Significance
Machine learning methodologies can be applied readily to biological problems, but standard training and testing methods are not designed to control for evolutionary relatedness or other biological phenomena. In this article, we propose, implement, and test two methods to control for and utilize evolutionary relatedness within a predictive deep learning framework. The methods are tested and applied within the context of predicting mRNA expression levels from whole-genome DNA sequence data and are applicable across biological organisms. Potential use cases for the methods include plant and animal breeding, disease research, gene editing, and others.
Keywords: machine learning, convolutional neural networks, regulation, RNA
Abstract
Deep learning methodologies have revolutionized prediction in many fields and show potential to do the same in molecular biology and genetics. However, applying these methods in their current forms ignores evolutionary dependencies within biological systems and can result in false positives and spurious conclusions. We developed two approaches that account for evolutionary relatedness in machine learning models: (i) gene-family–guided splitting and (ii) ortholog contrasts. The first approach accounts for evolution by constraining model training and testing sets to include different gene families. The second approach uses evolutionarily informed comparisons between orthologous genes to both control for and leverage evolutionary divergence during the training process. The two approaches were explored and validated within the context of mRNA expression level prediction and have the area under the ROC curve (auROC) values ranging from 0.75 to 0.94. Model weight inspections showed biologically interpretable patterns, resulting in the hypothesis that the 3′ UTR is more important for fine-tuning mRNA abundance levels while the 5′ UTR is more important for large-scale changes.
Machine and deep learning approaches such as Convolutional Neural Networks (CNNs) are largely responsible for a recent paradigm shift in image and natural language processing. These approaches are among the fundamental enablers of modern artificial intelligence advances such as facial recognition, speech recognition, and self-driving vehicles. The same deep learning approaches are beginning to be applied to molecular biology, genetics, agriculture, and medicine (1–7), but evolutionary relationships make properly training and testing models in biology much more challenging than the image or text classification problems mentioned above.
For example, if one wants to predict mRNA levels from DNA promoter regions (as we do here), the standard approach from image recognition problems would be to randomly split genes into training and testing sets (8). However, such a split will likely lead to dependencies between the sets because of shared evolutionary histories between genes (i.e., gene family relatedness, gene duplications, etc.) and may cause model overfitting and false-positive spurious conclusions. Models trained without properly accounting for the constraints imposed by evolutionary history (and perhaps other biological and technical factors specific to the modeling scenario) will likely memorize both the neutral and the functional evolutionary history, rather than learning only the functional elements, leading researchers to incorrect conclusions.
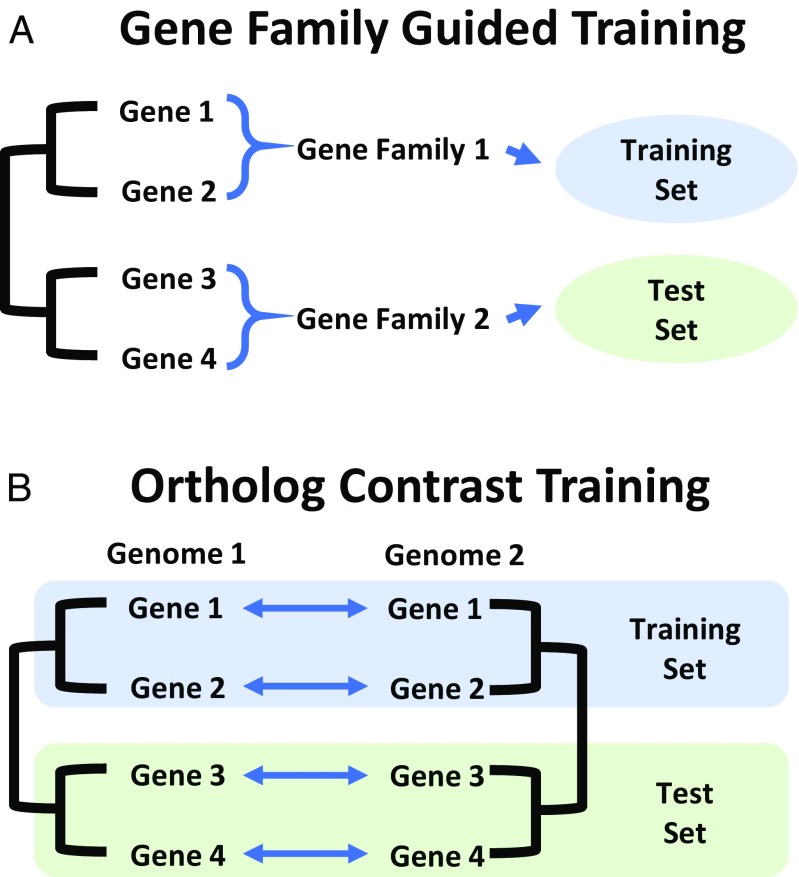
With these challenges in mind, we developed two CNN architectures for predicting mRNA expression levels from DNA promoter and/or terminator regions. These include models that predict the following: (i) if a given gene is highly or lowly expressed and (ii) which of two compared gene orthologs has higher mRNA abundance. The architectures are built around two methods developed here for properly structuring the model training and testing process to avoid the issues of training-set contamination by evolutionary relatedness. The first training method, which we call “gene-family guided splitting,” uses gene-family relationships to ensure that genes within the same family are not split between the training and testing sets. In this way, the model never sees a gene family in the testing set that it has already seen during the training process (Fig. 1A). The second training method uses what we call “ortholog contrasts” (comparisons between pairs of orthologs) to eliminate evolutionary dependencies (Fig. 1B). In addition to controlling for evolutionary relatedness, this method actually allows evolution to become an asset in the training process by leveraging whole-genome duplication events and/or genetic differences between species, two things that would normally be a hindrance to such models. Using evolutionary relatedness is powerful because it allows one to understand and train on what has survived selection. Considering deeper evolutionary divergence, between species rather than just within species, allows for sampling thousands of years of mutagenesis and selective pressures.
Fig. 1.
Evolutionarily informed strategies for deep learning. (A) For prediction tasks involving a single species, genes are grouped into gene families before being further divided into a training and a test set to prevent deep learning models from learning family-specific sequence features that are associated with target variables. (B) For prediction tasks involving two species, orthologs are paired before being divided into a training and a test set to eliminate evolutionary dependencies.
Results
Differentiating Between Expressed and Unexpressed Genes Based on DNA Sequence.
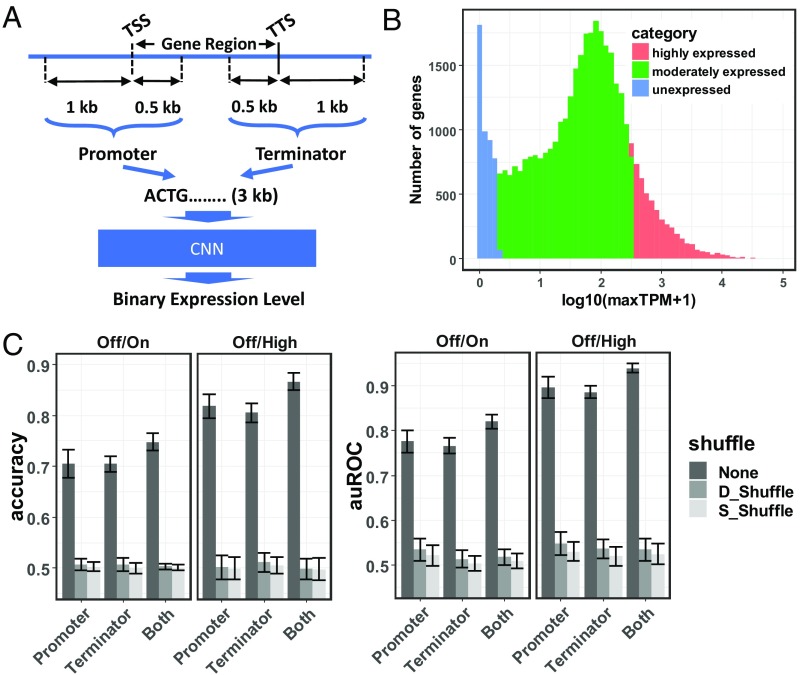
The first model was developed for the purpose of classifying genes as being expressed or not expressed (zero or near-zero expression level). This model has been named the “pseudogene model” because of its ability to predict genes that are potentially pseudogenized and therefore lack expression. The pseudogene model also serves as a simple use case for the gene-family–guided splitting approach. It requires as input the promoter and/or terminator sequences (defined in Materials and Methods and illustrated in Fig. 2A). To generate the output of the model (i.e., a binary value representing whether a gene is expressed or unexpressed), a comprehensive atlas of gene expression in maize covering major tissues at various developmental stages was generated by applying a unified pipeline on 422 tissues from seven RNA-Seq studies (9–15) (for full details, see Materials and Methods and Datasets S1 and S2). The distribution of the maximum log-transformed Transcripts Per Million (logTPM) revealed a peak at the lower tail comprising unexpressed genes (4,562 genes with maximum logTPM ≤ 1) along with normally distributed expressed genes (34,907 genes with maximum logTPM > 1) (Fig. 1B).
Fig. 2.
The architecture and performance of the pseudogene model. (A) A schematic representation of the architecture of the pseudogene model. The model takes promoter and/or terminator sequences as the predictor to predict binary expression levels. (B) A unified RNA-Seq data analysis pipeline is applied on 422 samples from seven references (9–15) representing a comprehensive collection of maize tissues at diverse developmental stages. The log-transformed maximum TPM over all samples is calculated for each gene and used to represent the strength of the corresponding predictor sequence. Shown is the distribution of log-transformed maximum TPMs for all maize genes. Genes are categorized into unexpressed genes (blue), moderately expressed genes (green), and highly expressed genes (red). (C) The accuracy and auROC of the pseudogene model trained on the Off/On gene set and the Off/High gene set, using promoters, terminators, or both promoter and terminator sequences as predictors. Models evaluated on test sets are either not shuffled (None) or shuffled while maintaining their di- or single-nucleotide composition (denoted as D_Shuffle and S_Shuffle, respectively). Error bars represent mean ± SD from gene-family–guided 10 times fivefold cross-validation.
As paralogous genes derived from more recent gene duplication events often share highly similar promoters or terminators, overfitting may potentially occur when highly similar paralogs are separated into training and testing sets. Moreover, as paralogs are often similar in their expression levels, separation of highly similar paralogs may force neural networks to learn gene-family–specific sequence features, rather than sequence features that determine expression levels per se. To solve these problems, genes were divided into gene families. The pseudogene model was trained on randomly selected families and tested on the remaining families not present in the training set (Dataset S3).
The number of expressed genes (34,907) and unexpressed genes (4,562) were highly imbalanced. Two approaches were used to handle the imbalance. First, expressed genes were divided into 4,562 highly expressed (maximum TPM ≥ 342.9) genes and 25,783 intermediately expressed genes (1 < maximum TPM < 342.9), and the model was trained to distinguish the 4,562 unexpressed genes from the 4,562 highly expressed genes (Off/High in Fig. 2C). The performance of the pseudogene model was evaluated using a 10 times fivefold cross-validation procedure, and the Off/High version achieved an average predictive accuracy of 86.6% (the area under the ROC curve, auROC = 0.94) when both promoters and terminators were used as the predictor. The average accuracy of the model reached 81.6% (auROC = 0.89) and 80.6% (auROC = 0.89) for promoters and terminators, respectively (Fig. 2C). Second, all expressed genes were randomly down-sampled to make them balanced with unexpressed genes. Using this approach, the model achieved an average predictive accuracy of 74.8% (auROC = 0.82), 70.1% (auROC = 0.77), and 70.6% (auROC = 0.77) for both promoters and terminators, promoters only, and terminators only, respectively (Off/On in Fig. 2C). The models learned higher-level features than single- or dinucleotide composition, since shuffling test set sequences while maintaining single- or dinucleotide composition abolished the predictive accuracy of these models (Fig. 2C) (16).
Random allocation of genes to training/test sets without considering their evolutionary relatedness led to significantly higher performance (in terms of auROC and accuracy) of our models on test-set genes than those obtained by family-guided training/test splitting (SI Appendix, Fig. S1). We further categorized the test-set genes into two groups: genes with homologs in the training set and genes without homologs in the training set, and the performance of our models was evaluated on the two groups separately. Interestingly, our models perform significantly more poorly on the latter group than the former group (SI Appendix, Fig. S1). Taken together, these results indicate that the evolutionary relatedness between training and test sets, if left uncontrolled, leads to overfitting on gene families present in both training and test sets.
Predicting Which of Two Genes Is More Highly Expressed Using Ortholog Contrasts.
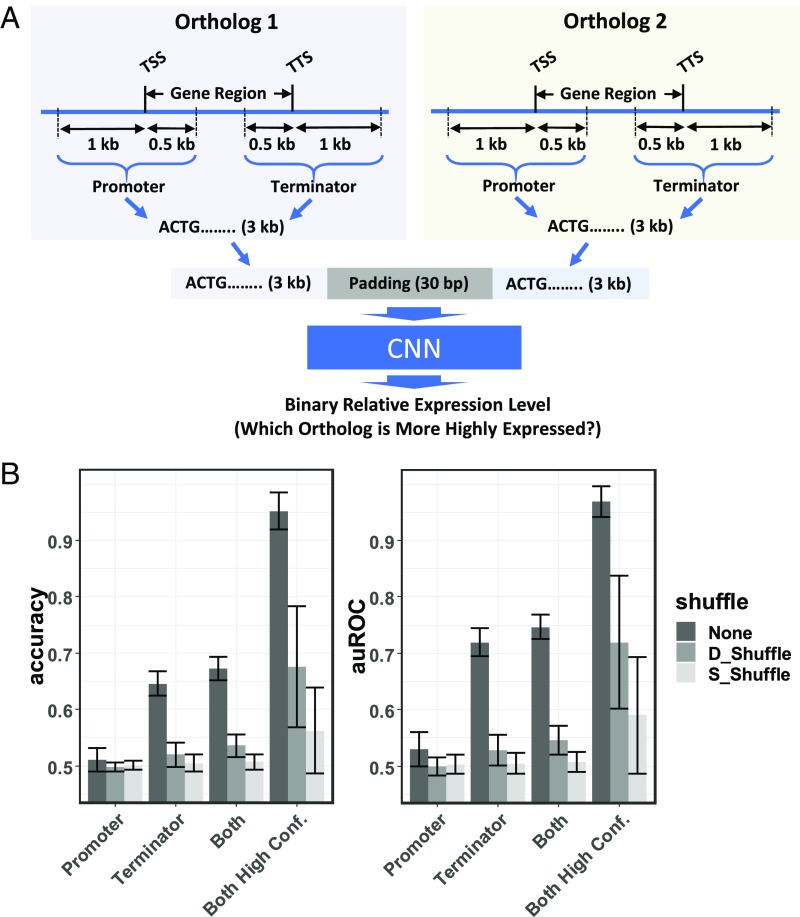
The ortholog contrast model follows a simple approach derived from phylogenetics, where the most recent common ancestor of two closely related genes can be represented as a contrast between the two (16, 17). Contrasting genes in this manner directly accounts for statistical dependencies between the genes that would otherwise hamper comparison with other genes (18). Building on this idea, the ortholog contrast method compares two genes from different genomes (or alleles from the same species) to each other and predicts the difference between the expression levels of the two (Fig. 3A). When each gene is compared directly to its ortholog, one can then compare that contrast value to the contrast values from other ortholog pairs without evolutionary dependence between them, hence enabling training and testing sets that are evolutionarily independent (Figs. 1B and 3A). To further simplify the contrast model, the values (the difference between the transcript abundance levels of the two compared genes) were converted to binary form: zero if the first gene is more highly expressed than the second, and 1 in the opposite case. Orthologs with no expression difference between them were excluded. This simplification results in a model where the CNN is asked to determine which of two orthologs is most highly expressed. In reality, this question of deciding between two genes or alleles is actually what is most needed in applications like plant breeding and medicine.
Fig. 3.
The architecture and performance of the ortholog contrast model. (A) A schematic representation of the architecture of the contrast model. The model takes promoter and/or terminator sequences from two orthologous genes as the predictor and predicts the binary difference in expression level between the two. (B) The accuracy and auROC of the ortholog contrast model trained using promoters, terminators, or both promoter and terminator sequences as predictors. Sequences in the test sets are either not shuffled (None) or shuffled while maintaining their di- or single-nucleotide composition (denoted as D_Shuffle and S_Shuffle, respectively). Error bars represent mean ± SD from 10 times fivefold cross-validation. The “Both High Conf.” bars represent the performance of the models when genes for which the model has less than 0.8 confidence are dropped.
However, there are a few challenges to applying this model to prediction. First, orthologs have highly correlated expression levels. Expression levels of the Sorghum bicolor by Zea mays orthologs used here had a Pearson correlation coefficient of 0.78 (P value < 0.001). This means that many of the genes that the model is predicting have very similar (perhaps functionally identical) expression levels and are simply noise in the data. Removing some of these similarly expressed orthologs from the data before training results in higher prediction accuracies, but it also lowers the number of ortholog pairs in the training set. This results in the second problem: low sample size. The nature of CNN models requires them to include many parameters (thousands generally), making high sample sizes of great importance to model training. In this case, the number of S. bicolor by Z. mays single-copy syntenic orthologs for the model is ∼13,000, leaving little room for filtering out ortholog pairs with similar expression levels.
To overcome these two challenges, a multistep strategy was adopted. To begin with, the S. bicolor by Z. mays ortholog training set was filtered to include only ortholog pairs with expression-level differences between them of 2,000 rank changes or more (∼0.54 logtwofold change; see Materials and Methods for explanation of rank change methodology). Higher-rank change filters resulted in untrainable models, presumably due to low sample size. Models trained with the 2,000 rank-change filtered set were able to predict never-before-seen pairs with an average auROC value of 0.75 across all fivefold 10-replicate testing sets (Fig. 3B). This moderate improvement over random guessing may have important utility in breeding, medical applications, and for hypothesis generation, but it is still based on many genes that are similarly expressed adding extra noise to the data.
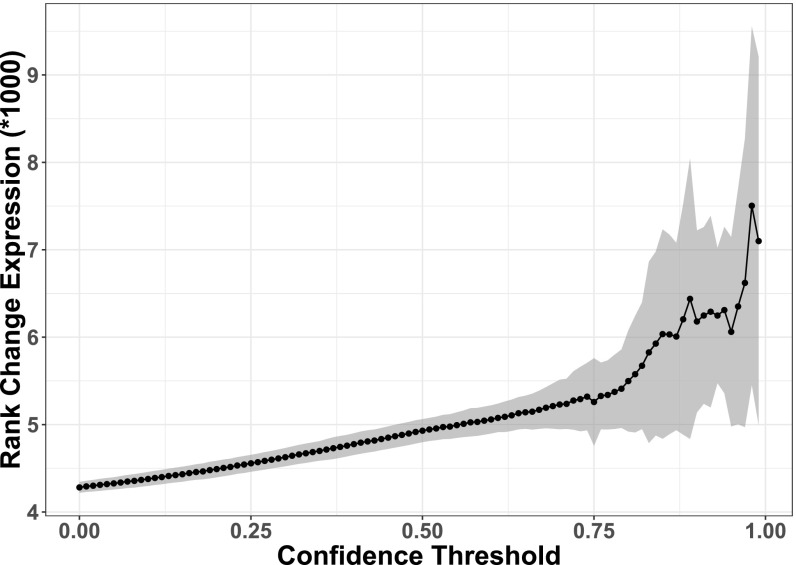
To further investigate the model’s potential to predict ortholog pairs with greater expression differences between them, a second filtering approach was applied. When the model is tested on never-before-seen ortholog pairs (test set), it outputs not only a binary prediction of which ortholog is most highly expressed, but also a value indicating how much confidence one should place in the model’s prediction of that ortholog pair. By setting a threshold confidence value, one can discard predictions for which the model is less confident and focus on only those pairs the confidence values of which are within a range deemed acceptable by the user (19). The entire range of threshold confidence values (0–1) was examined along with the average rank changes in expression levels (Fig. 4) and average auROC scores (SI Appendix, Fig. S2) that these threshold values produced. There is a clear and statistically significant (r = 0.92 with P value < 0.001) correlation between higher confidence threshold values and higher differences in expression between the two orthologs in the pair (rank-change expression values). The graph (Fig. 4) also has an inflection point at a confidence threshold value of ∼0.8. After this point, even small changes in the confidence threshold result in gene sets with much larger expression differences between the orthologs. This 0.8-threshold value was taken as a reasonable cutoff at which the testing set is composed of ortholog pairs that are different enough in expression level to be predicted accurately. Applying this threshold to the model resulted in an average auROC value of 0.97 across fivefold 10-replicate cross-validation (Fig. 3B, “Both High Conf.”). Similar approaches for focusing on values in which one is most confident have been commonly applied to other prediction problems (19, 20). Interestingly, while there is no indication that dinucleotide content contributes to the pseudogene model’s accuracy, it does appear to play a role in the contrast model’s predictions for at least some of the high-confidence ortholog pairs (Fig. 3B).
Fig. 4.
Model confidence threshold by mean expression rank change between ortholog pairs. A range of model confidence thresholds (model confidence value below which the ortholog pair is excluded from the test set) plotted against the average rank change in expression within each filtered test set across all 10-replicate fivefold validation sets. Pearson correlation coefficient between the values is r = 0.92 with a P value < 0.001.
The ortholog contrast model was also trained and tested to predict between the two Z. mays subgenomes. From the outset, this was assumed to be a more challenging task as the number of usable single-copy syntenic pairs between the two subgenomes is just above 4,000. This means that no training-set filtering could be done to limit the number of similarly expressed pairs. Without any filtering, this model performed with an average auROC value of 0.63. Using the same 0.8 confidence threshold as described above, the model achieved an average auROC score of 0.77. While these low prediction values are likely due in part to the low sample size and the filtering limitation that it imposes, neofunctionalization within the maize subgenomes is also probably an important factor. The Pearson correlation coefficient for expression levels between the two Z. mays subgenomes was 0.59 (compared with 0.78 for between S. bicolor and Z. mays). This, along with multiple studies, indicates that many of the genes within the Z. mays subgenomes are neofunctionalized or are undergoing neofunctionalization (21, 22). Many neofunctionalization-related expression changes are likely tissue-specific and due to distant enhancer elements or transacting factors that can be far outside the 3-kb regions used here for prediction (23). Taken together, the Z. mays subgenomes contrast model probably performs poorly because many important regulatory elements are not captured by the model. Because the S. bicolor by Z. mays model uses only single-copy syntenic orthologs, most of its genes are unlikely to have undergone neofunctionalization.
Interpretation of CNN Models Reveals Elements and Motifs Important for Transcript Abundance.
Transcript abundance is concertedly determined by its synthesis and degradation. Our current models cannot discriminate between these causes but could potentially do so in the future by training on Global Run-On followed by high-throughput sequencing of RNA (GRO-seq) (24), Precision Run-On followed by sequencing (PRO-seq) (25), or transcriptome-wide mRNA half-life data (26, 27).
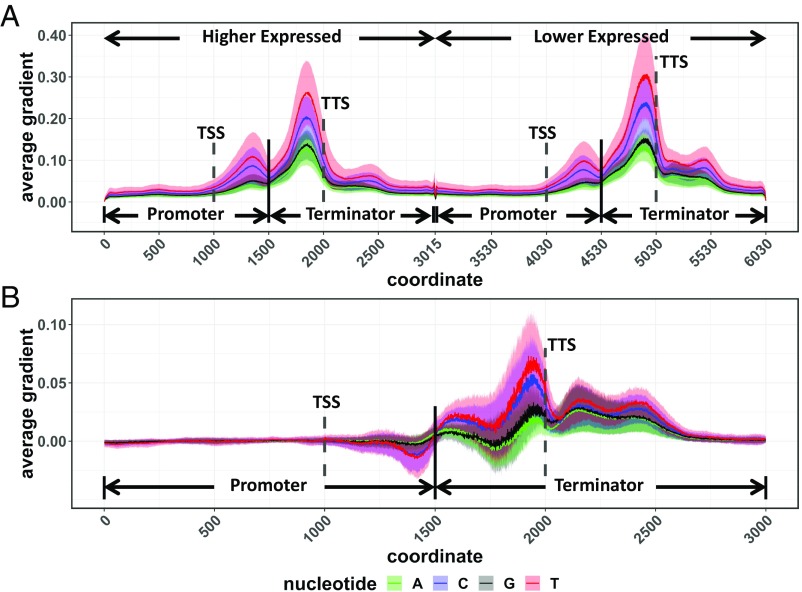
It has long been established that genomic sequences flanking coding sequences harbor important cis elements that determine the transcription rate and/or the stability of transcripts (28, 29). Many methods have been developed for interpreting and understanding CNNs (2, 30–32), but significant challenges and the possibility of misinterpretation remain (33–35). To identify motifs/putative cis elements, two gradient-based methods [Saliency and DeepLIFT (Deep Learning Important Features)] and one perturbation-based method (Occlusion) (31, 36) were applied to each of the models (Figs. 5 and 6 and SI Appendix, Figs. S3 and S4). The maps are based on the average values for all included genes across all 10 times fivefold cross-validations. Interestingly, the different training methods (gene-family–guided vs. orthologs contrasts) resulted in very different but potentially complementary results as to which regions of the sequence were most important for the prediction task.
Fig. 5.
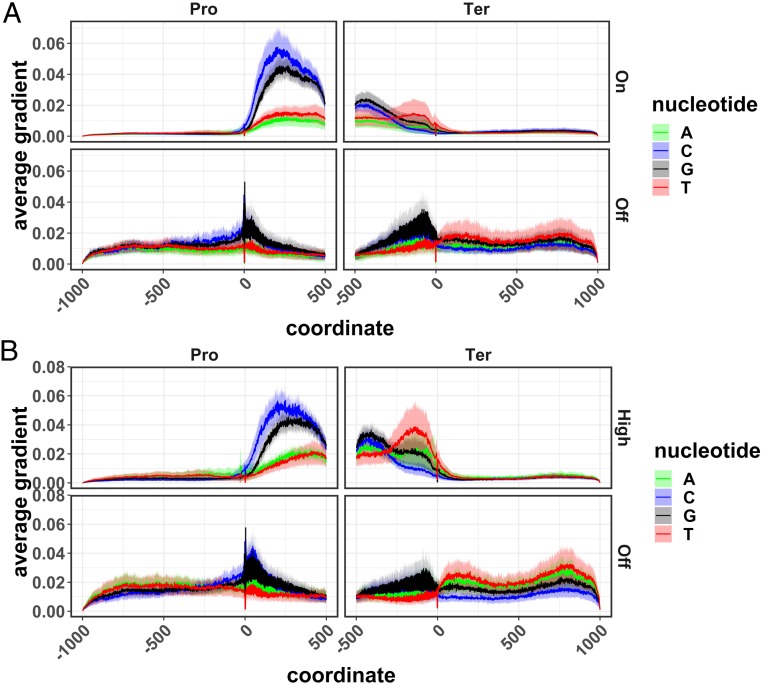
Averaged saliency map from the pseudogene model. Saliency map was calculated for the pseudogene model trained on either the Off/On gene set (A) or the Off/High gene set (B). Saliency was averaged over nonpseudogenes (Upper) and pseudogenes (Lower), respectively. Only genes with correctly predicted expression levels were used for the calculation of saliency maps. This figure is based on the average values over 10 times fivefold cross-validation, with solid lines representing the mean and shaded areas representing the SD.
Fig. 6.
Averaged saliency maps from the ortholog contrast model. Saliency maps were calculated for the ortholog contrast model. (A) True positive with ortholog 1 more highly expressed than ortholog 2. (B) True positive lower expressed orthologs minus true positive higher expressed orthologs. This figure is based on the average values from all genes over 10 times fivefold cross-validation, with solid lines representing the mean and shaded areas representing the SD.
In all tested models, the 5′ and 3′ UTR regions of what we call promoter and terminator sequences were much more important than the regions just outside of the gene models (Figs. 5 and 6). Pseudogene models trained on the off/on data set (Fig. 5A and SI Appendix, Fig. S3) and off/high data set (Fig. 5B and SI Appendix, Fig. S3) resulted in similar saliency maps. In both cases, nonpseudogenes showed stronger signals in the promoter regions than in the terminator regions. This is in agreement with the accuracy values for the models shown above (Fig. 2) where predictions based on the promoter sequences alone outperform those based on the terminator sequences alone. These results make sense based on what is currently known about cis regions that are important to gene expression (37).
For models trained using the ortholog contrast method, the results are somewhat different. In this case, the most heavily weighted areas of the input sequences were found in the terminator region (Fig. 6A and SI Appendix, Fig. S4). This is consistent whether the gene in the first position is more highly expressed than the gene in the second position or vice versa. The greater importance of terminator regions is further shown by the fact that models run with only the terminator sequence perform better than those with only promoter sequences (Fig. 3). The differences between expression values of the compared genes in the contrast model ranged from Log10-transformed TPM values of 0.12–2.80 with an average of 0.66. Log10-transformed TPM values for the off/high gene set in the pseudogene model ranged from 0 to 0.301 (with an average of 0.101) for the “off” gene set and ranged from 2.536 to 4.925 (with an average of 2.959) for the “high” gene set.
While pseudogenes (not expressed) in the family-guided approach showed very little signal in the promoter and terminator regions, the lower expressed genes in the contrast model actually showed very strong signals in these regions (at least for the saliency and occlusion methods). In the case of the terminator region, lower expressed genes showed higher values than highly expressed genes (Fig. 6B) for the saliency method, while the promoter regions showed the opposite trend with higher saliency, occlusion, and DeepLIFT values in the more highly expressed gene. All three feature-importance methods indicate that the terminator region is more important than the promoter region in the contrast model, giving us high confidence in this conclusion. Conversely, one of the three methods (saliency) shows higher values for the lower expressed gene, although these values are within one SD of the mean. The other two methods show similar values for both high and low (occlusion) or the opposite trend with higher expressed genes having higher values and lower expressed genes having very low values (DeepLIFT). This result is inconsistent across the three methods, making it harder to interpret with confidence.
Discussion
3′ UTR Potentially More Important for Small-Scale Changes in RNA Abundance.
There are a number of factors that might explain the differences in promoter and terminator importance between the pseudogene and ortholog contrast models. First, the method of controlling for evolutionary relatedness differs between the models. The grouping of genes into families for the gene-family–guided method relies on sequence similarity scores with subjectively defined cutoffs. This method is therefore potentially over- or under-controlling (or both in the case of different genes) for evolutionary relationships. The ortholog contrast model, on the other hand, fully controls for these relationships. Second, the pseudogene model is restricted to a single species while the contrast was applied both within species (although between subgenomes) and across species. Finally, and perhaps most likely, is that the two models are focused on different categories of gene expression and genes that have experienced different types of evolutionary constraint. The pseudogene model includes a bimodal distribution of genes that are expressed (highly or moderately) and genes that are not expressed, while the contrast model mostly contains genes that are expressed at some level (that likely does not include many pseudogenes). Given that small mutations in the promoter region could conceivably inhibit protein:DNA interactions and be responsible for drastic changes in gene expression (such as turning expression on or off), perhaps these types of mutations are responsible for the high predictive importance of promoter regions in the pseudogene model. The contrast model, on the other hand, was limited to genes with matching single-copy syntenic orthologs between distinct genomes to ensure reliable normalization and a balanced dataset for training and testing. These genes will be highly conserved (by definition) and likely under strong purifying selection to have remained conserved since the maize/sorghum split, meaning that large-scale expression changes are unlikely to be tolerated. Small-scale, fine-tuning adjustments of expression level, on the other hand, are more likely to be present in these highly conserved genes. We therefore hypothesize that the terminator region (particularly the 3′ UTR) plays a more important role in small-scale fine-tuning of RNA abundance levels than does the promoter (particularly the 5′ UTR) region. While the promoter and 5′ UTR regions are often thought of as important to transcriptional regulation, it has been known for some time that the 3′ UTR also plays an important role (38, 39). The 3′ UTR has been shown to regulate transcription via diverse mechanisms such as alternative polyadenylation, riboswitching, Nonsense-mediated decay, and alternative splicing (37). Which of these or other possible mechanisms may be at play here is not obvious, but the results presented here strongly implicate the 3′-UTR region in determining expression differences between syntenic orthologs across genomes.
Strengths and Weaknesses of Different Models and Training Approaches.
We have demonstrated the utility of two different approaches for mRNA expression prediction. Both approaches incorporate methods for dealing with evolutionary relatedness between genes within a predictive framework. Each approach has strengths and weaknesses, and which approach one choses will depend on the datasets available and the types of predictions/biological insights desired. For datasets or questions that are limited to a single sample from a single diploid species, the gene-family–guided approach is the most suitable option because ortholog contrasts are not possible. This method may also be most appropriate in situations where one wants to specifically identify genes that are unlikely to be expressed. The gene-family–guided method also has the benefit of being somewhat simpler to understand and interpret; however, one must define gene families, and that process is subjective and sensitive to parameter choices. In situations where multiple species or genotypes are involved, the contrast method is likely to be the method of choice. The contrast method is also the better choice when one wishes to compare orthologs/alleles that are both expressed at a moderate to high level.
Conclusions and Future Applications.
In the current study, CNN models have been successfully applied to the prediction of mRNA abundance under several distinct scenarios. The models are able to predict a gene’s On/Off state as well as which of two compared orthologs is more highly expressed. These models feature two very different approaches for handling evolutionary relationships between genes, gene-family–guided splitting, and ortholog contrasts. While not demonstrated here, the contrast model in theory should be extendable to determining which of two alleles in a population is more highly expressed, an application with clear utility in breeding and medicine.
In the future, it is hoped that gene-family–guided splitting, ortholog contrasts, and other potential strategies will be applied to deep learning models in various areas of biology. With larger datasets, the current models could also be built upon and further interpreted through partitioning genes by functional classes or predicting temporal-spatial expression patterns. The contrast model, in particular, could likely have increased performance if more than two genomes (or subgenomes) could be included in one model. Such a development would require novel strategies to properly control for dataset imbalances. Fundamentally, these models suffer from having more parameters than samples. Current technologies are making it possible to generate data from hundreds to thousands of individuals with many tissue types for each. As these large-scale datasets become available, models such as the ones developed here will become increasingly accurate and useful. The potential of deep learning to increase our understanding of and prediction within biological systems is enormous. Further creative strategies for focusing these methods on biologically relevant information and controlling for confounding biological factors will be critical to the success of deep learning in biology.
Materials and Methods
DNA Sequence Encoding and RNA-Seq Data Collection and Processing.
The Z. mays B73 reference genome (40, 41) was downloaded from Ensembl Plant Release 31 (plants.ensembl.org). The newest B73 reference genome, AGPv4 (42), was not used in this study due to known issues with the 3′-UTR annotations. Version 3.1.1 of the S. bicolor genome (43) was downloaded from Phytozome (https://phytozome.jgi.doe.gov). The transcription start site (TSS) and transcription termination site (TTS) are not explicitly annotated in these genomes, so the TSS was taken to be the start coordinate of the gene and the TTS the end coordinate of the gene. DNA sequences were transformed into one hot form using custom scripts (see Bitbucket repository, ref. 16).
To train models predicting unexpressed genes, 422 samples (from a total of 452) from seven references (9–15) representing a comprehensive collection of maize tissues at diverse developmental stages were used. The samples were downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), quality-trimmed, and checked using Sickle (version 1.33, https://github.com/najoshi/sickle) and FastQC (version 0.11.5, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The cleaned reads were aligned to the maize genome with HISAT2 (version 2.1.0) (44) and read counts were normalized to TPM by Stringtie (version 1.3.3) (45). Among the 452 samples, 26 with fewer than 5 million reads and four with a less than 50% alignment rate were excluded from downstream analysis, leaving 422 samples. Samples are summarized in Dataset S1, and gene expression levels are listed in Dataset S2.
Some gene models in the V3 annotation do not contain 5′ or 3′ UTRs, leaving discernible start and stop codons at TSS and TTS. To circumvent the model learning from these simple sequence features, the first three nucleotides downstream of the TSS and the last three nucleotides upstream of the TTS were masked. In both off/high and off/on comparisons, using longer promoters (from −2,500 to +500 bp with respect to the TSS) or terminators (from −500 to +2,500 bp with respect to TTS) did not improve predictive accuracy.
The Z. mays samples used for testing the ortholog contrast model come from the B73 Shoot data published by Kremling et al. (46). The corresponding S. bicolor data were generated and processed as described in that paper and can be found on the NCBI SRA website under project number PRJNA503076 (17). Both Z. mays and S. bicolor data were additionally processed together using the DESeq2 fragments per million normalization. All scripts associated with the analyses were deposited on BitBucket (https://bitbucket.org/bucklerlab/p_strength_prediction/) (16).
Categorizing Maize Genes into Gene Families and Family-Guided Splitting of Training and Testing Data Sets.
Z. mays genes were divided into gene families using a previously described pipeline (47) with modifications. An all-by-all BLAST was conducted on maize proteome sequences to evaluate pairwise similarity between maize proteins. As one gene may encode multiple protein isoforms, the result of the BLAST search was collapsed to gene-level similarity by an in-house R script. Then, an in-house python script was used to build a graph with nodes representing genes and edges connecting paralogous genes. This graph was further divided into clusters (i.e., gene families) by the Markov Clustering Algorithm implemented in the markov_clustering package in Python with default parameters except that inflation was set to 1.1. If a gene was not assigned to any gene family, it was considered as a family that contains only a single member. Each gene family was assigned an index (Dataset S3). For family-guided cross-validation, gene families were randomly partitioned into five subsamples with equal numbers of families. In each iteration, one subsample was retained as the test data, while the remaining four subsamples were used as training data (Dataset S3).
Model Architecture.
All models presented here use the same architecture. This architecture was originally determined for the pseudogene model based on a grid search. Multiple architectures were also tested for the ortholog contrast model (including the final pseudogene architecture). As the pseudogene architecture performed similarly to other tested architectures, it was determined to use it for both models for the sake of simplicity. All models were constructed in Python 2 using Keras 2 with a Tensorflow back end. The final architecture consisted of three groups of two convolutional layers, with each group of layers followed by a maximum pooling and a dropout layer, followed by two fully connected layers, each followed by a dropout layer, and a final prediction layer (see model code in Bitbucket repository for more details, ref. 16). A “relu” activation function was used for each layer in the model (except for the final prediction layer, which used a softmax or sigmoid activation function depending on the model).
To pick the final values of the 11 hyperparameters in the model, all hyperparameter combinations (1,344 combinations in total) were evaluated on the pseudogenes vs. expressed genes dataset using fivefold cross-validation (results are summarized in Dataset S4). Among them, most combinations achieved accuracies around 75%. Tukey’s Honestly Significant Difference test indicated that 1,294 of the 1,344 combinations are not significantly different (adjusted P value > 0.05) from the best-performing combination (with an accuracy of 77.7%). The Student’s t test indicated that 600 of the 1,344 combinations are not significantly different from the best-performing combination (false discovery rate adjusted P value > 0.05). Therefore, a single combination was randomly chosen from these 600 combinations (with an accuracy of 76.4%).
Syntenic Ortholog Contrasts.
Syntenic orthologs between the Z. mays and S. bicolor reference genomes were obtained from a previous publication (14). The training, validation, and testing sets were also divided by gene family in the same way as described for the pseudogene model above. To feed two genes at a time to the model, the gene sequences were first converted into one hot form. Each base pair, and a missing base-pair character, were included in the encoding. A column of all zeros was used as an additional class specific to padding characters. The two ortholog sequences being compared were then concatenated together with a block of all zero columns equivalent to 30 bp in between them. Different lengths of padding between the two sequences were tried, but 30 bp worked well and was used for all reported analyses. To control for the possibility of the network learning gene order (i.e., the first gene is always more highly expressed than the second), all gene pairs where fed to the model in both possible orders.
Transcript abundance data used in the contrast model included only one tissue type (shoot tissue) based on at least two replicates (distinct libraries created from pooled plants) for both maize and sorghum as described in ref. 46. Fragment Per Million values were log2-scaled and then normalized by percentage rank based only on genes with single-copy syntenic orthologs in both genomes. In cases where multiple transcripts had the same expression values, the average rank value was assigned to all. Orthologs were then paired and filtered to exclude pairs with a less than 2,000 rank expression difference between them (∼0.12 log10 TPM) and down-sampled to ensure an equal number of sorghum and maize winners. This resulted in 3,094 ortholog pairs for use in model training, validation, and testing.
Supplementary Material
Acknowledgments
We thank James Schnable for maize and sorghum ortholog lists. This material is based upon work supported by the NSF Postdoctoral Research Fellowship in Biology under Grant 1710618 (to J.D.W.); the NSF Plant Genome Research Program under Grant 1238014 (to E.S.B.); and the Tang Cornell-China Scholars Program (H.W.). Additional support comes from the United States Department of Agriculture, Agricultural Research Service.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive database (accession no. PRJNA503076) and the Bitbucket repository (https://bitbucket.org/bucklerlab/p_strength_prediction/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814551116/-/DCSupplemental.
References
- 1.Quang D, Xie X. 2017. FactorNet: A deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. bioRxiv, 10.1101/151274.
- 2.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 3.Ching T, et al. 2018. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 15:pii: 20170387.
- 4.Demirci S, Peters SA, de Ridder D, van Dijk ADJ. DNA sequence and shape are predictive for meiotic crossovers throughout the plant kingdom. Plant J. 2018 doi: 10.1111/tpj.13979. [DOI] [PubMed] [Google Scholar]
- 5.Webb S. Deep learning for biology. Nature. 2018;554:555–557. doi: 10.1038/d41586-018-02174-z. [DOI] [PubMed] [Google Scholar]
- 6.Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: A review of computational problems and data sets. Proc IEEE. 2016;104:176–197. [Google Scholar]
- 7.Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat Biotechnol. 2018;36:829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Li Y, Narayan R, Subramanian A, Xie X. Gene expression inference with deep learning. Bioinformatics. 2016;32:1832–1839. doi: 10.1093/bioinformatics/btw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 10.Davidson RM, et al. Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome J. 2011;4:191–203. [Google Scholar]
- 11.Chettoor AM, et al. Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 2014;15:414. doi: 10.1186/s13059-014-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelpflug SC, et al. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome. 2016 doi: 10.3835/plantgenome2015.04.0025. [DOI] [PubMed] [Google Scholar]
- 13.Bolduc N, et al. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012;26:1685–1690. doi: 10.1101/gad.193433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell. 2017;29:1938–1951. doi: 10.1105/tpc.17.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston R, et al. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell. 2014;26:4718–4732. doi: 10.1105/tpc.114.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn JD, Wang H. 2019 Data from “P_strength_prediction.” Bitbucket. Available at https://bitbucket.org/bucklerlab/p_strength_prediction/. Deposited July 6, 2018.
- 17.Washburn JD, Kremling KA, Valluru R, Buckler ES, Wang H. 2019 doi: 10.1073/pnas.1814551116. Evolutionarily informed deep learning methods: Predicting relative transcript abundance from DNA sequence. National Center for Biotechnology Information: Sequence Read Archive. Available at www.ncbi.nlm.nih.gov/bioproject/PRJNA503076. Deposited October 30, 2018. [DOI] [PMC free article] [PubMed]
- 18.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 19.Ketkar N. Deep Learning with Python: A Hands-On Introduction. Apress; New York: 2017. [Google Scholar]
- 20.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes TE, Langdale JA, Kelly S. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 2014;24:1348–1355. doi: 10.1101/gr.172684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnable JC, Freeling M. Polyploidy and Genome Evolution. Springer; Berlin: 2012. Maize (Zea mays) as a model for studying the impact of gene and regulatory sequence loss following whole-genome duplication; pp. 137–145. [Google Scholar]
- 23.Lu Z, Ricci WA, Schmitz RJ, Zhang X. Identification of cis-regulatory elements by chromatin structure. Curr Opin Plant Biol. 2018;42:90–94. doi: 10.1016/j.pbi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lugowski A, Nicholson B, Rissland OS. DRUID: A pipeline for transcriptome-wide measurements of mRNA stability. RNA. 2018;24:623–632. doi: 10.1261/rna.062877.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lugowski A, Nicholson B, Rissland OS. Determining mRNA half-lives on a transcriptome-wide scale. Methods. 2018;137:90–98. doi: 10.1016/j.ymeth.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 29.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrikumar A, Greenside P, Kundaje A. Learning important features through propagating activation differences. In: Precup D, Teh YW, editors. Proceedings of the 34th International Conference on Machine Learning, Proceedings of Machine Learning Research. PMLR, International Convention Centre; Sydney: 2017. pp. 3145–3153. [Google Scholar]
- 32.Zintgraf LM, Cohen TS, Adel T, Welling M. 2017. Visualizing deep neural network decisions: Prediction difference analysis. arXiv, 1702.04595.
- 33.Doshi-Velez F, Kim B. 2017. Towards a rigorous science of interpretable machine learning. arXiv, 1702.08608.
- 34.Choromanska A, Henaff M, Mathieu M, Ben Arous G, LeCun Y. 2014. The loss surfaces of multilayer networks. arXiv, 1412.0233.
- 35.Dinh L, Pascanu R, Bengio S, Bengio Y. Sharp minima can generalize for deep nets. In: Precup D, Teh YW, editors. Proceedings of the 34th International Conference on Machine Learning, Proceedings of Machine Learning Research. PMLR, International Convention Centre; Sydney: 2017. pp. 1019–1028. [Google Scholar]
- 36.Simonyan K, Vedaldi A, Zisserman A. 2013. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv, 1312.6034.
- 37.Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018;23:248–259. doi: 10.1016/j.tplants.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Hunt AG. Messenger RNA 3′ end formation in plants. Curr Top Microbiol Immunol. 2008;326:151–177. doi: 10.1007/978-3-540-76776-3_9. [DOI] [PubMed] [Google Scholar]
- 40.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 41.Wei F, et al. The physical and genetic framework of the maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao Y, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCormick RF, et al. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018;93:338–354. doi: 10.1111/tpj.13781. [DOI] [PubMed] [Google Scholar]
- 44.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremling KAG, et al. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature. 2018;555:520–523. doi: 10.1038/nature25966. [DOI] [PubMed] [Google Scholar]
- 47.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.