Significance
The filamentous bacteriophage IKe is one of many nonenveloped bacterial viruses that encapsulate a circular single-stranded viral genome in the phage capsid shell. The shell of IKe is comprised of about 3,100 copies of the IKe major coat protein. Atomic-resolution structures of filamentous phages are scarce, and the structure of the single-stranded DNA is a matter of debate. Our cryo-electron microscopy structure of the filamentous bacteriophage IKe at a resolution of 3.4 Å provides atomic details on the structure of the major coat protein, the symmetry of the capsid shell, and the key interactions driving its assembly. We propose a model for the conformation of the circular single-stranded DNA core and the interactions between the capsid shell and inner DNA core.
Keywords: filamentous virus, cryo-EM structure, symmetry mismatch, single-stranded circular DNA, helical reconstruction
Abstract
The filamentous bacteriophage IKe infects Escherichia coli cells bearing IncN pili. We report the cryo-electron microscopy structure of the micrometer-long IKe viral particle at a resolution of 3.4 Å. The major coat protein [protein 8 (p8)] consists of 47 residues that fold into a ∼68-Å-long helix. An atomic model of the coat protein was built. Five p8 helices in a horizontal layer form a pentamer, and symmetrically neighboring p8 layers form a right-handed helical cylinder having a rise per pentamer of 16.77 Å and a twist of 38.52°. The inner surface of the capsid cylinder is positively charged and has direct interactions with the encapsulated circular single-stranded DNA genome, which has an electron density consistent with an unusual left-handed helix structure. Similar to capsid structures of other filamentous viruses, strong capsid packing in the IKe particle is maintained by hydrophobic residues. Despite having a different length and large sequence differences from other filamentous phages, π–π interactions were found between Tyr9 of one p8 and Trp29 of a neighboring p8 in IKe that are similar to interactions observed in phage M13, suggesting that, despite sequence divergence, overall structural features are maintained.
Filamentous bacteriophages (phages) are a group of nonenveloped bacterial viruses with a unique morphology of ∼65–70 Å in diameter and 800–2,000 nm in length. Filamentous phages have similar life cycles, and virions probably have similar organization, although not many phage structures have been characterized to sufficiently high resolution to support this hypothesis. The capsid shells of phages are helical arrays of protein molecules assembled from 2,000–8,000 copies of a single major coat protein subunit. The exact number of the subunits depends on the length of the genome, and on the ratio between the number of the nucleotides (nts) and the number of the subunits (1, 2). In general, the encapsulated circular single-stranded DNA (ssDNA) genome of the phage consists of ∼5,000–8,000 nt that encode ∼10 proteins including the major coat protein, four minor coat proteins present in only a few copies and attached to the distal ends of the virion, and other nonstructural proteins that participate in virus genome replication and virus assembly and secretion (3).
The capsid structure and symmetry of filamentous phage was revealed by early X-ray fiber diffraction studies. Based on two distinct diffraction patterns (4), filamentous phages were classified into two groups (5). One group, which includes fd, M13, IKe, and If1, called symmetry class I, has diffraction patterns that are consistent with a fivefold symmetry (making the basic unit a pentamer) and an approximate 21 screw axis. The basic pentamer unit, made up of the major coat proteins, is related to the next unit by 16–17 Å in translation along the virion axis and ∼180° in rotation around the virion axis (C5S∼2). The two structures representative of class I filamentous phages are those of the Y21M mutant of fd phage (Tyr21 to Met), with a helical rise of 16.15 Å and a helical twist of 36.00° (6), and of the M13 phage, having a helical rise of 16.60 Å and a helical twist of 36.40° (7). The former structure is based on X-ray fiber diffraction and aligned solid-state NMR and the latter on magic-angle spinning (MAS) NMR. In the class II symmetry group of filamentous phages, which includes Pf1, Pf3, and Xf, the coat proteins are arranged in a helix with a rise per monomer of about 3.00 Å and a rotation of 66–67°. The only high-resolution structure that represents class II was obtained for Pf1 by using X-ray fiber diffraction (8) and aligned solid-state NMR (9). The consensus structure for Pf1 at a temperature of below 10 °C refined with respect to both methods (10) has a helical twist of 65.90° and a rise per subunit of 3.05 Å. The high-temperature form of Pf1 has a helical twist of 66.70° and a rise of 2.90 Å. This arrangement is probably similar in other class II phages [e.g., Pf3 (11)] but can have small variations.
The genomes of filamentous phages are circular, starting from one end of the filament, extending to the opposite end, and turning back again. Since the genome is single stranded, the two opposite strands do not have significant Watson–Crick base-pairing potential (12, 13). Details on the ssDNA structure in filamentous phage were obtained using various techniques. It was established by aligned solid-state NMR studies that the phosphate linkages (O–P–O angles) are disordered in fd but ordered in Pf1 (14). Measurements of the sugar pucker in fd by MAS NMR are indicative of a C2′-endo conformation (15), whereas Raman spectroscopy suggests that deoxyguanosines are C3′-endo and dexoythymidines can have either arrangement (16). The handedness of the DNA structure in various filamentous phages was deduced indirectly by circular dichroism measurements of silver-titrated particles. The similar spectrum of phage DNA to that of double-stranded B-DNA is suggestive of a right-handed helix in fd, Xf, and IKe with a bases-in and phosphates-out arrangement (17, 18).
The current study reveals the structure of the class I filamentous phage IKe, which infects Escherichia coli cells carrying the N conjugative pili (N-pili) (19). The wild-type viral genome has ∼6,880 nt and encodes 10 proteins. Similar to other filamentous phages, the encoded protein 3 (p3) is essential for host cell recognition and virus entry as it binds to a retracting pilus and the TolA receptor of the host cell through its two functional domains D1 and D2, respectively (20). Upon infection, the major capsid protein is inserted into the host cell membrane while changing its conformation (21), and the genome is released into the host cell cytoplasm. The viral ssDNA genome of IKe, as in other filamentous phage, is then converted into a double-stranded DNA and is replicated in a rolling circle manner (3). The replicated genome forms an elongated nucleoprotein complex with the viral replication-assembly protein [protein 5 (p5)] (22). The nucleoprotein complex moves close to the cell membrane where p5 is replaced by the major coat protein [protein 8 (p8)] that is preinserted into the membrane after synthesis (23). The minor coat proteins 9 (p9) and 7 (p7) attach to the tip of the filament to mediate virion secretion from host cells without killing the cells (24).
Fiber diffraction patterns of IKe phage have been obtained, and, although a complete atomic structure has not been deposited, IKe was reported to have a canonical C5S∼2 symmetry, similar to other class I phages (25). As was shown in the studies of M13, a minimal set of side chains is required to pack the ssDNA genome efficiently (26), yet any amino acid substitution in the capsid protein may affect the capsid assembly at the atomic level. It has been shown that the structures of M13 and fd-Y21M, which differ by only two amino acids on the coat protein, have different symmetries and rotation parameters (7). BLAST alignment of the major coat proteins of IKe and fd shows 47% sequence identity, and their lengths differ by three amino acids. We therefore decided to elucidate a high-resolution structure of IKe using cryo-electron microscopy (cryo-EM) to explore the differences and similarities of class I phages.
Here, we report an atomic model of the capsid of intact filamentous bacteriophage IKe, which was built based on the calculated 3.4-Å resolution electron density map. In addition, asymmetric reconstruction of the virion showed that the ssDNA genome encapsulated in the particle adopts a left-handed helix structure. Two of the four positively charged residues in the C terminus of the major capsid protein interact closely with and stabilize the inner DNA core structure.
Results
Structure Determination of the IKe Capsid Shell.
We prepared a vitrified cryo-EM sample of IKe phage and analyzed the sample using cryo-EM (SI Appendix, Fig. S1 A and B). The initial helical parameters were adapted from a low-resolution cryo-EM model of the bacteriophage fd (27) (twist, 37.40°; rise, 17.40 Å). The helical parameters were refined to be 38.52° for the helical twist and 16.77 Å for the helical rise with Relion2 (28) (SI Appendix, Fig. S2A). The final reconstruction, calculated with the fivefold and helical symmetry applied, had an overall resolution of 3.4 Å, and 112,808 selected segments were used of the total of 253,156 boxed segments (SI Appendix, Figs. S1 C–E, S2B, and S3A and Table S1). The density of the inner DNA core was smeared due to the symmetry mismatch between the capsid and the DNA core (Fig. 1A).
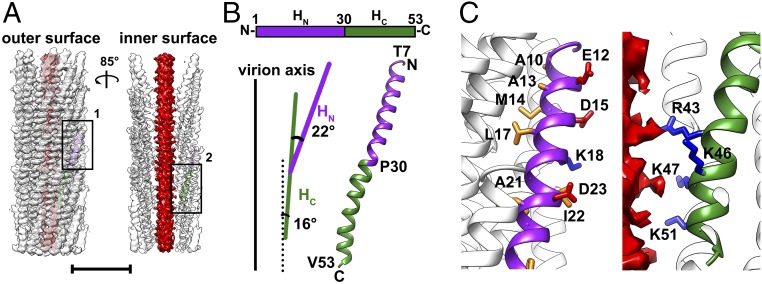
Fig. 1.
Structure of the filamentous phage IKe. (A) Ribbon and surface shadowed diagrams showing the cryo-EM structure of the filamentous phage IKe. The helical capsid shell is semitransparent and colored white. The inner smeared DNA core is colored red. (Scale bar, 5 nm.) (B, Top) A schematic diagram showing the polypeptide chain of the IKe major coat protein p8. (B, Bottom) Ribbon diagrams showing the backbone of a single p8 monomer structure. The HN and HC of p8 are colored purple and green, respectively. The angle between HC and the virion axis and the hinge angle between HN and HC were measured, and values are shown in the schematic diagrams. (C) Ribbon and surface shadowed diagrams showing the hydrophobic and hydrophilic residues of HN (Left, position 1 on A) and the four positively charged residues of HC (Right, position 2 on A). The side chains of the hydrophobic, positively charged, and negatively charged residues are shown and colored orange, blue, and red, respectively. Close interactions with the DNA of positively charged R43 and K51 are apparent from the image.
Structure of the Major Coat Protein.
The precursor of the IKe major coat protein p8 contains 82 amino acids; the first 29 N-terminal residues function as a signal peptide (29). The signal peptide helps to anchor the precursor protein of p8 onto the cell membrane, and the precursor is cleaved during virus assembly by the bacterial leader peptidase (30), leaving the C-terminal 53 residues to form the helical capsid shell. Forty-seven of these 53 residues (residues 7–53) were recognized in the calculated cryo-EM map and were built in the final atomic model (SI Appendix, Fig. S1E). The density around the missing six N-terminal residues of p8 is disordered, probably due to dynamics, as observed in other filamentous phage viruses (31).
Overall, p8 adopts an elongated right-handed α-helix structure (Fig. 1B). A kink introduced by residue Pro30 in the middle of the helix divides the 68-Å-long helix into two parts, HN and HC (Fig. 1B). The long helix bends in the middle with the distal end of HC in proximity to the viral axis, and the distal end of HN extends away from the viral axis. The hinge angle between HN and HC is ∼22° (Fig. 1B). HN contains 10 hydrophobic residues and four charged residues (Glu12, Asp15, Lys18, and Asp23). The 10 hydrophobic residues are located at one side of HN and are buried in the contact interfaces with neighboring protein subunits. The four charged residues are located on the side of HN that is completely exposed on the outer surface of the virion (Fig. 1C). HC includes 14 hydrophobic residues and four positively charged residues. The C-terminal distal end of HC is completely buried and has the four positively charged residues exposed on the inner surface of the capsid shell. Two of them point toward the encapsulated DNA (Fig. 1C).
Assembly of the Capsid Shell.
The p8 molecules assemble to form right-handed spirals in a symmetric cylindrical shell that has an outer diameter of 64 Å (excluding the disordered N terminus) and an inner diameter of about 22 Å (Fig. 2 A and B). Five p8 molecules (Fig. 2B, positions 1, 2, 3, 4, and 5) are symmetrically distributed around the viral axis to form a pentameric protein layer that is related to the five p8 molecules of the upper neighboring layer (Fig. 2B, positions 1′, 2′, 3′, 4′, and 5′) by a translation of 16.77 Å along and a rotation of 38.52° around the viral long axis (Fig. 2B). The 68-Å-long p8 molecule at position 1 of the pentameric protein layer n (n P1) interacts with additional three pentameric protein layers in each direction: its N-terminal distal end covers the distal HC ends of n + 3 P1′ and n + 2 P1 (Fig. 2A, part 1), its central portion interacts with the distal HN ends of n-1 P5′ and the HC of n + 1 P1′ (Fig. 2A, part 2), and its C-terminal portion interacts with the HN parts of n-2 P1 and n-3 P5′ (Fig. 2A, part 3). The C-terminal portions of the n + 1 p8 molecules and the N-terminal portions of the n-1 p8 molecules fill in the gaps between the five p8 molecules of the n pentameric protein layer (Fig. 2A).
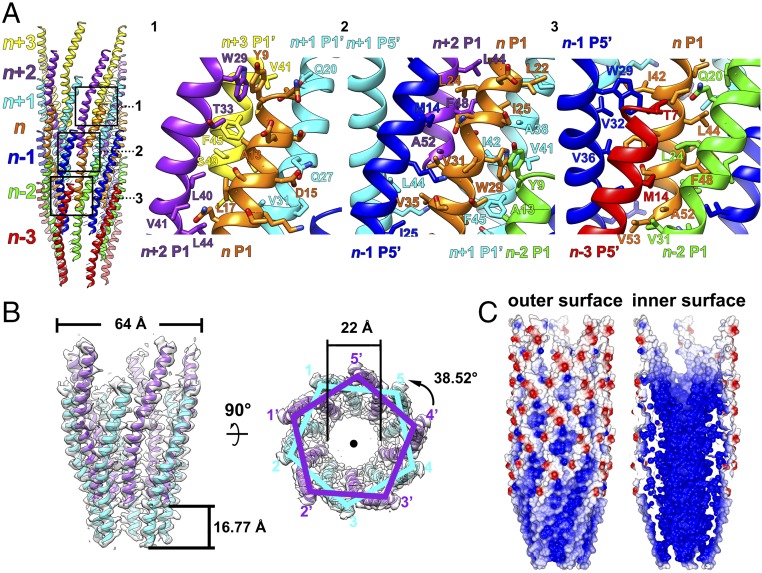
Fig. 2.
Helical assembly of the IKe capsid. (A, Left) Ribbon diagram showing an IKe capsid fragment of seven pentameric protein layers (n + 3, n + 2, n + 1, n, n − 1, n − 2, n − 3) in different colors. (A, Right) Ribbon and stick diagrams of numbered boxed areas in the Left panel showing the interactions of one p8 monomer (n P1, in orange) with neighboring subunits. The major coat proteins from the same pentameric protein layer are colored identically. (B) Ribbon and surface shadowed diagrams showing side (Left) and top (Right) views of two pentameric protein layers and their relationship via helical symmetry operators. (C) Surface electrostatic potential of the p8 helical shell. Negative and positive electrostatic potentials are colored red and blue, respectively.
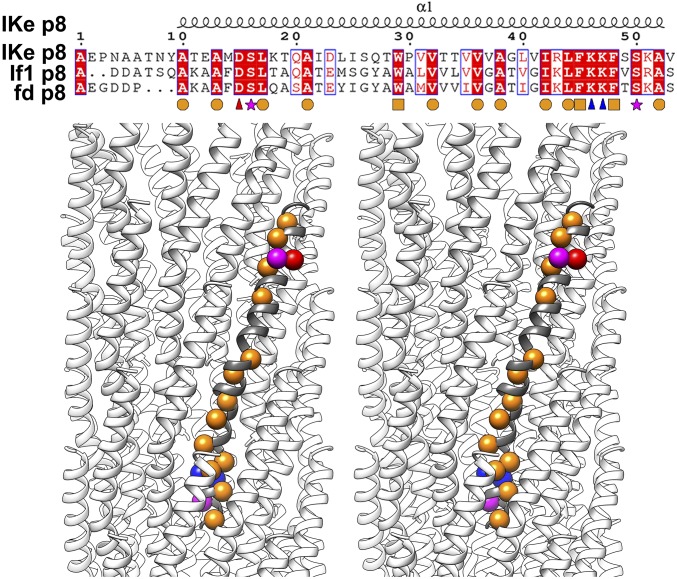
The interactions between the molecules are mediated mainly through hydrophobic residues, which are highly conserved among filamentous phages (Fig. 3). Of these conserved residues, mutational and structural studies suggested that in Ff phage (a common name for the family of three almost identical phages M13, fd, and f1) the single aromatic ring of Trp26 contacts neighboring hydrophobic residues to play a critical role in the capsid assembly (7, 32). In the capsid protein assembly of IKe, the aromatic ring of Trp29 forms a π–π interaction with the aromatic ring of residue Tyr9 from a neighboring molecule (Fig. 2A). Residue Tyr9 is not conserved in other filamentous bacteriophages (Fig. 3). However, in M13, the aromatic ring of Trp26 interacts with Pro6 in a manner similar to the Trp29 interaction with Tyr9, and the M13 Trp26 has additional contacts with two other hydrophobic residues, Ala7 and Ile39 (7, 32). Such proline–aromatic interactions are known to stabilize structures of many proteins (33). The interaction between the aromatic ring of Trp29 and that of Phe45 in IKe is geometrically similar to the corresponding Trp26–Phe42 interaction in M13 (7, 32). As we observed for M13, the stacking of aromatic rings in IKe could further enhance the packing and stability of the capsid shell.
Fig. 3.
Sequence alignment of three filamentous bacteriophage major coat proteins. (Top) Sequence alignments of the major coat proteins of IKe, If1, and fd. The secondary structure is shown above the alignments. Completely conserved residues are shown in white on a red background. Conserved residues are boxed. Completely conserved hydrophobic residues with short side chains, aromatic residues, positively charged residues, negatively charged residues, and polar residues are marked by orange circles, orange cubes, blue triangles, red triangles, and magenta stars, respectively. (Bottom) Stereo ribbon diagrams showing the locations of the completely conserved residues in the IKe p8 structure. The conserved residues are represented by balls that are colored as described in the legend for the Top panel.
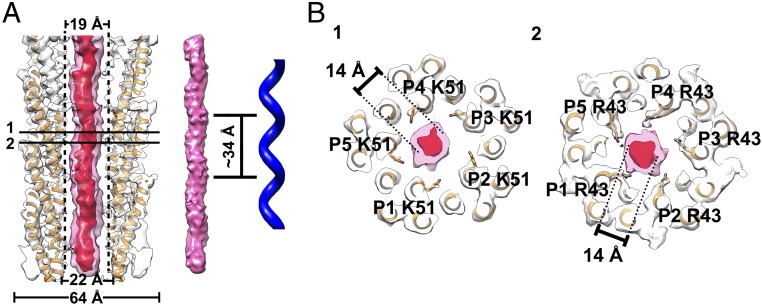
In addition to the hydrophobic interactions, two intermolecular hydrogen bonds between the subunits further stabilize the architecture of the capsid shell (Nζ of n P1 Lys46 to O of n + 1 P5′ Lys51, 2.8 Å; N ζ of n P1 Lys46 to O of n + 1 P5′ Val53, 2.6 Å). The outer surface of the IKe capsid shell is largely neutral and sparsely decorated with negatively and positively charged residues, whereas the inner surface is largely composed of positively charged residues (Fig. 2C). The side chains of Arg43 and Lys51 point to the inner lumen of the shell and mediate the interactions with the inner DNA core (Fig. 1C and SI Appendix, Fig. S4), and the side chains of Lys46 and Lys47 point more sideways. Interestingly, in IKe, the terminal lysine (Lys51) plays an important role in DNA binding but can be replaced by an alanine in M13 (26).
Structural Comparisons with Other Filamentous Bacteriophages.
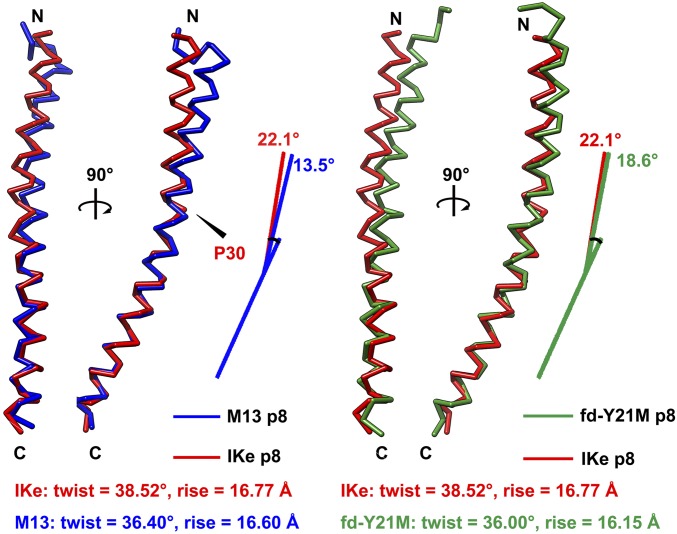
Structural comparisons of the filamentous bacteriophage major coat proteins show that the structure of IKe p8 is very similar to that of M13, despite large sequence deviations and a difference in length, which is primarily due to the longer N-terminal region of IKe. Structure superimposition of a single p8 subunit from IKe with that of M13 (PDB entry 2MJZ) shows an average root-mean-square deviation (rmsd) of 1.1 Å between the aligned Cα atoms (residues 8–53 of IKe p8 were compared with residues 5–50 of the three best M13 p8 NMR models; the individual rmsd values are 1.2, 1.3, and 0.9 Å). The major structural difference was caused by Pro30 of IKe p8, which introduces a kink in the helix and results in a larger hinge angle between HN and HC of the major coat protein of IKe compared with M13 (22.1° vs. 13.5°) (Fig. 4). Interestingly, in IKe several key residues, for example, W26, I42, F45, and F48, allow it to pack in a similar manner to other class I phages by adjusting its symmetry. Thus, in the currently very few structurally characterized members of the class I group differences are mainly in their helical parameters. The recently determined filamentous phage M13 structure (PDB entry 2MJZ) showed a twist of 36.40° and a rise of 16.60 Å (7) and fd-Y21M (PDB entry 2C0X) has a capsid arrangement with a twist of 36.00° and rise of 16.20 Å (6) (Fig. 4). Of the three, IKe has the largest twist angle of 38.52°, which is probably a consequence of the large hinge angle between the HN and HC of the IKe p8 and its slightly longer sequence.
Fig. 4.
Structural comparisons of the major coat proteins of class I phages IKe, M13, and the fd-Y21M mutant. (Left) Structural superimpositions of the IKe major coat protein (red) with that of M13 (PDB entry 2MJZ, blue) based on the HC regions. (Right) Structure superimpositions of the IKe major coat protein (red) with that of phage fd Y21M mutant (PDB entry 2C0X, green) based on the HC regions. The helical parameters are given for each phage structure.
Structure of the Inner DNA Core.
To determine the structure of the inner DNA core, densities of the capsid shell were subtracted from the images and an asymmetric reconstruction with the capsid shell-subtracted images was performed essentially as described previously (34). By assuming a fixed relationship between the capsid and the inner ssDNA, orientations of the inner ssDNA should be one of the five symmetry-related equivalent positions of the capsid shell (34). Thus, for each particle, five symmetry-related equivalent orientations of the capsid shell were generated (C5 expanded to C1, C5–C1). Orientations of the inner ssDNA were determined through 3D classifications using Relion2 performed with the equivalent orientations without doing ab initio particle orientation searches (SI Appendix, Fig. S3B). The classification results showed clear helical structures in the inner shell for some classes. Further analysis showed that the helical structures in different classes are equivalent and related to each other through a rotation along the helical axis by a multiple of 72° (SI Appendix, Fig. S3B).
Next, simulations were performed using the capsid structure model and a randomly generated inner helical object (we chose a regular B-DNA) that was either in a fixed or nonfixed position relative to the capsid (SI Appendix, Fig. S5). The results of the simulations showed that an inner helical structure could be obtained only when the helical object was in a fixed position to the capsid (SI Appendix, Fig. S5 A and B). This is consistent with previous asymmetric reconstruction studies for objects with mixed symmetries (34–36). In addition, the results suggested that the relative rotation of the inner helical structure among different classes was a result of an incorrectly assigned az angle that is off from the real value by n × 72° for most particles in one class (n is a constant for each class) (SI Appendix, Figs. S3B and S5A).
Particles in one of the classes were selected by their maximal likelihood values, and the orientations determined for the inner ssDNA core were used for the final asymmetric reconstruction with either the original raw images (Fig. 5A, Left) or the capsid subtracted images (Fig. 5A, Right, and SI Appendix, Fig. S3B). Reconstruction of the particles was also performed without imposing any symmetry (C1 reconstructions; SI Appendix, Fig. S6 A and B). Both the capsid and the ssDNA core were observed in the resulting C1 reconstruction map, although the resolution of the map is lower than that of the C5–C1 asymmetric reconstruction (orientation obtained from the symmetric reconstructions) (SI Appendix, Fig. S6 A and B), further confirming the fixed relationship between the capsid and the ssDNA core.
Fig. 5.
Structure of the inner ssDNA core. (A, Left) Ribbon and surface shadowed diagram showing the asymmetric reconstruction of the filamentous bacteriophage IKe calculated from the original raw images with the orientations determined for the inner DNA core. The helical shell is colored white, and the DNA core density contoured at a level of 17 σ is transparent and colored pink. The DNA core density contoured at a level of 26 σ is colored red. The model of the capsid is fitted in the map. (Middle and Right) Surface shadowed diagram showing the helical features of the inner DNA core and the left-handed helix. The left-handed helical pitch of the inner DNA core is ∼34 Å. (B) Ribbon and surface shadowed diagrams showing thin sections of the virus. The interactions between the capsid shell and the inner DNA core are mainly mediated through residues Lys51 and Arg43. The Arg43 residues are likely in contact with the DNA backbone. The densities from the shells are transparent and colored as described in A. The structure of IKe capsid shell is colored orange.
The final asymmetric reconstruction map clearly showed that the ssDNA adopts a left-handed helical structure (Fig. 5A). The diameter of this helix, when contoured at a level of 17 σ, is ∼19 Å. The pitch of the reconstructed structure is ∼34 Å (Fig. 5A), which is about twice the rise of the capsid protein layer. Since the ssDNA is not expected to show significant base pairing, it is most probably stabilized by asymmetric interactions (the nucleotide-to-protein subunit ratio is ∼2.4) with the capsid shell through the sidechains of two positively charged residues (Arg43 and Lys51) (Fig. 5B and SI Appendix, Fig. S4). These electrostatic interactions must play an important role in maintaining a stable conformation of the ssDNA core. Consistent with the modeled ssDNA core structure, power spectrum analysis of class-averaged images showed a peak around R = 0.06 Å−1 of the 1/34 Å−1 layer line (SI Appendix, Figs. S6D and S7).
Central sections of the ssDNA helix showed two separated peaks, suggesting that the two strands of the DNA are facing each other, as in double-stranded DNA structures (SI Appendix, Fig. S6C). However, the resolution of the inner DNA core is not high enough to allow us to build an atomic-resolution model of the DNA at this stage.
Summary and Discussion
Two main symmetry classes that describe the capsid organization of filamentous bacteriophages have been identified by X-ray fiber diffraction studies, and IKe belongs to the class I symmetry group. For this class, the capsid is organized as a helical arrangement of symmetric pentamers, each consisting of five symmetrically related and equivalent coat protein subunits, with twist and rise depending on the phage. Compared with the capsid structures of class I phages M13 and fd-Y21M, IKe has a larger twist angle between consecutive pentamers, probably resulting from the longer length of the main capsid protein and the presence of Pro30 that introduces a large curvature in the helix. In each of the class I structures, the capsid structure is maintained by strong hydrophobic interactions (Fig. 2A).
The structure of the ssDNA within the phage viral particle has been a topic of debate. Similarly to fd and M13 (1, 7), the ratio between nucleotides to subunits of IKe is noninteger (∼2.4). Base pairing is presumably sparse due to the nature of a ssDNA, and the phosphates were previously shown to be disordered (14). Attempts to unravel the DNA structure have been indirect. For example, a very early fiber diffraction study of fd pointed to “faint layer lines at … 26.7 Å,” which was suggested to be the pitch of the DNA helix, and a Z-DNA–like helical arrangement was not ruled out (37). Such reflections were not observed in the later study on M13, however (38). Circular dichroism spectroscopy has been used to study class I phages IKe, fd, and If1. Similarities of the spectra of fd phage, DNA isolated from fd, and a B-DNA in response to silver titrations lead to the hypothesis that the phage genome adopts a right-handed helix structure (18). Recent NMR studies of phage fd suggested strong electrostatic interactions between the C-terminal positively charged residues of the major coat protein and the negatively charged phosphate backbones of the ssDNA genome (12). On the other hand, NMR, X-ray fiber diffraction, and polarized Raman studies of the bacteriophage Pf1 suggested that the ssDNA genome of Pf1 may form an unusual, stretched conformation (13, 39) similar to that of the Pauling-like DNA structure (P-DNA) (40).
It was clear from previous studies that the structure of the genome in filamentous phage is unique and that the ssDNA may adopt non–B-form conformations. From the limited resolution we obtained from cryo-EM data and from a careful analysis of the electron density maps, we deduce that the ssDNA of the bacteriophage IKe adopts an unexpected left-handed helical structure that resembles a double-stranded helix but lacks base pairing. The ssDNA genome of IKe is a circular DNA. A double-stranded structure is likely to form when the genome is packed into the capsid shell. However, unlike the double-helix structure of genomic DNA, which is maintained by Watson–Crick base pairing, the two strands of the IKe ssDNA are held together via interactions with the capsid proteins (Figs. 1C and 2C, and SI Appendix, Fig. S4).
The left-handed helix structure of the IKe genome DNA core should not be a complete surprise. During the assembly of the virion, the newly replicated viral genome forms a complex with the viral replication-assembly protein p5. Early electron microscope measurements showed that the ssDNA–p5 complex adopts a left-handed conformation with a diameter of about 80 Å (41–43). Furthermore, circular dichroism and fluorescence spectroscopy studies reported that the left-handed nucleoprotein complex could be assembled at various stoichiometries of p5, depending on the genome length, and was retained regardless of the preparation techniques (44, 45). The p5 proteins in the nucleoprotein complex are replaced by p8 during virus secretion from the cell membrane, and it would be reasonable for the ssDNA genome to stay as in a left-handed helical conformation, since additional energy would be required for the conformation transition. The ssDNA helix might undergo a small conformational change upon replacement of p5 with the major coat protein p8 but not a change of handedness. Since a high-resolution structure of a full-length p5-ssDNA nucleoprotein complex is not available, the exact conformation of the p5 dimer, the interaction pattern between protein and ssDNA, and their relation to the intact phage structure requires further investigation.
Experimental Procedures
Sample Preparation.
We obtained from Prof. Marjorie Russel (The Rockefeller University, New York, NY) initial stocks of a hybrid phage, fIKe, in which wild-type p2 was replaced by p2 originating from f1 phage, thus allowing for significantly higher yields (46). The phage preparation protocol is based on previous reports (47) and is described in detail in SI Appendix, Materials and Methods.
Cryo-EM Grid Preparation and Data Collection.
The IKe samples were vitrified over 200-mesh Quantifoil grids (1.2-μm hole size) using a Vitrobot Mark III (FEI Company). Cryo-EM data of the bacteriophage IKe were collected manually as movie frame stacks at a nominal magnification of 22,500 (an effective pixel size of 1.32 Å) on an FEI Titan Krios electron microscope operating at 300 kV and equipped with a K2 Summit camera (Gatan). Details of cryo-EM grid preparation and data collection are provided in SI Appendix, Materials and Methods.
Image Processing.
Data analysis was performed by following the routine helical reconstruction procedure of Relion2 (28). The helical parameters from the PDB model 2HI5 of phage fd (twist, 37.40°; rise, 17.40 Å) were used for initial data processing. The final refined helical parameters were 38.52° for the twist and 16.77 Å for the rise. A subset of data that consists of 112,808 segmented particles was used in the final symmetric reconstruction. The final resolution of the entire segment was 3.4 Å after postprocessing (SI Appendix, Figs. S1 and S2B). To determine the structure of the inner DNA core, asymmetric reconstructions of IKe were performed by assuming a fixed relationship between the capsid and the DNA with the particle orientations determined in the fivefold symmetric reconstruction (C5 to C1). Alternatively, the particles used in shell reconstruction were subjected to reconstruction using Relion2 without imposing any symmetry (C1). Details of the reconstruction procedures are listed in SI Appendix, Materials and Methods.
Structural Modeling and Validation.
The model of the major coat protein was built and refined against the density map (SI Appendix, Table S1). Details are listed in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jingren Zhang, Jianlin Lei, and Nieng Yan for their support. We thank the Cryo-EM and Computing Platforms of the Tsinghua University Branch of the National Center for Protein Sciences (Beijing) for facility support. We thank Dr. Matthew Craddock for careful reading of the manuscript. This work was supported by funds from the Ministry of Science and Technology of China (Grant 2016YFA0501100), the National Natural Science Foundation of China (Grants 31861143027 and 31470721), the 973 Program (Grant 2015CB910102), the Junior Thousand Talents Program of China, the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, and the Beijing Advanced Innovation Center for Structural Biology (to Y.X.). This study was further supported by the joint Natural Science Foundation of China–Israel Science Foundation Grant 2423/18 (awarded to Y.X. and A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6A7F), and the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb/ (EMDB ID codes EMD-6993 and EMD-6994).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811929116/-/DCSupplemental.
References
- 1.Marzec CJ, Day LA. DNA and protein lattice-lattice interactions in the filamentous bacteriophages. Biophys J. 1983;42:171–180. doi: 10.1016/S0006-3495(83)84383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day LA, Marzec CJ, Reisberg SA, Casadevall A. DNA packing in filamentous bacteriophages. Annu Rev Biophys Biophys Chem. 1988;17:509–539. doi: 10.1146/annurev.bb.17.060188.002453. [DOI] [PubMed] [Google Scholar]
- 3.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr Issues Mol Biol. 2011;13:51–76. [PubMed] [Google Scholar]
- 4.Marvin DA. X-ray diffraction and electron microscope studies on the structure of the small filamentous bacteriophage fd. J Mol Biol. 1966;15:8–17. doi: 10.1016/s0022-2836(66)80205-x. [DOI] [PubMed] [Google Scholar]
- 5.Marzec CJ, Day LA. A theory of the symmetries of filamentous bacteriophages. Biophys J. 1988;53:425–440. doi: 10.1016/S0006-3495(88)83119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marvin DA, Welsh LC, Symmons MF, Scott WRP, Straus SK. Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J Mol Biol. 2006;355:294–309. doi: 10.1016/j.jmb.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 7.Morag O, Sgourakis NG, Baker D, Goldbourt A. The NMR-Rosetta capsid model of M13 bacteriophage reveals a quadrupled hydrophobic packing epitope. Proc Natl Acad Sci USA. 2015;112:971–976. doi: 10.1073/pnas.1415393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh LC, Symmons MF, Marvin DA. The molecular structure and structural transition of the alpha-helical capsid in filamentous bacteriophage Pf1. Acta Crystallogr D Biol Crystallogr. 2000;56:137–150. doi: 10.1107/s0907444999015334. [DOI] [PubMed] [Google Scholar]
- 9.Thiriot DS, Nevzorov AA, Zagyanskiy L, Wu CH, Opella SJ. Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. J Mol Biol. 2004;341:869–879. doi: 10.1016/j.jmb.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 10.Straus SK, Scott WRP, Schwieters CD, Marvin DA. Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR. Eur Biophys J. 2011;40:221–234. doi: 10.1007/s00249-010-0640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsh LC, Symmons MF, Sturtevant JM, Marvin DA, Perham RN. Structure of the capsid of Pf3 filamentous phage determined from X-ray fibre diffraction data at 3.1 Å resolution. J Mol Biol. 1998;283:155–177. doi: 10.1006/jmbi.1998.2081. [DOI] [PubMed] [Google Scholar]
- 12.Morag O, Abramov G, Goldbourt A. Complete chemical shift assignment of the ssDNA in the filamentous bacteriophage fd reports on its conformation and on its interface with the capsid shell. J Am Chem Soc. 2014;136:2292–2301. doi: 10.1021/ja412178n. [DOI] [PubMed] [Google Scholar]
- 13.Tsuboi M, Tsunoda M, Overman SA, Benevides JM, Thomas GJ., Jr A structural model for the single-stranded DNA genome of filamentous bacteriophage Pf1. Biochemistry. 2010;49:1737–1743. doi: 10.1021/bi901323a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross TA, Tsang P, Opella SJ. Comparison of protein and deoxyribonucleic acid backbone structures in fd and Pf1 bacteriophages. Biochemistry. 1983;22:721–726. doi: 10.1021/bi00273a002. [DOI] [PubMed] [Google Scholar]
- 15.Abramov G, Morag O, Goldbourt A. Magic-angle spinning NMR of a class I filamentous bacteriophage virus. J Phys Chem B. 2011;115:9671–9680. doi: 10.1021/jp2040955. [DOI] [PubMed] [Google Scholar]
- 16.Wen ZQ, Overman SA, Bondre P, Thomas GJ., Jr Structure and organization of bacteriophage Pf3 probed by Raman and ultraviolet resonance Raman spectroscopy. Biochemistry. 2001;40:449–458. doi: 10.1021/bi0018887. [DOI] [PubMed] [Google Scholar]
- 17.Casadevall A, Day LA. DNA packing in the filamentous viruses fd, Xf, Pf1 and Pf3. Nucleic Acids Res. 1982;10:2467–2481. doi: 10.1093/nar/10.7.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casadevall A, Day LA. Silver and mercury probing of deoxyribonucleic acid structures in the filamentous viruses fd, If1, IKe, Xf, Pf1, and Pf3. Biochemistry. 1983;22:4831–4842. doi: 10.1021/bi00289a033. [DOI] [PubMed] [Google Scholar]
- 19.Bradley DE, Coetzee JN, Hedges RW. IncI2 plasmids specify sensitivity to filamentous bacteriophage IKe. J Bacteriol. 1983;154:505–507. doi: 10.1128/jb.154.1.505-507.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakob RP, et al. Structural and energetic basis of infection by the filamentous bacteriophage IKe. Mol Microbiol. 2012;84:1124–1138. doi: 10.1111/j.1365-2958.2012.08079.x. [DOI] [PubMed] [Google Scholar]
- 21.Williams KA, Farrow NA, Deber CM, Kay LE. Structure and dynamics of bacteriophage IKe major coat protein in MPG micelles by solution NMR. Biochemistry. 1996;35:5145–5157. doi: 10.1021/bi952897w. [DOI] [PubMed] [Google Scholar]
- 22.Stassen APM, Folmer RHA, Hilbers CW, Konings RNH. Single-stranded DNA binding protein encoded by the filamentous bacteriophage M13: Structural and functional characteristics. Mol Biol Rep. 1994–1995;20:109–127. doi: 10.1007/BF00990543. [DOI] [PubMed] [Google Scholar]
- 23.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: Type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 24.Endemann H, Model P. Location of filamentous phage minor coat proteins in phage and in infected cells. J Mol Biol. 1995;250:496–506. doi: 10.1006/jmbi.1995.0393. [DOI] [PubMed] [Google Scholar]
- 25.Marvin DA, Hale RD, Nave C, Helmer-Citterich M. Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe. J Mol Biol. 1994;235:260–286. doi: 10.1016/s0022-2836(05)80032-4. [DOI] [PubMed] [Google Scholar]
- 26.Roth TA, Weiss GA, Eigenbrot C, Sidhu SS. A minimized M13 coat protein defines the requirements for assembly into the bacteriophage particle. J Mol Biol. 2002;322:357–367. doi: 10.1016/s0022-2836(02)00769-6. [DOI] [PubMed] [Google Scholar]
- 27.Wang YA, et al. The structure of a filamentous bacteriophage. J Mol Biol. 2006;361:209–215. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 28.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashima Y, Frangione B, Wiseman RL, Konigsberg WH. Primary structure of the major coat protein of the filamentous bacterial viruses, If1 and Ike. J Biol Chem. 1981;256:5792–5797. [PubMed] [Google Scholar]
- 30.Wickner W. Mechanisms of membrane assembly: General lessons from the study of M13 coat protein and Escherichia coli leader peptidase. Biochemistry. 1988;27:1081–1086. doi: 10.1021/bi00404a001. [DOI] [PubMed] [Google Scholar]
- 31.Aharoni T, Goldbourt A. Dynamics and rigidity of an intact filamentous bacteriophage virus probed by magic angle spinning NMR. Chemistry. 2018;24:8737–8741. doi: 10.1002/chem.201800532. [DOI] [PubMed] [Google Scholar]
- 32.Williams KA, et al. Packing of coat protein amphipathic and transmembrane helices in filamentous bacteriophage M13: Role of small residues in protein oligomerization. J Mol Biol. 1995;252:6–14. doi: 10.1006/jmbi.1995.0469. [DOI] [PubMed] [Google Scholar]
- 33.Zondlo NJ. Aromatic-proline interactions: Electronically tunable CH/π interactions. Acc Chem Res. 2013;46:1039–1049. doi: 10.1021/ar300087y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morais MC, et al. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Y, et al. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banner DW, Nave C, Marvin DA. Structure of the protein and DNA in fd filamentous bacterial virus. Nature. 1981;289:814–816. doi: 10.1038/289814a0. [DOI] [PubMed] [Google Scholar]
- 38.Glucksman MJ, Bhattacharjee S, Makowski L. Three-dimensional structure of a cloning vector. X-ray diffraction studies of filamentous bacteriophage M13 at 7 Å resolution. J Mol Biol. 1992;226:455–470. doi: 10.1016/0022-2836(92)90960-r. [DOI] [PubMed] [Google Scholar]
- 39.Liu DJ, Day LA. Pf1 virus structure: Helical coat protein and DNA with paraxial phosphates. Science. 1994;265:671–674. doi: 10.1126/science.8036516. [DOI] [PubMed] [Google Scholar]
- 40.Allemand JF, Bensimon D, Lavery R, Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci USA. 1998;95:14152–14157. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray CW. Three-dimensional structure of complexes of single-stranded DNA-binding proteins with DNA. IKe and fd gene 5 proteins form left-handed helices with single-stranded DNA. J Mol Biol. 1989;208:57–64. doi: 10.1016/0022-2836(89)90087-9. [DOI] [PubMed] [Google Scholar]
- 42.Olah GA, et al. Structures of fd gene 5 protein.nucleic acid complexes: A combined solution scattering and electron microscopy study. J Mol Biol. 1995;249:576–594. doi: 10.1006/jmbi.1995.0320. [DOI] [PubMed] [Google Scholar]
- 43.King GC, Coleman JE. Two-dimensional 1H NMR of gene 5 protein indicates that only two aromatic rings interact significantly with oligodeoxynucleotide bases. Biochemistry. 1987;26:2929–2937. doi: 10.1021/bi00384a039. [DOI] [PubMed] [Google Scholar]
- 44.Bulsink H, Harmsen BJ, Hilbers CW. DNA-binding properties of gene-5 protein encoded by bacteriophage M13. 2. Further characterization of the different binding modes for poly- and oligodeoxynucleic acids. Eur J Biochem. 1988;176:597–608. doi: 10.1111/j.1432-1033.1988.tb14319.x. [DOI] [PubMed] [Google Scholar]
- 45.Sang BC, Gray DM. CD measurements show that fd and IKe gene 5 proteins undergo minimal conformational changes upon binding to poly(rA) Biochemistry. 1989;28:9502–9507. doi: 10.1021/bi00450a038. [DOI] [PubMed] [Google Scholar]
- 46.Russel M. Interchangeability of related proteins and autonomy of function. The morphogenetic proteins of filamentous phage f1 and IKe cannot replace one another. J Mol Biol. 1992;227:453–462. doi: 10.1016/0022-2836(92)90900-5. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.