Significance
Permanent infections with beneficial bacteria are widespread in nature and are believed to play a pivotal role in evolution. How hosts’ immune functions are regulated to maintain cooperative bacteria while preventing them from constantly activating the host immune system is a key question in understanding host–bacterial associations’ sustainability. In insects, beneficial bacteria are often confined within specialized host cells, the bacteriocytes, which avoid direct contact between the host’s immune system and bacteria. Here, we report an additional mechanism that prevents bacteria-released immunogenic molecules (i.e., peptidoglycan fragments) from escaping the bacteriocytes. We show that two peptidoglycan-cleaving enzymes are specifically produced in the bacteriocytes, where they cleave bacterial peptidoglycan into nonimmunogenic fragments, thereby preventing continuous and damageable host systemic immune activation.
Keywords: endosymbiosis, insect, innate immunity, PGRP, coevolution
Abstract
Long-term intracellular symbiosis (or endosymbiosis) is widely distributed across invertebrates and is recognized as a major driving force in evolution. However, the maintenance of immune homeostasis in organisms chronically infected with mutualistic bacteria is a challenging task, and little is known about the molecular processes that limit endosymbiont immunogenicity and host inflammation. Here, we investigated peptidoglycan recognition protein (PGRP)-encoding genes in the cereal weevil Sitophilus zeamais’s association with Sodalis pierantonius endosymbiont. We discovered that weevil pgrp-lb generates three transcripts via alternative splicing and differential regulation. A secreted isoform is expressed in insect tissues under pathogenic conditions through activation of the PGRP-LC receptor of the immune deficiency pathway. In addition, cytosolic and transmembrane isoforms are permanently produced within endosymbiont-bearing organ, the bacteriome, in a PGRP-LC–independent manner. Bacteriome isoforms specifically cleave the tracheal cytotoxin (TCT), a peptidoglycan monomer released by endosymbionts. pgrp-lb silencing by RNAi results in TCT escape from the bacteriome to other insect tissues, where it chronically activates the host systemic immunity through PGRP-LC. While such immune deregulations did not impact endosymbiont load, they did negatively affect host physiology, as attested by a diminished sexual maturation of adult weevils. Whereas pgrp-lb was first described in pathogenic interactions, this work shows that, in an endosymbiosis context, specific bacteriome isoforms have evolved, allowing endosymbiont TCT scavenging and preventing chronic endosymbiont-induced immune responses, thus promoting host homeostasis.
Virtually all eukaryotes associate with mutualistic bacteria. It has been argued that such symbiotic associations are among the main evolutionary driving forces, notably by creating diversity, improving species adaptation, and generating biological novelty (1–4). One of the most critical challenges associated with housing mutualistic symbionts is discriminating between beneficial bacterial partners and pathogenic intruders. Both mutualistic and pathogenic bacteria share similar microbial-associated molecular patterns (MAMPs), including peptidoglycan (PG) and its derivatives, which trigger host innate immunity (5). How host–symbiont coevolution has shaped host immune genes, allowing chronic symbiont presence while retaining defensive efficiency against pathogens, remains a key issue.
One evolutionary strategy commonly observed in insects that house beneficial symbionts is bacterial seclusion in specialized host cells, called bacteriocytes, which can group together in an organ, the bacteriome (6). The intracellular compartmentalization of bacteria (endosymbionts) within bacteriocytes enables specific immune responses adapted to endosymbiont maintenance and control, while preventing symbionts from triggering the host systemic immune responses, which could be detrimental for both partners (7). In this way, the bacteriome immunity must not only avoid endosymbiont escape, but also prevent MAMP release and dissemination throughout the host tissues.
To understand bacteriome immune adaptation to endosymbiont presence, we investigated the association between the cereal weevil Sitophilus zeamais and the Gram-negative bacterium Candidatus Sodalis pierantonius, hereafter referred to as S. pierantonius (8–10) (Fig. 1 A and B). S. pierantonius was acquired recently (28,000 y ago), following symbiont replacement within the Dryophthoridae family, and retains similar genomic features with free-living bacteria (11–13). It encodes genes necessary for MAMP synthesis, including PG (9), and its experimental injection in the insect hemolymph elicits a systemic immune response (14), making this model system particularly relevant to study host immune adaptations to endosymbiont housing. We previously showed that the bacteriome selectively produces one antimicrobial peptide (AMP), coleoptericin A (ColA), which targets endosymbionts and prevents their escape from the bacteriocytes (15). Nonetheless, bacterial PG fragments could exit the bacteriocytes and trigger a systemic immune response. While it has been demonstrated that PG is able to cross membranes and epithelial barriers in other organisms (16–21), mechanisms that avoid PG release from insect bacteriomes remain unknown. PG is detected through host PG recognition proteins (PGRPs), a family of conserved proteins believed to have evolved from bacteriophage T7 lysozymes able to cleave PG (22). In animals, PGRPs have evolved toward three main functions in host–bacteria interactions: immunity activator upon PG recognition; immune pathway modulator, notably through PG cleavage into nonimmunogenic fragments; and immune effector through bactericidal activity (23, 24).
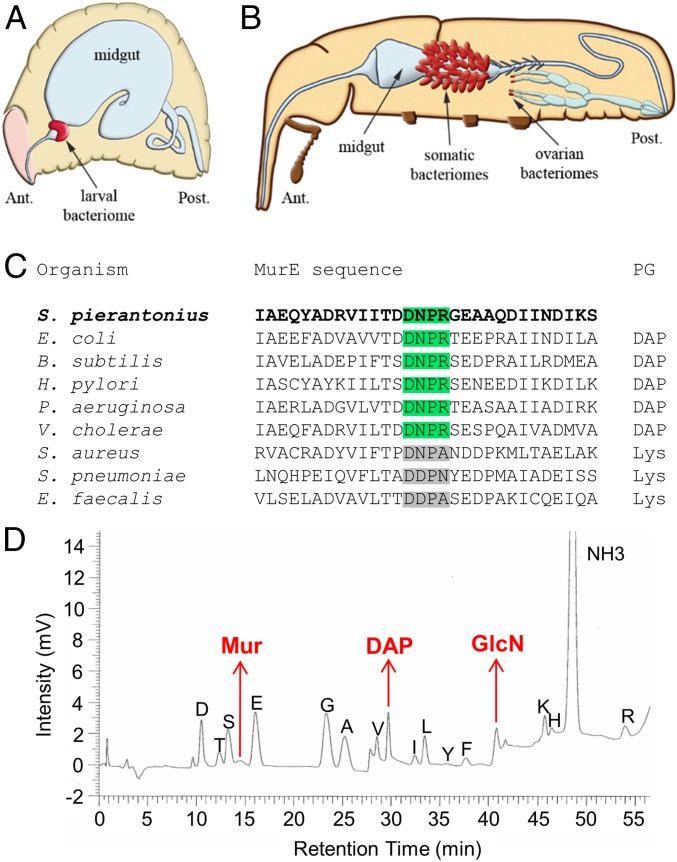
Fig. 1.
S. pierantonius produces a DAP-type peptidoglycan. (A and B) Schematic representation of endosymbiont localization in S. zeamais. In larvae (A), a unique bacteriome is located at the foregut–midgut junction. In adults (B), multiple bacteriomes are present at the apex of mesenteric caeca (“somatic bacteriomes”), as well as at the apex of the ovaries. Adapted with permission from ref. 7, and from ref. 37, with permission from Elsevier. Ant., anterior; Post., posterior. (C) Multiple protein sequence alignment of bacterial MurE. The DNPR motif found in S. pierantonius is found in DAP-adding MurE. (D) Biochemical analysis of S. pierantonius PG. PG extracted from larval bacteriome was hydrolyzed and inspected with an amino acid analyzer. Components in red are typically found in DAP-type PG.
In Drosophila, PGRPs are essential in the activation and regulation of the immune deficiency (IMD) pathway, in response to Gram-negative bacteria (24). In particular, the transmembrane receptor PGRP-LC recognizes circulating polymeric PG as well as a monomeric fragment, the tracheal cytotoxin (TCT), from Gram-negative bacteria (25, 26), while TCT located within insect cells is detected by the intracellular PGRP-LE (21, 27, 28) (SI Appendix, Fig. S1A). PG detection activates the IMD pathway, resulting in the nuclear translocation of NF-κB transcription factor Relish and AMP-encoding gene up-regulation (29). PGRP-LB negatively modulates the IMD pathway by cleaving bacterial PG into nonimmunogenic fragments (20, 30) (SI Appendix, Fig. S1A). We recently demonstrated that imd and relish are required for immune activation upon TCT injection in cereal weevils, and for endosymbiont-specific immune response within the bacteriome (31). However, the mechanism by which S. pierantonius activates the host local immunity in the bacteriome, without chronically triggering a systemic immune response, remains unknown.
Here, we have studied the function of pgrp genes with regards to symbiosis and host immunity in the cereal weevil S. zeamais. We show that pgrp-le is absent, and that pgrp-lc is essential for circulating-PG recognition and consequent IMD pathway activation. Remarkably, pgrp-lc is not involved in endosymbiont recognition within the bacteriome. Molecular investigations showed, however, that two bacteriocyte-expressed isoforms of the pgrp-lb gene specifically cleave within the bacteriome symbiont-released TCT, preventing its dissemination to insect tissues and subsequent chronic immune activation via PGRP-LC, which is detrimental to the weevil host but not to its associated endosymbionts.
Results and Discussion
S. pierantonius Produces a Diaminopimelic Acid-Type PG That Triggers the Weevil Systemic Immune Response.
We first sought to understand how S. pierantonius activates the host systemic immune response when injected into the insect hemolymph. Because S. pierantonius has conserved the genes required for PG synthesis (9), we hypothesized that such a molecule might mediate endosymbiont recognition through host immune receptors. PG is composed of glycan chains, formed by an alternation of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), and cross-linked by short peptides. Peptide composition changes between bacteria, depending on the amino acid being incorporated in the third position by the MurE enzyme [a lysine in Gram-positive cocci, and a diaminopimelic acid (DAP) in Gram-positive bacilli and Gram-negative bacteria (32, 33)]. S. pierantonius MurE possesses a four-amino acid consensus sequence found in DAP-incorporating MurE (Fig. 1C) (34). Biochemical analyses confirmed that S. pierantonius does synthesize DAP-type PG (Fig. 1D and SI Appendix, Table S1).
DAP-type PG is an elicitor of the IMD pathway in insects, so we wondered if such an activation occurs in S. zeamais. We injected larvae with polymeric DAP-type PG, TCT, or MurNAc-tripeptide-DAP purified from Escherichia coli (SI Appendix, Fig. S1B), and monitored the transcript steady-state levels of three AMP-encoding genes commonly used as an IMD pathway readout in Sitophilus (31, 35, 36), namely colA, coleoptericin B (colB), and sarcotoxin. We chose to study the larval instar in which, in contrast to adults, the endosymbiont load remains relatively stable and the bacteriocytes do not exhibit any cellular changes (37). Both polymeric DAP-type PG and TCT strongly induced systemic AMP-encoding gene expression compared with PBS or MurNAc-tripeptide-DAP injection (Fig. 2A and SI Appendix, Fig. S1C). This induction peaks at 6-h postinjection, a time point we used for further experiments. Induction of a systemic immune response by polymeric DAP-type PG and monomeric TCT is consistent with previous studies in cereal weevils (31) and Drosophila (38). We therefore hypothesized that S. pierantonius TCT could trigger the S. zeamais immune system. Indeed, in Gram-negative bacteria, PG is—most of the time—hidden from host immune receptors under the lipopolysaccharide-containing outer membrane layer, while the monomeric TCT is accessible due to its release during bacterial proliferation and cell wall recycling (39). Moreover, TCT is involved in several pathogenic infections, including with Bordetella pertussis and Neisseria gonorrhoeae, where it triggers epithelial shedding (40, 41). Finally, TCT was shown to be essential for symbiont colonization in the mutualistic association between the squid Euprymna scolopes and Vibrio fischeri (42).
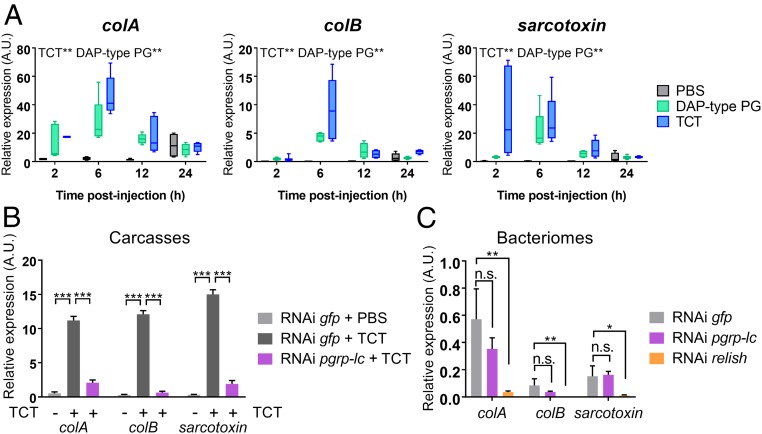
Fig. 2.
Circulating TCT, but not intracellular S. pierantonius, triggers a pgrp-lc–dependent immune response. (A) Systemic AMP-encoding gene expression kinetics following PG injection. colA, colB, and sarcotoxin expression were measured in whole larvae by qRT-PCR 2, 6, 12, or 24 h after injections of either PBS (gray), E. coli polymeric DAP-type PG (green), or E. coli-derived TCT (blue). TCT and polymeric DAP-type PG both significantly induce AMP expression in weevil larvae, based on the analysis of a generalized linear model. (B and C) AMP-encoding gene expression following pgrp-lc inhibition by RNAi. colA, colB, and sarcotoxin expression was measured in carcasses by qRT-PCR 6 h after either PBS or TCT injection (B), or in the bacteriome under standard conditions (C) 6 d following pgrp-lc or relish dsRNA injection. AMP expression does not increase following TCT injection when pgrp-lc is inhibited. However, AMP-encoding gene basal, symbiont-dependent expression in the bacteriome is not affected by pgrp-lc inhibition. By comparison, relish inhibition abolishes AMP-encoding gene expression in the bacteriome, as was previously shown (31). Asterisks indicate a significant difference between two conditions based on a Welch’s t test. The mean and SE for five independent replicates are represented. A.U., arbitrary units. n.s., nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001.
PGRP-LC Regulates Systemic Immunity, but Not the Bacteriome Immune Program.
To understand how TCT is recognized by the weevil’s immune system, we next studied the PGRP receptors in S. zeamais, PGRP-LC and -LE (SI Appendix, Fig. S1A). Remarkably, no pgrp-le transcript has been identified in S. zeamais transcriptomic data. Interestingly, pgrp-le is also absent from the genome of the tsetse fly (43), another symbiotic insect that harbors the endosymbiont Wigglesworthia glossinidia. The lack of intracellular TCT receptors in insects harboring endosymbionts may be a common feature that avoids the induction of local immune responses against endosymbionts. However, we did identify in S. zeamais a pgrp-lc ortholog and assessed its function with RNA interference (RNAi). To avoid interference with the bacteriome-specific immunity (7, 35), we separated larvae into two different samples: the bacteriome and the rest of the body, hereafter referred to as the “carcass.” Induction of systemic AMP-encoding gene expression by TCT was significantly lower in RNAi pgrp-lc carcasses, compared with controls (Fig. 2B). Thus, PGRP-LC is necessary for extracellular TCT detection and the ensuing systemic immune response in S. zeamais. Similar results were obtained in dissected bacteriomes (SI Appendix, Fig. S1D), suggesting that the bacteriome immunocompetency to exogenous challenges reported in a previous study (35) is also PGRP-LC–dependent. Intriguingly, AMP-encoding gene expression in the bacteriome, without TCT injection, did not significantly change following pgrp-lc inhibition (Fig. 2C), suggesting that the bacteriome immune response to intracellular stimuli (i.e., endosymbionts) is not initiated by PGRP-LC, despite being IMD/Relish-dependent (Fig. 2C) (31). Hence, S. zeamais does not seem to recognize intracellular TCT within the bacteriome, all the while being able to detect circulating TCT outside of the bacteriome, both systemically and locally, enabling the insect to mount an efficient immune response against bacterial intruders. Nonetheless, even in the absence of intracellular TCT detection inside the bacteriome, based on the strong metabolic exchanges between S. pierantonius and S. zeamais (9, 37, 44–46), we hypothesized that endosymbiont TCT might hitchhike with metabolites during their export from the bacteriome. Thus, symbiont-released TCT may exit the bacteriome and induce a systemic PGRP-LC–dependent immune response.
Weevil pgrp-lb Expresses Two Bacteriocyte Isoforms and a Secreted One Through Alternative Splicing.
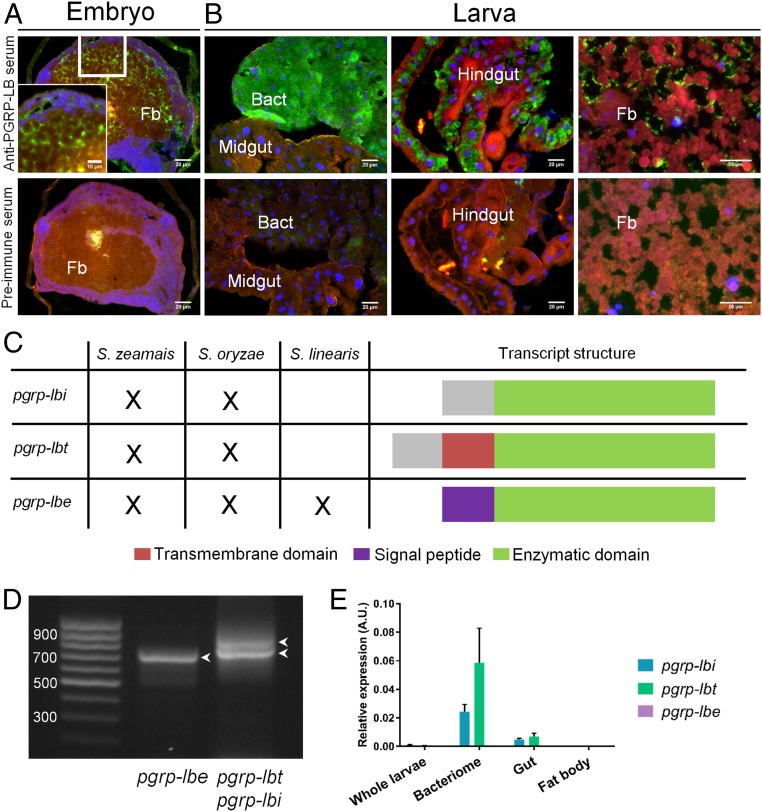
In Drosophila, down-regulation of PGRP-LC–mediated IMD pathway activation is notably accomplished by PGRP-LB (SI Appendix, Fig. S1A), which cleaves DAP-type PG into nonimmunogenic fragments through an amidase activity, hence, modulating the response to both pathogens and gut commensals (20, 30, 47). Interestingly, a pgrp-lb ortholog in S. zeamais, named wpgrp1 in previous studies, is highly expressed in the bacteriome (45, 48). Immunostaining experiments during weevil development showed that PGRP-LB could be detected both intra- and extracellularly (Fig. 3 A and B). During the embryonic stages, PGRP-LB seems to be extracellular in the fat body (Fig. 3A). At the larval and adult stages, PGRP-LB signal seems to be found extracellular in the fat body and, remarkably, intracellular within gut epithelial cells and the bacteriocytes (Fig. 3B and SI Appendix, Fig. S2). The strong production of PGRP-LB in the bacteriocytes presumes a potential function linked to endosymbiosis. We sequenced the transcriptome of S. zeamais whole adults and subsequently discovered three pgrp-lb splice variants, a finding that was further confirmed by RT-PCR (Fig. 3 C and D and SI Appendix, Supplementary Sequences). These transcripts encode three different proteins that share a predicted amidase domain. Interestingly, these isoforms are predicted to have distinct subcellular locations, according to which we named them PGRP-LBi for intracellular, PGRP-LBt for transmembrane, and PGRP-LBe for extracellular (Fig. 3C and SI Appendix, Supplementary Sequences).
Fig. 3.
S. zeamais pgrp-lb encodes two symbiosis related protein isoforms and one secreted isoform. (A and B) Immunostaining of PGRP-LB in embryos (A) and larval tissues (B). A preimmune serum was used as negative control (Lower). PGRP-LB–associated signal seems extracellular in the embryo fat body (A), and the larval fat body (B, Right). Intracellular signal is observed strongly in the bacteriome (B, Left), and in the gut (B, Center). Blue: DAPI; green: PGRP-LB; red: autofluorescence. Bact, bacteriome; fb, fat body. (C) Schematic representation of the three alternatively spliced pgrp-lb transcripts and their occurrence in three weevil species: S. zeamais and S. oryzae that house the endosymbiont S. pierantonius, and S. linearis that is naturally asymbiotic. Three transcripts are detected in the symbiotic species, while only pgrp-lbe (encoding the secreted isoform) is found in S. linearis. The green color indicates the common sequence between all three transcripts and includes the predicted enzymatic domain. The purple color indicates the sequence encoding the predicted signal peptide, specific to pgrp-lbe. The red color indicates the sequence encoding the predicted transmembrane domain, specific to pgrp-lbt. The gray color indicates the common sequence between pgrp-lbi and -lbt. The specific PGRP-LBi and -LBt N-terminal sequences (gray and red) do not show similarity with any sequence of the National Center for Biotechnology Information (NCBI) database. (D) Agarose gel electrophoresis showing the three pgrp-lb transcripts amplified by RT-PCR from S. zeamais’ total cDNA. Arrowheads point at the three bands correponding to the three pgrp-lb transcripts. (E) pgrp-lb transcript quantification in larval tissues. pgrp-lbi, pgrp-lbt, and pgrp-lbe transcripts were quantified by qRT-PCR with specific sets of primers. The mean and SE of five independent replicates are represented. pgrp-lbi levels are 5, 340, and 27 times higher in the bacteriome than in the gut, the fat body, and the whole body, respectively; pgrp-lbt levels are 8, 1,100, and 100 times higher in the bacteriome than in the gut, the fat body, and the whole body, respectively.
Consistent with the immunochemistry data, quantitative RT-PCR experiments revealed high amounts of pgrp-lbi and -lbt transcripts in the bacteriocytes, contrasting with the low amounts of pgrp-lbe in all tissues (Fig. 3E). To address the intracellular and transmembrane nature of the bacteriocyte PGRP-LB variants, we transfected Drosophila S2 cells with V5-tagged PGRP-LBi and PGRP-LBt. Immunostaining showed intracellular versus transmembrane signal for PGRP-LBi and -LBt, respectively (SI Appendix, Fig. S3). Using a double GFP-V5–tagged construction and immunostaining with or without membrane permeabilization, we further showed that PGRP-LBt is inserted in both orientations within Drosophila cell membranes (SI Appendix, Fig. S3). PGRP-LBt could either be initially inserted in both orientations, or it could be able to change its topology within membranes, as was notably described in E. coli following membrane lipid composition changes (49). This further strengthens the transmembrane localization of PGRP-LBt, and suggests that its predicted amidase activity may occur on both sides of the membranes, although the situation might be different in the bacteriocyte, where interactions with weevil proteins expressed in this specialized cell could stabilize PGRP-LBt orientation.
Intriguingly, N-terminal sequences specific to PGRP-LBi and -LBt do not show similarity with any known sequence (Fig. 3C and SI Appendix, Supplementary Sequences). To further understand the origin of these transcripts within the Sitophilus genus, and to determine the impact of endosymbiosis in pgrp-lb evolution, we sequenced the transcriptome of an additional symbiotic species, the cereal weevil Sitophilus oryzae that also houses S. pierantonius, and of a naturally nonsymbiotic species, Sitophilus linearis (13). While all three pgrp-lb transcripts were expressed in S. oryzae, only the pgrp-lbe transcript was found in the nonsymbiotic S. linearis transcriptome (Fig. 3C and SI Appendix, Supplementary Sequences). Thus, pgrp-lbi and -lbt splice variants might be the result of an endosymbiosis-driven evolution of the pgrp-lb gene structure or regulation. These findings point toward a specific function for PGRP-LBi and -LBt in host–symbiont interactions. It is noteworthy that three PGRP-LB isoforms with distinct localization (extracellular or intracellular) have also recently been reported in the Drosophila model, in which they show distinct local and systemic immune modulation to the presence of gut commensal bacteria (47). Therefore, pgrp-lb subfunctionalization through alternatively spliced variants and differential subcellular localization could be viewed as an evolutionary molecular shaping of this gene family to the distinct constraints present in fruit flies and weevils (i.e., gut extracellular commensals versus endosymbionts residing inside specialized cells).
Weevil pgrp-lb Transcripts Are Differentially Regulated.
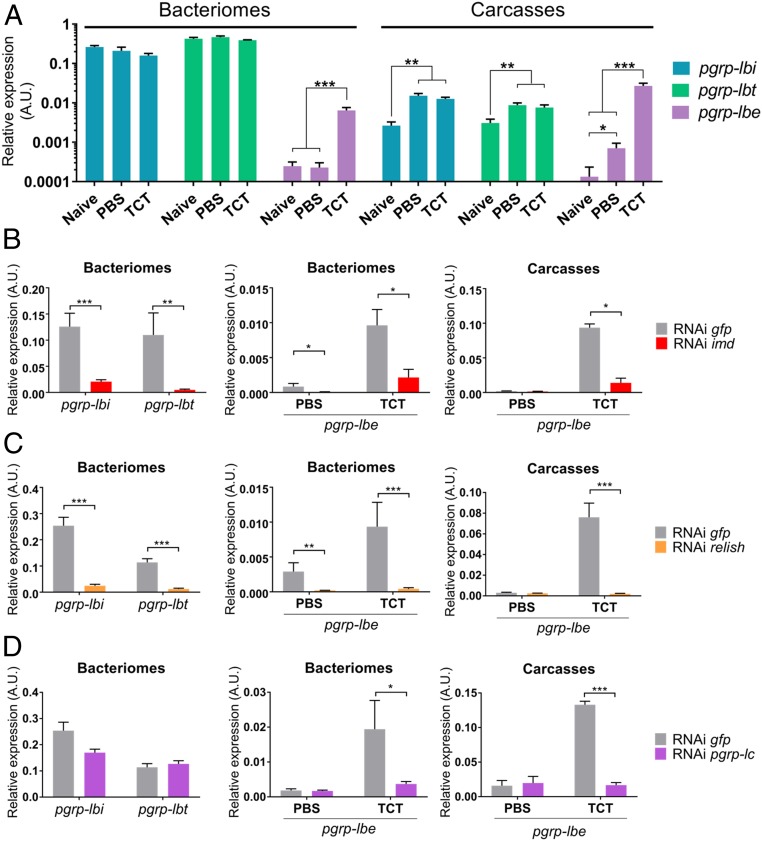
Because pgrp-lbi and -lbt are specifically expressed in bacteriocytes, we next explored how these variants are locally regulated within the bacteriome organ, compared with insect systemic immunity. We show that pgrp-lbi and -lbt transcript levels are not affected in the bacteriome following PBS or TCT injection (Fig. 4A). In the carcass, however, both pgrp-lbi and -lbt transcript levels increased similarly following either PBS or TCT injection (Fig. 4A), indicating that pgrp-lbi and -lbt transcript levels are affected by stress injury due to insect pricking and injection rather than by TCT. Conversely, pgrp-lbe transcript levels increased significantly in the carcass as well as in the bacteriome following TCT injection, indicating that pgrp-lb generates pgrp-lbe transcripts but not -lbi and -lbt transcripts following an immune challenge (Fig. 4A). RNAi inhibiting imd or relish expression significantly decreased pgrp-lbi and -lbt levels in the bacteriome, as well as the pgrp-lbe TCT-dependent levels in the bacteriome and carcass (Fig. 4 B and C), suggesting an IMD-dependent expression of all three transcripts. Pgrp-lc silencing also inhibited TCT-dependent up-regulation of pgrp-lbe levels in the bacteriome and carcass, but not pgrp-lbi and -lbt transcript levels in the bacteriome (Fig. 4D). These results point to a differential regulation of pgrp-lbe and pgrp-lbi/-lbt splicing. Indeed, pgrp-lbe transcript levels are regulated by extracellular TCT recognition through PGRP-LC and the IMD pathway. As all PGRP-LB proteins possess an amidase domain, we speculate that PGRP-LBe may participate in the modulation of the systemic immune response to pathogens, similarly to Drosophila PGRP-LB (20). In contrast, the transcript levels of pgrp-lbi/-lbt in the bacteriocytes are IMD/Relish-dependent but PGRP-LC–independent. This suggests that these isoforms are under the regulation of a yet unknown bacteriocyte-specific intracellular receptor, acting upstream of the IMD pathway, which could also regulate colA IMD-dependent (31), but PGRP-LC–independent expression (Fig. 2C). In the tsetse fly, pgrp-lb expression was shown to be symbiont-dependent but IMD-independent (50). While endosymbiont TCT could be a potential candidate for symbiont recognition inside the bacteriocytes, the IMD-independent pgrp-lb expression in the tsetse fly, and the PGRP-LC/-LE–independent expression of colA and pgrp-lbi/-lbt in Sitophilus, suggest that other bacterial molecules may trigger local gene expression in these model systems.
Fig. 4.
pgrp-lbi and pgrp-lbt transcripts are differentially regulated from pgrp-lbe. (A) Quantification of pgrp-lbi (blue), pgrp-lbt (green), and pgrp-lbe (purple) transcripts in bacteriomes and carcasses by qRT-PCR. Quantification was performed on naive individuals or 6 h after PBS or TCT injection. pgrp-lbe transcript levels are increased in all tissues following TCT injection, while pgrp-lbi and -lbt transcript levels remain stable. Arbitrary units are expressed as log-linear values. (B–D) Assessment of the IMD pathway implication in the regulation of pgrp-lb expression. Six days following imd (B), relish (C), or pgrp-lc (D) dsRNA injection, pgrp-lbi and pgrp-lbt transcript levels were quantified in the bacteriome (Left), and pgrp-lbe transcript level was quantified in the bacteriome and carcass, 6 h after PBS or TCT (Center and Right). pgrp-lbe transcript levels are IMD-, Relish-, and PGRP-LC–dependent. pgrp-lbi and -lbt expression transcript levels are IMD- and Relish-dependent, but not PGRP-LC-dependent. The mean and SE for five independent replicates are represented. Asterisks indicate a significant difference between two conditions based on a Welch’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Weevil PGRP-LB Enzymatic Domain Has Evolved Specificity Toward TCT.
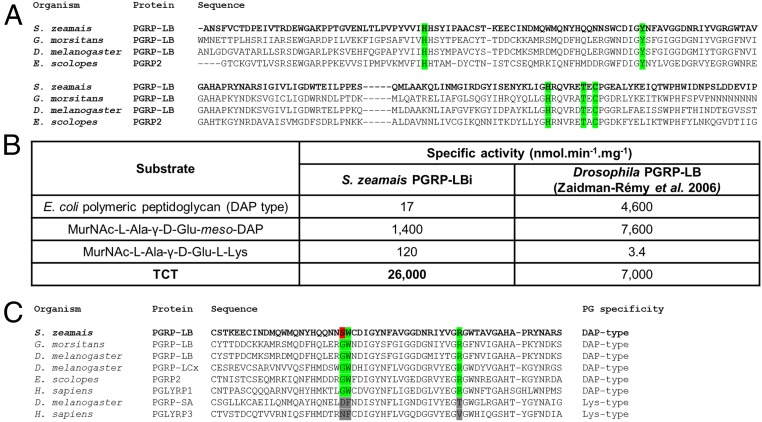
Because PGRP-LBi and -LBt are strongly produced in the bacteriocytes, we sought to understand their precise role by characterizing their enzymatic activity. The prediction of an N-acetylmuramoyl-l-alanine amidase activity that cleaves PG relies on the presence of five amino acid residues, conserved across catalytic PGRPs and required for the zinc-binding–dependent amidase activity (Fig. 5A) (51). To determine whether S. pierantonius DAP-type PG is cleaved by the weevil PGRP-LB enzymatic domain, we analyzed the amidase activity of one isoform, PGRP-LBi, against polymeric DAP-type PG, MurNAc-tripeptide-Lys, MurNAc-tripeptide-DAP, and TCT purified from culturable bacteria (SI Appendix, Fig. S1B). We show that the weevil PGRP-LB enzymatic domain displays a strong specificity toward TCT (Fig. 5B). Activities against DAP-type PG, MurNAc-tripeptide-Lys, and MurNAc-tripeptide-DAP were at least 18 times lower (Fig. 5B). These data contrast with the Drosophila PGRP-LB activity, which is versatile for polymeric and monomeric DAP-type PG, and suggest that weevil PGRP-LB has diverged, probably under the evolutionary pressure of endosymbiosis. Recognition of PG by the amidase activity relies on three conserved amino acid residues, which differ between Lys- and DAP-type binding PGRPs (24). Interestingly, one of these amino acids is not conserved in the weevil PGRP-LB, which could account for TCT specificity (Fig. 5C). TCT is a mediator of host–bacteria interactions in pathogenesis and mutualism. Modification of the bacterial TCT recycling machinery is known to modulate TCT release into the host. For example, in Neisseria species, the inner membrane permease AmpG ensures PG fragment recycling and limits the amount of TCT secreted (52, 53). Impaired AmpG activity correlates with a stronger virulence (53). Remarkably, we found that ampG is among the genes that were shown to be pseudogenized in S. pierantonius (9), suggesting that it may shed high quantities of TCT into the host. The PGRP-LB–specific amidase activity toward TCT could have emerged as an adaptive feature along host–symbiont coevolution, allowing the host to cope efficiently with symbiont-released TCT.
Fig. 5.
PGRP-LB efficiently and specifically cleaves TCT in vitro. (A) Multiple sequence alignment of the enzymatic domains of S. zeamais, Glossina morsitans, Drosophila melanogaster, and E. scolopes PGRPs. Residues critical to PGRP enzymatic activity are highlighted in green, and are conserved in S. zeamais PGRP-LB enzymatic domain. (B) In vitro enzymatic activity of recombinant PGRP-LBi toward different PG substrates (see SI Appendix, Fig. S1B for PG structures), assessed by HPLC. Drosophila PGRP-LB enzymatic activity (20) is indicated for comparison. PGRP-LBi enzymatic activity against TCT is at least 18 times higher than against other substrates. (C) Multiple sequence alignment of the PGRP domains of S. zeamais PGRP-LB and those of other organisms. Residues critical to DAP-type PG binding are highlighted in green. One of them (S103, red) is not conserved in S. zeamais PGRP-LB.
Weevil pgrp-lb Prevents Systemic Immune Activation by Endosymbionts.
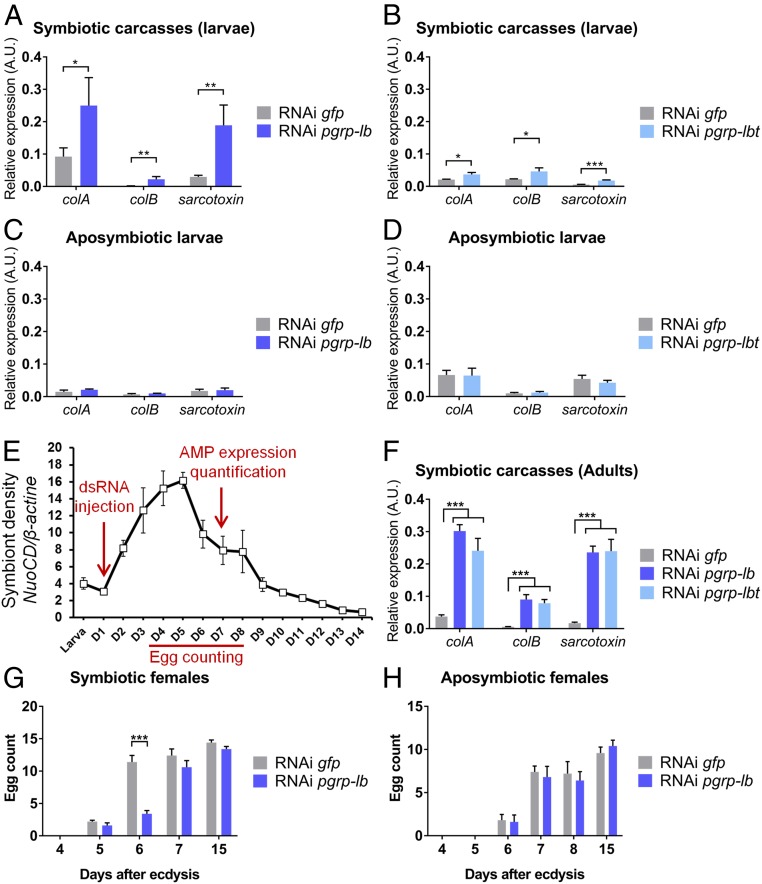
Our data strongly point to an endosymbiosis-driven evolution of pgrp-lb, notably in terms of gene regulation and protein enzymatic activity; hence, we next evaluated pgrp-lb function in host–symbiont interactions. To this end, we inhibited simultaneously all three pgrp-lb transcripts by RNAi in larvae and quantified AMP-encoding gene expression in the carcass, in the absence of any exogenous challenge. Interestingly, all AMP-encoding genes tested showed a strong induction upon RNAi (Fig. 6A). This induction likely relies on pgrp-lbi and pgrp-lbt inhibition because these two transcripts are at least 1,000-fold more abundant than the pgrp-lbe transcript within the bacteriome and carcass in standard conditions (Fig. 3E). To support this hypothesis, we chose to specifically inhibit pgrp-lbt. Because the transmembrane domain-encoding cDNA of pgrp-lbt is too short, we have generated a double-strand (ds) RNA harboring two concatenated cDNA sequences. Pgrp-lbt specific inhibition was sufficient to observe a phenotype similar to pgrp-lb inhibition (Fig. 6B), attesting the crucial function of this isoform in avoiding a systemic immune response. To confirm that the systemic immune activation following weevil pgrp-lb extinction is related to endosymbiont presence, we repeated these experiments with insects artificially depleted of their endosymbionts (aposymbiotic) (54). We show that pgrp-lb inhibition did not generate any immune activation in these insects (Fig. 6 C and D). Taken together, these results indicate that the immune response following pgrp-lb or specific pgrp-lbt extinction is related to symbiont presence, similarly to Drosophila, the tsetse fly, and the red palm weevil (20, 30, 47, 50, 55, 56). It is noteworthy that similar mechanisms have evolved in both extracellular and intracellular symbioses, highlighting the similar immune adaptations to associations with bacteria.
Fig. 6.
PGRP-LB prevents chronic immune systemic activation and detrimental effect on host physiology. (A–D) Systemic AMP-encoding gene expression in symbiotic (A and B) or aposymbiotic (C and D) larvae after pgrp-lb (A and C) or pgrp-lbt (B and D) RNAi inhibition. colA, colB, and sarcotoxin expression was measured in symbiotic larval carcasses or whole aposymbiotic larvae by qRT-PCR 8 d following pgrp-lb or pgrp-lbt RNAi treatment. AMP gene expression in the carcass increases following pgrp-lb or specific pgrp-lbt inhibition in symbiotic larvae, but not in aposymbiotic larvae. (E) Experimental timing for pgrp-lb inhibition in adults, based on symbiont density in emerging S. zeamais adults. (D) Days after final ecdysis. dsRNA was injected at day 1 after final ecdysis. Six days after dsRNA injection, individuals were dissected and AMP expression was quantified in carcasses. Eggs were counted from day 4 to day 8, and at day 15. Adapted from ref. 37, with permission from Elsevier. (F) Systemic AMP-encoding gene expression in adults after pgrp-lb or pgrp-lbt inhibition. colA, colB, and sarcotoxin expression was measured in adult carcasses by qRT-PCR and strongly increases following pgrp-lb or specific pgrp-lbt RNAi inhibition in adults. (G and H) Egg count in ovaries from symbiotic (G) or aposymbiotic (H) females following pgrp-lb RNAi treatment. At day 6, egg load in RNAi pgrp-lb females is significantly lower than in RNAi gfp females. The mean and SE for five independent replicates are represented. Asterisks indicate a significant difference between two conditions based on a Welch’s t test (A, D, and F) or based on a Student’s t test (G and H) (*P < 0.05; **P < 0.01; ***P < 0.001).
While in larvae endosymbiont load remains relatively stable and low, emerging adults experience a drastic and rapid increase of endosymbiont load (37). Therefore, to understand to which extent PGRP-LB isoforms avoid systemic immune activation and sustain host homeostasis, we studied the impact of pgrp-lb suppression in emerging adults (Fig. 6E). AMP-encoding gene expression also significantly increased in carcasses of RNAi pgrp-lb or RNAi pgrp-lbt adults (Fig. 6F), but in a much stronger extent than in larvae, consistent with the higher symbiont load. Beyond immunity, we hypothesized that such strong deregulations during a crucial developmental period might affect insect biological processes. Consequently, we analyzed the impact of pgrp-lb inhibition on female sexual maturation, a process that occurs at the first days following adult emergence, concomitantly with endosymbiont population increase (Fig. 6E). By counting the egg load in ovaries, we observed that sexual maturation takes place between days 5 and 6 in control females (Fig. 6G). In pgrp-lb RNAi females, however, egg load at day 6 was significantly lower (Fig. 6G), and sexual maturation took one additional day compared with control females. This delay was symbiont-dependent because pgrp-lb–silenced aposymbiotic females did not display any difference with the controls (Fig. 6H). Taken together, these data show that symbiotic homeostasis disruption, through pgrp-lb inhibition, impacts the weevil’s overall physiology. The sexual maturation delay could be interpreted as a resource allocation shift from egg production to a costly immune response, including AMP production. In several insects, overstimulated immunity negatively impacts reproductive traits, and reversely, active reproduction reduces immune abilities (57). The impact observed on egg production could also be a direct deleterious effect of IMD pathway overactivation, which promotes apoptosis in Drosophila’s wing discs (58) and gut (59).
Weevil immune overactivation following pgrp-lb inhibition impaired host physiological functions but did not impact endosymbiont load (SI Appendix, Fig. S4A). We therefore conclude that in S. zeamais, PGRP-LB bacteriocyte-isoforms do not act per se in endosymbiont preservation from the host immunity (i.e., what some authors refer to as “tolerance”), but in preventing chronic host inflammation due to endosymbiont presence. This contrasts with the tsetse fly in which pgrp-lb inhibition leads to endosymbiont load decrease (50, 55), attesting to the crucial role of PGRP-LB in endosymbiont immune tolerance in this model. pgrp-lb inhibition also results in an impaired fertility in female tsetse (55), which in this model could be the consequence of either resource competition with the overly activated immune system, or more probably of the decreased symbiotic population that is essential for host fertility (60).
PGRP-LB Isoforms Prevent Endosymbiont TCT from Reaching the Host Systemic Immune System.
To pinpoint the molecular basis of AMP up-regulation in RNAi pgrp-lb individuals, we simultaneously inhibited pgrp-lb and pgrp-lc expression, because PGRP-LC regulates AMP expression in the carcass (Fig. 2B). We show that RNAi against pgrp-lc rescued the pgrp-lb RNAi phenotype (Fig. 7A), indicating that the systemic immune activation upon pgrp-lb inhibition is mediated by the receptor PGRP-LC. The presence of PG in the carcass could result from either endosymbionts or PG fragments escaping the bacteriome. However, FISH experiments with a specific probe directed against S. pierantonius did not reveal any symbiont outside of the bacteriome in RNAi pgrp-lb insects (SI Appendix, Fig. S4B). To strengthen these histological results, we show that PGRP-LBi did not display any bactericidal activity against the Gram-negative bacterium E. coli in vitro (SI Appendix, Fig. S4C), and that, as mentioned above, global endosymbiont density was not affected by pgrp-lb RNAi (SI Appendix, Fig. S4A), indicating that PGRP-LB does not directly control symbiont populations. Furthermore, colA expression, the disruption of which results in endosymbiont escape (15, 31), was not affected in the bacteriome by pgrp-lb inhibition (SI Appendix, Fig. S4 D and E), suggesting that PGRP-LB does not indirectly control endosymbionts either. Overall, we demonstrate that systemic immune activation following pgrp-lb inhibition is not due to symbiont escape from the bacteriome.
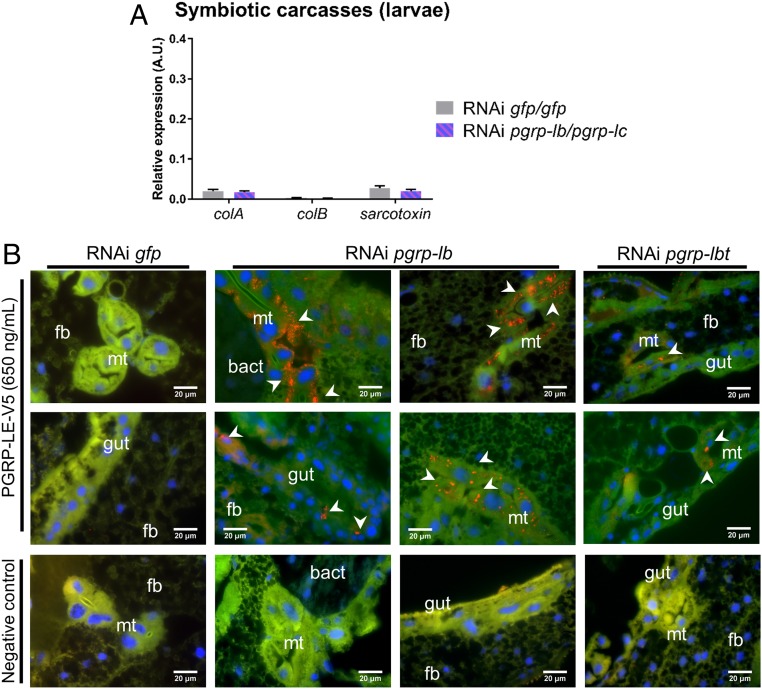
Fig. 7.
PGRP-LB prevents symbiotic TCT escape from the bacteriome and recognition by PGRP-LC. (A) Systemic AMP-encoding gene expression in symbiotic larvae after pgrp-lb and pgrp-lc RNAi treatment. colA, colB, and sarcotoxin expression was measured in carcasses by qRT-PCR 8 d after simultaneous pgrp-lb/pgrp-lc RNAi treatment. AMP expression does not vary following simultaneous pgrp-lb and pgrp-lc inhibition. The mean and SE for five independent replicates are represented. No significant difference was found between two conditions based on a Welch’s t test. (B) TCT localization in symbiotic larvae 8 d following gfp (column 1), pgrp-lb (columns 2–3), or pgrp-lbt (column 4) RNAi treatment. TCT was identified using recombinant Drosophila PGRP-LE (SI Appendix, Fig. S5). A negative control without PGRP-LE was included (Bottom). In RNAi pgrp-lb or pgrp-lbt, TCT is seen in gut cells and in Malpighian tubules (white arrowheads). bact, bacteriome; fb, fat body; Mt, Malpighian tubules. Blue: DAPI; green: autofluorescence; red: TCT.
To attest that the immune activation following pgrp-lb inhibition is due to TCT escape from the bacteriome organ, we developed a tool for in situ TCT detection. Based on the specific TCT binding ability of the Drosophila PGRP-LE recognition domain (27, 61), we constructed a V5-tagged recombinant PGRP-LE to use as a TCT detector by immunostaining (SI Appendix, Fig. S5). Remarkably, individuals treated with RNAi pgrp-lb or RNAi pgrp-lbt exhibited strong punctuated signal in the gut epithelia and Malpighian tubules (Fig. 7B and SI Appendix, Fig. S6 and Table S2). Malpighian tubules endorse detoxifying and excretory functions in insects, and are known to internalize TCT via oligopeptide carriers in Drosophila (16, 27).
These findings show that PGRP-LB specifically cleaves symbiotic TCT, preventing its dissemination out of the bacteriome and the subsequent activation of a PGRP-LC–dependent systemic immune response. Several mechanisms allowing PG to enter eukaryotic cells have been described in pathogenic interactions, including bacterial pore-forming toxins (62), outermembrane vesicles (63), and oligopeptide carriers (16). We hypothesize that similar mechanisms may permit PG released by intracellular symbionts to exit bacteriocytes. The strong vesicular network previously described in bacteriocytes and suggested to mediate host–symbiont metabolic exchanges (45, 64) could also participate in TCT transfer outside of the bacteriocytes. Based on the subcellular locations of PGRP-LBi and -LBt, we propose that these bacteriome-specific isoforms cleave TCT both in the cytosol and in export vesicles, preventing direct contact between TCT and the systemic immunity receptor PGRP-LC. We also speculate that PGRP-LBe is able to cleave any TCT that might still escape the bacteriome, as is suggested by pgrp-lbe transcript level increase following specific pgrp-lbt transcript inhibition (SI Appendix, Fig. S7A).
Since TCT identification as the main toxin released by B. pertussis and N. gonorrhoeae, and as the causative agent of the whooping cough and gonorrhea (40, 41), TCT has been shown to be a major microbe–host signaling molecule both in pathogenic and mutualistic interactions. Not only does TCT trigger host immunity, but it also widely affects host behavior (65) and morphogenetic processes (42, 66). Notably, in the beneficial association between the squid E. scolopes and V. fischeri, a TCT-cleaving PGRP is produced only after the symbiont-released TCT has triggered the symbiotic light organ morphogenesis (67). TCT could therefore impact other functions than immunity in the association between cereal weevils and S. pierantonius, functions that remain to be explored.
In conclusion, we have characterized host-specific local immune barriers that complement morphogenetic adaptations (bacteriocytes) and ensure immune homeostasis by preventing a chronic and detrimental host immune activation. While S. pierantonius ColA-dependent seclusion in bacteriocytes prevents endosymbionts from directly triggering host immunity, the PGRP family plays an equally important role by limiting symbiotic TCT-mediated local and systemic immune activation. We show that TCT is most likely not detected within the bacteriome, due to the absence of appropriate receptors, while its dissemination throughout host tissues and subsequent recognition by systemic receptors is limited through PGRP-LB local action in the bacteriome organ. Weevil pgrp-lb has evolved several features specific to endosymbiont-derived MAMP tolerance, including an increased enzymatic specificity against TCT, and the specific subcellular distribution of two isoforms within the bacteriocytes via the acquisition of novel sequences. This indicates how the PG-cleaving function has evolved from a presumed offensive role in phage T7 lysozymes to sustaining host homeostasis in symbiotic relationships. These local regulations do not seem to affect the host’s ability to properly respond to pathogens, through PGRP-LC and PGRP-LBe systemic functions. This work shows how host–symbiont coevolution not only promotes biological novelty, for example through the emergence of dedicated symbiotic structures, but also shapes existing host immune genes and implements novel mechanisms that favor symbiont persistence without compromising the host’s defenses against pathogens.
Materials and Methods
See SI Appendix for additional materials and methods.
Biological Material and Sample Preparation.
S. zeamais and S. oryzae weevils are reared on wheat grains at 27.5 °C and at 70% relative humidity. The Lagoa (S. zeamais) strains were chosen in this work because they are free of any facultative symbionts, including Wolbachia, and harbor only S. pierantonius. S. linearis individuals were collected in Niamey (Niger) and reared on tamarind seeds at 27.5 °C and at 70% relative humidity. Aposymbiotic insects were obtained as previously described (54). The aposymbiotic status was confirmed by PCR and histology experiments.
Insect organs (bacteriomes, guts, ovaries) were dissected in diethylpyrocarbonate-treated Buffer A (25 mM KCl, 10 mM MgCl2, 250 mM Sucrose, 35 mM Tris⋅HCl, pH = 7.5). For RNA extraction, at least five organs or whole individuals per condition were pooled and stored at −80 °C and each sampling was independently repeated five times. For DNA extraction, single whole individuals were stored at −80 °C and each sampling was independently repeated eight times.
PG Injections.
PG fragments were produced and purified from E. coli as previously described (38). For TCT and MurNAc-tripeptide-DAP, 55 nL at a 0.2 mM concentration were injected into the hemolymph using a Nanoject II (Drummond). For whole DAP-type PG, after 5 min of sonication, 55 nL at 1 mM were injected. Sterile PBS was used as a negative control. Individuals were recovered 2, 6, 12, or 24 h after injection.
dsRNA Synthesis and Injection.
dsRNA was prepared as described previously (68). Primers used for T7 DNA fragments are listed in SI Appendix, Table S3 and were designed to amplify a fragment from 200 bp to 300 bp. Next, 50 ng of dsRNA were injected into the hemolymph of third-instar larvae or 1-d-old adults with a Nanoject II (Drummond). dsRNA targeting the gfp gene was injected as a negative control. When two genes were targeted at once, 50 ng of each dsRNA were injected, and 100 ng of gfp dsRNA were injected as a negative control. Individuals were then kept on wheat flour, for 6 or 8 d, at a controlled temperature (27.5 °C) and relative humidity (70%). Efficiency of the RNAi was then checked by qRT-PCR (SI Appendix, Fig. S7A) and by immunostaining in the case of pgrp-lb inhibition (SI Appendix, Fig. S7B).
Total RNA Extraction and Reverse-Transcription.
Total RNA from whole larvae or carcasses was extracted with TRIzol reagent (ThermoFisher Scientific) following the manufacturer’s instructions. RNA was incubated with 1 U/µg of RQ1 RNase-free DNase (Promega) for 30 min at 37 °C. Total RNA from bacteriomes, guts and fat body was extracted using RNAqueous Micro (Ambion), which allows for a better RNA yield from small tissue samples. After purification, the RNA concentration was measured with a Nanodrop spectrophotometer (ThermoFisher Scientific) and RNA quality was checked using agarose gel electrophoresis. Reverse-transcription into the first strand cDNA was carried out using the iScript cDNA Synthesis Kit (Bio-Rad).
Real-Time qRT-PCR Transcript Quantification.
The quantification was performed with a LightCycler instrument using the LightCycler Fast Start DNA Master SYBR Green I kit (Roche Diagnostics). Data were normalized using the ratio of the target cDNA concentration to that of two housekeeping genes: ribosomal protein L29 (rpL29, both larvae and adults), glyceraldehyde 3-phosphate dehydrogenase (gapdh, adults only), and malate dehydrogenase (mdh, larvae only). Gene expression is therefore represented as the relative expression of the gene of interest compared with normalizing genes. As such, this ratio of two concentrations is expressed in arbitrary units. Primers were designed to amplify fragments of ∼200 bp. A complete list of the primers can be found in SI Appendix, Table S3. The PCR reactions were carried out as previously described (31).
Biochemical Assays.
PGRP-LBi enzymatic activity was measured in 50-µL reaction mixtures containing 50 mM ethanolamine buffer, pH 9.6, 2.5 mM ZnSO4, and unlabeled or radiolabeled forms of DAP-type PG, TCT or Lys- and DAP-containing MurNAc-tripeptide as substrates (0.1–0.5 mM range). Radiolabeled PG polymer and fragments were obtained as previously described (20, 38). After incubation for 30 min (PG fragments) or several hours (PG) at 37 °C, the substrates and reaction products were separated by HPLC on a Nucleosil 100 C18 5µ column (250 × 4.6 mm; Alltech France) using a gradient of methanol from 0 to 20% in 50 mM sodium phosphate buffer, pH 4.3, for elution, at a flow rate of 0.6 mL/mL Peaks were detected at 207 nm and the radioactivity was detected and quantitated with a flow detector as described in Zaidman-Rémy et al. (20). Digestion of the whole PG polymer by PGRP-LBi released tetra- and octapeptides as main products and digestion of its MurNAc-tripeptide and TCT fragments released tri- and tetrapeptide, respectively. The amounts of peptides thus released were quantitated and used to determine the specific enzymatic activity of the PGRP-LBi.
Construction, Expression and Purification of a Drosophila PGRP-LE Recognition Domain Fusion with V5 Epitope.
The gene segment coding for the PGRP domain (residues 173–345) of Drosophila PGRP-LE (Accession no. Q9VXN9) was chemically synthesized with optimization for E. coli expression (ProteoGenix). The sequences coding for a 6His-tag and a V5-tag were added in 5′/Nter position in fusion with the ORF. This cDNA sequence was cloned in an E. coli pT7 expression vector. The sequence of the expected protein produced is illustrated in SI Appendix, Fig. S5 A and B.
The target protein was produced in a 1 L E. coli culture at 37 °C, with an induction of 1 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) during 4 h, and after centrifugation the pellet of bacterial culture was solubilized with a denaturing buffer with 8 M urea.
The denatured protein extract was applied to a His-Tag on nickel Sepharose. The column was equilibrated with TBS Buffer pH 7.5 and washed, then eluted by imidazole shift. The final buffer is TBS pH 7.5 supplemented with urea 4 M. The protein of interest is refolded by dialysis method in a buffer with 50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 5 mM GSH, and 0.5 mM GSSG. The final samples were then buffer-exchanged by a standard dialysis method versus PBS pH 7.5 at a final concentration of 0.13 mg/mL (purity >90%) (ProteoGenix).
TCT Localization Using a Drosophila PGRP-LE Recognition Domain Fusion with V5 Epitope.
The same protocol as the one used for PGRP-LB immunostainings was used, using the fusion PGRP-LE-V5 diluted either 1:100 or 1:200 incubated overnight at 4 °C, a mouse anti-V5 antibody diluted 1:500 (ThermoFisher Scientific) incubated for 1 h at room temperature, and an Alexa Fluor 647 goat anti-mouse IgG diluted 1:1,000 (ThermoFisher Scientific) incubated for 1 h at room temperature.
Aposymbiotic larvae were used as a negative control to find the dilution that yielded the least possible background signal in individuals that do not contain any bacteria that potentially release TCT (SI Appendix, Fig. S5D). TCT-injected larvae were used as a positive control: 55 nL of a 0.2 mM purified TCT solution were injected, and larvae were recovered, fixed 1 h later and processed as described in SI Appendix, Supplementary Materials and Methods. The signal observed in the fat body and its decrease proportional to PGRP-LE-V5 concentration confirmed the capacity of our construction to detect TCT in situ (SI Appendix, Fig. S5C). Images were acquired the same way as described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. Fiol, S. López-Madrigal, and C. Vargas-Chávez for their valuable help on animal rearing and sampling; R. Rebollo for her critical reading of the manuscript; S. Hughes and B. Gillet (Institut de Génomique Fonctionnelle de Lyon) for RNA sequencing; N. Silverman (University of Massachusetts) for the fruitful discussion on novel ways to detect tracheal cytotoxin; B.-H. Oh (Korea Advanced Institute of Science and Technology) for providing Drosophila’s pgrp-le sequence; T. Touzé (Université Paris Sud) for trying to produce recombinant PGRP-LBt; A. Khila’s team (Institut de Génomique Fonctionnelle de Lyon) for providing Drosophila S2 cells; J.-Y. Rasplus for collecting Sitophilus linearis individuals from Niger; and G. Gabant and V. Aucagne (Center for Molecular Biophysics, Orléans) for mass spectrometry experiments. qPCR was performed at Développement de Techniques et Analyse Moléculaire de la Biodiversité, from the Bio Environnement et Santé Research Federation. Confocal microscopy was performed at the Centre Technologique des Microstructures. Funding for this project was provided by Institut National de la Recherche Agronomique, Institut National des Sciences Appliquées-Lyon, the French ANR-13-BSV7-0016-01 (IMetSym), the French ANR-17-CE20-0031-01 (GREEN), and a grant from La Région Rhône-Alpes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra [transcriptome datasets (accession nos. SRP149413 and SRP149424); cDNA sequences (accession nos. MH423625 and MH423627–MH423629)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821806116/-/DCSupplemental.
References
- 1.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heddi A, Zaidman-Rémy A. Endosymbiosis as a source of immune innovation. C R Biol. 2018;341:290–296. doi: 10.1016/j.crvi.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medzhitov R, Janeway CA. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 6.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. Interscience Publishers/John Wiley; New York: 1965. [Google Scholar]
- 7.Masson F, Zaidman-Rémy A, Heddi A. Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Philos Trans R Soc Lond B Biol Sci. 2016;371:403–411. doi: 10.1098/rstb.2015.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: A peculiar G + C content of an endocytobiotic DNA. J Mol Evol. 1998;47:52–61. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 9.Oakeson KF, et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 2014;6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles H, Heddi A, Rahbe Y. A putative insect intracellular endosymbiont stem clade, within the Enterobacteriaceae, infered from phylogenetic analysis based on a heterogeneous model of DNA evolution. C R Acad Sci III. 2001;324:489–494. doi: 10.1016/s0764-4469(01)01328-2. [DOI] [PubMed] [Google Scholar]
- 11.Clayton AL, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 2012;8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conord C, et al. Long-term evolutionary stability of bacterial endosymbiosis in curculionoidea: Additional evidence of symbiont replacement in the dryophthoridae family. Mol Biol Evol. 2008;25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- 13.Lefèvre C, et al. Endosymbiont phylogenesis in the dryophthoridae weevils: Evidence for bacterial replacement. Mol Biol Evol. 2004;21:965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- 14.Anselme C, et al. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 16.Paik D, et al. SLC46 family transporters facilitate cytosolic innate immune recognition of monomeric peptidoglycans. J Immunol. 2017;199:263–270. doi: 10.4049/jimmunol.1600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendrin M, Welchman DP, Poidevin M, Hervé M, Lemaitre B. Long-range activation of systemic immunity through peptidoglycan diffusion in Drosophila. PLoS Pathog. 2009;5:e1000694. doi: 10.1371/journal.ppat.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arentsen T, et al. The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol Psychiatry. 2017;22:257–266. doi: 10.1038/mp.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaidman-Rémy A, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol. 2012;189:1886–1897. doi: 10.4049/jimmunol.1201022. [DOI] [PubMed] [Google Scholar]
- 22.Montaño AM, Tsujino F, Takahata N, Satta Y. Evolutionary origin of peptidoglycan recognition proteins in vertebrate innate immune system. BMC Evol Biol. 2011;11:79. doi: 10.1186/1471-2148-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat Rev Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 24.Royet J, Dziarski R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 26.Leulier F, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 28.Bosco-Drayon V, et al. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. 2012;12:153–165. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 30.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Maire J, Vincent-Monégat C, Masson F, Zaidman-Rémy A, Heddi A. An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp. Microbiome. 2018;6:6. doi: 10.1186/s40168-017-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith CA. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol. 2006;362:640–655. doi: 10.1016/j.jmb.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 33.El Zoeiby A, Sanschagrin F, Levesque RC. Structure and function of the Mur enzymes: Development of novel inhibitors. Mol Microbiol. 2003;47:1–12. doi: 10.1046/j.1365-2958.2003.03289.x. [DOI] [PubMed] [Google Scholar]
- 34.Patin D, et al. Unusual substrate specificity of the peptidoglycan MurE ligase from Erysipelothrix rhusiopathiae. Biochimie. 2016;121:209–218. doi: 10.1016/j.biochi.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Masson F, et al. Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis. J Innate Immun. 2015;7:290–301. doi: 10.1159/000368928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Madrigal S, Maire J, Balmand S, Zaidman-Rémy A, Heddi A. Effects of symbiotic status on cellular immunity dynamics in Sitophilus oryzae. Dev Comp Immunol. 2017;77:259–269. doi: 10.1016/j.dci.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Vigneron A, et al. Insects recycle endosymbionts when the benefit is over. Curr Biol. 2014;24:2267–2273. doi: 10.1016/j.cub.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 38.Stenbak CR, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- 39.Park JT. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 40.Goldman WE, Klapper DG, Baseman JB. Detection, isolation, and analysis of a released Bordetella pertussis product toxic to cultured tracheal cells. Infect Immun. 1982;36:782–794. doi: 10.1128/iai.36.2.782-794.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melly MA, McGee ZA, Rosenthal RS. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J Infect Dis. 1984;149:378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- 42.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 43.Attardo GM, et al. Genome sequence of the tsetse fly (Glossina morsitans): Vector of African Trypanosomiasis. Science. 2014;344:380–386. doi: 10.1126/science.1249656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci USA. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heddi A, et al. Molecular and cellular profiles of insect bacteriocytes: Mutualism and harm at the initial evolutionary step of symbiogenesis. Cell Microbiol. 2005;7:293–305. doi: 10.1111/j.1462-5822.2004.00461.x. [DOI] [PubMed] [Google Scholar]
- 46.Heddi A, Lefebvre F, Nardon P. Effect of endocytobiotic bacteria on mitochondrial enzymatic-activities in the weevil Sitophilus oryzae (Coleoptera, Curculionidae) Insect Biochem Mol Biol. 1993;23:403–411. [Google Scholar]
- 47.Charroux B, et al. Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe. 2018;23:215–228.e4. doi: 10.1016/j.chom.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Anselme C, Vallier A, Balmand S, Fauvarque M-O, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol. 2006;72:6766–6772. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitrac H, MacLean DM, Jayaraman V, Bogdanov M, Dowhan W. Dynamic membrane protein topological switching upon changes in phospholipid environment. Proc Natl Acad Sci USA. 2015;112:13874–13879. doi: 10.1073/pnas.1512994112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106:12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M-S, Byun M, Oh B-H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- 52.Garcia DL, Dillard JP. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J Bacteriol. 2008;190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JM, Dillard JP. Neisseria gonorrhoeae crippled its peptidoglycan fragment permease to facilitate toxic peptidoglycan monomer release. J Bacteriol. 2016;198:3029–3040. doi: 10.1128/JB.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nardon P. Obtention d’une souche aposymbiotique chez le charançon Sitophilus sasakii Tak : Différentes méthodes d’obtention et comparaison avec la souche symbiotique d’origine. C R Acad Sci Paris. 1973;227D:981–984. [Google Scholar]
- 55.Wang J, Aksoy S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc Natl Acad Sci USA. 2012;109:10552–10557. doi: 10.1073/pnas.1116431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dawadi B, et al. PGRP-LB homolog acts as a negative modulator of immunity in maintaining the gut-microbe symbiosis of red palm weevil, Rhynchophorus ferrugineus Olivier. Dev Comp Immunol. 2018;86:65–77. doi: 10.1016/j.dci.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Schwenke RA, Lazzaro BP, Wolfner MF. Reproduction-immunity trade-offs in insects. Annu Rev Entomol. 2015;18:16. doi: 10.1146/annurev-ento-010715-023924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bischoff V, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryu J-H, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 60.Pais R, Lohs C, Wu Y, Wang J, Aksoy S. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol. 2008;74:5965–5974. doi: 10.1128/AEM.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim J-H, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 62.Hruz P, et al. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci USA. 2009;106:12873–12878. doi: 10.1073/pnas.0904958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaparakis M, et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12:372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 64.Vigneron A, et al. Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol. 2012;12:S14. doi: 10.1186/1471-2180-12-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurz CL, Charroux B, Chaduli D, Viallat-Lieutaud A, Royet J. Peptidoglycan sensing by octopaminergic neurons modulates Drosophila oviposition. eLife. 2017;6:1059–1067. doi: 10.7554/eLife.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 67.Troll JV, et al. Taming the symbiont for coexistence: A host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallier A, Vincent-Monégat C, Laurençon A, Heddi A. RNAi in the cereal weevil Sitophilus spp: Systemic gene knockdown in the bacteriome tissue. BMC Biotechnol. 2009;9:44. doi: 10.1186/1472-6750-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.