Significance
Preterm birth (PTB) complications are the leading cause of long-term morbidity and mortality in children. The genetic and molecular characteristics of PTB and related disorders remain unclear. In this study, a family-based cohort of 791 family trios, including 270 PTB and 521 control families, was investigated by using whole-genome sequencing, RNA sequencing, and DNA methylation data. Integrative analysis identified 160 genomic variants associated with PTB-related phenotypes and led to the discovery of 72 candidate biomarker genes for very early PTB (VEPTB). The genes associated with VEPTB involve growth signaling and inflammation- and immunity-related pathways. With these data, and by stratifying PTB by subphenotype, we have identified PTB genes and pathways that can be used as a starting point in further clinical studies.
Keywords: preterm birth, whole genome sequencing, family trios, integrative computational analysis, genomic variants
Abstract
Preterm birth (PTB) complications are the leading cause of long-term morbidity and mortality in children. By using whole blood samples, we integrated whole-genome sequencing (WGS), RNA sequencing (RNA-seq), and DNA methylation data for 270 PTB and 521 control families. We analyzed this combined dataset to identify genomic variants associated with PTB and secondary analyses to identify variants associated with very early PTB (VEPTB) as well as other subcategories of disease that may contribute to PTB. We identified differentially expressed genes (DEGs) and methylated genomic loci and performed expression and methylation quantitative trait loci analyses to link genomic variants to these expression and methylation changes. We performed enrichment tests to identify overlaps between new and known PTB candidate gene systems. We identified 160 significant genomic variants associated with PTB-related phenotypes. The most significant variants, DEGs, and differentially methylated loci were associated with VEPTB. Integration of all data types identified a set of 72 candidate biomarker genes for VEPTB, encompassing genes and those previously associated with PTB. Notably, PTB-associated genes RAB31 and RBPJ were identified by all three data types (WGS, RNA-seq, and methylation). Pathways associated with VEPTB include EGFR and prolactin signaling pathways, inflammation- and immunity-related pathways, chemokine signaling, IFN-γ signaling, and Notch1 signaling. Progress in identifying molecular components of a complex disease is aided by integrated analyses of multiple molecular data types and clinical data. With these data, and by stratifying PTB by subphenotype, we have identified associations between VEPTB and the underlying biology.
Preterm birth (PTB) is defined as a live birth occurring before 37 weeks (wk) of completed gestation. Every year, ∼15 million infants worldwide are born preterm (1). PTB-related complications result in 1 million deaths yearly (2); surviving preterm infants have a significantly increased risk of developmental and medical disabilities (3). PTB is the endpoint of a complex process spanning fetal development and is influenced by fetal and maternal factors (4–8). In approximately half of PTB cases, there are no apparent known risk factors; in the other half, known risk factors or clinical antecedents such as preeclampsia lead to early delivery (9–11). There is a racial disparity in preterm and previable birth rates that may result from genetic, epidemiological, or other factors (12–14). PTB heritability is estimated to be 15–35% based on twin and family studies (15–17). Genomewide association studies (GWASs) and candidate gene studies have implicated genes (18–21) that illuminate several systems involved in PTB, including immune response, inflammation, coagulation factors, and connective tissue remodeling (19). The most comprehensive GWAS performed to date for PTB identified replicable loci in six genes (EBF1, EEFSEC, AGTR2, WNT4, ADCY5 and RAP2C) associated with gestational duration, with three of these six genes (EBF1, EEFSEC and AGTR2) strongly associated with PTB in a European ancestry cohort of 43,568 women (22). Furthermore, Zhang et al. (22) determined that common maternal SNPs could explain 23% of the phenotypic variance in PTB observed in this cohort, suggesting that other sources that could explain PTB phenotypic variation need to be investigated further. Gene expression and DNA methylation studies have also identified biomarkers (23–25), often in the same systems illuminated by the candidate genes.
Current knowledge of PTB environmental and genetic factors is incomplete, and little is known of the strength of these factors or how they interact. Integrative systems biology approaches can be applied to overcome the challenges in studying pregnancy-related complications (26). To characterize the molecular mechanisms that may be altered in PTB, we integrated molecular and clinical data for 270 PTB and 521 control families. We performed whole-genome sequencing (WGS) of maternal, paternal, and neonatal DNA for each family trio. We carried out mRNA sequencing (RNA-seq) and miRNA sequencing and DNA methylation profiling of maternal whole blood. We employed an integrative approach to characterize PTB at the genomic, transcriptomic, and epigenomic levels. We identified dozens of PTB candidate genes and provide further evidence for previously reported PTB genes and networks. This type of approach can be used to analyze other etiologically complex conditions for which very large cohorts are not yet available.
Results
Study Population.
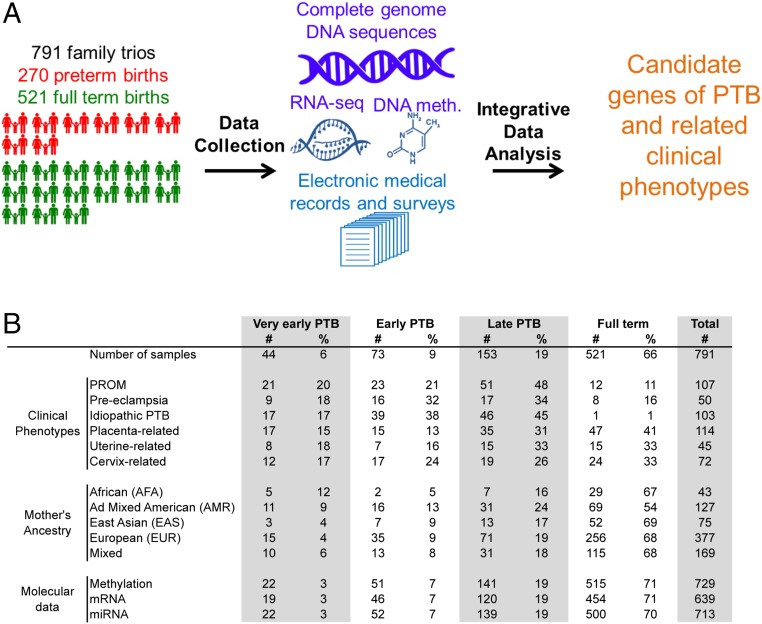
The study population consisted of 791 family trios (father, mother, and newborn), of which 270 involved deliveries of live births before 37 wk of gestation (Fig. 1A). Families were recruited at the time of delivery at the Inova Fairfax Medical Center from 2011 to 2013. The recruitment was performed to enrich for cases of PTB, early PTB (EPTB), and very early PTB (VETBP). Clinical information was obtained by interview, electronic medical records (EMRs), and self-reported data from a study-specific case report form. Because of potential biases in self-reported ancestry, we computed admixture coefficients from WGS data by using the 1000 Genomes reference ancestries (27). The maternal cohort included African (n = 43), East Asian (n = 75), European (n = 377), admixed American (n = 127), and mixed individuals in cases in which inferred admixture coefficients were inconclusive (n = 169; Fig. 1B).
Fig. 1.
Study overview. (A) Graphical overview of the study described in this report. We collected peripheral blood samples from 791 family trios, of which 270 represented PTBs. We carried out WGS of DNA for each member of the family trio, i.e., the father, mother, and newborn. We profiled mRNA and miRNA expression by using RNA-seq as well as DNA methylation in the maternal samples. Extensive clinical information was captured by using EMRs and study-specific patient surveys. All these data were integrated in an analytical framework to characterize the genomic and molecular associations with PTB and related clinical phenotypes. (B) Summary of distribution of family trios across clinical phenotypes and ancestries broken down by PTB categories based on gestational age. Molecular data indicate the number of maternal samples profiled for DNA methylation and mRNA and miRNA expression. Numbers cited indicate samples that passed stringent quality-control criteria for inclusion in this report.
PTB-Related Clinical Phenotypes.
For our primary analysis, we tested associations with the PTB phenotype (gestation <37 wk) by using all full-term births (FTBs; gestation ≥37 and <42 wk) as controls. We defined eight additional phenotypes (Table 1) for secondary case-control analyses: (i) VEPTB <28 wk, (ii) EPTB <34 wk (including VEPTB cases), (iii) premature rupture of membranes (PROM), (iv) preeclampsia (encompassing eclampsia), (v) placenta-related (e.g., placental previa/abruption), (vi) uterine-related (e.g., uterine anomalies and endometriosis), (vii) cervix-related, and (viii) idiopathic (no evidence of infection, cervical insufficiency, uterine abnormalities, placentation abnormalities, or maternal disease known to cause premature delivery). These phenotypes are not mutually exclusive. Cases and controls for each secondary analysis are depicted in SI Appendix, Fig. S1. It is important to note that the case and control definitions refer to an individual such as the mother or newborn or refer to the family trio. This highlights a unique challenge in analyzing disorders that affect pregnancy, in which multiple persons or a group of persons (i.e., family trio) can be viewed as the “case” in an analysis. Detailed descriptions of these phenotypes are given in the SI Appendix.
Table 1.
Summary of genomic association tests across clinical phenotypes
| Clinical phenotype | No. of Cases | No. of Controls | FBAT | EIGENSTRAT | ||||||
| Family trio | Father | Mother | Newborn | |||||||
| No. genes | No. igr | No. genes | No. igr | No. genes | No. igr | No. genes | No. igr | |||
| Preterm | 270 | 521 | — | — | — | — | — | — | — | — |
| Early preterm | 117 | 521 | 1 | 1 | — | — | 1 | 1 | — | — |
| Very early preterm | 44 | 521 | 3 | 1 | 1 | 2 | 3 | 6 | 3 | 5 |
| PROM | 107 | 684 | 3 | 1 | 1 | — | 1 | — | — | — |
| Pre-eclampsia | 50 | 741 | 11 | 5 | 3 | 4 | 7 | 5 | 6 | 10 |
| Idiopathic PTB | 103 | 520 | — | 1 | — | — | — | — | — | — |
| Placenta-related | 114 | 677 | 2 | 1 | 1 | — | — | — | 1 | 1 |
| Uterine-related | 45 | 746 | 9 | 6 | 1 | 5 | 2 | 4 | 7 | 7 |
| Cervix-related | 72 | 719 | 4 | 3 | 1 | 2 | — | 2 | 2 | 1 |
Number of statistically significant variants associated with a given clinical phenotype at the P value threshold 10−8 for the family-based test FBAT and the EIGENSTRAT test, which was performed for the maternal, paternal, and neonatal genomes separately. igr, intergenic region.
Genomewide Statistical Tests Identify Candidate PTB Genes.
WGS was performed by Complete Genomics on 791 mothers, 791 fathers, and 839 newborns. In total, there were 784 complete family trios, i.e., the genomes from the mother, father, and newborn were successfully sequenced. Sixty-three families had twins and one family had triplets. For our analyses, only one newborn was (randomly) chosen from each multiple-birth family. After filtering for quality and minor allele frequency (MAF), the total number of variants with MAF ≥ 1% across all individuals was 6,987,906 (Dataset S1). We applied two complementary statistical tests for genomic associations: EIGENSTRAT (28) and the family-based association test (FBAT) (29). EIGENSTRAT is an individual-based test that corrects for population stratification. FBAT is robust to population stratification and quantifies the disequilibrium of transmission of alleles from parents to offspring. We performed EIGENSTRAT for each of the nine phenotypes (Table 1) and each of the three sample groups: mothers, fathers, and newborns. We performed FBAT under the additive genetic model for each of the nine phenotypes separately. FBAT was run for the 784 family trios with complete WGS. To avoid inflated P values caused by small sample sizes, we did not test rare variants (MAF < 5%) for FBAT and required at least 10 cases and 10 controls with nonreference alleles for EIGENSTRAT. Further details are provided in SI Appendix.
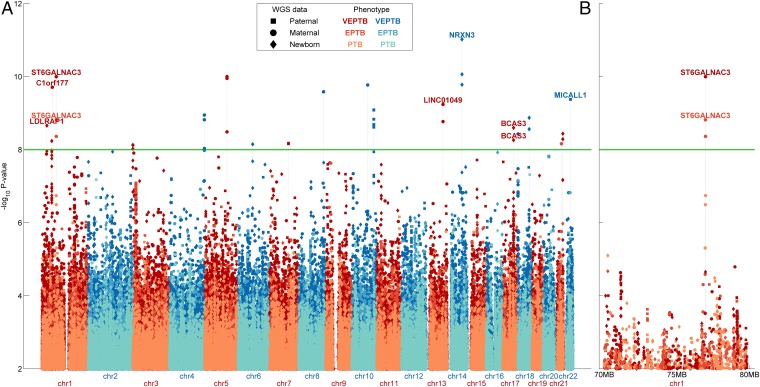
When considering PTB as a single cohort (our primary analysis), we found no significant variant associations by EIGENSTRAT or FBAT. However, application of these tests in our eight secondary analyses (EPTB, VEPTB, PROM, preeclampsia, placenta-related, uterine-related, cervix-related, and idiopathic) identified 160 variants with moderately significant associations at an uncorrected P value cutoff of 1 × 10−8 (Table 1 and Dataset S2). Of these variants, 27 were discovered by using FBAT and 133 were discovered by using EIGENSTRAT; we found no overlap between significant EIGENSTRAT and FBAT variants for any phenotype, but several identified variants showed associations across multiple phenotypes (Dataset S3). Of the 160 variants, 66 were within genes, none of which led to coding changes, and 94 were intergenic, including 9 variants in long intergenic noncoding RNAs. Importantly, we found no highly significant variants, but many moderately significant variants, as can be seen by the large number of small peaks in the corresponding Manhattan plots (Fig. 2A and SI Appendix, Fig. S2). Notably, 48 were associated with VEPTB and EPTB. For some variants, including one in the ST6GALNAC3 gene, we observed progressively increasing statistical significance for the PTB, EPTB, and VEPTB phenotypes (Fig. 2B). Fifty-two variants were associated with preeclampsia, 7 with PROM, and 1 with idiopathic PTB; 6 were placenta-related, 59 were uterine-related, and 17 were cervix-related (Table 1 and Dataset S2). Of the associations identified by EIGENSTRAT, 40 were to variants in the maternal genome, 57 to the newborn genome, and 36 to the paternal genome. These sets of significant variants were mutually exclusive among the maternal, newborn, and paternal cohorts. After collapsing correlated and nearby variants, i.e., those that are in linkage disequilibrium, we identified variants in 8, 19, and 14 genes from the paternal, newborn, and maternal genomes, respectively (Table 1). These results are consistent with a larger role for the maternal genome in PTB (7), as well as a role for the fetal genome (with paternal contributions). These trends are maintained when analyzing the variants at higher or lower levels of statistical significance (Dataset S2). The maternal and paternal contributions are corroborated by FBAT analysis, which demonstrated significant over- or undertransmission of nonreference alleles from mothers and fathers to their prematurely born offspring. At a less stringent level of statistical significance, i.e., a false discovery rate (FDR) < 10%, our analyses reproduced many reported PTB gene associations, e.g., AGT, COL5A2, CRHR2, FLT1, MMP2, and NAT1 (Dataset S4). Moreover, the genes identified at FDR < 10% were enriched in published lists of PTB genes (Dataset S5). We also detected associations, such as a variant in PIN4 that was associated with EPTB and VETPB according to FBAT (P = 1.2 × 10−9 and P = 2.5 × 10−13, respectively). Although PIN4 has already been associated with human spontaneous labor without histologic chorioamnionitis based on gene-expression studies (30), genomic correlations were not previously identified.
Fig. 2.
Manhattan plot of genomic associations in PTB. (A) Genomewide significance values (−log10 P values) for all variants tested for association with PTB, EPTB, and VEPTB. Association tests were performed by using EIGENSTRAT on the paternal, maternal, or neonatal genomes separately. The green horizontal line represents the global P value threshold of 10−8. Stacked points represent variants within close proximity of one another. (B) Zoomed-in view of chr1 from 70,000,000 bps to 80,000,000 bps, which includes the ST6GALNAC3 locus.
To validate significant findings, we used an independent cohort of more than 1,300 family trios ascertained through the First 1,000 Days of Life Study (31). We used the validation cohort to specifically test the significant variants. In this cohort, we observed a small number of variants with statistical significance (FDR < 10%). A variant in IFN Lambda Receptor 1 (IFNLR1) showed a particularly strong association with the PTB phenotype. The details of this validation study are included in SI Appendix, Supplementary Note 1 and Fig. S3 and Datasets S6–S8.
Statistical Tests for Single-Gestation Families Highlight Variability in Genomic Associations but Confirm Strong Hits.
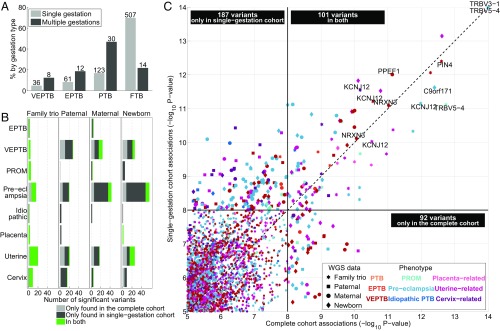
Our study population included families from different ancestries and with various known risk factors of PTB. Of these factors, multiple-gestation pregnancy is well known to be strongly related to gestational age, i.e., spontaneous PTB occurs much more frequently in twin and triplet pregnancies than in singleton pregnancies (32). Indeed, of the 64 multiple-gestations families (63 twins, 1 triplet), 50 (78%) were PTB (including EPTB and VEPTB; Fig. 3A). To assess the statistical significance of the PTB-associated genomic variants when excluding the effect of multiple-gestation pregnancies, we removed the 64 families with twins and triplets from our study cohort and repeated the same statistical tests. We observed that associations around the global threshold of 1 × 10−8 were quite variable, i.e., of the 160 unique variants that were discovered using the complete cohort, 71 (44%), were identified as statistically significant in the single-gestation cohort. Conversely, 169 unique variants were identified when excluding twin and triplet families that were not significant previously (Fig. 3 B and C). Importantly, the variants that were statistically most strongly related to PTB, i.e., P values lower than 1 × 10−10 in the complete cohort, were identified at a similar level of statistical significance for the single-gestation cohort (Fig. 3C). None of these variants were paternal. Related, significant paternal variants showed the smallest agreement between the complete and single-gestation cohort (Fig. 3B). Overall, we observed more variants that were significantly related to PTB-related phenotypes in the single-gestation cohort (Fig. 3 B and C and Dataset S9), including maternal variants in EBF1 and WNT4, two of the six genes identified by Zhang et al. (22), associated with VEPTB at the less stringent statistical threshold of FDR < 10%. However, the P values were not substantially lower for the strongest associations compared with the complete cohort. The largest number of associations was detected for the phenotypes with the smallest number of cases, i.e., VEPTB, preeclampsia, and uterine-related (Fig. 3B and Table 1). For the single-gestation cohort, the number of cases is even smaller, leading to slightly more biased P values and potentially more false-positive findings (SI Appendix, Figs. S4 and S5).
Fig. 3.
Genomic associations when excluding multiple-gestation families. (A) Bar plot showing the distribution of single- and multiple-gestation families across the four term categories. The light gray bars for single gestations and dark gray bars for multiple gestations each add up to 100%. The numbers above the bars indicate the number of family trios. (B) Bar plots indicating the number of genomic variants associated with PTB-related phenotypes (stratified vertically) across genomic tests (stratified horizontally) at P < 10−8, divided into (i) variants found only in the complete cohort (light gray), (ii) variants found only in the single-gestation families (dark gray), and (iii) variants found in both (green). (C) Scatter plot displaying P values for variants that were statistically associated with the nine PTB-related phenotypes across the four genomic tests (indicated by various markers and colors) in the complete cohort (x axis) and the single-gestation cohort (y axis). Gene names are printed for variants with P < 10−10 in both cohorts that were in a gene. The black boxes indicate the number of variants observed at P < 10−8 in only single-gestation families (top left box), in only the complete cohort (bottom right box), or in both (center top box). Note that these are not numbers of unique variants; a variant may be represented multiple times if significant for multiple tests or phenotypes.
Integrative Analysis Highlights Known and PTB Genes and Pathways.
We performed an integrative analysis of WGS, mRNA, miRNA, and DNA methylation data from samples of maternal blood drawn after birth. For this analysis, we used maternal samples from the complete cohort, i.e., including multiple-gestation families. mRNA, miRNA, and DNA methylation measurements were all obtained from maternal whole blood samples. Totals of 684 RNA-seq and 766 miRNA samples were sequenced on an Illumina HiSeq2000 system. We identified miRNA targets with multiMiR (33). A total of 784 DNA methylation samples were analyzed on the Illumina Methylation 450K platform by using probes previously identified as high in quality (34). All four data types (WGS, RNA-seq, miRNA, and DNA methylation) were obtained for 629 families. For the majority of mothers (71%), whole blood samples were taken 1 d after birth. For 93% of the mothers, blood was drawn within the first 4 d after birth. Molecular data derived from samples taken more than 4 d after birth were not used in further analyses. Fig. 1B shows an overview of the samples in this study.
We developed a unified workflow for the identification of significant differences in the levels of expression or methylation by using a generalized linear model framework (35). Age, admixture, and blood draw dates (as integers with day of birth as zero) were used as covariates in the analysis (SI Appendix). We identified known PTB genes from the literature (Dataset S4) and used the hypergeometric test to test significance of overlap between gene lists (i.e., those newly identified in our work compared with existing lists). For the expression and methylation quantitative trait loci (QTL) analyses, we used MatrixEQTL (36) to identify genes and methylation probes that were within (or overlapped with) candidate genes. Covariates included maternal age, admixture coefficients, and blood draw date.
We observed differential gene expression and DNA methylation across five phenotypes studied (Table 2 and Datasets S2, S10, and S11), including our primary PTB phenotype, as well as EPTB, VEPTB, preeclampsia, and idiopathic PTB. There were 215 differentially expressed genes (DEGs) and two differentially methylated probes (DMPs) between PTB and FTB. There were even more differences between FTB and each of the two more extreme gestational age phenotypes: 650 DEGs and 273 DMPs for EPTB and 838 DEGs and 811 DMPs for VEPTB. We did not identify any significantly differentially expressed miRNAs. Differentially expressed miRNAs have been reported in gestational tissues such as cervix and placenta (37–39). However, we found that this observation does not extend to peripheral blood, similar to findings by Elovitz et al. (40).
Table 2.
Summary of genomic and molecular associations across clinical phenotypes
| Clinical phenotype | Genomic | Molecular | |||||
| FBAT | EIGENSTRAT | DNA methylation | mRNA | miRNA | |||
| Family trio | Maternal | Union of FBAT and EIGENSTRAT | Maternal | ||||
| No. of variants | No. of variants | No. of genes | No. of probes | No. of genes | No. of genes | No. of miRNAs | |
| PTB | — | — | — | 2 | 2 | 215*,†,‡ | — |
| EPTB | 3 | 42 | 7 | 273 | 258* | 650*,†,‡,§ | — |
| VEPTB | 7 | 960 | 217*,†,§ | 811 | 735 | 838*,†,‡ | — |
| PROM | 23 | 3 | 12 | — | — | — | — |
| Pre-eclampsia | 78 | 1046 | 312*,§ | — | — | 8 | — |
| Idiopathic PTB | 1 | 10 | 3 | 11 | 11 | 17 | — |
| Placenta-related | 13 | — | 10 | — | — | — | — |
| Uterine-related | 105 | 276 | 132§ | — | — | — | — |
| Cervix-related | 28 | 16 | 18 | — | — | — | — |
Overview of statistically significant genomic associations (FDR <10%) and differentially expressed [FDR <10% and absolute log2(FC) >0.5] and methylated genes (FDR <10%) between cases and controls for each phenotype.
Statistically significant overlap with gene lists from dbPTB (41).
Statistically significant overlap with candidate PTB genes (McElroy_PTB, ref. 19).
Statistically significant overlap with Pre-Eclampsia SNP Resource (59).
Statistically significant overlap with genes involved in human birth timing (Plunkett_HBT, ref. 60).
Enrichment tests showed overlaps between the DEGs and DMPs and previously reported PTB genes we obtained from a variety of metastudies and databases (Dataset S4). Specifically, DEGs from PTB, EPTB, and VEPTB were strongly enriched in the Database for Preterm Birth (dbPTB) (41) (P = 6.2 × 10−4, P = 7.7 × 10−15, and P = 8.0 × 10−12, respectively; Table 2 and Dataset S5) as well as the genes reported in the work of McElroy et al. (19) (P = 2.4 × 10−3, P = 2.1 × 10−7, and P = 3.0 × 10−8, respectively). For VEPTB, we also detected significant overlap between the DEGs and DMPs (P = 4.2 × 10−4; Dataset S5). The substantial overlap between our results and those already published demonstrate the value of integrating the genomic and molecular data in this cohort, even though the previously reported PTB genes were obtained from studies with quite different experimental setups and analyses.
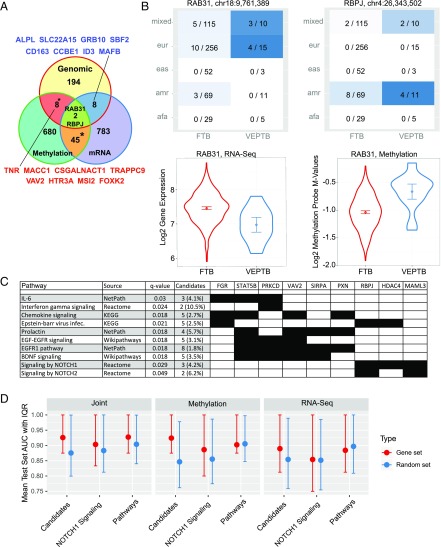
Of all phenotypes tested, we observed a large number of associated variants, DEGs, and DMPs in only VEPTB (Table 2). To further study VEPTB, we created a set of 72 “VEPTB genes” (Dataset S12). These genes had a strong genomic association with the VEPTB phenotype or were associated with VEPTB in at least two of the three data types (WGS, DNA methylation, mRNA expression). Of these, 16 genes (TNR, MACC1, CSGALNACT1, TRAPPC9, VAV2, HTR3A, MSI2, FOXK2, ALPL, SLC22A15, GRB10, SBF2, CD163, CCBE1, ID3, and MAFB) had variants significantly associated with VEPTB and were differentially methylated or differentially expressed (Fig. 4A). In particular, these 16 are not in dbPTB and therefore represent implicated genes. Only three VEPTB candidate genes are found in dbPTB (IL10, FGD4, and STAT5B). When looking across all PTB-related gene sets (Dataset S4), 92% (66 of 72) of the VEPTB candidate genes have not been previously reported to be associated with PTB. Of the six previously reported genes, IL-10 was the most commonly reported (five of seven gene sets), followed by TNR and TIMP2 (two of seven gene sets). Two genes, RAB31 and RBPJ, had significant associations found for all three data types (WGS, DNA methylation, and mRNA expression). A variant at chr18:9,761,389 (rs117652912) in an intron of RAB31 was present in mothers from European and mixed ancestry (Fig. 4B). RAB31 is a member of the RAS oncogene family coding for a small GTPase-binding protein involved in vesicle and granule targeting (42). RAB31 is up-regulated at term relative to midgestation (43) and has been implicated in a GWAS of placental abruption (44). (In this study, variants in RAB31 were not significantly associated with the placenta-related phenotype.) A variant in RBPJ at chr4:26,343,502 was found associated with VEPTB in the American and mixed populations (Fig. 4B). RBPJ is up-regulated and hypermethylated in VEPTB. RBPJ is a transcriptional regulator in the Notch signaling pathway (45). Heterozygous mutations in RBPJ, resulting in impaired DNA binding of the encoded protein, are one of the causes of the rare developmental disorder Adams–Oliver syndrome, which can involve dermatologic, limb, and vascular anomalies (46).
Fig. 4.
Integrative analysis of genomic and molecular data for VEPTB families uncovers candidate genes. (A) Venn diagram of the overlap between genes with significant variants associated with VETPB and differentially expressed and methylated genes. * Indicates statistically significant overlap between gene sets (hypergeometric test P < 0.05). (B) Heat maps depicting the distribution of variants in RAB31 (Upper Left) and RBPJ (Upper Right) across different ancestries for FTB and VEPTB mothers. In each heat map panel, the ratio is the number of mothers who have the minor allele (homozygous or heterozygous) over the total number of mothers from that ancestry group. Ancestries are represented by using the 1000 Genomes super populations notation. (Lower) Violin plots of differential gene expression (Left) and differential DNA methylation (Right) of RAB31 between FTB and VEPTB. (C) Overview of pathways that were significantly enriched with genes in the VEPTB candidate list of 72 genes. This overview is a selection of all significant pathways (listed in Dataset S14). The selection was performed manually with the goal of including pathways related to immune and growth factor signaling, which formed the large majority of the enriched pathways, yet avoiding redundancy among the selected pathways, i.e., excluding pathways with similar names and gene membership. (D) Mean area under the curve (AUC) and associated interquartile range of VEPTB class prediction using a random forest classifier with different data types including RNA-seq data, DNA methylation data, and a joint set of RNA-seq and methylation data. Prediction was performed with the 72 VEPTB genes (candidate); the 1,324 VEPTB pathway genes, i.e., the full set of genes in associated pathways excluding the 72 VEPTB genes (pathway genes); and on each candidate pathway individually (one example shown, i.e., the Notch1 pathway). Sets of random genes with identical set sizes are shown for comparison. Each mean AUC was computed by using cross-validation on a test set.
As further supporting evidence for these VEPTB genes, we observed within the validation cohort more variants in and around the 72 VEPTB genes that were statistically associated with PTB, EPTB, or VEPTB than in a comparable randomly selected set of 72 genes (SI Appendix, Supplementary Note 1).
To determine whether our identified genomic variants are responsible for differential expression and/or methylation, we performed cis-expression QTL (eQTL) and cis-methylation QTL (mQTL) analysis for all 72 VEPTB candidate genes. Specifically, we performed the e/mQTL analysis by associating any variant with a MAF > 1% in or near each of the VEPTB genes with the expression and methylation levels of that gene. We found 2,328 eQTL and 403 mQTL pairs at an FDR < 10% (Dataset S13). These pairs provide evidence that these genomic variations may influence expression of these nearby genes. However, we identified only one significant eQTL relationship for a variant (chr20:1,825,838, rs73584704) that was itself statistically associated with the VEPTB phenotype. This variant was correlated with the expression of SIRPA (SI Appendix, Fig. S6).
PTB is thought to result from aberrations in systems such as inflammatory, immune-related, and hormone regulation pathways (47). We used ConsensusPathDB (48), an interaction database that integrates multiple public resources, to determine whether the VEPTB candidate genes are enriched in particular pathways. Our results indicate that VEPTB candidate genes are involved in systems including EGFR (q-value 2.4 × 10−4) and prolactin signaling pathways (q-value 2.4 × 10−4), inflammation- and immunity-related pathways such as the IL-6 pathway (q-value 0.030), chemokine signaling (q-value 0.018), IFN-γ signaling (q-value 0.024), and Notch1 signaling (q-value 0.029; Fig. 4C, SI Appendix, Fig. S7, and Dataset S14). Although only 3 of the 72 VEPTB genes were previously implicated in PTB, the identified pathways included many pathways that were previously associated with PTB. For example, the IL-6 signaling pathway and inflammation-related processes have been previously implicated in PTB (49–52). Analysis of GWAS data also implicated the RAS pathway, which is downstream of EGFR (and part of the MAPK/ERK pathway) with PTB (53). Notch signaling pathways, which have a role in innate and adaptive immunity, were significantly perturbed in our results and have recently been linked to PTB in mouse models (54).
To further validate the association between VEPTB and these pathways, we performed additional analyses of the genes in these pathways, excluding, however, the 72 VEPTB genes that were used to identify them. Specifically, we looked for association between VEPTB and these pathways outside of the identified 72 VEPTB genes. For this analysis, we defined a set of 1,324 “VEPTB pathway genes” across 27 enriched pathways. Reexamination of the genomic data showed a statistically significant enrichment of variants in or around these VEPTB pathway genes that were associated with VEPTB. Finally, we trained random forest machine learning models to predict VEPTB status by using the gene-expression and DNA methylation data for all genes in the associated pathways, excluding the 72 VEPTB genes. The machine learning analyses showed that VEPTB status could be predicted by the VEPTB pathway genes with similar accuracy compared with the original 72 VEPTB genes, and, in both cases, much better than with random gene sets (Fig. 4D, SI Appendix, Fig. S8, and Dataset S15). Detailed results of the analyses on the VEPTB pathways are presented in SI Appendix, Supplementary Note 2.
Discussion
We have described a study of PTB using a cohort of 791 family trios from various ancestries, of which 270 had PTB. We obtained and integrated WGS data from the fathers, mothers, and newborns in these family trios and RNA-seq gene expression and DNA methylation data from maternal blood samples and gathered comprehensive clinical information concerning pregnancy, delivery, and newborn health. By using these clinical data, we nonexclusively assigned the family trios into clinical groups that are likely to have different molecular causes underlying PTB. These groups included three based on gestational age, delineating PTB (<37 wk; n = 270), EPTB (<34 wk; n = 117), VEPTB (<28 wk; n = 44), and known clinical antecedents of PTB, such as PROM and structural issues with the placenta, uterus, or cervix. Stratification of the cohort in case and control sets for each of the PTB-related clinical phenotypes enabled the discovery of genomic and molecular correlates that were specific to phenotypes across our heterogeneous cohort.
By using genomic statistical tests, we identified many variants that were associated with the various PTB-related phenotypes, albeit at moderate levels of statistical significance. Although we found genes previously implicated in PTB, many previously identified genes were not retrieved by our analyses. For example, our study did not replicate the findings by Zhang et al. (22) at the genome-wide significance level of 10−8, although, at a less stringent statistical threshold, i.e., FDR < 10%, we did identify four of the six genes described by Zhang et al. (22), albeit in different SNPs (loci) associated with these genes. This could be explained in part by our relatively lower sample count and ethnically diverse cohort. In general, published PTB association studies report different variants and genes. Differences in study design, cohort definition, and statistical data-analysis methods certainly play a role in explaining this lack of agreement among the various studies. We further evaluated the uncovered genomic associations in two ways, first by removing families with twins and triplets and second by employing an independent validation cohort of more than 1,300 family trios. Exclusion of multiple-gestation pregnancies, a well-known risk factor for PTB, may lead to a “cleaner” study design. Indeed, we identified more significant variants when using the single-gestation cohort, although these associations were not stronger than those found by using the complete cohort. Further, the most significant variants identified in the complete cohort were also found when families with twins and triplets were removed. In the validation cohort, we were able to reproduce a small number of variants. Our validation exercises may be considered as partially successful, as individual variants were hard to reproduce. More large-scale genetic association studies with deep clinical phenotypic data will be required to fully ascertain the genomic contribution of this multifactorial syndrome.
By using an integrative approach whereby we combined the WGS data with RNA-seq gene expression and DNA methylation data from maternal blood samples, we identified PTB genes and pathways. Specifically for VEPTB, we uncovered various significantly associated variants as well as differentially expressed and methylated genes, many of which are involved in growth factor signaling and inflammation- and immunity-related pathways. The finding that inflammatory pathways are significant is not unexpected based on previous biomarker studies, but our comprehensive approach with the use of WGS, RNA-seq, and methylation data provides specificity in terms of gene involvement and suggests generalizability. Implicated genes included RAB31 and RBPJ, members of the RAS oncogene family and Notch signaling pathway, respectively, which had significant associations found for all three data types. VEPTB is associated with higher risks of infant death and disabilities. Therefore, our identification of genes and pathways for VEPTB may be of particular clinical relevance. In addition, dissecting the causes of VEPTB may provide a window to a more broad understanding of PTB. Interestingly, the genes identified in this study showed marginal overlap with published biomarkers for PTB (SI Appendix, Supplementary Note 3). This implies that our study potentially identified PTB-related associations or biomarkers. Our observations could also be explained, at least in part, by inconsistent reports of genetic associations within this heterogeneously defined phenotype of PTB-like syndromes. On the functional level of biological pathways, we found a greater agreement of our primary results with published literature and within the validation cohort. Twenty of the VEPTB genes we uncovered, as well as the previously known genes, are involved in the immune response, inflammation, and growth factor signaling pathways. Integrative analysis with gene expression and DNA methylation data strengthened these findings. The expression and methylation data were derived from maternal blood samples taken after birth, and may not be useful as predictors of PTB. Specifically, samples taken in the immediate postpartum period may reflect the transcriptomics of (preterm) labor rather than the differences that are the underlying causes of PTB (55). Additional gene-expression data around the time of birth from placenta, uterine tissue, and/or blood will be informative to further interpret our results (11). Still, in our integrative approach, the molecular data played an important role in implicating factors involved in the biology of PTB. Importantly, the identified pathways, or gene signatures derived from these pathways, may be more robust biomarkers of VEPTB than individual genes or variants, and could be replicated in future studies. Similar study designs would be useful in addressing the limited published studies with multiomic characterization in the same individuals, e.g., Eidem et al. (55) report that 18% of published PTB-related studies include transcriptomic profiling. The integration of other data types, such as from microbiome-based studies as well as blood-based metabolites and proteins, could also be an important adjunct to further analysis (56).
Our WGS focus allowed us to study rare variants. The importance of rare variants is likely to have been missed by previous studies. Variants uncovered in GWAS studies are common variants, generally with an MAF of at least 1%. In our primary analysis, we focused on common variants; however, rare variants may play a crucial role for complex diseases such as PTB (57). For example, McElroy et al. (58) looked at rare variants in the context of PTB by examining 10 exomes, including 2 mother–daughter pairs, and found a potential link to the complement/coagulation factor pathway. We also investigated rare variants in the set of 72 VEPTB genes (SI Appendix, Supplementary Note 4 and Fig. S9 and Datasets S16–S18). This analysis led to several moderately statistically significant genes. The function of these genes and associations shown in other studies suggests that they could play potential roles in PTB. However, validation of these results will need to be carried out in an independent cohort with a larger number of VEPTB samples.
The WGS data also enabled us to explore the relationship between the prevalence of PTB with genetic risks of other diseases through a comorbidity analysis by using published GWAS markers for 56 common diseases. We observed that variation in genetic risk varied substantially with ancestry, but not significantly across PTB categories (SI Appendix, Supplementary Note 5 and Fig. S10 and Datasets S19 and S20).
Our study is an important step forward in understanding PTB by using WGS-based clinical genomics. The comprehensive and integrative approach we developed provided genomic information at an unprecedented scale and resolution. This approach will aid in the search for genetic contributors to the complex and heterogeneous condition of PTB. Such knowledge of the biology of PTB should lead to better prediction and prevention.
Supplementary Material
Acknowledgments
We acknowledge Dr. Alison Paquette for a critical reading of the manuscript and for useful suggestions.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716314116/-/DCSupplemental.
References
- 1.World Health Organization 2015. Preterm Birth Fact Sheet (Geneva, WHO)
- 2.Hamilton BE, Martin JA, Ventura SJ. Births: Preliminary data for 2012. Natl Vital Stat Rep. 2013;62:1–20. [PubMed] [Google Scholar]
- 3.Mwaniki MK, Atieno M, Lawn JE, Newton CR. Long-term neurodevelopmental outcomes after intrauterine and neonatal insults: A systematic review. Lancet. 2012;379:445–452. doi: 10.1016/S0140-6736(11)61577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrman RE, Butler AS. 2007. Preterm Birth: Causes, Consequences, and Prevention, eds Behrman RE, Butler AS. The National Academies Collection: Reports Funded by National Institutes of Health (National Institutes of Health, Washington, DC)
- 5.York TP, Eaves LJ, Neale MC, Strauss JF., 3rd The contribution of genetic and environmental factors to the duration of pregnancy. Am J Obstet Gynecol. 2014;210:398–405. doi: 10.1016/j.ajog.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith R. Parturition. N Engl J Med. 2007;356:271–283. doi: 10.1056/NEJMra061360. [DOI] [PubMed] [Google Scholar]
- 7.Plunkett J, et al. Mother’s genome or maternally-inherited genes acting in the fetus influence gestational age in familial preterm birth. Hum Hered. 2009;68:209–219. doi: 10.1159/000224641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd HA, et al. Maternal contributions to preterm delivery. Am J Epidemiol. 2009;170:1358–1364. doi: 10.1093/aje/kwp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362:529–535. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 10.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero R, Dey SK, Fisher SJ. Preterm labor: One syndrome, many causes. Science. 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai HJ, et al. Role of African ancestry and gene-environment interactions in predicting preterm birth. Obstet Gynecol. 2011;118:1081–1089. doi: 10.1097/AOG.0b013e31823389bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeFranco EA, Hall ES, Muglia LJ. Racial disparity in previable birth. Am J Obstet Gynecol. 2016;214:394.e1-7. doi: 10.1016/j.ajog.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 14.Menon R. Spontaneous preterm birth, a clinical dilemma: Etiologic, pathophysiologic and genetic heterogeneities and racial disparity. Acta Obstet Gynecol Scand. 2008;87:590–600. doi: 10.1080/00016340802005126. [DOI] [PubMed] [Google Scholar]
- 15.Wu W, et al. The heritability of gestational age in a two-million member cohort: Implications for spontaneous preterm birth. Hum Genet. 2015;134:803–808. doi: 10.1007/s00439-015-1558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kistka ZA, et al. Heritability of parturition timing: An extended twin design analysis. Am J Obstet Gynecol. 2008;199:43.e1-5. doi: 10.1016/j.ajog.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 17.York TP, et al. Fetal and maternal genes’ influence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. Am J Epidemiol. 2013;178:543–550. doi: 10.1093/aje/kwt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plunkett J, Muglia LJ. Genetic contributions to preterm birth: Implications from epidemiological and genetic association studies. Ann Med. 2008;40:167–195. doi: 10.1080/07853890701806181. [DOI] [PubMed] [Google Scholar]
- 19.McElroy JJ. 2013. Genetics of spontaneous idiopathic preterm birth: Exploration of maternal and fetal genomes. PhD dissertation (Vanderbilt University, Nashville, TN)
- 20.Crider KS, Whitehead N, Buus RM. Genetic variation associated with preterm birth: A HuGE review. Genet Med. 2005;7:593–604. doi: 10.1097/01.gim.0000187223.69947.db. [DOI] [PubMed] [Google Scholar]
- 21.Uzun A, et al. Targeted sequencing and meta-analysis of preterm birth. PLoS One. 2016;11:e0155021. doi: 10.1371/journal.pone.0155021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang G, et al. Genetic associations with gestational duration and spontaneous preterm birth. N Engl J Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enquobahrie DA, et al. Early pregnancy peripheral blood gene expression and risk of preterm delivery: A nested case control study. BMC Pregnancy Childbirth. 2009;9:56. doi: 10.1186/1471-2393-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heng YJ, Pennell CE, Chua HN, Perkins JE, Lye SJ. Whole blood gene expression profile associated with spontaneous preterm birth in women with threatened preterm labor. PLoS One. 2014;9:e96901. doi: 10.1371/journal.pone.0096901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsuya K, Singh N, Sooranna SR, Johnson MR, Myatt L. Epigenetics of human myometrium: DNA methylation of genes encoding contraction-associated proteins in term and preterm labor. Biol Reprod. 2014;90:98. doi: 10.1095/biolreprod.113.113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eidem HR, McGary KL, Capra JA, Abbot P, Rokas A. The transformative potential of an integrative approach to pregnancy. Placenta. 2017;57:204–215. doi: 10.1016/j.placenta.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 29.Horvath S, Xu X, Laird NM. The family based association test method: Strategies for studying general genotype–Phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 30.Haddad R, et al. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol. 2006;195:394.e1-24. doi: 10.1016/j.ajog.2005.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavey AR, et al. Utilization of genomic sequencing for population screening of immunodeficiencies in the newborn. Genet Med. 2017;19:1367–1375. doi: 10.1038/gim.2017.57. [DOI] [PubMed] [Google Scholar]
- 32.National Collaborating Centre for Women's and Children's Health 2011. Multiple pregnancy: The management of twin and triplet pregnancies in the antenatal period (RCOG, London)
- 33.Ru Y, et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42:e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naeem H, et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics. 2014;15:51. doi: 10.1186/1471-2164-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elovitz MA, et al. Distinct cervical microRNA profiles are present in women destined to have a preterm birth. Am J Obstet Gynecol. 2014;210:221.e1-11. doi: 10.1016/j.ajog.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 38.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18:46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu P, et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 2014;63:1276–1284. doi: 10.1161/HYPERTENSIONAHA.113.02647. [DOI] [PubMed] [Google Scholar]
- 40.Elovitz MA, Anton L, Bastek J, Brown AG. Can microRNA profiling in maternal blood identify women at risk for preterm birth? Am J Obstet Gynecol. 2015;212:782.e1-5. doi: 10.1016/j.ajog.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Uzun A, et al. dbPTB: A database for preterm birth. Database (Oxford) 2012;2012:bar069. doi: 10.1093/database/bar069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao X, Faris AE, Jang EK, Haslam RJ. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur J Biochem. 2002;269:259–271. doi: 10.1046/j.0014-2956.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 43.Winn VD, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 44.Denis M, et al. Placental genome and maternal-placental genetic interactions: A genome-wide and candidate gene association study of placental abruption. PLoS One. 2014;9:e116346. doi: 10.1371/journal.pone.0116346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 46.Hassed SJ, et al. RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am J Hum Genet. 2012;91:391–395. doi: 10.1016/j.ajhg.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capece A, Vasieva O, Meher S, Alfirevic Z, Alfirevic A. Pathway analysis of genetic factors associated with spontaneous preterm birth and pre-labor preterm rupture of membranes. PLoS One. 2014;9:e108578. doi: 10.1371/journal.pone.0108578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prins JR, Gomez-Lopez N, Robertson SA. Interleukin-6 in pregnancy and gestational disorders. J Reprod Immunol. 2012;95:1–14. doi: 10.1016/j.jri.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Brou L, et al. Dysregulated biomarkers induce distinct pathways in preterm birth. BJOG. 2012;119:458–473. doi: 10.1111/j.1471-0528.2011.03266.x. [DOI] [PubMed] [Google Scholar]
- 51.Velez DR, et al. Preterm birth in Caucasians is associated with coagulation and inflammation pathway gene variants. PLoS One. 2008;3:e3283. doi: 10.1371/journal.pone.0003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menon R, Camargo MC, Thorsen P, Lombardi SJ, Fortunato SJ. Amniotic fluid interleukin-6 increase is an indicator of spontaneous preterm birth in white but not black Americans. Am J Obstet Gynecol. 2008;198:77.e1-7. doi: 10.1016/j.ajog.2007.06.071. [DOI] [PubMed] [Google Scholar]
- 53.Uzun A, Dewan AT, Istrail S, Padbury JF. Pathway-based genetic analysis of preterm birth. Genomics. 2013;101:163–170. doi: 10.1016/j.ygeno.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaiswal MK, et al. Notch signaling in inflammation-induced preterm labor. Sci Rep. 2015;5:15221. doi: 10.1038/srep15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eidem HR, Ackerman WE, 4th, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: A systematic review and meta-analysis. BMC Med Genomics. 2015;8:27. doi: 10.1186/s12920-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiGiulio DB, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA. 2015;112:11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibson G. Rare and common variants: Twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McElroy JJ, et al. Maternal coding variants in complement receptor 1 and spontaneous idiopathic preterm birth. Hum Genet. 2013;132:935–942. doi: 10.1007/s00439-013-1304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuteja G, Cheng E, Papadakis H, Bejerano G. PESNPdb: A comprehensive database of SNPs studied in association with pre-eclampsia. Placenta. 2012;33:1055–1057. doi: 10.1016/j.placenta.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 60.Plunkett J, et al. An evolutionary genomic approach to identify genes involved in human birth timing. PLoS Genet. 2011;7:e1001365. doi: 10.1371/journal.pgen.1001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.