Significance
Reptiles show an amazing color diversity based on variation in melanins, carotenoids, and pterins. This study reveals genes controlling differences between three color morphs (white, orange, and yellow) in the common wall lizard. Orange pigmentation, due to high levels of orange/red pterins in skin, is caused by genetic changes in the sepiapterin reductase gene. Yellow skin, showing high levels of yellow carotenoids, is controlled by the beta-carotene oxygenase 2 locus. Thus, the color polymorphism in the common wall lizard is associated with changes in two small regions of the genome containing genes with crucial roles in pterin and carotenoid metabolism. These genes are likely to have pleiotropic effects on behavior and other traits associated with the different color morphs.
Keywords: Podarcis muralis, carotenoid pigmentation, pterin pigmentation, balanced polymorphism, introgression
Abstract
Reptiles use pterin and carotenoid pigments to produce yellow, orange, and red colors. These conspicuous colors serve a diversity of signaling functions, but their molecular basis remains unresolved. Here, we show that the genomes of sympatric color morphs of the European common wall lizard (Podarcis muralis), which differ in orange and yellow pigmentation and in their ecology and behavior, are virtually undifferentiated. Genetic differences are restricted to two small regulatory regions near genes associated with pterin [sepiapterin reductase (SPR)] and carotenoid [beta-carotene oxygenase 2 (BCO2)] metabolism, demonstrating that a core gene in the housekeeping pathway of pterin biosynthesis has been coopted for bright coloration in reptiles and indicating that these loci exert pleiotropic effects on other aspects of physiology. Pigmentation differences are explained by extremely divergent alleles, and haplotype analysis revealed abundant transspecific allele sharing with other lacertids exhibiting color polymorphisms. The evolution of these conspicuous color ornaments is the result of ancient genetic variation and cross-species hybridization.
Color morphs can be found in thousands of species in nature. In many animals, color morphs persist over extended periods of time; co-occur across large geographic regions; and differ in a range of key morphological, physiological, or behavioral traits (1–5). These features offer an exceptional framework with which to investigate central mechanisms of phenotypic diversity and innovation, with potential insights into a variety of processes, including adaptation, convergent evolution, sexual selection, and the early stages of sympatric speciation (1, 6). It is thus of special interest to understand the evolutionary and mutational mechanisms promoting the emergence and long-term persistence of sympatric color polymorphisms in nature.
Reptiles are among the most colorful animals. They achieve this striking color variation in different ways, but their vibrant colors spanning the gradient of hues between yellow and red are generated primarily by carotenoid and/or pterin pigments (7). The colors produced by these pigments serve many biological functions and are thought to be of key importance in intra- and interspecific communication (8). Striking colors often differ between closely related species and have been gained and lost across the reptilian phylogeny, demonstrating that coloration driven by these types of pigments can change over short time scales and presumably evolved multiple times independently. However, despite intense study of the biochemical basis and ecology of carotenoid- and pterin-based pigmentation in reptiles (3, 7, 9–11), we have a limited understanding regarding the genetic changes and molecular pathways that govern differences among individuals and species.
To investigate the genetic and evolutionary bases of the vivid colors displayed by reptiles, and to test hypothesis about how and why color polymorphisms and correlated trait variation persist within populations, we studied the European common wall lizard (Podarcis muralis; Fig. 1A): a polymorphic lizard in which the ventral scales of males and females exhibit one of three distinct colors (orange, yellow, and white) or a mosaic pattern combining two colors (orange-yellow and orange-white) (12, 13). These five color morphs can be found throughout most of the broad geographic distribution of the species (Fig. 1B), and are shared by intraspecific sublineages thought to have diverged up to 2.5 Mya (14). Whereas the white morph is typically the most common (>50%), the relative frequency of morphs is highly variable even at small regional scales and the yellow or orange morph may occasionally prevail (15–17) (SI Appendix, Fig. S1). The widespread distribution and persistence of color variation is thought to be due to balancing selection and the product of an interplay between natural and sexual selection (15, 17). Previous work has shown that morphs mate assortatively with respect to ventral color (∼75% of pairs) and differ in additional traits, including morphology, behavior, physiology, immunology, and reproduction (12, 18–22). The mode of inheritance of the color morphs is unknown.
Fig. 1.
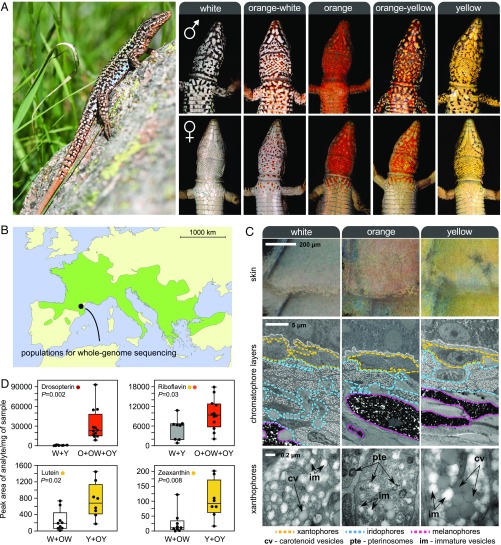
Color polymorphism in the European common wall lizard, P. muralis. (A, Left) Common wall lizard. (A, Right) Illustrations of the five discrete ventral morphs. These conspicuous colors likely function as visual ornaments implicated in sexual signaling. The yellow and orange colors are restricted to the ventral surface, and males and females exhibit marked differences in the extent of pigmentation in some populations. (B) Geographic distribution of the species in Europe (light green). (C) Ultrastructure of the ventral skin of the three pure morphs. (Top) Close-up view of the ventral scales of each morph under a light microscope. (Middle) TEM of the three chromatophore layers [xantophores (yellow), iridophores (blue), and melanophores (pink)]. (Bottom) Electromicrographs detailing the structure of xanthophores. Examples of pterinosomes (pte), carotenoid vesicles (cv), and immature vesicles (im) are highlighted. (D) Levels of colored pterin and carotenoid compounds in the ventral skin of the different morphs obtained by HPLC-MS/MS. O, orange; W, white; Y, yellow.
Results
Carotenoid and Pterin Pigments Underlie Pigmentation Differences.
We began by determining the biochemical and cellular basis of pigmentation differences among morphs. Using transmission electron microscopy (TEM), we found that the ventral skin of all morphs contained the same set of dermal pigment cells arranged as three superimposed layers (xantophores, iridophores, and melanophores; Fig. 1C). The iridophore layer was thinner in orange individuals compared with yellow and white individuals, but the most noticeable difference among morphs was observed in the xantophore layer, which usually contains pterins and carotenoid pigments in reptiles (7). Although two types of morphologically distinct pigment-containing organelles (pterinosomes and carotenoid vesicles) coexisted within many xantophores irrespective of skin color, their relative abundance varied drastically. Pterinosomes were abundant in xantophores from orange skin, whereas carotenoid vesicles of a large size were abundant in yellow skin. The xantophores of individuals exhibiting white skin were characterized by low numbers of pterinosomes and carotenoid vesicles, as well as by large numbers of immature vesicles of small size and low electron density under TEM.
Next, we used high-performance liquid chromatography–mass spectrometry (HPLC-MS/MS) to quantify individual carotenoid and pterin derivatives (Fig. 1D and SI Appendix, Fig. S2A). In agreement with microscopy, carotenoids and pterins were present in skin of all colors, but the relative proportions of specific metabolites varied according to color morph. Both lutein and zeathanxin, two yellow-colored carotenoids, were significantly more abundant in yellow skin relative to white skin (Mann–Whitney U test, P < 0.05 for both tests; Fig. 1D). By quantifying nine colored and colorless pterin metabolites, we found no evidence for substantial changes in the overall pterin profiles between white and yellow individuals (SI Appendix, Fig. S2 A and B). However, the skin of individuals expressing orange pigmentation (pure and mosaic) contained significantly higher levels of orange/red pterins (riboflavin and drosopterin) compared with nonorange individuals (Mann–Whitney U test, P < 0.05 for both tests; Fig. 1D). The observed variation in colored pterins and carotenoids among morphs provides a biochemical basis for the interindividual differences in orange and yellow coloration, respectively. Together, the microscopy and biochemical analyses support the hypothesis that chromatic variation is due to alterations in the metabolism, transport, or deposition of carotenoids and pterins, or to some combination of these processes.
A Highly Contiguous Genome Sequence for the Common Wall Lizard.
As a backbone for our genetic and evolutionary studies, we sequenced and assembled a reference genome for the common wall lizard (PodMur1.0). The assembly was primarily based on Pacific Biosciences (PacBio) long-read sequences (∼100-fold coverage) and complemented with Illumina sequencing (30-fold coverage). To aid in the assembly of the genome, we generated chromosome conformation capture sequencing data using CHICAGO and HiC libraries (23, 24). The combination of these methodologies resulted in a high-quality, chromosome-scale, genome assembly of 1.51 Gb with a contig N50 of 714.6 kb and a scaffold N50 of 92.4 Mb (SI Appendix, Table S1). Our assembly produced 19 scaffolds of large size (18 larger than 40 Mb and one larger than 10 Mb; SI Appendix, Table S2), which matches well with the karyotype of the species (2n = 38) (25). A search for conserved single-copy orthologs revealed that the assembly included a large percentage of full-length genes (93.2%; SI Appendix, Table S1), indicating that the sequence is both highly contiguous and accurate.
Karyotypic analysis demonstrated that the common wall lizard possesses a ZW/ZZ sex determination system and that the two sex chromosomes are of similar size and shape (26). To identify the Z chromosome in our assembly, we compared sequence coverage between DNA pools of males and females (SI Appendix, Fig. S3). We found a single chromosome, which we named Z, for which sequence coverage in females was roughly half that of males throughout most of the chromosome, as expected from reads mapping on the Z, but not on the W, chromosome. The results show that Z and W, although similar in size, have likely evolved independently for a long time and have diverged extensively in sequence.
Finally, the genome assembly was annotated using in silico predictions and transcriptome data from five tissues and one embryonic stage (SI Appendix, Table S3), which predicted 24,656 protein-coding genes (SI Appendix, Table S1). This high-quality lizard genome provides an important resource for comparative genomic analysis within squamate reptiles.
The Distinct Morphs Have Near-Identical Genome Sequences.
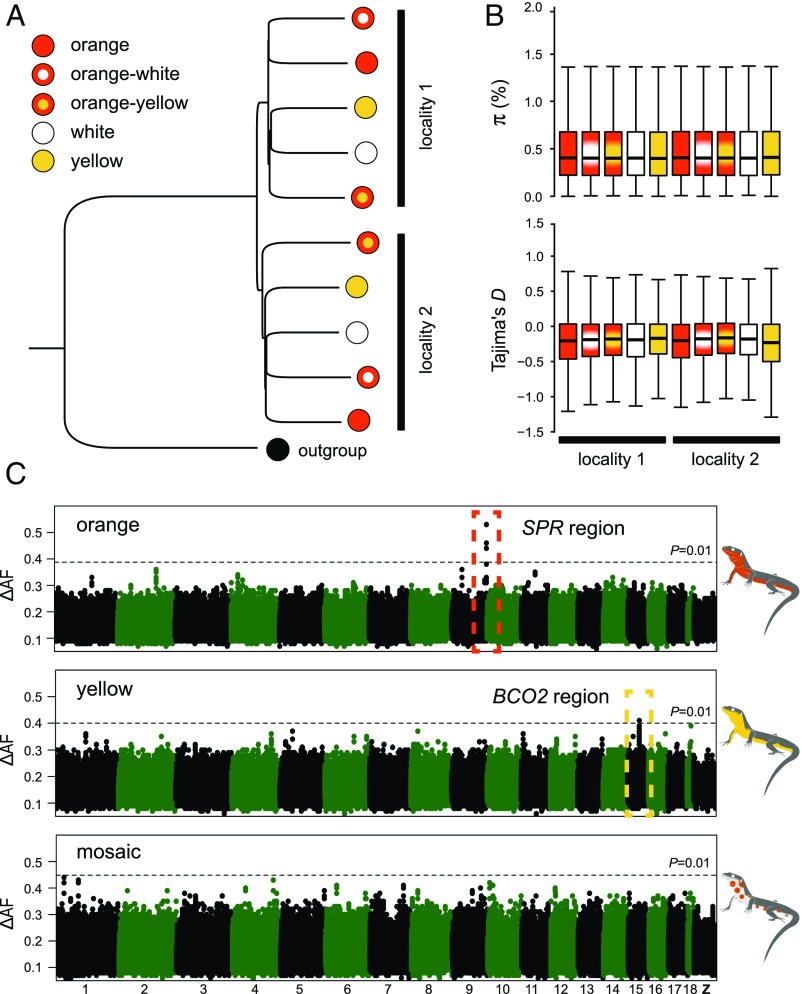
To obtain genome-wide polymorphism data, we sampled 154 individuals from two neighboring localities in the eastern Pyrenees and performed whole-genome sequencing (Fig. 1B). We sequenced 10 DNA pools of individuals grouped by color morph and locality (mean coverage = ∼16-fold; SI Appendix, Table S4). The short-sequence reads were aligned to the reference genome for variant identification and allele frequency estimation, yielding a total of 12,066,526 variants (SNPs and indels). A neighbor-joining tree based on these genome-wide data revealed that the overall genetic structure was predominantly influenced by sampling site as opposed to color phenotype (Fig. 2A). The low differentiation between morphs is well demonstrated by an average fixation index (FST) value close to 0 (0.02) and by a paucity of variants displaying high allele frequency differences in pairwise comparisons (SI Appendix, Fig. S4). The different morphs also displayed similar levels of nucleotide diversity (π) and nearly identical allele frequency spectra as measured by Tajima’s D (Fig. 2B), as expected from individuals drawn from a single population. These results demonstrate that the genome-wide impacts of assortative mating are minor and that rates of gene flow between morphs are sufficiently high to prevent the buildup of strong genetic differentiation.
Fig. 2.
Population structure and genetic basis of color polymorphism in the common wall lizard. (A) Neighbor-joining tree summarizing genetic distance among individual pools using 250,000 randomly chosen SNPs. The tree was rooted with a DNA pool of individuals sampled from Italy and belonging to a different intraspecific sublineage. (B) Nucleotide diversity (π) and Tajima’s D estimated for each morph. Both statistics were calculated in 10-kb nonoverlapping windows, and a genome-wide estimate was obtained by averaging all windows across the genome. (C) Genetic mapping based on differences in allele frequencies (ΔAF) for the orange, yellow, and mosaic phenotypes. The Manhattan plots show the median value of 20-SNP windows (five-SNP overlap) across the reference genome. The dashed lines represent a 1% significance cutoff based on 1,000 permutations conducted for each dataset.
High-Resolution Mapping of Genomic Regions Underlying Differences in Coloration.
Encouraged by the overall low levels of genetic differentiation among morphs, we next carried out population genomic analysis to identify chromosome regions associated with color variation. We calculated allele frequency differentiation (ΔAF) averaged in sliding windows, and guided by our phenotypic characterization, we contrasted morphs by the presence/absence of specific colors (orange and yellow) or by their patterning (mosaic). For both orange and yellow coloration, we found in each case a single genomic region showing significantly high ΔAF compared with an empirical null distribution generated by permutation (Fig. 2C). These regions were small, located on the autosomes, and embedded within an otherwise undifferentiated genome (Fig. 3A). A Cochran–Mantel–Hanzel (CMH) test, configured to identify consistent changes in allele frequency between different samples, corroborated the ΔAF analysis and revealed the same two regions as the top outliers for orange and yellow coloration (SI Appendix, Fig. S5).
Fig. 3.
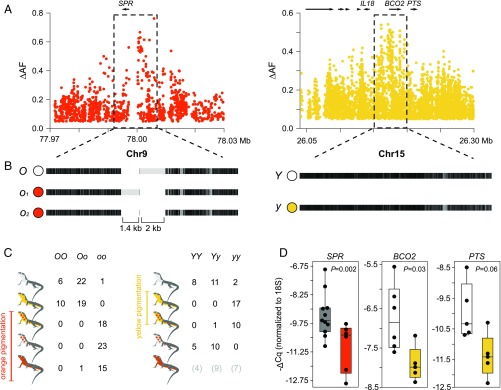
Regulatory variation explains color polymorphism in the common wall lizard. (A) Differences in allele frequencies (ΔAF) for the orange and yellow phenotypes around SPR and BCO2 (each dot represents an SNP). (B) Haplotype structure for the same two regions based on the alignment of our reference genome sequence to consensus sequences of the alternative haplotypes obtained using Nanopore and Sanger sequencing. Black indicates homology, and light gray indicates mismatches that can originate from point mutations or indel variants. (C) Individual genotypes for the SPR and BCO2 loci among the five morphs based on high ΔAF variants selected from the whole-genome data. (D) qPCR measurements of SPR, BCO2, and PTS expression in ventral skin.
The region significantly associated with orange coloration was located near the SPR gene [chromosome 9 (Chr9); Fig. 3A], which encodes sepiapterin reductase, a key enzyme in pterin metabolism (SI Appendix, Fig. S6A). The presence/absence of yellow coloration mapped to the genomic region around the BCO2 gene (Chr15; Fig. 3A). BCO2 encodes the beta-carotene oxygenase 2 that oxidizes colorful carotenoids to colorless apocarotenoids during the biosynthesis of vitamin A (SI Appendix, Fig. S6B). In light of the phenotypic differences between morphs, PTS is another gene of interest that was located near the BCO2 region (Fig. 3A). PTS encodes the 6-pyruvoyltetrahydropterin synthase, which is also a key enzyme in pterin metabolism (SI Appendix, Fig. S6A). Although evolutionary theory predicts that alleles encoding sexually selected traits, such as color ornaments, should accumulate preferentially on sex chromosomes (27, 28), particularly for ZZ/ZW systems, our results demonstrate that the two genomic regions most strongly associated with orange and yellow pigmentation are not sex-linked.
To confirm the association and to gain additional insight about the genetic architecture of orange and yellow coloration, we obtained individual genotypes for high ΔAF variants selected from the resequencing data (Fig. 3C). This genotyping revealed that in 56 of 57 cases, orange and mosaic individuals were homozygous for one allele (hereafter o) at the SPR locus, whereas yellow and white individuals, with one exception, were either heterozygous (n = 41) or homozygous (n = 16) for the alternative allele (hereafter O): a highly significant association (recessive model, P = 9.3 × 10−29). The association at the BCO2 locus was also highly significant (recessive model, P = 1.2 × 10−15), with most individuals displaying yellow coloration (yellow and orange-yellow) being homozygous (27 of 28 individuals) for one allele (hereafter y) and 34 of 36 individuals displaying white coloration (white and orange-white) being heterozygous (n = 21) or homozygous (n = 13) for the alternative allele (hereafter Y). The few discordant individuals in both loci can be explained by incomplete linkage between the genotyped and causal variants, incomplete penetrance due to environmental effects, or other interacting genetic factors. We found the three different genotypic classes among individuals of the pure orange morph at the BCO2 locus, suggesting that yellow carotenoids, if present, are likely masked by the stronger pterin-based orange coloration. Whereas other loci not revealed in our genomic analysis might play a minor role, these loci explain a large component of the differences in pterin and carotenoid pigmentation among morphs.
Subsequently, we extended the genotyping to a larger cohort of common wall lizards covering the broad geographic range of the species and belonging to several of its intraspecific sublineages (14). This analysis confirmed that SPR is the primary locus explaining orange coloration across the species’ range (SI Appendix, Table S5). In contrast, the variant near BCO2, which was strongly associated with yellow pigmentation in the Pyrenees, was either not present in other sublineages or not evidently associated with yellow coloration (SI Appendix, Table S5). The absence of an association might be explained by some of the same reasons as above, or by the possibility that yellow coloration might have evolved convergently more than once, either through independent mutation events near BCO2 or through a different gene.
By contrast, our genomic analysis (ΔAF and CMH test) did not reveal any genetic candidate associated with mosaic morphs (Fig. 2C and SI Appendix, Fig. S5). The mosaic morphs are also not explained by any combination of alleles between SPR and BCO2 (Fig. 3C). The lack of genetic signal could be explained by a polygenic architecture consisting of a few loci with small to moderate effects that our sequencing efforts might not be powerful enough to detect. Alternatively, this phenotype might not be directly under genetic control; rather, it might reflect a temporary ontogenetic stage or phenotypic plasticity. In fact, a long-term study of a natural population showed that many subadult individuals displaying isolated orange scales, identical to the orange-white morph, subsequently develop pure orange coloration (12). Additional work is needed to resolve the genetic basis (or lack thereof) of the mosaic morphs.
Synteny and Gene Content Are Preserved Between Haplotypes Associated with Coloration.
Structural changes could be responsible for the observed genetic differentiation associated with each color morph. To explore this possibility, we sequenced an additional individual homozygous for the haplotypes not represented in the reference sequence (o and Y) using Nanopore long reads (9.7-fold coverage). We found that the region immediately upstream of SPR (∼7 kb) contained two medium-sized indel polymorphisms compared with the reference sequence (1.4 kb and 2 kb) (Fig. 3B). PCR amplification of this region in a larger number of orange individuals (n = 15) revealed two orange alleles of different size (o1 and o2) that shared the 2-kb indel (Fig. 3B). However, we failed to identify homologous sequences to the two indels in the chicken or the anole lizard; thus, they are likely derived insertions lacking any strongly conserved sequence. In the region associated with yellow coloration surrounding BCO2, the y and Y haplotypes were identical in their general structure (Fig. 3B). Taken together, we found no evidence for the existence of large-scale structural differences, copy number variants, or translocations in both loci. Thus, gene content is preserved between the haplotypes that control pigmentation differences, demonstrating that the described multitrait divergence between morphs is not explained by a supergene organization implicating many genes.
Cis-Regulatory Sequence Variation Underlies the Color Polymorphism.
Next, we examined in greater detail the region of association around SPR and BCO2. The signal of association for orange coloration was restricted to a noncoding region immediately upstream of SPR (Fig. 3A). The association for yellow coloration in BCO2 was strong both upstream and overlapping part of the coding region (Fig. 3A); however, we found no mutations strongly associated with yellow coloration that could alter the protein structure of BCO2 (nonsynonymous, stop, splicing, or frameshift). We thus hypothesized that the pigmentation differences are controlled by regulatory sequence variation at these loci.
To examine the role of regulatory variation in pigmentation differences, we studied gene expression in several tissues (ventral skin, brain, muscle, and liver) harvested from orange, yellow, and white lizards using quantitative PCR (qPCR) (Fig. 3D and SI Appendix, Fig. S6C). SPR expression was significantly lower in orange skin relative to other colors (Mann–Whitney U test, P = 0.002), whereas the same was found for BCO2 in yellow skin relative to white skin (Mann–Whitney U test, P = 0.03). Low BCO2 expression is consistent with reduced activity of this enzyme that presumably leads to accumulation of colorful carotenoids, similar to what has been shown for carotenoid-based yellow coloration in birds (29). Despite the fact that the strongest signal of association was located near BCO2 (Fig. 3A), the expression of PTS was also higher in white individuals compared with yellow individuals, although not significantly so (Mann–Whitney U test, P = 0.06). Given that we found no obvious differences in pterin content between white and yellow individuals (SI Appendix, Fig. S2 A and B), it is unclear whether PTS could play a role in pigmentation differences. In the other tissues, we found no significant differences between morphs for BCO2, whereas SPR was slightly up-regulated in the muscle and brain of orange individuals (opposite to the pattern that was observed in the skin), and the differences were significant for the muscle and marginally significant for the brain (SI Appendix, Fig. S6C).
We next examined the expression differences in BCO2 and SPR by measuring allele-specific expression in the skin. If cis-acting regulatory mutations are driving expression differences between morphs, then one haplotype should preferentially be expressed in the shared cellular environment of individuals heterozygous for the Y/y or O/o alleles. Among the individuals used in the qPCR analysis, we found for each gene one heterozygous individual containing a polymorphism overlapping the coding region that could be used to quantify allelic imbalance using cDNA sequencing. Corroborating the observed differences in the qPCR experiments, we found a strong preferential expression of one allele over the other for both SPR (81% vs. 19%; n = 8,210 reads; χ2, P < 10−16) and BCO2 (72% vs. 28%; n = 25,026 reads; χ2, P < 10−16). Overall, our expression studies provide evidence that pigmentation differences in the skin evolved through one or more cis-acting mutations affecting the activity of SPR and BCO2, and possibly PTS. They further suggest that gene expression of SPR in other tissues might also differ between morphs.
Genetic Variation Upstream of SPR and BCO2 Is Shared Among Divergent Species.
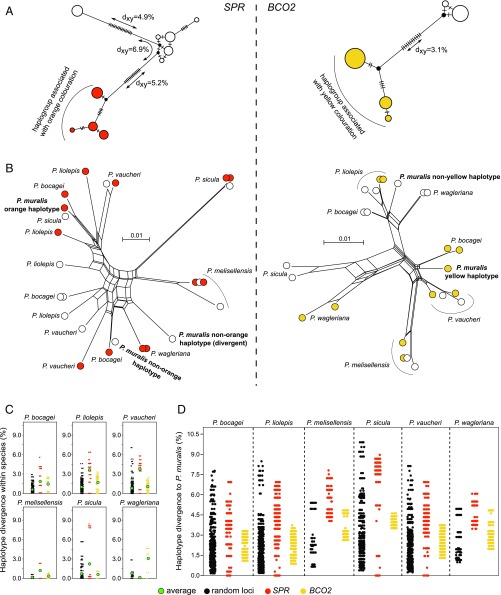
To gain insight into the evolutionary history of the two regions that explain pterin- and carotenoid-based pigmentation, we sequenced amplicons (∼550 bp) overlapping the strong signals of association upstream of SPR and BCO2 in individuals from the Pyrenees, and used the sequence data to construct haplotype trees (Fig. 4A). In both BCO2 and SPR, the haplotypes were typically associated with the presence or absence of yellow (y and Y) and orange (o and O) coloration clustered into extremely divergent haplogroups (Fig. 4A). In SPR, we also uncovered two divergent haplotypes associated with nonorange coloration. The average number of pairwise differences (dXY) between the orange and nonorange haplogroups in SPR (dXY = 5.2–6.9%) and between the yellow and nonyellow haplogroups in BCO2 (dXY = 3.1%) was approximately eight- to 17-fold higher than the average number of pairwise differences between sequences within the French population (π ≈ 0.4%; Fig. 2B). To put this into context, the average human–chimpanzee nucleotide divergence in noncoding regions is ∼1.2% (30). The magnitude of divergence between haplotypes associated with pigmentation differences shows that the alleles controlling the presence or absence of orange and yellow coloration are evolutionary old and not the result of recent mutational events. Tajima’s D values were also high for both loci (DSPR = 2.1 and DBCO2 = 2.3), likely as a consequence of the highly differentiated haplotypes and consistent with a molecular signature of balancing selection. However, since our sequencing data do not originate from a random sampling of morphs with respect to their observed frequencies, this should be interpreted with caution.
Fig. 4.
Evolution of the regulatory regions associated with pterin- and carotenoid-based coloration in the genus Podarcis. (A) Median-joining genealogies of the genomic regions associated with coloration upstream of SPR (Left) and BCO2 (Right) from common wall lizards from the Pyrenees. The dashes on branches indicate the number of observed mutations. (B) NeighborNet trees of the genomic regions associated with coloration upstream of SPR (Left) and BCO2 (Right) combining common wall lizards from the Pyrenees and six other species in the genus Podarcis. Haplotypes are colored orange, yellow, or white, indicating the color morph of the individual. For representation purposes, only a subset of the sequences is presented. (C) Pairwise nucleotide differences between haplotypes within species for SPR (orange), BCO2 (yellow), and 31 random loci (black). The average is indicated by a green circle. (D) Pairwise nucleotide differences between haplotypes of the common wall lizard and other Podarcis species for SPR (orange), BCO2 (yellow), and 31 random loci (black). The contrasts involving other species are presented in SI Appendix, Fig. S7.
Other European lacertids exhibit intraspecific color polymorphisms resembling those of the common wall lizard (12). To study the evolutionary history of the regulatory regions associated with pigmentation in an extended phylogenetic context, we sequenced the same amplicons for six additional species of the genus Podarcis showing color polymorphism. In both SPR and BCO2, we found a slight nonrandom clustering with respect to color morph across species. However, assuming the same recessive pattern of inheritance inferred for the common wall lizards, we would not expect to see haplotypes derived from orange and yellow individuals clustering together with haplotypes from nonorange and nonyellow individuals. The discrepancy could be explained by a combination of incomplete linkage between the sequenced amplicons and the causative mutations, as well as independent molecular bases for orange and yellow coloration in some species. Notably, the genealogies at both loci revealed abundant transspecies polymorphisms, in which haplotypes from a particular species were more closely related to haplotypes from a different species (Fig. 4B). For both loci, the average number of pairwise differences between haplotypes within taxa for several Podarcis species was also elevated compared with a set of 31 amplicons randomly distributed across the genome (average size = 410 bp; Fig. 4C). This pattern was particularly strong at SPR, where a subset of the comparisons exceeds all values for the random loci, demonstrating that highly divergent haplotypes also co-occur in these species.
The patterns of deep haplotype divergence and allele sharing between species at both pigmentation loci could be explained by ancient alleles that predate speciation events and/or by interspecific hybridization. These scenarios generate different predictions regarding patterns of molecular variation. For example, introgressed haplotypes are expected to show reduced sequence divergence between species compared with the genome-wide average, whereas the maintenance of ancestral polymorphisms, compatible with a balancing selection scenario or incomplete lineage sorting, predicts no such reduction. We compared pairwise haplotype differences across species for SPR, BCO2, and the 31 random loci (Fig. 4D and SI Appendix, Fig. S7). In contrasts between the common wall lizard and other species, the distribution of haplotype divergence for BCO2 was within the distribution of values calculated for the random loci, consistent with a scenario of ancestral genetic variation preserved between species. In the case of SPR, however, we found identical haplotypes between the common wall lizard and Podarcis bocagei, Podarcis liolepis, Podarcis sicula, and Podarcis vaucheri, a pattern never observed for the random loci (Fig. 4D). The same pattern was observed in other interspecific contrasts (SI Appendix, Fig. S7). Given that these species have diverged from the common wall lizard for millions of years (>10 Mya) (31), these identical sequences strongly suggest that introgression via hybridization occurred with these or related species. Collectively, these results suggest that patterns of molecular evolution at the pigmentation loci are explained by a combination of ancestral genetic variation, likely maintained through balancing selection, and introgression.
Discussion
Reptiles exhibit striking variation in color within and between species. Here, we combined whole-genome sequencing with gene expression, biochemical, and microscopy analyses to dissect the molecular basis of the coloration differences and correlated traits among morphs of the common wall lizard. Our analysis revealed genes responsible for production of carotenoid- and pterin-based pigmentation in reptiles, providing the basis to unravel the evolutionary processes that influenced the emergence of the broad palette of colors and visual ornaments so prevalently found in the reptilian world.
Co-option, the recruitment of genes and gene pathways to serve a different function, is emerging as a major evolutionary force underlying pigmentation novelty (32). Examples include a cytochrome P450 enzyme required for color vision and implicated in carotenoid-based red coloration in birds (33, 34), a polyketide synthase essential for producing yellow psittacofulvins in parrots (35), and early developmental genes governing wing patterns in butterflies (36). Pterins are produced through ancient multistep biosynthetic pathways, and orthologs of SPR exist throughout the animal kingdom. The SPR enzyme is required to catalyze one of the three key steps in the synthesis of tetrahydrobiopterin (SI Appendix, Fig. S6A), an essential cofactor for a vast range of enzymatic reactions of key metabolic importance, including degradation of the amino acid phenylalanine and biosynthesis of the neurotransmitters dopamine, serotonin, melatonin, noradrenalin, and adrenalin (37). For example, defects in pterin metabolism in humans are associated with many neurological, behavioral, and movement disorders (38). Given that SPR has housekeeping functions and is expressed constitutively across the skin and other tissues in vertebrates that do not use pterin-based compounds in pigmentation (39), our results show that a key enzyme in pterin metabolism, such as SPR and possibly PTS, has been co-opted for color variation through changes in gene regulation.
This study further expands our understanding of the genetic mechanisms linking color polymorphism to other phenotypic and fitness-related traits. It is well established that chromosomal rearrangements, such as inversions, can facilitate the evolution of multitrait differences by suppressing recombination and preserve tight physical linkage over extended genomic segments (40–43). In contrast, genetic divergence between color morphs in the common wall lizard was restricted to very small and localized genomic intervals adjacent to pigmentation genes. This important result raises an immediate question: How do the described differences in morphology, physiology, and behavior between morphs arise? One plausible explanation is pleiotropic effects of the loci controlling skin pigmentation on other biological processes. The alleles associated with pigmentation differences were characterized by striking sequence divergence, and it is therefore possible that each variant allele carries multiple mutations affecting the expression of SPR, BCO2, and possibly PTS differently in the skin and other tissues, for instance, in brain regions controlling behavioral differences between morphs. Carotenoid and pterin compounds are known to be involved in a wide range of vital metabolic processes (37, 44); thus, alterations in the regulation of their biosynthetic pathways may form the basis of the multitrait divergence often observed between color morphs in many species of reptiles.
Finally, we show that transspecific allele sharing in both pigmentation loci was frequent among several species harboring color polymorphisms identical to those of the common wall lizard. Our results indicate that a combination of balancing selection and introgressive hybridization played a role in the evolution of coloration in the genus Podarcis and led to an unusually long-term maintenance of genetic variation at these pigmentation loci. Other examples of the combination of these two processes in reptiles include highly divergent haplotypes underlying alternate venom types in rattlesnakes (45) and signals of positive selection and introgression at the MHC locus in green lizards (46). Since polymorphic ventral coloration is common in lacertid lizards from multiple genera other than Podarcis, our results raise the intriguing possibility that hybridization might be a frequent mechanism promoting cross-species transfer of visual ornaments and associated traits, which are subsequently kept by balancing selection within species. This fits the emerging paradigm that hybridization and transfer of ancient alleles contribute to adaptive evolution, as shown by a growing list of studies on multiple traits and taxonomic groups, such as the venom types in rattlesnakes (45), wing coloration in Heliconius butterflies (47), seasonal coat color change in hares (48), and beak shape in Darwin’s finches (49). As genomic data accumulate, the molecular determinants of color variation in other polymorphic systems and the role of hybridization and balancing selection in the evolution of visual ornaments can be evaluated systematically.
Materials and Methods
Fully detailed and referenced methods are available in SI Appendix. Permits for sampling and killing these lizards were provided by the Prefecture des Pyrénées Orientales (arrêté no. 2016-2-09) and the Servei de Biodiversitat i Protecció dels Animals (SF/474).
Phenotypic Characterization.
We characterized the phenotype in detail using both TEM and biochemical analyses of pigments, using ventral skin harvested from individuals of all morphs of P. muralis (n = 22). TEM was performed following standard procedures in skin portions collected from animals exhibiting different color morphs. Carotenoid and pteridine content in the skin of all morphs was determined using HPLC-MS/MS.
Reference Genome Sequencing, Assembly, and Annotation.
To generate a reference genome sequence for the common wall lizard, we sequenced a yellow male individual from the Pyrenees region using PacBio single-molecule real-time sequencing at ∼100-fold coverage. To improve the accuracy of the genome sequence, we sequenced the same individual at 30-fold coverage using Illumina reads. Proximity ligation data (CHICAGO and HiC libraries) from an additional individual from the same location were used for scaffolding the draft assembly. To identify the Z chromosome, we compared sequence coverage at each scaffold between DNA pools of males and females. For the annotation of the genome, we obtained RNA-sequencing (RNA-seq) data of five tissues (brain, duodenum, muscle, skin, and testis) using Illumina technology. RNA-seq data from two embryos at the 31-somite stage, incubated at 15 °C or 24 °C, were obtained from a previous publication (50) and combined with the newly generated data. The annotation was performed using an ab initio-based annotation strategy combining gene predictions with the available evidence data.
Population Genomics and Association Mapping.
Genome-wide polymorphism data were obtained using whole-genome resequencing of DNA pools (Illumina). We sampled male individuals of the five color morphs at two localities in the eastern Pyrenees (France). Individuals were pooled by morph and locality in equimolar concentrations and sequenced (10 pools, plus one pool from a location in northern Italy to serve as an outgroup). The number of individuals included in each pool varied from nine to 21, and each pool was sequenced to an effective coverage of 15- to 18-fold. We constructed a neighbor-joining tree based on Nei’s standard genetic distances to infer patterns of population structure among sampled localities and among color morphs, and compared levels and patterns of genetic diversity between localities and morphs using Tajima’s D and genetic diversity using nucleotide diversity (π). Genetic differentiation was summarized using FST. To test for the association between each color morph and specific genomic regions, we calculated the median value of the allele frequency difference (ΔAF) at each SNP between each relevant comparison based on the phenotypic data: (i) presence or absence of orange pigmentation, (ii) presence or absence of yellow pigmentation, and (iii) mosaic patterning or uniform coloration. We then used a sliding window approach (20 SNPs and steps of five SNPs) to identify regions with consistent differentiation across many SNPs. To estimate whether windows displayed higher ΔAF than expected by chance, we conducted 1,000 permutation tests for each contrast. We further conducted a CMH test, which assesses consistent allele frequency changes across biological or technical replicates. To combine the results for each CMH test, we summarized P values using Fisher’s combined probability test, and we performed the same sliding window analysis as for the ΔAF analysis. We confirmed the genotype/phenotype associations in individuals from the Pyrenees through individual-based genotyping using Sanger sequencing. To investigate structural variation around the candidate loci, we prepared Oxford Nanopore libraries of an orange-white individual for sequencing in a MinION device (9.7-fold coverage) and mapped the reads to our reference assembly. We retrieved reads that mapped within and around SPR and BCO2, and manually realigned the reads to the reference (a yellow individual) for analysis. Additionally, for the pool-sequencing dataset, we further performed functional annotation of the variants to check for SNP and indel variants with potential functional significance around the candidate regions.
Gene Expression and Allelic Imbalance Analysis.
Gene expression for SPR, BCO2, PTS, and 18S (a housekeeping gene for expression normalization) was quantified using qPCR. We sampled 26 individuals belonging to all five morphs. After dissection, several organs (skin, brain, liver, and muscle) were harvested and stored for RNA extraction and cDNA synthesis. For the analysis on the skin, we focused only on the pure morph animals so as to avoid possible variation in expression across skin patches of mosaic animals. Quantification cycle (Cq) values of three technical replicates were averaged, and the expression of each focal gene for each sample was normalized to the expression of 18S using a −ΔCq approach.
We also assessed levels of gene expression by analyzing allele imbalance. For all individuals among our cohort of samples used for the qPCR analysis that were heterozygous O/o and Y/y, we screened part of the coding sequence to detect polymorphisms that could be used to quantify the relative expression of each allele. We then designed primers to amplify small fragments from cDNA overlapping these polymorphisms and sequenced these amplicons on a MiSeq system. To calculate the relative proportion of alleles expressed in the skin of each individual for each transcript, we counted the number of reads corresponding to the reference and alternative alleles.
Amplicon Sequencing Overlapping Regions of Association.
We sequenced amplicons (∼550 bp) overlapping the regions of association for the two loci recovered in the genome-wide association mapping for a set of samples (n = 48) that included common wall lizard samples from our primary study location in the eastern Pyrenees (n = 16) and other Podarcis species (n = 32) that are known to exhibit ventral color polymorphism. After individual amplification and barcoding, samples were pooled and sequenced on a MiSeq system. We constructed median-joining haplotype networks and calculated values of dxy and Tajima’s D for each locus. To explore patterns of sequence evolution in a broader phylogenetic context, we constructed neighbor-net networks using an extended dataset with additional sequences from other Podarcis species. Finally, we estimated pairwise sequence divergence between haplotypes of the SPR and BCO2 amplicons within P. muralis and between P. muralis and six additional Podarcis species. The same calculations were also performed for 31 amplicons randomly distributed in the genome (255–667 bp).
Supplementary Material
Acknowledgments
We thank Javier Ábalos, Arnaud Badiane, and Senda Reguera for assistance during field work, and Alvarina Couto, Sara Rocha, Carla Luís, Carolina Pereira, and Joana Mendes for preliminary analyses of the 31 random amplicons. We thank the National Genomics Infrastructure (NGI)/Uppsala Genome Center and Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) for providing assistance in massive parallel sequencing and computational infrastructure. This work was supported by research grants from the Knut and Alice Wallenberg Foundations (to L.A.); by the Swedish Research Council (Grant 2017-02907 to L.A. and Grant E0446501 to T.U.); by the Fundação para a Ciência e Tecnologia (FCT) through POPH-QREN funds from the European Social Fund and Portuguese MCTES (FCT Investigator Grants IF/00283/2014/CP1256/CT0012 to M.C. and IF/01597/2014/CP1256/CT0009 to C.P. and Postdoctoral Fellowships SFRH/BPD/99138/2013 to S.J.S. and SFRH/BPD/94582/2013 to G.P.i.d.L.); by a Wallenberg Academy Fellowship (to T.U.); by Research Fellowships attributed to P.A. (PD/BD/114028/2015) and P.P. (PD/BD/128492/2017) in the scope of the Biodiversity, Genetics, and Evolution PhD Program at InBIO/Centro de Investigação em Biodiversidade e Recursos Genéticos (CIBIO) and the University of Porto; by the project “PTDC/BIA-EVL/30288/2017 - NORTE -01-0145-FEDER-30288”, cofunded by NORTE2020 through Portugal 2020 and FEDER Funds and by National Funds through the FCT; by the projects “Genomics and Evolutionary Biology” and “Genomics Applied to Genetic Resources” cofinanced by North Portugal Regional Operational Programme 2007/2013 (ON.2 – O Novo Norte) under the National Strategic Reference Framework and the European Regional Development Fund; and by a European Union FP7 REGPOT grant (CIBIO-New-Gen, 286431). M.A.C. was supported by the project NORTE-01-0145-FEDER-000007. D.S. is currently supported by the program “Rita Levi Montalcini” (Ministero dell’Istruzione dell’Università e della Ricerca) for the recruitment of young researchers at the University of L’Aquila. Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and the Science for Life Laboratory, Sweden.
Footnotes
The authors declare no conflict of interest.
Data deposition: The reference genome sequence, annotation, and all high-throughput sequencing data reported in this article have been deposited to National Center for Biotechnology Information (BioProject PRJNA515813). Pterin and carotenoid quantification data, quantitative PCR results, and amplicon sequence alignments have been deposited in the Dryad Digital Repository (doi:10.5061/dryad.j307mv3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820320116/-/DCSupplemental.
References
- 1.McKinnon JS, Pierotti MER. Colour polymorphism and correlated characters: Genetic mechanisms and evolution. Mol Ecol. 2010;19:5101–5125. doi: 10.1111/j.1365-294X.2010.04846.x. [DOI] [PubMed] [Google Scholar]
- 2.Wellenreuther M, Svensson EI, Hansson B. Sexual selection and genetic colour polymorphisms in animals. Mol Ecol. 2014;23:5398–5414. doi: 10.1111/mec.12935. [DOI] [PubMed] [Google Scholar]
- 3.Sinervo B, Lively CM. The rock-paper-scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. [Google Scholar]
- 4.Sinervo B, Svensson E. Correlational selection and the evolution of genomic architecture. Heredity (Edinb) 2002;89:329–338. doi: 10.1038/sj.hdy.6800148. [DOI] [PubMed] [Google Scholar]
- 5.Roulin A. The evolution, maintenance and adaptive function of genetic colour polymorphism in birds. Biol Rev Camb Philos Soc. 2004;79:815–848. doi: 10.1017/s1464793104006487. [DOI] [PubMed] [Google Scholar]
- 6.Cuthill IC, et al. The biology of color. Science. 2017;357:eaan0221. doi: 10.1126/science.aan0221. [DOI] [PubMed] [Google Scholar]
- 7.Olsson M, Stuart-Fox D, Ballen C. Genetics and evolution of colour patterns in reptiles. Semin Cell Dev Biol. 2013;24:529–541. doi: 10.1016/j.semcdb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Cooper WE, Jr, Greenberg N. Reptilian coloration and behavior. In: Gans C, Crews D, editors. Biology of the Reptilia. Vol 18. Univ Chicago Press; Chicago: 1992. pp. 298–422. [Google Scholar]
- 9.McLean CA, Lutz A, Rankin KJ, Stuart-Fox D, Moussalli A. Revealing the biochemical and genetic basis of color variation in a polymorphic lizard. Mol Biol Evol. 2017;34:1924–1935. doi: 10.1093/molbev/msx136. [DOI] [PubMed] [Google Scholar]
- 10.Rankin KJ, McLean CA, Kemp DJ, Stuart-Fox D. The genetic basis of discrete and quantitative colour variation in the polymorphic lizard, Ctenophorus decresii. BMC Evol Biol. 2016;16:179. doi: 10.1186/s12862-016-0757-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitze PS, et al. Carotenoid-based colours reflect the stress response in the common lizard. PLoS One. 2009;4:e5111. doi: 10.1371/journal.pone.0005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez i de Lanuza G, Font E, Carazo P. Color-assortative mating in a color-polymorphic lacertid lizard. Behav Ecol. 2013;24:273–279. [Google Scholar]
- 13.Pérez i de Lanuza G, Ábalos J, Bartolomé A, Font E. Through the eye of a lizard: Hue discrimination in a lizard with ventral polymorphic coloration. J Exp Biol. 2018;221:jeb169565. doi: 10.1242/jeb.169565. [DOI] [PubMed] [Google Scholar]
- 14.Salvi D, Harris DJ, Kaliontzopoulou A, Carretero MA, Pinho C. Persistence across Pleistocene ice ages in Mediterranean and extra-Mediterranean refugia: Phylogeographic insights from the common wall lizard. BMC Evol Biol. 2013;13:147. doi: 10.1186/1471-2148-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez i de Lanuza G, Sillero N, Carretero MA. Climate suggests environment-dependent selection on lizard colour morphs. J Biogeogr. 2018;45:2791–2802. [Google Scholar]
- 16.Sacchi R, et al. Microgeographic variation of colour morph frequency and biometry of common wall lizards. J Zool (Lond) 2007;273:389–396. [Google Scholar]
- 17.Pérez I de Lanuza G, Carretero MA, Font E. Intensity of male-male competition predicts morph diversity in a color polymorphic lizard. Evolution. 2017;71:1832–1840. doi: 10.1111/evo.13256. [DOI] [PubMed] [Google Scholar]
- 18.Calsbeek B, Hasselquist D, Clobert J. Multivariate phenotypes and the potential for alternative phenotypic optima in wall lizard (Podarcis muralis) ventral colour morphs. J Evol Biol. 2010;23:1138–1147. doi: 10.1111/j.1420-9101.2010.01978.x. [DOI] [PubMed] [Google Scholar]
- 19.Sacchi R, Mangiacotti M, Scali S, Ghitti M, Zuffi MAL. Effects of colour morph and temperature on immunity in males and females of the common wall lizard. Evol Biol. 2017;44:496–504. [Google Scholar]
- 20.Abalos J, Pérez i de Lanuza G, Carazo P, Font E. The role of male coloration in the outcome of staged contests in the European common wall lizard (Podarcis muralis) Behaviour. 2016;153:607–631. [Google Scholar]
- 21.Galeotti P, et al. Colour polymorphism alternative breeding strategies: Effects of parent’s colour morph on fitness traits in the common wall lizard. Evol Biol. 2013;40:385–394. [Google Scholar]
- 22.Sacchi R, et al. Seasonal variations of plasma testosterone among colour-morph common wall lizards (Podarcis muralis) Gen Comp Endocrinol. 2017;240:114–120. doi: 10.1016/j.ygcen.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Putnam NH, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016;26:342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vujošević M, Blagojević J. The distribution of constitutive heterochromatin and nucleolus organizers in lizards of the family Lacertidae (Sauria) Genetika. 1999;31:269–276. [Google Scholar]
- 26.Olmo E, Odierna G, Capriglione T. Evolution of sex-chromosomes in lacertid lizards. Chromosoma. 1987;96:33–38. [Google Scholar]
- 27.Reeve HK, Pfennig DW. Genetic biases for showy males: Are some genetic systems especially conducive to sexual selection? Proc Natl Acad Sci USA. 2003;100:1089–1094. doi: 10.1073/pnas.0337427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirkpatrick M, Hall DW. Sexual selection and sex linkage. Evolution. 2004;58:683–691. doi: 10.1111/j.0014-3820.2004.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson J, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4:e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazave E, Marqués-Bonet T, Fernando O, Charlesworth B, Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaliontzopoulou A, Pinho C, Harris DJ, Carretero MA. When cryptic diversity blurs the picture: A cautionary tale from Iberian and North African Podarcis wall lizards. Biol J Linn Soc Lond. 2011;103:779–800. [Google Scholar]
- 32.Nordén KK, Price TD. Historical contingency and developmental constraints in avian coloration. Trends Ecol Evol. 2018;33:574–576. doi: 10.1016/j.tree.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Mundy NI, et al. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr Biol. 2016;26:1435–1440. doi: 10.1016/j.cub.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 34.Lopes RJ, et al. Genetic basis for red coloration in birds. Curr Biol. 2016;26:1427–1434. doi: 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooke TF, et al. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell. 2017;171:427–439.e21. doi: 10.1016/j.cell.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshmukh R, Baral S, Gandhimathi A, Kuwalekar M, Kunte K. Mimicry in butterflies: Co-option and a bag of magnificent developmental genetic tricks. Wiley Interdiscip Rev Dev Biol. 2018;7:e291. doi: 10.1002/wdev.291. [DOI] [PubMed] [Google Scholar]
- 37.Thöny B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347:1–16. [PMC free article] [PubMed] [Google Scholar]
- 38.Longo N. Disorders of biopterin metabolism. J Inherit Metab Dis. 2009;32:333–342. doi: 10.1007/s10545-009-1067-2. [DOI] [PubMed] [Google Scholar]
- 39.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Tuttle EM, et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr Biol. 2016;26:344–350. doi: 10.1016/j.cub.2015.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014;24:R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 43.Lamichhaney S, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nat Genet. 2016;48:84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 44.Olson VA, Owens IPF. Costly sexual signals: Are carotenoids rare, risky or required? Trends Ecol Evol. 1998;13:510–514. doi: 10.1016/s0169-5347(98)01484-0. [DOI] [PubMed] [Google Scholar]
- 45.Dowell NL, et al. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr Biol. 2018;28:1016–1026.e4. doi: 10.1016/j.cub.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagonas K, et al. Selection, drift, and introgression shape MHC polymorphism in lizards. Heredity (Edinb) (September 26, 2018 doi: 10.1038/s41437-018-0146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones MR, et al. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science. 2018;360:1355–1358. doi: 10.1126/science.aar5273. [DOI] [PubMed] [Google Scholar]
- 49.Lamichhaney S, et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 50.Feiner N, Rago A, While GM, Uller T. Signatures of selection in embryonic transcriptomes of lizards adapting in parallel to cool climate. Evolution. 2018;72:67–81. doi: 10.1111/evo.13397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.