Significance
IL-37 is the newest member of the IL-1 family of cytokines that elicit diverse activities integrally related to the innate immune responses and inflammation. Whereas most members elicit proinflammatory responses; in contrast, IL-37 broadly down-regulates inflammation and exists in a unique monomer/dimer equilibrium. Experimental and theoretical analyses show that monomeric IL-37 results in more effective suppression of inflammatory markers on multiple cell types compared with native IL-37, whereas the IL-37 dimer functions to block the activity of the monomer. Considering the emerging efficacy of antagonists and antibodies that specifically target IL-1 in human disease, identifying the active form of IL-37 may lead to the clinical development of recombinant IL-37 to down-regulate inflammation.
Keywords: interleukin, inflammation, dimer, innate immunity
Abstract
Interleukin-37 (IL-37), a member of the IL-1 family of cytokines, is a fundamental suppressor of innate and acquired immunities. Here, we used an integrative approach that combines biophysical, biochemical, and biological studies to elucidate the unique characteristics of IL-37. Our studies reveal that single amino acid mutations at the IL-37 dimer interface that result in the stable formation of IL-37 monomers also remain monomeric at high micromolar concentrations and that these monomeric IL-37 forms comprise higher antiinflammatory activities than native IL-37 on multiple cell types. We find that, because native IL-37 forms dimers with nanomolar affinity, higher IL-37 only weakly suppresses downstream markers of inflammation whereas lower concentrations are more effective. We further show that IL-37 is a heparin binding protein that modulates this self-association and that the IL-37 dimers must block the activity of the IL-37 monomer. Specifically, native IL-37 at 2.5 nM reduces lipopolysaccharide (LPS)-induced vascular cell adhesion molecule (VCAM) protein levels by ∼50%, whereas the monomeric D73K mutant reduced VCAM by 90% at the same concentration. Compared with other members of the IL-1 family, both the N and the C termini of IL-37 are extended, and we show they are disordered in the context of the free protein. Furthermore, the presence of, at least, one of these extended termini is required for IL-37 suppressive activity. Based on these structural and biological studies, we present a model of IL-37 interactions that accounts for its mechanism in suppressing innate inflammation.
The discovery of IL-1 marked a revolution in biology with the identification of a single highly active protein that could induce inflammation at picamolar concentrations (1). In many ways, the discovery of IL-37 has led to a similar revolution whereby IL-37 can limit or even prevent a broad spectrum of inflammatory models now termed innate immunity. Specifically, IL-37 can reverse inflammation, diminish multiple phenotypes related to cancer, and even reverse age-related phenotypes (2–6). Much of the molecular details of IL-37 signaling has been surmised from animal models and cellular studies that include colocalization studies, which suggest that IL-37 engages both the IL-18Rα receptor and the previously orphaned IL-1R8 receptor (7–10). The elegance of this proposed signaling mechanism is that IL-37 could both block the proinflammatory-induced response by IL-18 and simultaneously signal to promote an antiinflammatory response. However, the molecular details of IL-37 remain elusive as IL-37 exhibits several unique features relative to other IL-1 family members, which include the formation of a dimer and the presence of extended termini.
Unlike other members of the IL-1 family that are monomeric, IL-37 self-associates with nanomolar affinity (11, 12) and comprises longer extended termini relative to other family members. ILs from different families have been shown to dimerize with a spectrum of functional consequences, which range from IL-10 family members that are fully active as swapped dimers (13, 14) to chemokines, such as IL-8 that engage their receptors primarily as monomers (15–17). Indeed, the recent crystal structure of IL-37 by Ellisdon et al. (12) has afforded the engineering of IL-37 mutant forms that disrupt dimer formation and simultaneously lead to higher IL-37 activities. However, the activity of these IL-37 mutant forms were compared well below their determined self-association affinities where dimer formation would be expected to be negligible, suggesting the presence of cellular determinants that further modulate dimerization analogous to an array of molecules that modulate chemokine dimerization (18). Considering the effectiveness of IL-1 blocking therapies already in use today (19), further elucidating the activities of these monomeric IL-37 forms and the molecular determinants of IL-37 activity may open a window to new therapies that utilize variations of IL-37 to block acute inflammation and potentially cancer progression. Thus, here we sought to address the solution behavior of IL-37 and its monomeric forms, determine whether the higher activities of IL-37 monomeric versions are a more general phenotype that would further support an inhibitory role for the IL-37 dimer, and determine the role of the extended IL-37 termini.
Results and Discussion
Only Mutations at the IL-37 Dimer Interface Specifically Disrupt Dimer Formation.
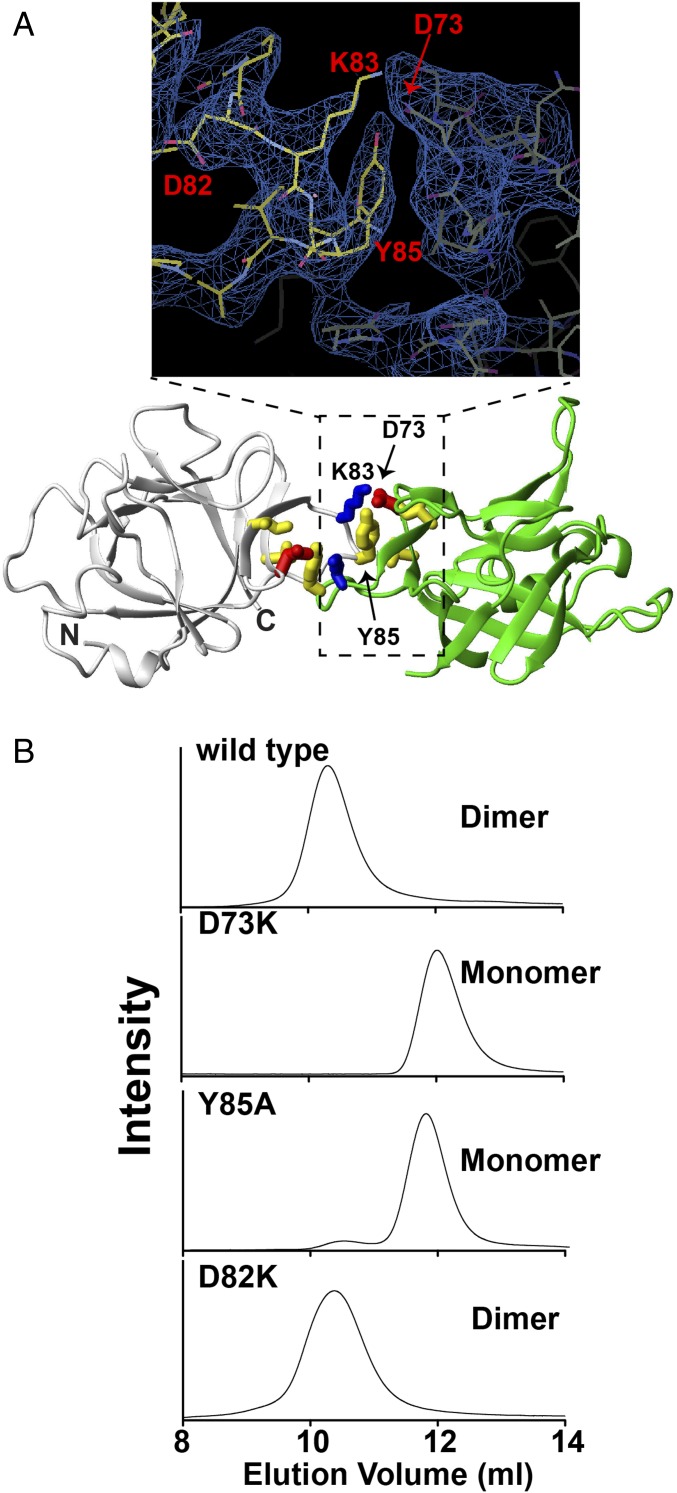
The biological form of secreted IL-37 comprises residues 46–218 (IL-3746–218), and terminal truncations together with NMR analysis described below identified a stable core of IL-3753–206 amenable to crystallization and subsequent structure determination. Whereas the recent X-ray crystal structure of a similar construct of IL-3750–206 was solved with the proposed dimer interface formed between IL-37 monomers within different asymmetric units (12), IL-3753–206 crystallized here was in the P3121 space group where each dimer was visible within each asymmetric unit [Protein Data Bank (PDB) accession number 6NCU]. Thus, despite crystallization within a completely different space group and relatively low resolution of these crystals (Table 1), density across symmetry mates clearly identifies the same dimer interface as this recent IL-37 structure (Fig. 1A). Specifically, Asp73 in one monomer forms a stabilizing ionic bond with Lys83 in another monomer, and Tyr85 forms a hydrophobic cluster with Val71 and Ile78 across this interface. As expected, mutation of either IL-37D73K (D73K) or IL-37Y85A (Y85A) results in stable monomeric forms as assessed by size-exclusion chromatography, whereas a control mutation of IL-37D82K retains dimer formation (Fig. 1B). Thus, IL-37 dimers can be converted into IL-37 monomers by specific point mutations to the dimer interface. NMR studies were also undertaken to probe the solution behavior of IL-37 and characterize the extended termini of IL-37 that are longer than other IL-1 family members.
Table 1.
Structural statistics for the IL-37 crystal structure
| Data collection | |
| Space group | P3121 |
| Cell dimensions | |
| a, b, c (Å) | 67.50, 67.510, 142.74 |
| α, β, γ (Å) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 3.5 |
| Rmerge (%) | 9.0 |
| I/αI | 16.6 |
| Completeness (%) | 99 |
| Redundancy | 19 |
| Refinement | |
| Resolution (Å) | 54.1–3.5 |
| Number of reflections | 5,091 |
| Rwork/Rfree | 34.0 (39.9)* |
| Number of atoms | 2385 |
| Protein | 2385 |
| B-factors | 108.8 |
| rms deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.0612 |
| Ramachandran | |
| Favored (%) | 83.8 |
| Allowed (%) | 12.84 |
| Outliers (%) | 3.4 |
| Clash score | 12 |
Values in the parenthesis refer to the highest-resolution shell.
Fig. 1.
IL-37 crystal structure and analytical sizing of IL-37 point mutations. (A) Electron density of the IL-3753–206 dimer interface (Top) and key interactions (Bottom). These include an ionic interaction among D73 (red) and K83 (blue) and Y85 mediated hydrophobic interactions (yellow). (B) Analytical size-exclusion chromatography (Superdex 75) of wild-type (WT) IL-37 and monomer-inducing point mutations D73K and Y85A along with a control mutation IL-37D82K. Analytical size exclusion was conducted in the context of the mature IL-37 (residues 46–218 with the WT sequence and the single point mutations).
The IL-37 Termini Are Disordered, Whereas the Dimer Interface Is Highly Ordered.
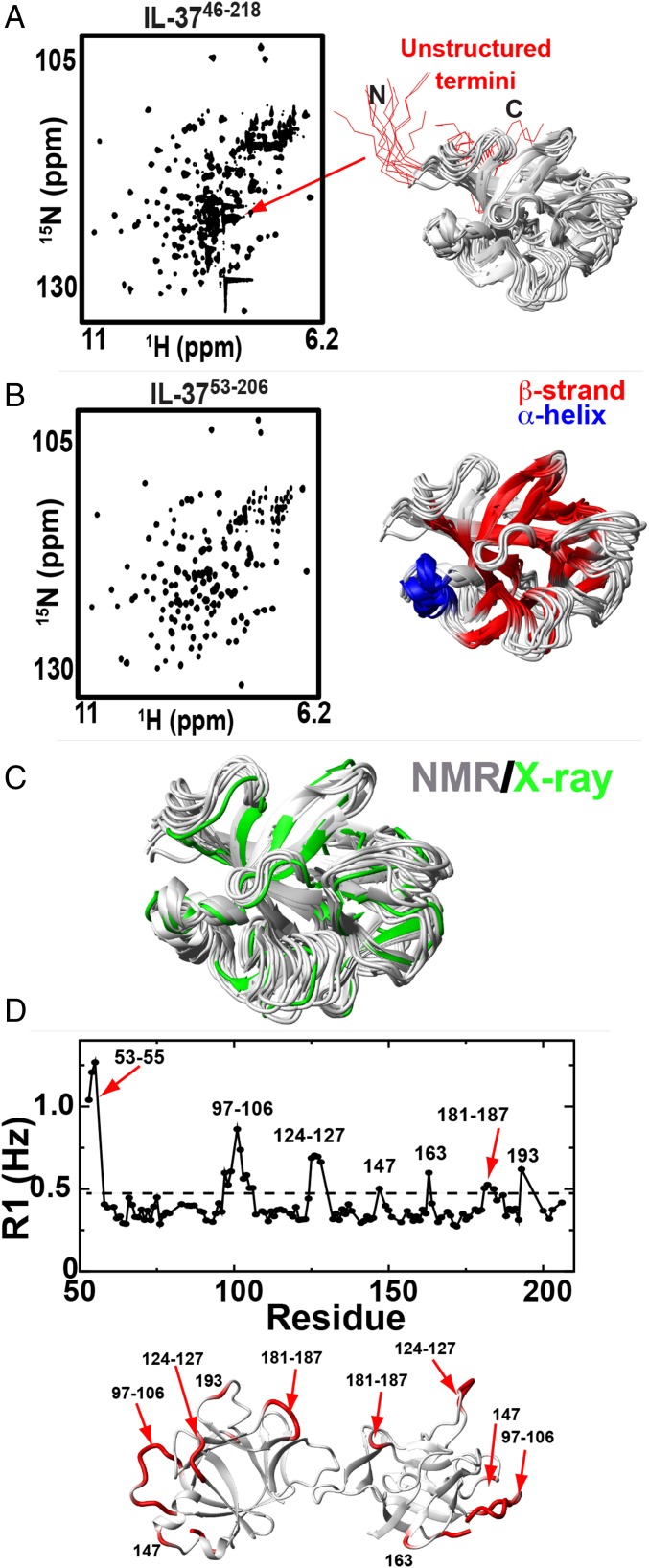
To address the solution behavior of IL-37 and provide a foundation for further comparative studies of the monomeric versions of IL-37, we exploited the high sensitivity of NMR. The WT biologically active form of IL-3746–218 gives rise to a well-dispersed 2D-heteronuclear single quantum coherence (HSQC) spectrum that is indicative of a well-folded protein (Fig. 2A). However, a significant overlap of highly intense peaks within the center of the spectrum is suggestive of unfolded regions, which we hypothesized are due to the presence of disordered termini. To test this hypothesis, we truncated both termini to the smallest soluble recombinant protein of IL3753–206, which results in a HSQC devoid of this spectral crowding (Fig. 2B). Moreover, these termini do not alter the chemical environment of the centrally folded region of IL3753–206 as the spectra are completely superimposable that suggests there is no structural change to the IL-37 core upon their removal. The determination of the low-resolution solution structure of the monomer guided by our chemical shift assignments (Biological Magnetic Resonance Bank accession number 27729) and experimental amide NOEs facilitated the low-resolution structure of IL3753–206 in the context of its native dimer that indicates the solution structure is identical to that determined within the crystal (Fig. 2C and Table 2). Thus, these solution studies provide compelling evidence to suggest that the extended termini of the atypical IL-1 family member are disordered in the context of the free protein.
Fig. 2.
NMR solution studies of IL-37. (A, Left) 15N-HSQC spectra of IL-3746–218. (A, Right) Structural ensemble of the 10 highest scoring structures showing residues 46–218 calculated from resolution adapted structural recombination (RASREC) Rosetta with the flexible termini shown (red). (B, Left) 15N-HSQC spectra of IL-3753–206. (B, Right) Structural ensemble of the same 10 highest scoring structures with a secondary structure of α-helices (blue) and β-strands (red) mapped onto the ensemble calculated using measured chemical shifts and predictions calculated in the chemical shift index (33). Calculated β-strands include residues 59–63, 69–80, 90–97, 109–113, 119–124, 131–137, 155–161, 164–169, 175–179, 189–190, and 198–203, and α-helices include residues 140–145 and 148–151. (C) Comparison between this solution ensemble (white) and one subunit within the X-ray crystal structure (green) of residues 53–206 shown. (D) R1 relaxation rates of IL-3753–206 with the average 0.41 ± 0.16 s−1 (dashed line) and residues greater than +0.5 SD mapped onto the X-ray crystal structure dimer (red).
Table 2.
Structural statistics for the IL-37 monomer 10 NMR solution structure determined using backbone chemical shifts with RASREC Rosetta
| NOE distance restraints | 234 |
| Short-range (intraresidue) | 22 |
| Sequential | 91 |
| Medium-range (|i-j|<=4) | 46 |
| Long-range (|i-j|>4) | 75 |
| Violations >0.5 Å of hard restraints | 23 ± 2 |
| Long-range violations >0.5 Å of hard restraints | 5 ± 1 |
| Average long-range violation (Å) | 0.59 |
| Average rmsd (over all β-strands) | |
| Backbone (Å) | 1.21 ± 0.3 |
| Heavy atom (Å) | 1.76 ± 0.39 |
| Ramachandran plot summary | |
| Most favored regions (%) | 83.1 |
| Allowed regions (%) | 15.7 |
| Generously allowed regions (%) | 0.2 |
| Disallowed regions (%) | 1.1 |
In addition, to determine that the IL-37 termini are disordered, the dynamic nature of the centrally folded IL-37 core was probed using R1 relaxation rates as sensitive markers for local flexibility (Fig. 2D). Overall, the R1 relaxation rates are consistent with the relatively large dimer with an average R1 relaxation rate of 0.41 ± 0.16 s−1. Relatively flexible regions exhibiting R1 relaxation rates can be identified by elevated R1 relaxation rates, shown as, at least, 0.5 SD above the average (Fig. 2C, dotted line on the left and shown in red mapped onto our X-ray structure on the right). The entire IL-37 dimer interface is relatively rigid, indicating that the loops at this interface are relatively rigid. Conversely, many loops on the periphery of the IL-37 dimer are disordered that include the relative large loop of residues 96–106 and residues 181–187. Whether these loops are “flexible for function” and comprise the binding epitopes will await future studies aimed at assessing IL-37 interactions with its identified receptors.
IL-37 Mutations at the Dimer Interface Remain Monomeric at Millimolar Concentrations.
In general, mutations of protein dimer interfaces may disrupt dimer association and thereby simply shift the equilibrium toward a monomeric form with continued association at higher concentrations, begging the question of whether the engineered IL-37 monomers weakly associate. For example, we have previously shown that CXCL8 mutants that disrupt dimer association nevertheless persist in dimer interactions in solution (20). Specifically, R1 relaxation was used to both probe for local disorder and was simultaneously used as a sensitive reporter of the overall tumbling time that reports on the oligomeric state (i.e., monomer versus dimer). R1 relaxation rates were determined for both D73K and Y85A, which were compared with WT IL-37 (all in the context of IL-3753–206).
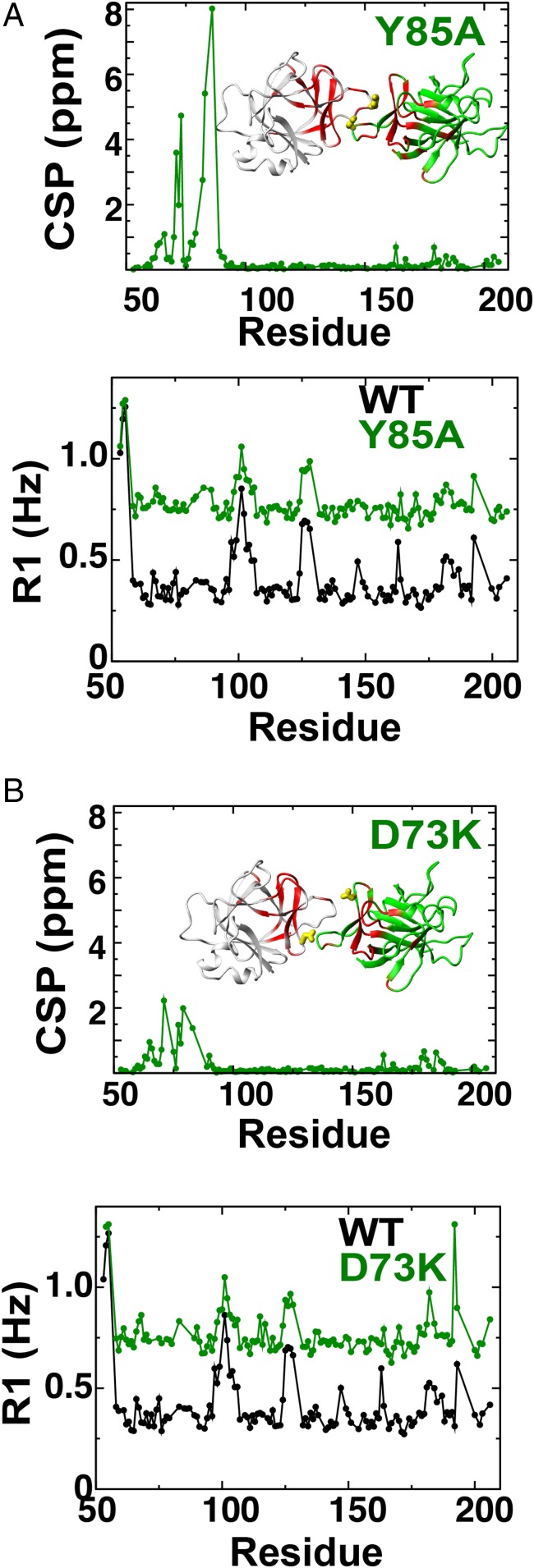
Using the central core of IL-37 residues 53–206 with point mutations of D73K and Y85A, chemical shift perturbations (CSPs) were induced only to the dimer interface (Fig. 3 A and B), suggesting that there are little to no changes to the global structure. These monomeric forms exhibit average R1 relaxation rates that are nearly twice as high as the WT dimer with average R1 rates of 0.77 ± 0.10 and 0.79 ± 0.10 s−1 for D73K and Y85A, respectively (Fig. 3B). Such relatively high R1 relaxation rates are consistent with these mutants retaining their oligomeric states as monomers down to the high concentrations necessary for NMR (near millimolar). Additionally, R1 relaxation rates of the individual mutants indicate that their internal dynamics are remarkably similar to the WT IL-37, indicating that these point mutations do not incur substantial global effects on the dynamics and that the dimer interface still remains relatively rigid.
Fig. 3.
Solution characterization of IL-37 monomeric mutants. Both amide CSPs and R1 relaxation rates are shown for (A) Y85A and (B) D73K. CSPs were determined for WTs and mutants in the context of IL-3753–206, and R1 relaxation rates are plotted for the WT IL-3753–206 together with each mutant form for comparison. Average R1 relaxation rates for Y85A and D73K are 0.79 ± 0.10 and 0.77 ± 0.10 s−1, respectively, consistent with monomer tumbling and compared with the slower average R1 relaxation rate of the WT IL-3753–206 dimer with an average of 0.41 ± 0.16 s−1.
IL-37 Exhibits a Unique Dose-Dependent Profile on Multiple Cell Types.
Dose dependencies that monitor the antiinflammatory response of recombinant IL-37 on primary cells elicit a distinct response (Fig. 4), which can be rationalized based on the unique dimerization property of this IL-1 family member. Specifically, increasing doses of IL-37 initially blocks the secretion of proinflammatory cytokines in both mouse and human primary cells induced by either PMA or LPS (Fig. 4 A and B). IL-37 antiinflammatory activity is observed to reach a maximum response after which increased doses of IL-37 become less effective. As a further corroboration of these results, a similar IL-37 dose–response was also observed using J774A.1 (J1) macrophages as an in vitro model system (Fig. 4 C and D). Collectively, these data further support a role in IL-37 dimerization in suppressing its antiinflammatory activity.
Fig. 4.
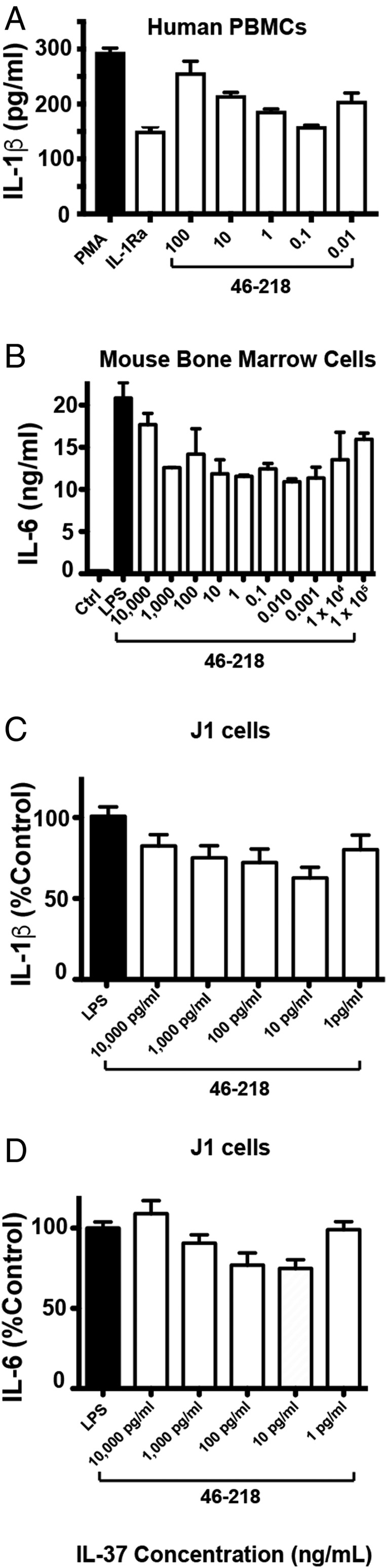
IL-37 elicits a unique (parabolic) dose dependence observed under multiple conditions. (A) Recombinant IL-37 suppression of phorbol 12-myristate 13-acetate (PMA)-induced IL-1β secretion in human peripheral blood mononuclear cells (PBMCs) pretreated with indicated doses of IL-37 2 h before treatment with 25 ng/mL of PMA for 24 h in the presence of 10% FCS. IL-1Ra was also used at 10 μg/mL (B) Recombinant IL-37 (IL-3746–218) suppression of LPS-induced IL-6 secretion in bone marrow cells of mice pretreated with indicated doses of IL-37 2 h before treatment with 100 ng/mL LPS. Assays were performed in quadruplicate in 96 well plates with 500,000 cells/mL. J1 murine macrophage cells were stimulated with LPS, and the indicated doses of IL-3746–218 and supernatants were probed for both (C) IL-1β and (D) IL-6 as described in Fig. 5.
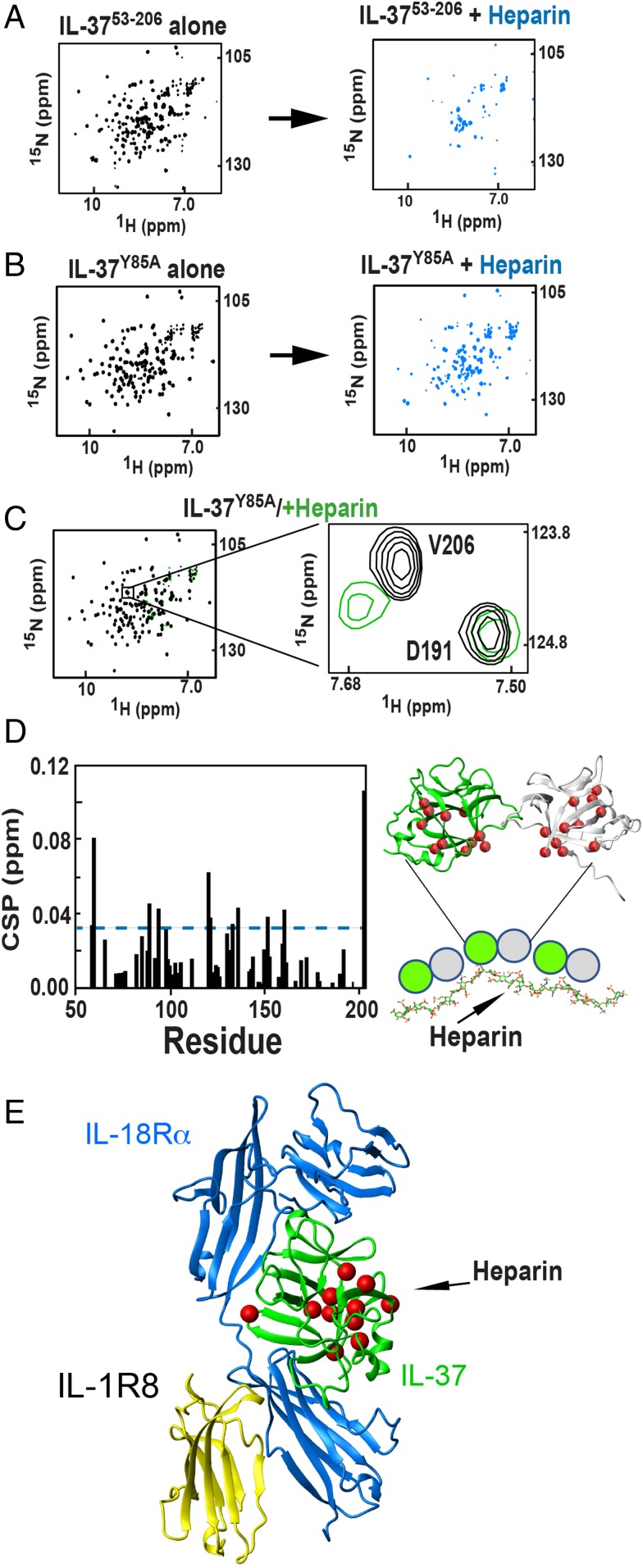
Mechanistically, these dose dependencies reveal surprising characteristics of IL-37 dimer association that include the role of the IL-37 dimer in blocking IL-37 activity induced by the IL-37 monomer. For example, a theoretical derivation of the IL-37 self-association binding isotherm shows that, although increasing concentrations of IL-37 result in the expected relative increase in IL-37 dimers (SI Appendix, Fig. S1A), the absolute concentration of IL-37 monomers still increases (SI Appendix, Fig. S1B). Collectively, these data suggest that the increase in IL-37 dimers inhibit the increase in IL-37 monomer activity, potentially by occluding monomer binding sites. Moreover, IL-37 dimer formation is likely mediated by endogenous molecules. For example, the self-association constant was initially reported as 5 nM (11), and an even weaker self-association constant was recently reported (12). However, the inflection point where IL-37 concentrations becomes inactive varies with different cell types and is well below the reported self-association constants. This modulation of IL-37 oligomerization may be reminiscent of chemokines that interact with a variety of glycosaminoglycans and may therefore be cell-type dependent. Such a possibility was directly assessed by NMR titrations with heparin below, which is a common glycosaminoglycan that does, indeed, engage IL-37 (see Fig. 8).
Fig. 8.
IL-37 interacts with heparin. Heparin induces severe line broadening in IL-37 15N-HSQC spectra as shown in a comparison of free protein and equimolar concentrations of heparin for the (A) IL-3753–206 dimer and (B) mutant Y85A. (C) 15N-HSQC spectra Y85A (300 μM) with substoichiometric heparin (50 μM), highlighting amide chemical shifts. (D) IL-37 residues that exhibit large CSPs in the presence of heparin are largely confined to one surface. (Left) Amide CSPs per residue induced by heparin binding within Y85A with substoichiometric heparin added. (Right) CSPs greater than 1.0 SD above the average (0.033 ppm) are mapped onto the structural model of the IL-37 dimer (red spheres) that illustrate how either a monomer or a dimer may engage heparin and a representative model of IL-37 interacting with a heparin chain. (E) Residues are mapped (red spheres) onto the ternary complex model of IL-37/IL-18Rα/IL-1R8. Mapped residues that exhibit large CSPs upon substoichiometric addition of heparin are K59, F60, I90, A95, C122, D123, L135, E138, F154, S163, and V206.
Either Termini of IL-37 Are Required for Activity and Monomeric Forms of IL-37 Increase the Anti-Inflammatory Activity.
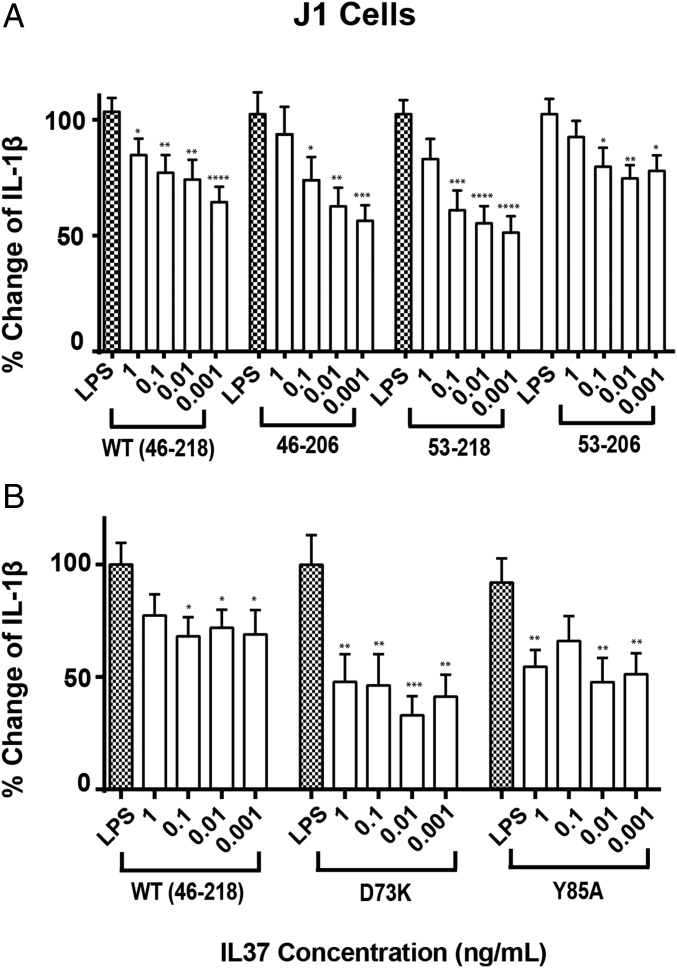
The lack of structured termini for IL-3746–218 and the suppressive effects of its dimerization facilitate multiple avenues to engineer variants of IL-37 to explore their relative contributions to function. To this end, we sought to directly test the contributions to the activity of the IL-37 termini as well as confirm that IL-37 monomeric mutants enhance IL-37 activity. As J1 cells were shown above to recapitulate the IL-37 activity profiles observed in primary cells, J1 cells were therefore utilized to test the activity of engineered IL-37 forms described herein.
Terminal truncations of the mature WT IL-3746–218 were constructed, which include the C-terminally truncated form (IL-3746–206), the N-terminally truncated form (IL-3753–218), and the central structured form that was subjected above to both X-ray crystallography and NMR studies (IL-3753–206). Activities of these terminal truncated forms were compared with WT IL-37 (Fig. 5A). Interestingly, the removal of either the N-terminal or the C-terminal residues leads to no measurable change to LPS-induced IL-1β secretion, yet the removal of both termini led to a recombinant form with slightly reduced activity compared with WT IL-37. These data indicate that the removal of the disordered termini of IL-37 do not simply remove regions that are sterically obstructing a single receptor interaction but that there is possibly a more complicated mechanism of engagement. For example, one possibility is that each terminus is critical for specific interactions for specific receptors whereby the other terminus is a negative regulator. Conversely, there may be nonspecific interactions that enhance receptor affinity. Either way, the removal of both termini renders IL-37 unable to bind both receptors.
Fig. 5.
IL-37-mediated antiinflammatory activity is not increased by terminal truncations but only mutations that disrupt dimer formation. (A) J1 murine macrophage cells were pretreated 1 h with the indicated doses of WT IL-3746–218 and engineered terminal mutants of IL-37, stimulated with 1 µg/mL LPS for 4 h and, subsequently, treated with 20 µM of nigericin for 1 h before monitoring IL-1β secretion. (B) WT IL-3746–218 along with both point mutations that specifically disrupt the IL-37 dimer, Y85A, and D73K, was assayed identically to A. Assays were performed in triplicate in a 96 well plate with 750,000 cells/mL Cytokine production was quantified by ELISA. All assays are representative of, at least, five independent experiments. Error bars represent the SEM; *P < 0.05, **P < 0.001, and ***P < 0.0001, statistical significance was assessed using the unpaired Student’s t test.
Both IL-37 monomeric mutations were also tested to confirm that disruption of the WT IL-37 dimer enhances IL-37 activity in the context of the fully mature IL-3746–218. Indeed, the activities of both D73K and Y85A are enhanced relative to WT IL-37 (Fig. 5B). These data support earlier important findings that indicate the antiinflammatory activity of IL-37 lies in its monomeric form (12), at least, with regard to macrophages that were also extended to primary cells below.
IL-37 Monomers Down-Regulate VCAM-1 Expression in a Dose-Dependent Manner as Opposed to WT IL-37.
Consistent with the data described above that utilized cell lines, we studied the dose–response of native IL-3746–218 in primary human aortic valve cells (AVICs). As shown in Fig. 6A, LPS-induced expression of VACM-1 is reduced from an eightfold increase over the baseline level (normalized to 1.0) to only 1.8-fold by IL-3746–218 at 0.05 nM. This low concentration of IL-3746–218 is typical of many in vitro studies. However, as shown in Fig. 6B, an increase in IL-3746–218 to 0.5 nM results in the loss of a significant ability to suppress VCAM-1 expression. In contrast, both mutant monomers in the same assay at 1.0 nM suppressed VCAM-1 expression much better than the WT form. The interpretations of these data are consistent with dimeric forms of WT IL-37 preventing the monomeric IL-37 from access to its receptors. With increasing doses of IL-37 WT and monomeric mutant forms, the higher activity of the IL-37 monomers become increasingly apparent (Fig. 6 C and D). For example, at 2.5 nM, there was a 50% reduction in VCAM-1 for WT IL-37, but the monomeric mutant D73K reduced expression from eightfold nearly back to the baseline level. Here, the monomeric D73K reduced VCAM-1 from eightfold to 1.8-fold, a 90% decrease. These data support the higher activity of the monomeric IL-37 forms shown above in the cell lines. Thus, using primary cells, we can appreciate the antiinflammatory properties of monomeric IL-37 as the effective form for limiting inflammation. Although monomeric mutants do not exist in nature, the data nevertheless convincingly reveal that the IL-37 monomer is the active form. As discussed below, the ability of low concentrations to be effective in suppressing innate inflammation likely requires a cofactor (s).
Fig. 6.
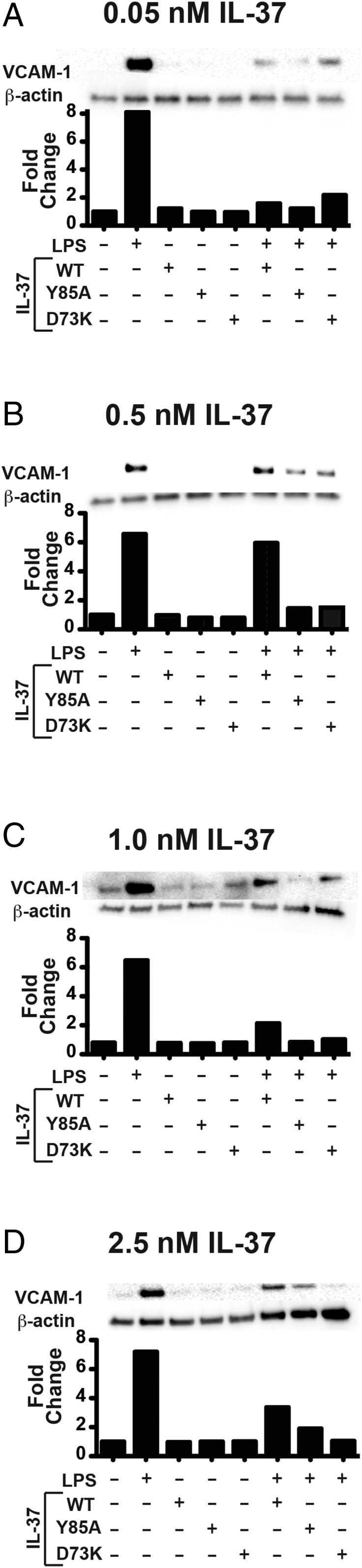
Suppression of LPS-induced VCAM-1 by high concentrations of IL-37 monomers in human AVICs. (A) Normal human AVICs were treated with 0.05 nM recombinant WT IL-3746–218, Y85A, and D73K or untreated before stimulation with LPS and then subsequently probed for VCAM-1. Identical assays were performed with (B) 0.05 nM, (C) 1.0 nM, and (D) 2.5 nM recombinant IL-37 proteins.
Implications for IL-37 Interactions.
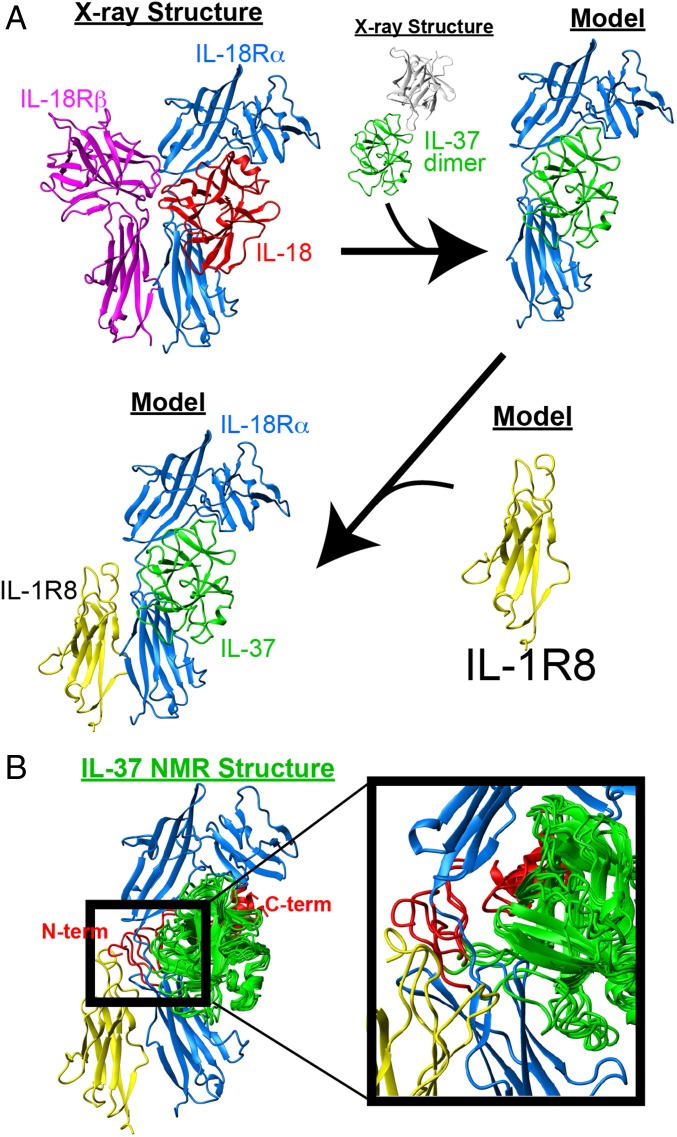
Whereas IL-37 has been shown to associate with both IL-18Rα and IL-1R8 (8, 9, 11), studies shown here and previous studies indicate that these interactions are likely more complicated than originally conceived. For example, cononical IL-1 family interactions, such as that of IL-18, first engage a primary receptor followed by the association with a second receptor as is the case for ternary complexes of IL-1/IL-1R/IL-1R3 (21) and IL-18/IL-18Rα/IL-18Rβ (22). Following such a canonical binding mechanism, a straightforward model would suggest that IL-37 first interacts with one receptor (IL-18Rα), and that this is followed by ternary complex formation with a second receptor (IL-1R8) as we have modeled here using the recently determined IL-18 ternary complex (Fig. 7A). The evidence for such a model is based on fluorescent studies that identified an IL-37/IL-18Rα/IL-1R8 complex in primary cells and cell lines (9). The elegance of such a model seems appealing as the IL-37 dimer must dissociate in order for the IL-37 monomer to interact with IL-18Rα, which is consistent with our data (Fig. 4), and the antiinflammatory activity of IL-37 would also inhibit the proinflammatory IL-18 activity. Moreover, superposition of the NMR solution structure ensemble of the monomer in this model suggests that the flexible N terminus would need to reorient to avoid steric clashes with IL-18Rα (Fig. 7B). This too may be consistent with the role of one of these termini in dictating IL-37 interactions and the role of the other that may negatively regulate binding (in Fig. 5).
Fig. 7.
Potential model of IL-37 interactions. (A) Utilizing the IL-18 (red) ternary complex with IL-18Rα (blue), IL-18Rβ (magenta) as a template (PDB accession 3WO4), and an IL-37 monomer (green) from the X-ray crystal structure determined here (green and white) is superimposed onto IL-18 with IL-18Rα and the homology model of IL-1R8 superimposed onto IL-18Rα. Note, the IL-37 dimer is shown in the same orientation of the IL-37/IL-18Ra complex to illustrate the occlusion of the interacting surface within the IL-37 dimer. (B) NMR-derived solution ensembles are superimposed onto this model with the mature IL-37 extended termini of residues 46–52 and 207–218 shown (red). A blowup of the N-terminal region of IL-3746–218 illustrates potential clashes with the receptor complex, suggesting that this region may need to reorient for interactions.
There are important problems with this model that should be addressed in the future. First, the relatively weak affinity of IL-37 to IL-18Rα previously determined as ∼130 nM (11) is inconsistent with the picomolar activity of IL-37 observed here and elsewhere, which may suggest that there are missing cofactors of this interaction. Second, the previously determined 5 nM affinity for IL-37 dimerization would preclude the weaker binding of IL-18Rα in the first place, which further suggests the existence of cofactors. Thus, although the presented canonical mode of IL-37 binding to IL-18Rα and IL-1R8 is consistent with signal transduction in the IL-1 family, there are likely missing cofactors that contribute to the molecular details of IL-37 signaling in full. Such a cofactor (s) may also explain how IL-1R8 functions, which contains only one Ig-like domain compared with, at least, two Ig-like domains for receptors within the canonical interactions.
Finally, to address the findings that IL-37 activity is diminished well below its in vitro determined self-association affinities (11, 12), we hypothesized that glycosaminoglycan interactions are likely to occur and thereby serve to modulate IL-37 interactions. Thus, we sought to directly address whether IL-37 interacts with such endogenous molecules. To test such a scenario, we utilized heparin as a representative of common glycosaminoglycans, which has been shown to modify the activity of several chemokines by stabilizing an oligomeric state (23–25). Indeed, heparin addition to both the WT IL-3753–206 dimer (Fig. 8A) and the monomeric mutant of Y85A (Fig. 8B) induced severe line broadening at stoichiometric concentrations. Although such line broadening may be induced by either oligomerization or binding to multiple interaction sites, the fact that specific CSPs can be observed while simultaneously being diminished in intensities indicates a preferred interaction site with sampling of larger oligomeric species. This simultaneous shifts with concomitant diminishment in intensity is exemplified using Y85A (Fig. 8C). A plot of these CSPs reveals a likely heparin binding surface within this monomer (Fig. 8D), which is largely distinct from any interaction surface within the modeled ternary complex of IL-37/IL-18Rα/IL-1R8 (Fig. 8E). As glycosaminoglycans are present within multiple surface exposed glycoproteins and are abundant within the extracellular matrix (26), these NMR studies identify IL-37 as a potential glycosaminoglycan binding protein. Such interactions may stabilize IL-37 oligomerization by either shifting the monomer/dimer equilibrium toward the dimer or promoting higher order oligomers of the dimer (Fig. 8D). Additionally, glycosaminoglycan could compete for an additional cofactor interaction (Fig. 8E). In either event, glycosaminoglycan may down-regulate IL-37 activity in accord with its inactivation at concentrations well below the self-association affinity.
Conclusion
Revealing the underlying molecular mechanism of IL-37 will identify promising pathways that may be exploited for controlling inflammation. The biophysical, biochemical, and biological studies here confirm that elevated IL-37 concentrations result in a dampening of its antiinflammatory activities whereby dimerization occludes the activity of the IL-37 monomer. Point mutations that specifically disrupt the IL-37 dimer remain stable monomers in vitro and provide greater suppression of inflammatory cytokines compared with native IL-37 on multiple cell types. Athough NMR solution studies have specifically shown that, in the absence of any other interaction, the IL-37 termini are unstructured, the presence of one extended terminus is necessary for activity. Such findings may be reconciled by the existence of multiple cell surface interactions and/or multiple mediators of IL-37 activity, which would also explain how IL-37 is active at picomolar concentrations despite its relatively weak interaction with IL-18Rα.
Materials and Methods
Protein Expression and Purification.
Recombinant IL-37 constructs with an N-terminal 6xHis tag and thrombin cleavage site were cloned into pET21 for subsequent expression in BL21/DE3 cells at room temperature. The soluble protein was purified using Ni affinity and subjected to sulphopropyl (SP) ion exchange (50 mM Tris, pH 7.2, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT) with bound IL-37 eluted with high salt (2 M NaCl). After removal of the 6xHis tag with thrombin, the protein was subjected to Superdex-75 size exclusion using the final NMR buffer (50 mM phosphate, 150 mM NaCl, and 1 mM DTT), crystallization buffer (20 mM Trizma, pH 7, 50 mM NaCl, and 1 mM DTT), or biological buffer (PBS). For 2H15N13C-labeled NMR samples, protein was grown at 37 °C and refolded as previously published to facilitate amide exchange (27, 28).
X-Ray Crystallography and NMR Relaxation.
Recombinant IL-3753–206 was used for all structural studies unless otherwise stated. For NMR assignments, standard BioPack sequences were collected on a 2H15N13C-labeled protein acquired on a 600 MHz Varian spectrometer along with 15N- and 13C-nuclear Overhauser effect spectroscopy (NOESY) spectra acquired on a 900 MHz Varian spectrometer and R1 relaxation data were collected at 900 MHz. Samples were ∼500 μM. Heparin sodium salt from porcine intestinal mucosa (average molecular weight 11–12.5 kDa; Sigma-Aldrich) was added as indicated. Data were processed using nmrPipe software (29) and analyzed using CCPNmr software (30). Chemical shifts and NOEs for the truncated IL-37 residues 53–206 were used with RASREC Rosetta to calculate solution ensembles of the IL-37 monomer (31) with distance restraints all set to 6 Å and IL-37 residues 1–218 used for all solution calculations. For crystallography, conditions were identified within JCSG Core I (Qiagen) condition 92 (0.1 M citric acid pH 2.5, 20% MPD), and crystals were collected at the Advanced Light Source beamline 8.2.1. For computational models, the ectodomain of IL-1R8 residues 1–118 was calculated with the Rosetta structural prediction, and all superpositions were calculated using Coot Software (32).
Cell Lines and in Vitro Assays.
The murine macrophage cell line J1 was purchased from American Type Culture Collection and cultured according to the distributor’s instructions in DMEM (Corning) supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C and 5% CO2. For in vitro assays, 75,000 cells per well were plated overnight or 1 h before pretreatment in a flat bottom 96 well plate. J1 cells were stimulated for 4 h with 1 µg/mL LPS; Escherichia coli 055:B5; Sigma-Aldrich. For inflammasome activation, 20 µM nigericin sodium salt dissolved in 100% ethanol (InvivoGen) was added for another 1 h. IL-1β and IL-6 concentrations were measured in the cell supernatants by specific ELISA (DuoSet, R&D Systems). Manufacturer’s instructions were strictly followed.
Isolation and Culture of Human AVICs.
All studies were approved by the Institutional Review Board of University of Colorado (COMIRB; Protocol 08-0280) and performed in accordance with the Declaration of Helsinki. All aortic valve donors gave their written informed consent before their inclusion in this study. The normal tricuspid aortic valves were collected from cardiomyopathy heart-transplant patients at the University of Colorado Hospital.
Aortic valve leaflets were excised, washed in PBS, and then incubated with collagenase solution (type I, 1.0 mg/mL) in hybridization ovens at 37 °C for 30 min to separate endothelial cells. Leaflets were further digested with a fresh solution of 1 mg/mL collagenase solution for 4–6 h at 37 °C to isolate human AVICs. After vortexing and aspirating repeatedly to break up the tissue mass, human AVIC suspensions were centrifuged at 1,000 rpm for 10 min. Pellets were resuspended and cultured in M199 growth medium (Lonza), supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin (Sigma-Aldrich) in an incubator with 5% CO2 at 37 °C. The medium was replaced every 3 d throughout the growth, differentiation, and experimental periods. Human AVICs were passaged three to six times and used at 80–90% confluence for all experiments. The AVICs were stimulated with LPS (200 ng/mL) for 24 h. In the experiments, the AVICs were pretreated with recombinant IL-3746–218, the single point mutation of D73K, or Y85A for 2 h before the addition of LPS. After 24 h, the cultures were processed for Western blotting with antihuman VCAM-1 antibodies.
Statistical Analysis.
Statistical significance was calculated using the two-tailed Student’s t test with GraphPad Prism 5.0. P values < 0.05 were considered to be significant with *P < 0.05, **P < 0.001, and ***P < 0.0001.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NCU) and in Biological Magnetic Resonance Data Bank (BMRB accession no. 27729).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819672116/-/DCSupplemental.
References
- 1.Auron PE, et al. Nucleotide sequence of human monocyte interleukin 1 precursor cDNA. Proc Natl Acad Sci USA. 1984;81:7907–7911. [Google Scholar]
- 2.Charrad R, et al. Anti-inflammatory activity of IL-37 in asthmatic children: Correlation with inflammatory cytokines TNF-α, IL-β, IL-6 and IL-17A. Immunobiology. 2016;221:182–187. doi: 10.1016/j.imbio.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Ding VA, et al. The role of IL-37 in cancer. Med Oncol. 2016;33:68. doi: 10.1007/s12032-016-0782-4. [DOI] [PubMed] [Google Scholar]
- 4.Ge G, et al. Interleukin-37 suppresses tumor growth through inhibition of angiogenesis in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:13. doi: 10.1186/s13046-016-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu WD, Zhao Y, Liu Y. Insights into IL-37, the role in autoimmune diseases. Autoimmun Rev. 2015;14:1170–1175. doi: 10.1016/j.autrev.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Zhang J, Tao J, Lu T. Elevated serum levels of interleukin-37 are associated with inflammatory cytokines and disease activity in rheumatoid arthritis. APMIS. 2015;123:1025–1031. doi: 10.1111/apm.12467. [DOI] [PubMed] [Google Scholar]
- 7.Nold MF, et al. CS05-3. Interleukin 37 exerts its anti-inflammatory functions by associating with IL-18Rα and SIGIRR. Cytokine. 2011;56:12. [Google Scholar]
- 8.Nold C, et al. Interleukin 37 employs the IL-1 family orphan receptor SIGIRR and the alpha chain of the IL-18 receptor to inhibit innate immunity. J Immunol. 2013;190:184.7. [Google Scholar]
- 9.Nold-Petry CA, et al. IL-37 requires the receptors IL-18Rα and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal transduction. Nat Immunol. 2015;16:354–365. doi: 10.1038/ni.3103. [DOI] [PubMed] [Google Scholar]
- 10.Garlanda C, Riva F, Bonavita E, Mantovani A. Negative regulatory receptors of the IL-1 family. Semin Immunol. 2013;25:408–415. doi: 10.1016/j.smim.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, et al. Interleukin-1F7B (IL-1H4/IL-1F7) is processed by caspase-1 and mature IL-1F7B binds to the IL-18 receptor but does not induce IFN-gamma production. Cytokine. 2002;18:61–71. doi: 10.1006/cyto.2002.0873. [DOI] [PubMed] [Google Scholar]
- 12.Ellisdon A, et al. Homodimerization attenuates the anti-inflammatory activity of interleukin-37. Sci Immunol. 2017;2:eaaj1548. doi: 10.1126/sciimmunol.aaj1548. [DOI] [PubMed] [Google Scholar]
- 13.de Oliveira Neto M, et al. Interleukin-22 forms dimers that are recognized by two interleukin-22R1 receptor chains. Biophys J. 2008;94:1754–1765. doi: 10.1529/biophysj.107.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zdanov A. Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 2010;21:325–330. doi: 10.1016/j.cytogfr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kufareva I, Salanga CL, Handel TM. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol Cell Biol. 2015;93:372–383. doi: 10.1038/icb.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravindran A, Joseph PRB, Rajarathnam K. Structural basis for differential binding of the interleukin-8 monomer and dimer to the CXCR1 N-domain: Role of coupled interactions and dynamics. Biochemistry. 2009;48:8795–8805. doi: 10.1021/bi901194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J Mol Biol. 2011;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson S, et al. Regulation of chemokine function: The roles of GAG-binding and post-translational nitration. Int J Mol Sci. 2017;18:E1692. doi: 10.3390/ijms18081692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinarello CA, van der Meer JWM. Treating inflammation by blocking interleukin-1 in humans. Semin Immunol. 2013;25:469–484. doi: 10.1016/j.smim.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendrick AA, et al. The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide. Protein Sci. 2014;23:464–480. doi: 10.1002/pro.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, et al. Structural insights into the assembly and activation of IL-1β with its receptors. Nat Immunol. 2010;11:905–911. doi: 10.1038/ni.1925. [DOI] [PubMed] [Google Scholar]
- 22.Tsutsumi N, et al. The structural basis for receptor recognition of human interleukin-18. Nat Commun. 2014;5:5340. doi: 10.1038/ncomms6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poluri KM, Joseph PRB, Sawant KV, Rajarathnam K. Molecular basis of glycosaminoglycan heparin binding to the chemokine CXCL1 dimer. J Biol Chem. 2013;288:25143–25153. doi: 10.1074/jbc.M113.492579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziarek JJ, et al. Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus. J Biol Chem. 2013;288:737–746. doi: 10.1074/jbc.M112.394064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph PRB, Sawant KV, Rajarathnam K. Heparin-bound chemokine CXCL8 monomer and dimer are impaired for CXCR1 and CXCR2 activation: Implications for gradients and neutrophil trafficking. Open Biol. 2017;7:170168. doi: 10.1098/rsob.170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjellén L, Lindahl U. Specificity of glycosaminoglycan-protein interactions. Curr Opin Struct Biol. 2018;50:101–108. doi: 10.1016/j.sbi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Schlegel J, et al. Solution characterization of the extracellular region of CD147 and its interaction with its enzyme ligand cyclophilin A. J Mol Biol. 2009;391:518–535. doi: 10.1016/j.jmb.2009.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel J, Armstrong GS, Redzic JS, Zhang F, Eisenmesser EZ. Characterizing and controlling the inherent dynamics of cyclophilin-A. Protein Sci. 2009;18:811–824. doi: 10.1002/pro.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 31.Lange OF. Automatic NOESY assignment in CS-RASREC-Rosetta. J Biomol NMR. 2014;59:147–159. doi: 10.1007/s10858-014-9833-3. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, et al. CS23D: A web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.