Google Scholar]
- 18.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol 15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon CA, Hodges NA, Marriott C. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J Antimicrob Chemother 22:667–674. [DOI] [PubMed] [Google Scholar]
- 20.Nichols WW, Dorrington SM, Slack MP, Walmsley HL. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother 32:518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MRW, Gilbert P. 1995. Microbiological quality assurance: a guide towards relevance and reproducibility of inocula. CRC Press, Boca Raton, FL. [Google Scholar]
- 22.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 25.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 26.Wolcott RD, Kennedy JP, Dowd SE. 2009. Regular debridement is the main tool for maintaining a healthy wound bed in most chronic wounds. J Wound Care 18:54–56. doi: 10.12968/jowc.2009.18.2.38743. [DOI] [PubMed] [Google Scholar]
- 27.Rediske AM, Roeder BL, Brown MK, Nelson JL, Robison RL, Draper DO, Schaalje GB, Robison RA, Pitt WG. 1999. Ultrasonic enhancement of antibiotic action on Escherichia coli biofilms: an in vivo model. Antimicrob Agents Chemother 43:1211–1214. doi: 10.1128/AAC.43.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänsch GM. 2012. Host defence against bacterial biofilms: “mission impossible”? Int Sch Res Notices 2012:853123. doi: 10.5402/2012/853123. [DOI] [Google Scholar]
- 29.Algburi A, Comito N, Kashtanov D, Dicks LMT, Chikindas ML. 2017. Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol 83:e02508-16. doi: 10.1128/AEM.02508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bechinger B, Gorr S-U. 2017. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res 96:254–260. doi: 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bionda N, Fleeman RM, de la Fuente-Núñez C, Rodriguez MC, Reffuveille F, Shaw LN, Pastar I, Davis SC, Hancock REW, Cudic P. 2016. Identification of novel cyclic lipopeptides from a positional scanning combinatorial library with enhanced antibacterial and antibiofilm activities. Eur J Med Chem 108:354–363. doi: 10.1016/j.ejmech.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traxler MF, Summers SM, Nguyen H-T, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock REW. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Götz F. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3:e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darouiche RO, Mansouri MD, Gawande PV, Madhyastha S. 2009. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother 64:88–93. doi: 10.1093/jac/dkp158. [DOI] [PubMed] [Google Scholar]
- 36.Izano EA, Wang H, Ragunath C, Ramasubbu N, Kaplan JB. 2007. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dent Res 86:618–622. doi: 10.1177/154405910708600707. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan JB, LoVetri K, Cardona ST, Madhyastha S, Sadovskaya I, Jabbouri S, Izano EA. 2012. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J Antibiot (Tokyo) 65:73–77. doi: 10.1038/ja.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones EM, Cochrane CA, Percival SL. 2015. The effect of pH on the extracellular matrix and biofilms. Adv Wound Care (New Rochelle) 4:431–439. doi: 10.1089/wound.2014.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Larose C, Della Porta AC, Schultz GS, Gibson DJ. 2017. A surfactant-based wound dressing can reduce bacterial biofilms in a porcine skin explant model. Int Wound J 14:408–413. doi: 10.1111/iwj.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clinton A, Carter T. 2015. Chronic wound biofilms: pathogenesis and potential therapies. Lab Med 46:277–284. doi: 10.1309/LMBNSWKUI4JPN7SO. [DOI] [PubMed] [Google Scholar]
- 42.Balaban N, Cirioni O, Giacometti A, Ghiselli R, Braunstein JB, Silvestri C, Mocchegiani F, Saba V, Scalise G. 2007. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob Agents Chemother 51:2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A 110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 45.Otto M, Süssmuth R, Vuong C, Jung G, Götz F. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett 450:257–262. [DOI] [PubMed] [Google Scholar]
- 46.Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 47.Paul D, Gopal J, Kumar M, Manikandan M. 2018. Nature to the natural rescue: silencing microbial chats. Chem Biol Interact 280:86–98. doi: 10.1016/j.cbi.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, Skindersoe ME, Rasmussen TB, Friedrich K, Uthe F, Jensen PØ, Moser C, Nielsen KF, Eberl L, Larsen TO, Tanner D, Høiby N, Bjarnsholt T, Givskov M. 2012. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother 56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jakobsen TH, Bragason SK, Phipps RK, Christensen LD, van Gennip M, Alhede M, Skindersoe M, Larsen TO, Høiby N, Bjarnsholt T, Givskov M. 2012. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl Environ Microbiol 78:2410–2421. doi: 10.1128/AEM.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter K, Van den Driessche F, Coenye T. 2017. Innovative approaches to treat Staphylococcus aureus biofilm-related infections. Essays Biochem 61:61–70. doi: 10.1042/EBC20160056. [DOI] [PubMed] [Google Scholar]
- 51.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, Palys TJ. 2014. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 30:17–28. doi: 10.1080/08927014.2013.832224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E56 Committee. 2006. Terminology relating to nanotechnology. ASTM International, West Conshohocken, PA. [Google Scholar]
- 54.Sun H, Gao N, Dong K, Ren J, Qu X. 2014. Graphene quantum dots—band-aids used for wound disinfection. ACS Nano 8:6202–6210. doi: 10.1021/nn501640q. [DOI] [PubMed] [Google Scholar]
- 55.Ashour AH, El-Batal AI, Maksoud MIAA, El-Sayyad GS, Labib S, Abdeltwab E, El-Okr MM. 2018. Antimicrobial activity of metal-substituted cobalt ferrite nanoparticles synthesized by sol-gel technique. Particuology 40:141–151. doi: 10.1016/j.partic.2017.12.001. [DOI] [Google Scholar]
- 56.Xiu Z-M, Ma J, Alvarez PJJ. 2011. Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ Sci Technol 45:9003–9008. doi: 10.1021/es201918f. [DOI] [PubMed] [Google Scholar]
- 57.Kalishwaralal K, BarathManiKanth S, Pandian SRK, Deepak V, Gurunathan S. 2010. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf B Biointerfaces 79:340–344. doi: 10.1016/j.colsurfb.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 58.Azam A, Ahmed AS, Oves M, Khan MS, Habib SS, Memic A. 2012. Antimicrobial activity of metal oxide nanoparticles against Gram-positive and Gram-negative bacteria: a comparative study. Int J Nanomedicine 7:6003–6009. doi: 10.2147/IJN.S35347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holder IA, Boyce ST. 1994. Agar well diffusion assay testing of bacterial susceptibility to various antimicrobials in concentrations non-toxic for human cells in culture. Burns 20:426–429. [DOI] [PubMed] [Google Scholar]
- 60.Balouiri M, Sadiki M, Ibnsouda SK. 2016. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J-H, Kim Y-G, Cho MH, Lee J. 2014. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol Res 169:888–896. doi: 10.1016/j.micres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Seil JT, Webster TJ. 2011. Reduced Staphylococcus aureus proliferation and biofilm formation on zinc oxide nanoparticle PVC composite surfaces. Acta Biomater 7:2579–2584. doi: 10.1016/j.actbio.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 63.El-Batal AI, Mosalam FM, Ghorab MM, Hanora A, Elbarbary AM. 2018. Antimicrobial, antioxidant and anticancer activities of zinc nanoparticles prepared by natural polysaccharides and gamma radiation. Int J Biol Macromol 107:2298–2311. doi: 10.1016/j.ijbiomac.2017.10.121. [DOI] [PubMed] [Google Scholar]
- 64.Mukha I, Eremenko А, Korchak G, Michienkova А. 2010. Antibacterial action and physicochemical properties of stabilized silver and gold nanostructures on the surface of disperse silica. J Water Resource Prot 02:131–136. doi: 10.4236/jwarp.2010.22015. [DOI] [Google Scholar]
- 65.Hernández-Sierra JF, Ruiz F, Pena DCC, Martínez-Gutiérrez F, Martínez AE, Guillén ADJP, Tapia-Pérez H, Castañón GM. 2008. The antimicrobial sensitivity of Streptococcus mutans to nanoparticles of silver, zinc oxide, and gold. Nanomedicine 4:237–240. doi: 10.1016/j.nano.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 66.Dwivedi S, Wahab R, Khan F, Mishra YK, Musarrat J, Al-Khedhairy AA. 2014. Reactive oxygen species mediated bacterial biofilm inhibition via zinc oxide nanoparticles and their statistical determination. PLoS One 9:e111289. doi: 10.1371/journal.pone.0111289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heng BC, Zhao X, Xiong S, Ng KW, Boey FY-C, Loo JS-C. 2010. Toxicity of zinc oxide (ZnO) nanoparticles on human bronchial epithelial cells (BEAS-2B) is accentuated by oxidative stress. Food Chem Toxicol 48:1762–1766. doi: 10.1016/j.fct.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 68.Vandebriel RJ, De Jong WH. 2012. A review of mammalian toxicity of ZnO nanoparticles. Nanotechnol Sci Appl 5:61–71. doi: 10.2147/NSA.S23932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loh JV, Percival SL, Woods EJ, Williams NJ, Cochrane CA. 2009. Silver resistance in MRSA isolated from wound and nasal sources in humans and animals. Int Wound J 6:32–38. doi: 10.1111/j.1742-481X.2008.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan Y, Neuss S, Leifert A, Fischler M, Wen F, Simon U, Schmid G, Brandau W, Jahnen-Dechent W. 2007. Size-dependent cytotoxicity of gold nanoparticles. Small 3:1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 71.Pan Y, Leifert A, Ruau D, Neuss S, Bornemann J, Schmid G, Brandau W, Simon U, Jahnen-Dechent W. 2009. Gold nanoparticles of diameter 1.4 nm trigger necrosis by oxidative stress and mitochondrial damage. Small 5:2067–2076. doi: 10.1002/smll.200900466. [DOI] [PubMed] [Google Scholar]
- 72.Shukla R, Bansal V, Chaudhary M, Basu A, Bhonde RR, Sastry M. 2005. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: a microscopic overview. Langmuir 21:10644–10654. doi: 10.1021/la0513712. [DOI] [PubMed] [Google Scholar]
- 73.Villiers C, Freitas H, Couderc R, Villiers M-B, Marche P. 2010. Analysis of the toxicity of gold nano particles on the immune system: effect on dendritic cell functions. J Nanopart Res 12:55–60. doi: 10.1007/s11051-009-9692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y-H, Tsai C-Y, Huang P-Y, Chang M-Y, Cheng P-C, Chou C-H, Chen D-H, Wang C-R, Shiau A-L, Wu C-L. 2007. Methotrexate conjugated to gold nanoparticles inhibits tumor growth in a syngeneic lung tumor model. Mol Pharm 4:713–722. doi: 10.1021/mp060132k. [DOI] [PubMed] [Google Scholar]
- 75.Wang F, Wang Y-C, Dou S, Xiong M-H, Sun T-M, Wang J. 2011. Doxorubicin-tethered responsive gold nanoparticles facilitate intracellular drug delivery for overcoming multidrug resistance in cancer cells. ACS Nano 5:3679–3692. doi: 10.1021/nn200007z. [DOI] [PubMed] [Google Scholar]
- 76.Wadhwani P, Heidenreich N, Podeyn B, Bürck J, Ulrich AS. 2017. Antibiotic gold: tethering of antimicrobial peptides to gold nanoparticles maintains conformational flexibility of peptides and improves trypsin susceptibility. Biomater Sci 5:817–827. doi: 10.1039/c7bm00069c. [DOI] [PubMed] [Google Scholar]
- 77.Iijima S. 1991. Helical microtubules of graphitic carbon. Nature 354:56–58. doi: 10.1038/354056a0. [DOI] [Google Scholar]
- 78.Yang K, Feng L, Hong H, Cai W, Liu Z. 2013. Preparation and functionalization of graphene nanocomposites for biomedical applications. Nat Protoc 8:2392–2403. doi: 10.1038/nprot.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng L, Yang X, Shi X, Tan X, Peng R, Wang J, Liu Z. 2013. Polyethylene glycol and polyethylenimine dual-functionalized nano-graphene oxide for photothermally enhanced gene delivery. Small 9:1989–1997. doi: 10.1002/smll.201202538. [DOI] [PubMed] [Google Scholar]
- 80.Purtov KV, Petunin AI, Burov AE, Puzyr AP, Bondar VS. 2010. Nanodiamonds as carriers for address delivery of biologically active substances. Nanoscale Res Lett 5:631–636. doi: 10.1007/s11671-010-9526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mochalin VN, Shenderova O, Ho D, Gogotsi Y. 2011. The properties and applications of nanodiamonds. Nat Nanotechnol 7:11–23. doi: 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X-Q, Chen M, Lam R, Xu X, Osawa E, Ho D. 2009. Polymer-functionalized nanodiamond platforms as vehicles for gene delivery. ACS Nano 3:2609–2616. doi: 10.1021/nn900865g. [DOI] [PubMed] [Google Scholar]
- 83.Gibney KA, Sovadinova I, Lopez AI, Urban M, Ridgway Z, Caputo GA, Kuroda K. 2012. Poly(ethylene imine)s as antimicrobial agents with selective activity. Macromol Biosci 12:1279–1289. doi: 10.1002/mabi.201200052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tran PA, Webster TJ. 2011. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int J Nanomedicine 6:1553–1558. doi: 10.2147/IJN.S21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Zhao Q, Han N, Bai L, Li J, Liu J, Che E, Hu L, Zhang Q, Jiang T, Wang S. 2015. Mesoporous silica nanoparticles in drug delivery and biomedical applications. Nanomedicine 11:313–327. doi: 10.1016/j.nano.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 86.Esmaeili F, Hosseini-Nasr M, Rad-Malekshahi M, Samadi N, Atyabi F, Dinarvand R. 2007. Preparation and antibacterial activity evaluation of rifampicin-loaded poly lactide-co-glycolide nanoparticles. Nanomedicine 3:161–167. doi: 10.1016/j.nano.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 87.Kwon MJ, An S, Choi S, Nam K, Jung HS, Yoon CS, Ko JH, Jun HJ, Kim TK, Jung SJ, Park JH, Lee Y, Park J-S. 2012. Effective healing of diabetic skin wounds by using nonviral gene therapy based on minicircle vascular endothelial growth factor DNA and a cationic dendrimer. J Gene Med 14:272–278. doi: 10.1002/jgm.2618. [DOI] [PubMed] [Google Scholar]
- 88.Canfarotta F, Poma A, Guerreiro A, Piletsky S. 2016. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat Protoc 11:443–455. doi: 10.1038/nprot.2016.030. [DOI] [PubMed] [Google Scholar]
- 89.Martin C, Low WL, Gupta A, Amin MCIM, Radecka I, Britland ST, Raj P, Kenward KMA. 2015. Strategies for antimicrobial drug delivery to biofilm. Curr Pharm Des 21:43–66. [DOI] [PubMed] [Google Scholar]
- 90.Rukavina Z, Vanić Ž. 2016. Current trends in development of liposomes for targeting bacterial biofilms. Pharmaceutics 8:E18. doi: 10.3390/pharmaceutics8020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sachetelli S, Khalil H, Chen T, Beaulac C, Senechal S, Lagace J. 2000. Demonstration of a fusion mechanism between a fluid bactericidal liposomal formulation and bacterial cells. Biochim Biophys Acta 1463:254–266. [DOI] [PubMed] [Google Scholar]
- 92.Dong D, Thomas N, Thierry B, Vreugde S, Prestidge CA, Wormald P-J. 2015. Distribution and inhibition of liposomes on Staphylococcus aureus and Pseudomonas aeruginosa biofilm. PLoS One 10:e0131806. doi: 10.1371/journal.pone.0131806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moghadas-Sharif N, Fazly Bazzaz BS, Khameneh B, Malaekeh-Nikouei B. 2015. The effect of nanoliposomal formulations on Staphylococcus epidermidis biofilm. Drug Dev Ind Pharm 41:445–450. doi: 10.3109/03639045.2013.877483. [DOI] [PubMed] [Google Scholar]
- 94.Ahmed K, Muiruri PW, Jones GH, Scott MJ, Jones MN. 2001. The effect of grafted poly(ethylene glycol) on the electrophoretic properties of phospholipid liposomes and their adsorption to bacterial biofilms. Colloids Surf A Physicochem Eng Asp 194:287–296. doi: 10.1016/S0927-7757(01)00817-2. [DOI] [Google Scholar]
- 95.Kania RE, Lamers GEM, Vonk MJ, Huy PTB, Hiemstra PS, Bloemberg GV, Grote JJ. 2007. Demonstration of bacterial cells and glycocalyx in biofilms on human tonsils. Arch Otolaryngol Head Neck Surg 133:115–121. doi: 10.1001/archotol.133.2.115. [DOI] [PubMed] [Google Scholar]
- 96.Galili U. 2017. α-Gal nanoparticles in wound and burn healing acceleration. Adv Wound Care (New Rochelle) 6:81–92. doi: 10.1089/wound.2016.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dobrovolskaia MA, McNeil SE. 2007. Immunological properties of engineered nanomaterials. Nat Nanotechnol 2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 98.Soliman S, Sant S, Nichol JW, Khabiry M, Traversa E, Khademhosseini A. 2011. Controlling the porosity of fibrous scaffolds by modulating the fiber diameter and packing density. J Biomed Mater Res A 96:566–574. doi: 10.1002/jbm.a.33010. [DOI] [PubMed] [Google Scholar]
- 99.Junker JPE, Kamel RA, Caterson EJ, Eriksson E. 2013. Clinical impact upon wound healing and inflammation in moist, wet, and dry environments. Adv Wound Care (New Rochelle) 2:348–356. doi: 10.1089/wound.2012.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Montaser AS, Abdel-Mohsen AM, Ramadan MA, Sleem AA, Sahffie NM, Jancar J, Hebeish A. 2016. Preparation and characterization of alginate/silver/nicotinamide nanocomposites for treating diabetic wounds. Int J Biol Macromol 92:739–747. doi: 10.1016/j.ijbiomac.2016.07.050. [DOI] [PubMed] [Google Scholar]
- 101.Hurler J, Sørensen KK, Fallarero A, Vuorela P, Škalko-Basnet N. 2013. Liposomes-in-hydrogel delivery system with mupirocin: in vitro antibiofilm studies and in vivo evaluation in mice burn model. Biomed Res Int 2013:498485. doi: 10.1155/2013/498485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang K, Han Q, Chen B, Zheng Y, Zhang K, Li Q, Wang J. 2018. Antimicrobial hydrogels: promising materials for medical application. Int J Nanomedicine 13:2217–2263. doi: 10.2147/IJN.S154748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Negre O, Eggimann A-V, Beuzard Y, Ribeil J-A, Bourget P, Borwornpinyo S, Hongeng S, Hacein-Bey S, Cavazzana M, Leboulch P, Payen E. 2016. Gene therapy of the β-hemoglobinopathies by lentiviral transfer of the βA(T87Q)-globin gene. Hum Gene Ther 27:148–165. doi: 10.1089/hum.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Finer M, Glorioso J. 2017. A brief account of viral vectors and their promise for gene therapy. Gene Ther 24:1–2. doi: 10.1038/gt.2016.71. [DOI] [PubMed] [Google Scholar]
- 105.Park S, Kim JI, Lee I, Bae J-Y, Hwang M-W, Kim D, Jang S-I, Kim H, Park MS, Kwon H-J, Song J-W, Cho YS, Chun W, Park M-S. 2014. Inhibition of Pseudomonas aeruginosa with a recombinant RNA-based viral vector expressing human β-defensin 4. BMC Microbiol 14:237. doi: 10.1186/s12866-014-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barker JC, Barker AD, Bills J, Huang J, Wight-Carter M, Delgado I, Noble DL, Huang LJ, Porteus MH, Davis KE. 2014. Genome editing of mouse fibroblasts by homologous recombination for sustained secretion of PDGF-B and augmentation of wound healing. Plast Reconstr Surg 134:389e–401e. doi: 10.1097/PRS.0000000000000427. [DOI] [PubMed] [Google Scholar]
- 107.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. 2013. The in vivo biofilm. Trends Microbiol 21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 108.Bouloussa H, Humblot V, Court C. 2017. Comparative description of in vitro and in vivo MRSA biofilms on titanium surfaces: why animal models still matter. Spine J 17:S167–S168. doi: 10.1016/j.spinee.2017.07.247. [DOI] [Google Scholar]
- 109.Hotterbeekx A, Kumar-Singh S, Goossens H, Malhotra-Kumar S. 2017. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol 7:106. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Otto M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Selle K, Barrangou R. 2015. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 112.Sismaet HJ, Banerjee A, McNish S, Choi Y, Torralba M, Lucas S, Chan A, Shanmugam VK, Goluch ED. 2016. Electrochemical detection of Pseudomonas in wound exudate samples from patients with chronic wounds. Wound Repair Regen 24:366–372. doi: 10.1111/wrr.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]