Significance
MicroRNAs (miRNAs) are indispensable regulators of normal development and mediators of diverse diseases, including cancer, and cardiovascular and neurodegenerative diseases. As a consequence, the biogenesis of miRNAs is a critical determinant in disease etiology and pathogenesis. Cytoplasmic processing by DICER and downstream modifications of miRNAs are crucial regulators of miRNA biogenesis. mda-7/IL-24, a cancer-selective apoptosis-inducing cytokine, specifically downregulates expression of DICER, thereby deregulating miRNA expression. A link among mda-7/IL-24, DICER, and reactive oxygen species in cancer cells is described that might be exploited to produce enhanced and effective therapies for cancer. Accordingly, targeting this pathway uncovers exceptional opportunities to potentially eradicate cancer. This study demonstrates that mda-7/IL-24 effectively downregulates a miRNA processing protein and highlights the importance of the mda-7/IL-24-MITF-DICER-miRNA axis in cancer.
Keywords: mda-7/IL-24, miRNA, ROS, DICER, MITF
Abstract
Melanoma differentiation-associated gene-7/interleukin-24 (mda-7/IL-24) is a multifunctional cytokine displaying broad-spectrum anticancer activity in vitro or in vivo in preclinical animal cancer models and in a phase 1/2 clinical trial in patients with advanced cancers. mda-7/IL-24 targets specific miRNAs, including miR-221 and miR-320, for down-regulation in a cancer-selective manner. We demonstrate that mda-7/IL-24, administered through a replication incompetent type 5 adenovirus (Ad.mda-7) or with His-MDA-7/IL-24 protein, down-regulates DICER, a critical regulator in miRNA processing. This effect is specific for mature miR-221, as it does not affect Pri-miR-221 expression, and the DICER protein, as no changes occur in other miRNA processing cofactors, including DROSHA, PASHA, or Argonaute. DICER is unchanged by Ad.mda-7/IL-24 in normal immortal prostate cells, whereas Ad.mda-7 down-regulates DICER in multiple cancer cells including glioblastoma multiforme and prostate, breast, lung, and liver carcinoma cells. MDA-7/IL-24 protein down-regulates DICER expression through canonical IL-20/IL-22 receptors. Gain- and loss-of-function studies confirm that overexpression of DICER rescues deregulation of miRNAs by mda-7/IL-24, partially rescuing cancer cells from mda-7/IL-24-mediated cell death. Stable overexpression of DICER in cancer cells impedes Ad.mda-7 or His-MDA-7/IL-24 inhibition of cell growth, colony formation, PARP cleavage, and apoptosis. In addition, stable overexpression of DICER renders cancer cells more resistant to Ad.mda-7 inhibition of primary and secondary tumor growth. MDA-7/IL-24-mediated regulation of DICER is reactive oxygen species-dependent and mediated by melanogenesis-associated transcription factor. Our research uncovers a distinct role of mda-7/IL-24 in the regulation of miRNA biogenesis through alteration of the MITF-DICER pathway.
RNAi-mediated gene regulation is an evolutionarily conserved pathway that regulates gene expression in a sequence-specific manner. MicroRNAs are noncoding 18–22-nucleotide-long RNAs, which either degrade RNA or translationally block protein synthesis (1). These small RNAs play a decisive role in evolution and disease development, including cancer (2). Biogenesis of miRNAs (microRNAs or miRs) is regulated by multiple steps including DROSHA-regulating nuclear generation of primary miRNA (pri-), Exportin 5 transferring pri-miRNA to the cytoplasm, and pri-miRNA maturation in the cytoplasm by DICER (3). Conventional miRNA biogenesis consists of nuclear primary miRNA generation to synthesis of cytoplasmic mature miRNA (3). Various RNA-induced silencing complex models have been proposed, and these are either DICER-dependent or DICER-independent (4). The DICER-independent miRNA, miR-451, uses Ago2 for maturation (4). The identification of different oncogenic and tumor suppressor miRNAs can be used as biomarkers that correlate with disease stage and type (5).
DICER is a RNase III endonuclease molecule localized in the cytoplasm, which provides key functions in miRNA processing (3). Expression of DICER in different cancer types varies (6). Expression is up-regulated in prostate cancers (7) and precursor lesions of lung adenocarcinomas (8), but reduced in ovarian (9) and advanced lung carcinomas (8). In addition, both high and low expression of DICER correlate with poor prognosis in distinct populations of patients with cancer (10).
Melanoma differentiation-associated gene-7/interleukin-24 (MDA-7/IL-24), a cancer-selective antitumor cytokine, is a member of the IL-10 gene superfamily (11). The gene and protein exhibit in vitro and in vivo antitumor activity (12). Elevated MDA-7/IL-24 expression suppresses tumor growth and promotes apoptosis and toxic autophagy in a broad array of cancer cells including osteosarcoma, melanoma, glioblastoma, neuroblastoma, and colorectal, liver, lung, breast, pancreatic, and prostate cancer (12). In a phase 1/2 clinical trial, delivery of MDA-7/IL-24 by replication-incompetent adenovirus showed clinical efficacy in patients with advanced cancers (12–16). MDA-7/IL-24 not only directly diminishes tumor growth but also blocks angiogenesis and activates the immune system (17), functioning as an anticancer double-edged sword. An additional attribute, and perhaps a key distinctive property, of MDA-7/IL-24 that contributes to its profound anticancer properties involves its ability to be secreted and induce its own synthesis through canonical IL-20/IL-22 receptors, thereby promoting both localized and distant tumor cell killing (i.e., “bystander anticancer activity”) (18–20).
We showed previously that MDA-7/IL-24 regulates expression of several miRNAs, including miR-221 (21). However, the mechanism of miRNA regulation by MDA-7/IL-24 was not resolved in this study. We now elucidate the primary underlying mechanism of regulation of miRNAs by MDA-7/IL-24. MDA-7/IL-24 down-regulates DICER in a reactive oxygen species (ROS)-dependent manner. This results in deregulation of miRNAs that are controlled by MDA-7/IL-24. Overexpression of DICER partially rescues MDA-7/IL-24-mediated cell death in cancer cells. Moreover, MDA-7/IL-24-mediated DICER deregulation occurs through regulation of melanogenesis-associated transcription factor (MITF). Our studies confirm a functional link between mda-7/IL-24 and specific miR regulation and show that this pathway is enabled by a complex loop consisting of MITF and DICER. These discoveries provide targets for potentially enhancing anticancer activity of the therapeutic cytokine MDA-7/IL-24.
Results
MDA-7/IL-24 Regulates Mature miR-221 and Not Pri-miR-221.
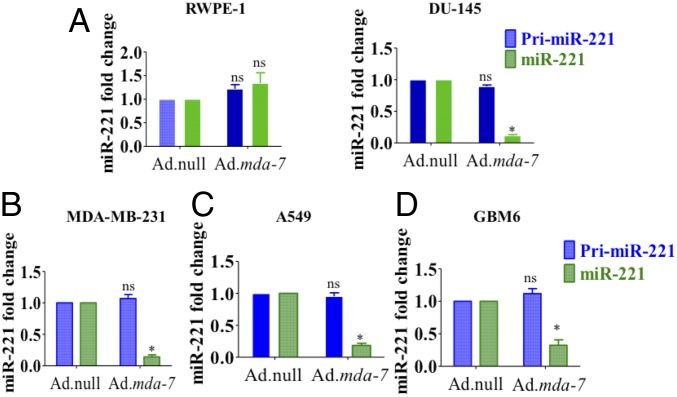
We demonstrated previously that mda-7/IL-24 down-regulated the oncogenic microRNA, miR-221, in a broad spectrum of cancer cell lines (21). Overexpression of miR-221 partially rescued cells from mda-7/IL-24-mediated cell death. To study the mechanism underlying this regulation, we first determined the level of primary miRNA (i.e., pri-miR-221 or premiR-221) in mda-7/IL-24-treated cells (Fig. 1). Cells were infected with Ad.null or Ad.mda-7 [2,000 viral particles (vp)/cell] for 72 h, and the levels of primary and mature miR-221 were determined by real-time PCR. As shown in Fig. 1, infection with Ad.mda-7 induced a significant down-regulation of mature miR-221 in a diverse array of cancer cell lines, supporting previous observations (21). However, no significant change was evident in pri-miR-221 levels in these different cancer cells (Fig. 1). In contrast, no alterations in pri-miR-221 or mature miR-221 were evident in RWPE-1 cells (Fig. 1A), which are primary immortal normal prostate epithelial cells. These results suggest that mda-7/IL-24-mediated miRNA regulation in cancer cells is posttranscriptional, which affects mature miRNA and not primary miRNA. In addition, our findings demonstrate a lack of effect on miRNA in normal prostate epithelial cells, confirming a cancer-selective effect of mda-7/IL-24 on miR-221 expression.
Fig. 1.
mda-7/IL-24 down-regulates mature miR-221, but not pri-miR-221. Real-time quantitative PCR of cancer cells [DU-145 (A), MDA-MB-231 (B), A549 (C), and GBM6 (D)] infected with Ad.mda-7 (2,000 vp/cell for 72 h) indicates significantly less miR-221. miR-221 expression does not change in RWPE-1 cells (A) after infection with Ad.mda-7. No significant change in primary-miR-221 occurs after Ad.mda-7 infection.
MDA-7/IL-24 Down-Regulates the miRNA-Processing Enzyme DICER.
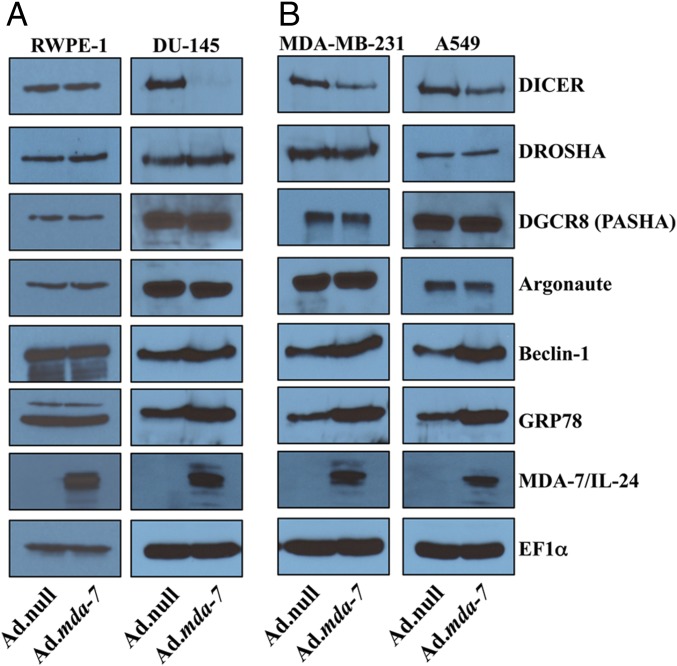
To scrutinize the mechanism of MDA-7/IL-24 regulation of miR-221, we examined the expression of the proteins involved in miRNA processing. miRNA processing is complex: mature miRNAs develop through a series of posttranscriptional steps, including production of premiRNA by Drosha/DGCR8, export into the cytoplasm by exportin-5, and digestion by DICER, an RNase III endonuclease. There are also reported miRNAs, which follow DICER-independent pathways (i.e., miR-451) (4). Moreover, there are groups of miRNAs called mirtrons, which use the splicing machinery and bypass Drosha-mediated cleavage. We checked the expression of regulatory molecules of the miRNA biogenesis pathway in a series of cancer cells of diverse origin including prostate (DU-145), breast (MDA-MB-231), and lung (A549) carcinomas and normal prostate epithelial cells (RWPE-1) after Ad.null or Ad.mda-7 infection (Fig. 2 and SI Appendix, Fig. S1). Cell lysates were collected 72 h after treatment, and Western blotting was done with the indicated antibodies. DICER expression decreased with mda-7/IL-24 overexpression, whereas DROSHA and DGCR8 (PASHA) expression remained unaffected in Ad.mda-7-infected cells (Fig. 2 and SI Appendix, Fig. S1). Ago2 also remained unaltered after overexpression of mda-7/IL-24. Interestingly, DICER expression remained unchanged in RWPE-1 cells supporting cancer-specific activity of mda-7/IL-24 (Fig. 2A). MDA-7/IL-24 overexpression after Ad.mda-7 infection was confirmed by Western blotting. To validate further the activity of mda-7/IL-24, we checked the expression of two established downstream targets of mda-7/IL-24; that is, BiP/GRP78 (22) and Beclin-1 (21, 23). As anticipated, expression of GRP78 and Beclin-1 were increased after mda-7/IL-24 treatment (Fig. 2 and SI Appendix, Fig. S1). The level of decrease in DICER expression by mda-7/IL-24 varied in different cancer cell lines, whereas no change in DICER (or GRP78 or Beclin-1) was observed in RWPE-1 cells after infection with Ad.mda-7 (Fig. 2A and SI Appendix, Fig. S1).
Fig. 2.
mda-7/IL-24 down-regulates DICER protein. Effect of mda-7/IL-24 on DICER, DROSHA, DGCR8 (PASHA), Ago2, Beclin-1, and GRP78 protein levels determined by Western blotting. RWPE-1 and DU-145 (A), MDA-MB-231 and A549 (B) cells were infected with Ad.null or Ad.mda-7 (2,000 vp/cell) for 72 h. Cells were lysed, and Western blotting was done. EF1α was used as a loading control.
To explore further the mechanistic details of DICER regulation by MDA-7/IL-24, we determined the level of DICER transcripts after mda-7/IL-24-treatment. Real-time PCR confirmed a consistent down-regulation of DICER mRNA after overexpression of mda-7/IL-24 in multiple cancer cell lines (SI Appendix, Fig. S2). In addition, mda-7/IL-24 significantly down-regulated DICER protein level in a dose- and time-dependent manner in both DU-145 and MDA-MB-231 cells (SI Appendix, Fig. S3).
MDA-7/IL-24 is a secreted protein that requires a complete set of dimeric cell surface receptors (IL-20R1/IL-20R2, IL-20R1/IL-22R1, or IL-20R2/IL-22R1) for internalization and signaling (24). We purified recombinant His tagged MDA-7/IL-24 protein, using a previously described protocol (24–26), and treated A549 cells (which lack the dimeric set of IL-20/IL-22 receptors) and DU-145 cells (with a complete set of IL-20/IL-22 receptors) with His-MDA-7/IL-24 and determined the DICER transcript levels. As shown in SI Appendix, Fig. S4A, expression of DICER significantly decreased in DU-145 cells, whereas the level remained unchanged in A549 cells. This validates previous data using viral-based delivery of MDA-7/IL-24 and confirms that MDA-7/IL-24 down-regulates DICER in DU-145 cells, which have a complete set of receptors for this cytokine, but not in A549 cells, which only express IL-20R1. However, when IL-22R1 or IL-20R2 receptors were introduced into A549 cells and these reconstituted A549 cells were treated with His-MDA-7/IL-24, DICER levels decreased to a comparable level, as seen in DU-145 cells (SI Appendix, Fig. S4A).
DICER Overexpression Rescues miRs Down-Regulated by MDA-7/IL-24.
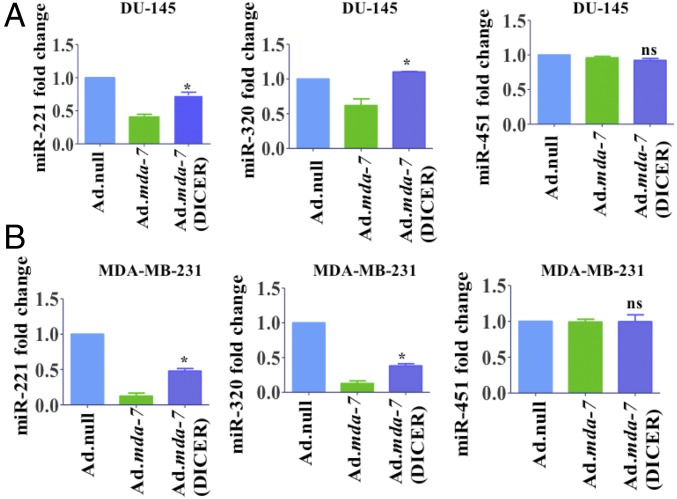
Because DICER expression decreased with mda-7/IL-24 treatment, we examined the expression of miRNAs that were deregulated by mda-7/IL-24 after overexpression of DICER. Whereas mda-7/IL-24 down-regulated miR-221 and miR-320 in control (nontransfected) cells, there was a significant rescue in the levels of miR-221 and miR-320 in DICER-overexpressing cells (Fig. 3). In contrast, miR-451, a DICER-independent miRNA (4), showed no significant change after overexpression of mda-7/IL-24 or DICER (Fig. 3). These results were observed both in DU-145 (Fig. 3A) and MDA-MB-231 cells (Fig. 3B). Overall, these studies support DICER-dependent regulation of miRNA by mda-7/IL-24.
Fig. 3.
DICER overexpression rescues miRs regulated by mda-7/IL-24. DU-145 (A) and MDA-MB-231 (B) cells were transfected with DICER overexpressing plasmid and infected with Ad.mda-7. miR-221, miR-320, and miR-451 levels were monitored by real-time quantitative PCR. Treatment with Ad.mda-7 significantly down-regulated miR-221 and miR-320, which were rescued in DICER-transfected cells. The level of miR-451, which is a DICER-independent miRNA, does not change with either Ad.mda-7 infection or DICER overexpression.
MDA-7/IL-24-Mediated DICER Regulation Is ROS-Dependent.
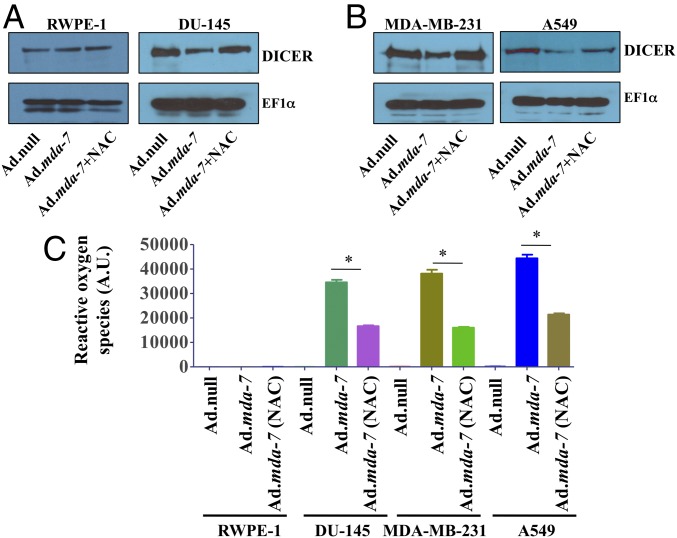
ROS are key contributors to mda-7/IL-24-mediated cell death uniquely in cancer cells (27). Prior studies have shown that mda-7/IL-24-mediated miR-221 regulation is ROS-dependent (21). Infection with Ad.mda-7 down-regulated DICER expression in multiple cancer cells, irrespective of their origin, whereas pretreatment of cells with N-acetyl cysteine (NAC), a ROS scavenger, rescued DICER expression (Fig. 4 and SI Appendix, Fig. S4B). Moreover, these changes in DICER levels were not observed in RWPE-1 cells after Ad.mda-7 infection. ROS levels were quantified with DCFD reagent, which showed that ROS levels increased after mda-7/IL-24 expression in cancer cells, and NAC treatment decreased ROS levels (Fig. 4C).
Fig. 4.
ROS-dependent regulation of DICER by mda-7/IL-24. RWPE-1 and DU-145 (A), MDA-MB-231, and A549 (B) cells were pretreated with 10 mM NAC and infected with either Ad.null or Ad.mda-7 (2,000 vp/cell for 72 h). Western blotting was done with DICER antibody. EF1α was used as a loading control. (C) Bar graph representation of quantitative ROS levels.
DICER Overexpression Rescues MDA-7/IL-24-Mediated Cell Death.
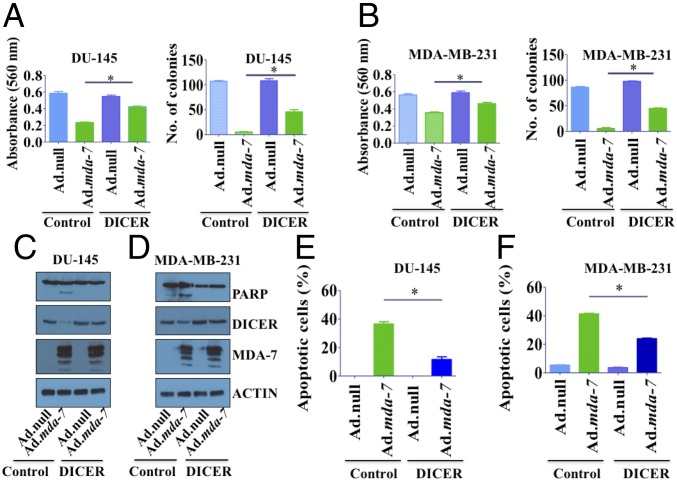
Because DICER is a downstream target of mda-7/IL-24, we determined whether DICER overexpression could mitigate mda-7/IL-24-mediated effects on cell proliferation and death. Cells were transfected with a vector control or DICER and infected with either Ad.null or Ad.mda-7. DICER-transfected cells were resistant to mda-7/IL-24-mediated inhibitory effects on cell proliferation, as monitored by MTT (3-4,5-dimethyl thiazol-2-yl-2, 5-diphenyl tetrazolium bromide) assays (Fig. 5). These antiproliferative effects were also evident in long-term colony-forming assays (Fig. 5). A strong inhibition in colony formation was seen in mda-7/IL-24-infected cells, which was partially but significantly rescued in DICER-overexpressing cells.
Fig. 5.
DICER overexpression rescues cancer cells from mda-7/IL-24-mediated cell death. DU-145 (A) and MDA-MB-231 (B) cells were transfected with pCDNA3.1 (control) or DICER and then infected with Ad.null or Ad.mda-7. MTT assays showed that treatment with mda-7/IL-24 resulted in an inhibition in proliferation of DU-145 and MDA-MB-231 cells, which is less in DICER-overexpressing cells. DICER overexpression also rescues inhibition in colony formation in DU-145 (A) and MDA-MB-231 (B) cells treated with Ad.mda-7. Data presented as mean ± SD. DU-145 (C) and MDA-MB-231 (D) cells were transfected with pCDNA3.1 (control) or DICER and then infected with Ad.null or Ad.mda-7 (2,000 vp/cell). After 72 h, cells were lysed and Western blotting was done with PARP, DICER, and MDA-7/IL-24 antibody. Actin was used as a loading control. DU-145 (E) and MDA-MB-231 (F) cells were transfected with pCDNA3.1 (control) or DICER and then treated with Ad.null or Ad.mda-7. After 72 h, cells were stained with Annexin-V/PI and then analyzed by flow cytometry.
We also studied apoptosis by monitoring cleaved PARP levels in control and DICER-transfected cells. There was significantly less cleaved PARP in DICER-overexpressing cells compared with vector-transfected cells after mda-7/IL-24 expression (Fig. 5 C and D and SI Appendix, Fig. S5). Annexin-V assays also confirmed similar results, with DICER-overexpressing cells being significantly more resistant to mda-7/IL-24-mediated cell death (Fig. 5 E and F).
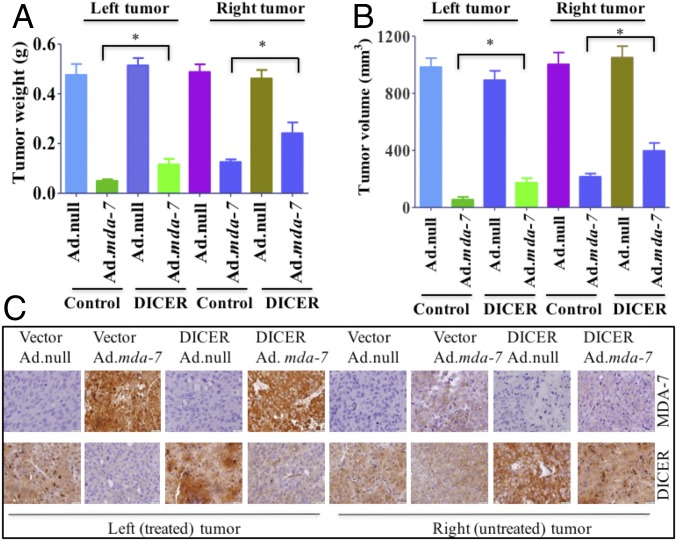
To evaluate the role of mda-7/IL-24 and DICER in vivo, DU-145 cells stably overexpressing vector or DICER were injected s.c. to establish tumor xenografts in male athymic nude mice. After a palpable tumor developed in ∼2 wk, the tumors were injected with eight intratumoral injections during a 3-wk period with 1 × 108 viral particles of Ad.null or Ad.mda-7. In control vector-transfected cells, significant growth inhibition was evident, but in DICER-overexpressing cells, the effect of Ad.mda-7 was diminished in both the injected left tumor and the uninjected right tumor, as previously observed in vitro (Fig. 6 A and B and SI Appendix, Fig. S6). The expression of MDA-7/IL-24 and DICER were confirmed by immunohistochemistry (Fig. 6C and SI Appendix, Fig. S7). Taken together, these experiments demonstrate the significance of DICER in mda-7/IL-24-mediated cancer cell death both in vitro and in vivo.
Fig. 6.
In vivo xenograft study showing DICER-mediated cell death by mda-7/IL-24. DU-145 cells, stably expressing a control pCDNA3.1 vector or DICER, were s.c. implanted in both flanks of athymic male nude mice. Left-sided tumors were injected with eight intratumoral injections of Ad.null or Ad.mda-7 (1 × 108 vp). Once the control tumors reached maximum allowable limits, animals were killed and tumors were isolated. (A) Graphical representation of the tumor weight on both flanks. (B) Tumor volumes on both flanks were measured, and results are presented in a graphical manner. (C) Immunohistochemical analysis of MDA-7/IL-24 and DICER in tumor sections (Left, injected/treated tumors; Right, uninjected/untreated tumors).
MDA-7/IL-24-Mediated DICER Regulation Is Mediated by the Transcription Factor MITF.
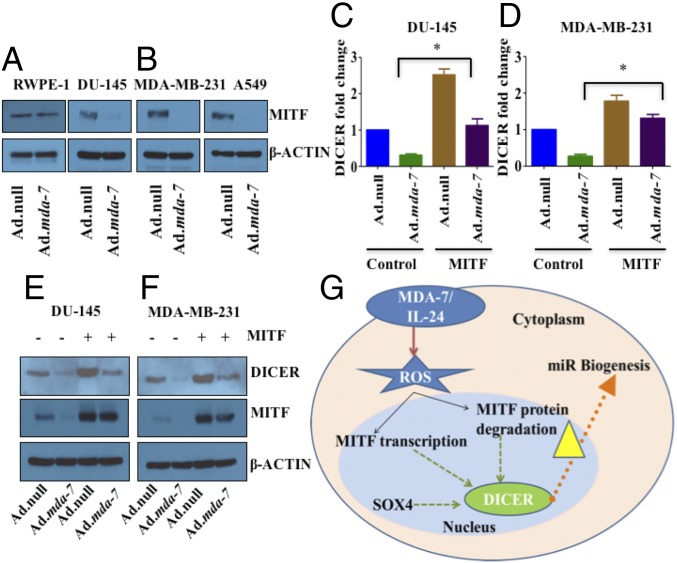
DICER plays a central regulatory role in miRNA processing and biogenesis pathways. However, little is known regarding the regulation of DICER. Conversion of pre- to mature-miRNA is dependent on the level of DICER expression. Accordingly, understanding the underpinnings of transcriptional regulation of DICER is relevant. Previous studies have shown that SOX4 (28) and MITF (29) can transcriptionally activate DICER. Infection with Ad.mda-7 dramatically inhibited MITF expression in all three cancer types (Fig. 7 A and B and SI Appendix, Fig. S8A). Also, the transcript levels of MITF decreased with mda-7/IL-24 expression (SI Appendix, Fig. S8B). In contrast, no significant changes in MITF occurred in RWPE-1 cells infected with Ad.mda-7. Also, treatment with NAC, the ROS scavenger, rescued MITF expression in Ad.mda-7 infected cells, suggesting a role of ROS in the regulation of MITF (SI Appendix, Fig. S9).
Fig. 7.
mda-7/IL-24-mediated DICER regulation is controlled by the transcription factor MITF. mda-7/IL-24 down-regulates MITF in different cancer cell lines (A and B), but not in normal RWPE-1 cells (A). DU-145 (C) and MDA-MB-231 cells (D) were transfected with vector or MITF, and then treated with Ad.null or Ad.mda-7. RNA was isolated 72 h postinfection, and real-time quantitative PCR was done to check the level of DICER. DU-145 (E) and MDA-MB-231 cells (F) were treated as described in C and D, total protein was isolated, and Western blotting was done with DICER and MITF antibodies. Actin was used as a loading control. (G) Schematic representation of regulation of the miRNA processing enzyme DICER by mda-7/IL-24. MDA-7/IL-24 down-regulates the transcription factor MITF in a ROS-dependent manner, which in turn down-regulates DICER.
Because secreted MDA-7/IL-24 protein induces cancer toxicity through a paracrine/autocrine loop through induction of endogenous MDA-7/IL-24 protein expression and secretion (18), we generated mda-7/IL-24 knockout clones (KO), using the CRISPR-Cas9 method in DU-145 cells to evaluate the effect of mda-7/IL-24 on MITF regulation. Both vector and Cas9 mda-7 KO cells were treated with recombinant His-MDA-7/IL-24 protein, and as reported earlier, MDA-7/IL-24 protein turned on its own transcript expression through an autocrine regulatory pathway (18) in control vector cells. In contrast, His-MDA-7 failed to turn on mda-7/IL-24 in Cas9 mda-7 KO cells (SI Appendix, Fig. S10). Also, in the vector group, there was a down-regulation in MITF transcript level after mda-7/IL-24 expression, which was abrogated in the mda-7-cas9 KO cells. These data confirm mda-7/IL-24 regulation of MITF expression.
Next, we monitored the expression of DICER in MITF-overexpressing cells and determined whether overexpressing MITF could diminish the suppressive effect of mda-7/IL-24 on DICER. Cells were transfected with vector or MITF plasmids and infected with Ad.null or Ad.mda-7. In MITF transfected cells, DICER expression was significantly enhanced (Fig. 7 C–F and SI Appendix, Fig. S11). As anticipated, DICER levels decreased in mda-7/IL-24-treated cells; however, in MITF-overexpressing cells, DICER expression was rescued after treatment with mda-7/IL-24 (Fig. 7 C–F and SI Appendix, Fig. S11). These results confirm MITF-dependent mda-7/IL-24-mediated DICER regulation. Taken together, these results delineate the significance of MITF-DICER and miRNAs in the cancer-specific cytotoxicity of mda-7/IL-24, establishing a pathway of mda-7/IL-24-mediated gene regulation in cancer cells.
To validate that the effect of mda-7/IL-24 on the transcriptional regulation of DICER involves MITF, we performed a luciferase reporter gene assay using a DICER promoter construct and MITF. Cells infected with Ad.mda-7 had significantly lower luciferase activity than the Ad.null infected controls (SI Appendix, Fig. S12). Also, as reported earlier, MITF induced DICER promoter activity (29), which decreased after mda-7/IL-24-treatment. These data confirm that MITF mediates DICER regulation by mda-7/IL-24.
Discussion
miRNAs are abundant in different cell types and in diverse organisms including plants, animals, and viruses. They resemble siRNAs and are part of the RNA interference pathway (30). However, they differ in that miRNAs are produced from RNA transcripts that fold on themselves, forming hairpins, whereas siRNAs are generated from double-stranded RNAs. miRNAs target about 60% of genes in humans and other mammals (30). They are evolutionarily conserved, implying their importance in many biological functions. Many miRNA genes are located in both exons and introns and are transcribed by RNA polymerase II (30). Our study reveals a previously unrecognized mechanism of DICER regulation by mda-7/IL-24 and the melanocytic transcription factor MITF. This regulation leads to noteworthy changes in downstream miRNAs. We also demonstrate that DICER plays a decisive role in mda-7/IL-24-mediated cell death that is cancer selective.
Although DICER is ubiquitously expressed in many cell types, its regulation of miRNA biogenesis in diverse and specific cellular and biological contexts requires clarification. DICER expression and its occurrence in cancer varies (31), with no clear correlation between DICER expression and disease stage or occurrence (32). DICER can function both as an oncogene and as a tumor suppressor gene in specific cancer indications. Although DICER is up-regulated in prostate cancer (33), it is down-regulated in breast cancer (34). Also, contradicting studies suggest that DICER protein levels do not always correlate with mature miRNA production (35).
Maturation of miRNA involves several steps and three central molecules, DROSHA, DGCR8, and DICER, which play crucial roles in maturation (36). DICER is recognized for its canonical role in the generation and maturation of miRNAs (37). This was traditionally thought to be a cytoplasmic process; however, recent evidence suggests that functional DICER can also localize to the nucleus (38). A DICER-independent pathway was described for the precursor of miR-451. miR-451 bypasses DICER, is loaded into Ago2 directly (39), and uses Ago2 for its maturation instead of DICER (39). mda-7/IL-24 does not regulate miR-451, but regulates other DICER-dependent miRs, emphasizing the specific role of mda-7/IL-24 on DICER-mediated miR regulation.
mda-7/IL-24 is a tumor suppressor gene displaying selective tumor inhibitory activity in a broad spectrum of cancer cells (12, 40). This IL-10 gene family cytokine targets several anti-apoptotic proteins for suppression, that is, Bcl-2, Bcl-xL (12), Mcl-1, and AIF (41), whereas it activates the tumor suppressors SARI (24, 42), Beclin-1 (21, 23), p27 (21), and PUMA (21). mda-7/IL-24 also regulates specific microRNAs including miR-221 (21). This immunomodulatory anticancer cytokine inhibits angiogenesis (43), and the secreted MDA-7/IL-24 protein has potent “bystander activity,” targeting both local and distant tumors (19, 44). We now uncover a pathway of mda-7/IL-24-mediated miRNA regulation that involves ROS, MITF, and DICER.
MITF, a microphthalmia family oncogenic transcription factor, transcriptionally activates many oncogenes and lysosomal genes (45). The underlying mechanisms of regulation of MITF are not well understood. It is an evolutionarily conserved transcription factor with homologs in Caenorhabditis elegans and Drosophila (46). MITF is mainly expressed in melanocytes and retinal pigment epithelial cells, but it is also expressed in other cell types (47). Genomic amplification of MITF is observed in cancer, where it can function as an oncogene (48). MITF promotes cellular growth and oncogenesis through regulation of genes involved in survival, proliferation, motility, oxidative stress, and DNA repair (45). Cancer cells depend on glucose for efficient growth, and glucose metabolism controls MITF expression (49). Restriction of glucose to physiological levels induces ROS production (49). Increased ROS levels lead to activation of ATF4, which in turn suppresses MITF expression by competing with CREB, an otherwise potent inducer of the MITF promoter (49). The ROS-ERK signaling pathway plays an essential role in cancer, promoting phosphorylation of Serine 73 on MITF and thereby accelerating its proteasomal degradation (50). We show that mda-7/IL-24 can specifically down-regulate MITF transcriptionally and translationally, which are mediated by ROS. Also, this regulation down-regulates the target of MITF, that is, DICER, culminating in a decrease in miRNA biogenesis (Fig. 7G). Wiesen and Tomasi have shown that ROS and HDAC inhibitors regulate DICER protein levels (51). As a result, the HDAC inhibitor, panobinostat, down-regulates DICER activity, which provides therapeutic opportunities in cancer. In these contexts, downregulating DICER can have direct therapeutic applications and may be an additional mechanism by which MDA-7/IL-24 exerts such potent anticancer activity in diverse cancers.
In conclusion, our study highlights a distinctive role of mda-7/IL-24 in miRNA processing by regulating the ROS/MITF/DICER pathway. Further detailed investigations are necessary to mechanistically define precisely how DICER is regulated by mda-7/IL-24 and MITF. Aberrant expression of miRNAs in different pathological conditions, including cancer, neurodegeneration, cardiovascular disease, and infectious diseases, represent important areas of health research, and consequently mda-7/IL-24-mediated DICER regulation may provide a target for therapeutic intervention in multiple diseases.
Materials and Methods
Plasmids, Cell Lines, Stable Clones, Adenoviruses, and Purified MDA-7/IL-24 Protein.
DICER, DICER promoter, and MITF plasmids were obtained from Addgene. The MDA-7/IL-24 CAS9 construct and its vector control were from Genecopoeia. Ad.mda-7, a replication incompetent Ad5 expressing mda-7/IL-24, and Ad.null, a replication incompetent Ad5 without a gene insert, were constructed and amplified as described (25–27). IM-PHFA cells were infected with Ad.5-His-mda-7, using a standard protocol. Cell supernatant was mixed with Ni-NTA slurry to bind MDA-7/IL-24 with Ni-NTA beads. Twenty-four hours after incubation, the Ni-NTA beads were collected, washed, and purified. MDA-7/IL-24 protein was eluted in imidazole buffer and validated by Western blotting. DU-145, MDA-MB-231, A549, and RWPE-1 cells were obtained from the American Type Culture Collection. They were cultured as per American Type Culture Collection instructions and regularly tested for mycoplasma contaminations, using mycoplasma detection kit (Sigma Aldrich). GBM6 cells were described (52) and provided by C. David James (University of California, San Francisco). The IM-PHFA cell line was established and maintained as described (19). DICER stable cell lines were generated using a standard protocol (19).
Transfection and Western Blotting.
Cells were transfected with Lipofectamine 2000 (Invitrogen), as suggested by the manufacturer. Plasmid concentrations vary in the size and area of the plates, as per the manufacturer’s instructions. Western blotting was done using standard protocols (21). Primary antibodies were DICER, DROSHA, MITF, DGCR8, Argonaute, Beclin-1, Actin (Cell Signaling Technology), MDA-7/IL-24 (Genhunter), BiP/GRP78 (Santacruz), and EF1α (Upstate biotechnology). Secondary antibodies were from Cell Signaling Technology.
Real-Time PCR.
Total RNA and miRNA-enriched fractions were isolated from cell lines using RNA and miRNA isolation kits from Qiagen. cDNA synthesis was done using the cDNA synthesis kit from Applied Biosystems. Real-time PCR was performed using the TaqMan master mix from Applied Biosystems. Data were analyzed using GraphPad Prism software.
Cell Proliferation Assays.
MTT was used to study cellular proliferation. Standard protocol was used (18).
Colony Formation Assays.
Clonal assays were performed as described (24). Briefly, ∼500 cells were plated and treated as described in figure legends. After 2 wk, colonies were stained and counted. Graphical representation was done for different groups in triplicates, with SD of the mean. Statistical analysis was done with P value < 0.05 considered significant.
Apoptosis Assays.
Cells were treated as indicated in the figure legends. Annexin-V/PI assay was used to study apoptosis as described in the manufacturer’s instructions (BD Biosciences). Flow cytometric analysis was done to quantify the apoptotic cells using BD FACS canto II and BDFACS DIVA software.
ROS Measurements.
Carboxy-2′,7′-dichloro dihydro fluorescein diacetate (Molecular Probes; Life Technologies) was used to measure the total amount of ROS produced by cells after treatment, as described in the figure legends (Fig. 4C).
Reporter Gene Assays.
For luciferase assays, cells were transfected with the DICER promoter construct and MITF overexpressing plasmid using lipofectamine. Cells were incubated for 72 h, and luciferase assays were performed with the dual-luciferase assay kit from Promega.
Xenograft Tumor Studies.
Prostate cancer cell lines, including DU-145 and DICER overexpressing stable clones of DU-145, were implanted in both flanks of 6–7-wk-old athymic nude mice. All animal experiments were performed as per guidelines approved by the American Association for Accreditation of Laboratory Animal Care and Institutional review board (IRB). After the tumor reached palpable size, they were injected with the adenoviruses 108 vp for eight doses. Once the control tumors reached a maximum allowable limit, the animals were killed, tumors were isolated, and tumor weight was measured. Also, tumor sizes were measured periodically, using calipers, and graphical representation was done. After completion of the experiment, tumors were fixed and the sections were evaluated immunohistochemically.
Statistical Analysis.
Data are presented as mean ± SD of at least three experiments, performed as a minimum in triplicate. Statistical analysis were done using Student t test. P < 0.05 was considered to be significant. Analysis and graphical representation were done using the GraphPad prism software.
Supplementary Material
Acknowledgments
We thank Xue-Ning Shen for outstanding technical assistance. The present study was supported in part by National Cancer Institute Cancer Center Support Grant to the VCU Massey Cancer Center P30 CA016059 (to P.B.F. and D. Sarkar), the National Foundation for Cancer Research (P.B.F. and W.K.C.), the VCU Institute of Molecular Medicine (P.B.F.), and the Genetics Enhancement Fund (to P.B.F., S.K.D., and L.E.). Support was also provided by a Sponsored Research Agreement from ILCT (L.E.). P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research at the Massey Cancer Center.
Footnotes
Conflict of interest statement: W.K.C. and P.B.F. are cofounders of InterLeukin Combinatorial Therapies, Inc. (ILCT). W.K.C., P.B.F., and Virginia Commonwealth University own stock in ILCT. L.E. is the principal investigator of a sponsored research agreement provided by ILCT to Virginia Commonwealth University. The other authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819869116/-/DCSupplemental.
References
- 1.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Ma L. MicroRNA and metastasis. Adv Cancer Res. 2016;132:165–207. doi: 10.1016/bs.acr.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamam R, et al. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stratmann J, et al. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer. 2011;11:345. doi: 10.1186/1471-2407-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bian X, et al. Expression of dicer and its related miRNAs in the progression of prostate cancer. PLoS One. 2015;10:e0120159. doi: 10.1371/journal.pone.0120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiosea S, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 9.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem. 2010;43:324–327. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Jakymiw A, et al. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–559. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher PB, et al. Melanoma differentiation associated gene-7/interleukin-24 (mda-7/IL-24): Novel gene therapeutic for metastatic melanoma. Toxicol Appl Pharmacol. 2007;224:300–307. doi: 10.1016/j.taap.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menezes ME, et al. MDA-7/IL-24: Multifunctional cancer killing cytokine. Adv Exp Med Biol. 2014;818:127–153. doi: 10.1007/978-1-4471-6458-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher PB, et al. mda-7/IL-24, a novel cancer selective apoptosis inducing cytokine gene: From the laboratory into the clinic. Cancer Biol Ther. 2003;2(4) Suppl 1:S23–S37. [PubMed] [Google Scholar]
- 14.Fisher PB. Is mda-7/IL-24 a “magic bullet” for cancer? Cancer Res. 2005;65:10128–10138. doi: 10.1158/0008-5472.CAN-05-3127. [DOI] [PubMed] [Google Scholar]
- 15.Tong AW, et al. Intratumoral injection of INGN 241, a nonreplicating adenovector expressing the melanoma-differentiation associated gene-7 (mda-7/IL24): Biologic outcome in advanced cancer patients. Mol Ther. 2005;11:160–172. doi: 10.1016/j.ymthe.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham CC, et al. Clinical and local biological effects of an intratumoral injection of mda-7 (IL24; INGN 241) in patients with advanced carcinoma: A phase I study. Mol Ther. 2005;11:149–159. doi: 10.1016/j.ymthe.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Menezes ME, et al. MDA-7/IL-24 functions as a tumor suppressor gene in vivo in transgenic mouse models of breast cancer. Oncotarget. 2015;6:36928–36942. doi: 10.18632/oncotarget.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauane M, et al. Autocrine regulation of mda-7/IL-24 mediates cancer-specific apoptosis. Proc Natl Acad Sci USA. 2008;105:9763–9768. doi: 10.1073/pnas.0804089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su Z, et al. Unique aspects of mda-7/IL-24 antitumor bystander activity: Establishing a role for secretion of MDA-7/IL-24 protein by normal cells. Oncogene. 2005;24:7552–7566. doi: 10.1038/sj.onc.1208911. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar D, et al. Dual cancer-specific targeting strategy cures primary and distant breast carcinomas in nude mice. Proc Natl Acad Sci USA. 2005;102:14034–14039. doi: 10.1073/pnas.0506837102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradhan AK, et al. mda-7/IL-24 mediates cancer cell-specific death via regulation of miR-221 and the Beclin-1 axis. Cancer Res. 2017;77:949–959. doi: 10.1158/0008-5472.CAN-16-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta P, et al. BiP/GRP78 is an intracellular target for MDA-7/IL-24 induction of cancer-specific apoptosis. Cancer Res. 2006;66:8182–8191. doi: 10.1158/0008-5472.CAN-06-0577. [DOI] [PubMed] [Google Scholar]
- 23.Bhutia SK, et al. Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer Res. 2010;70:3667–3676. doi: 10.1158/0008-5472.CAN-09-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dash R, et al. Novel mechanism of MDA-7/IL-24 cancer-specific apoptosis through SARI induction. Cancer Res. 2014;74:563–574. doi: 10.1158/0008-5472.CAN-13-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su ZZ, et al. The cancer growth suppressor gene mda-7 selectively induces apoptosis in human breast cancer cells and inhibits tumor growth in nude mice. Proc Natl Acad Sci USA. 1998;95:14400–14405. doi: 10.1073/pnas.95.24.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradhan AK, et al. Recombinant MDA-7/IL-24 suppresses prostate cancer bone metastasis through downregulation of the Akt/Mcl-1 pathway. Mol Cancer Ther. 2018;17:1951–1960. doi: 10.1158/1535-7163.MCT-17-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebedeva IV, et al. Melanoma differentiation associated gene-7, mda-7/interleukin-24, induces apoptosis in prostate cancer cells by promoting mitochondrial dysfunction and inducing reactive oxygen species. Cancer Res. 2003;63:8138–8144. [PubMed] [Google Scholar]
- 28.Jafarnejad SM, Ardekani GS, Ghaffari M, Martinka M, Li G. Sox4-mediated Dicer expression is critical for suppression of melanoma cell invasion. Oncogene. 2013;32:2131–2139. doi: 10.1038/onc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradhan AK, Emdad L, Das SK, Sarkar D, Fisher PB. The enigma of miRNA regulation in cancer. Adv Cancer Res. 2017;135:25–52. doi: 10.1016/bs.acr.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Swahari V, Nakamura A, Deshmukh M. The paradox of dicer in cancer. Mol Cell Oncol. 2016;3:e1155006. doi: 10.1080/23723556.2016.1155006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurzynska-Kokorniak A, et al. The many faces of dicer: The complexity of the mechanisms regulating dicer gene expression and enzyme activities. Nucleic Acids Res. 2015;43:4365–4380. doi: 10.1093/nar/gkv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 34.Grelier G, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffend NC, Magner WJ, Tomasi TB. The epigenetic regulation of Dicer and microRNA biogenesis by Panobinostat. Epigenetics. 2017;12:105–112. doi: 10.1080/15592294.2016.1267886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song MS, Rossi JJ. Molecular mechanisms of dicer: Endonuclease and enzymatic activity. Biochem J. 2017;474:1603–1618. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menezes ME, et al. Role of MDA-7/IL-24 a multifunction protein in human diseases. Adv Cancer Res. 2018;138:143–182. doi: 10.1016/bs.acr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhoopathi P, et al. mda-7/IL-24 induces cell death in neuroblastoma through a novel mechanism involving AIF and ATM. Cancer Res. 2016;76:3572–3582. doi: 10.1158/0008-5472.CAN-15-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su ZZ, et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN) Proc Natl Acad Sci USA. 2008;105:20906–20911. doi: 10.1073/pnas.0807975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishikawa T, Ramesh R, Munshi A, Chada S, Meyn RE. Adenovirus-mediated mda-7 (IL24) gene therapy suppresses angiogenesis and sensitizes NSCLC xenograft tumors to radiation. Mol Ther. 2004;9:818–828. doi: 10.1016/j.ymthe.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Chada S, et al. Bystander activity of Ad-mda7: Human MDA-7 protein kills melanoma cells via an IL-20 receptor-dependent but STAT3-independent mechanism. Mol Ther. 2004;10:1085–1095. doi: 10.1016/j.ymthe.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Ploper D, De Robertis EM. The MITF family of transcription factors: Role in endolysosomal biogenesis, Wnt signaling, and oncogenesis. Pharmacol Res. 2015;99:36–43. doi: 10.1016/j.phrs.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Slade L, Pulinilkunnil T. The MiTF/TFE family of transcription factors: Master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15:1637–1643. doi: 10.1158/1541-7786.MCR-17-0320. [DOI] [PubMed] [Google Scholar]
- 47.Maruotti J, Thein T, Zack DJ, Esumi N. MITF-M, a ‘melanocyte-specific’ isoform, is expressed in the adult retinal pigment epithelium. Pigment Cell Melanoma Res. 2012;25:641–644. doi: 10.1111/j.1755-148X.2012.01033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartman ML, Czyz M. Pro-survival role of MITF in melanoma. J Invest Dermatol. 2015;135:352–358. doi: 10.1038/jid.2014.319. [DOI] [PubMed] [Google Scholar]
- 49.Ferguson J, Smith M, Zudaire I, Wellbrock C, Arozarena I. Glucose availability controls ATF4-mediated MITF suppression to drive melanoma cell growth. Oncotarget. 2017;8:32946–32959. doi: 10.18632/oncotarget.16514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim ES, et al. Mitochondrial dynamics regulate melanogenesis through proteasomal degradation of MITF via ROS-ERK activation. Pigment Cell Melanoma Res. 2014;27:1051–1062. doi: 10.1111/pcmr.12298. [DOI] [PubMed] [Google Scholar]
- 51.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yacoub A, et al. MDA-7/IL-24 plus radiation enhance survival in animals with intracranial primary human GBM tumors. Cancer Biol Ther. 2008;7:917–933. doi: 10.4161/cbt.7.6.5928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.