Significance
Many enzymes harness the energy of molecular oxygen to catalyze diverse transformations. However, reaction with oxygen is kinetically challenging and must be precisely controlled to prevent cellular damage. The formylglycine-generating enzyme activates its targets by utilizing a mononuclear copper center to facilitate oxidation of a substrate cysteine. We reveal the structure of its atypical copper active site and discover that substrate binding directly to copper precedes and initiates reactivity toward oxygen. We furthermore identify a transient intermediate that may provide evidence for the involvement of an elusive catalytic species. This work uncovers a distinct strategy among copper enzymes that exploits a dynamic binding site to tightly couple the activation of oxygen with selective substrate oxidation.
Keywords: formylglycine, copper oxidase, metalloenzyme, X-ray spectroscopy, bioinorganic chemistry
Abstract
The formylglycine-generating enzyme (FGE) is required for the posttranslational activation of type I sulfatases by oxidation of an active-site cysteine to Cα-formylglycine. FGE has emerged as an enabling biotechnology tool due to the robust utility of the aldehyde product as a bioconjugation handle in recombinant proteins. Here, we show that Cu(I)–FGE is functional in O2 activation and reveal a high-resolution X-ray crystal structure of FGE in complex with its catalytic copper cofactor. We establish that the copper atom is coordinated by two active-site cysteine residues in a nearly linear geometry, supporting and extending prior biochemical and structural data. The active cuprous FGE complex was interrogated directly by X-ray absorption spectroscopy. These data unambiguously establish the configuration of the resting enzyme metal center and, importantly, reveal the formation of a three-coordinate tris(thiolate) trigonal planar complex upon substrate binding as furthermore supported by density functional theory (DFT) calculations. Critically, inner-sphere substrate coordination turns on O2 activation at the copper center. These collective results provide a detailed mechanistic framework for understanding why nature chose this structurally unique monocopper active site to catalyze oxidase chemistry for sulfatase activation.
The formylglycine-generating enzyme (FGE) catalyzes the cotranslational or posttranslational activation of type I sulfatases in eukaryotes and aerobic microbes (1). This is accomplished by oxidation of a sulfatase active-site cysteine residue located in the minimum consensus sequence CXPXR to Cα-formylglycine (fGly), a critical cofactor for the hydrolysis of sulfate ester substrates (Fig. 1). Dysfunction of FGE in humans results in a congenital disease, multiple sulfatase deficiency, which originates from a global decrease of sulfatase function (2, 3). The promiscuity of FGE has enabled its use in biotechnology and therapeutic applications, such as site-specific drug attachment to fGly in monoclonal antibodies (4–6).
Fig. 1.
The O2-dependent conversion of cysteine to Cα-formylglycine (fGly) catalyzed by FGE.
Initially, FGE was thought to catalyze Cys-to-fGly conversion using an unprecedented cofactorless oxidase/monooxygenase mechanism, relying on an essential redox-active cysteine pair (7). However, in 2015, Holder et al. (8) provided evidence for activation and stoichiometric Cu binding in Streptomyces coelicolor and human FGEs, implicating FGE as a Cu-dependent metalloenzyme. Further investigation was carried out by Knop et al. (9), detailing that high-affinity (Kd ∼10−17 M) copper binding is dependent on the aforementioned active-site cysteine pair (10). This was unexpected because typical mononuclear copper oxidases contain N- and O-ligands at higher coordination states (11). FGE crystal structures bound to catalytically inactive Ag(I) and Cd(II) have provided structural evidence for a dicysteine binding motif reminiscent of known Cu(I) binding proteins (12). Likely owing to the difficulty of isolating metallated Cu(I) protein crystals (13), structural characterization of an authentic Cu–FGE complex has remained elusive. Additionally, the manner in which the monocopper site reacts with O2, and the reactive intermediates responsible for substrate oxidation are unclear. Therefore, elucidation of the catalytic mechanism of FGE is fundamental to a broader understanding of copper-mediated biotransformations.
Here, we present the X-ray crystal structure of Cu-bound FGE at a resolution of 2.2 Å. The linear cuprous dithiolate ligand system observed in the holoenzyme structure is unusual for a copper oxidase and is superficially similar to well-characterized Cu(I) transporters and chaperones (14). Additionally, we report the electronic absorption and electron paramagnetic resonance (EPR) features of a transient cupric-FGE complex, providing direct evidence that FGE can bind both copper redox states with the same active-site ligands. By directly interrogating Cu(I)–FGE using X-ray absorption spectroscopy (XAS), the linear two-coordinate crystallographic model was confirmed in solution. Moreover, these studies reveal a shift to a three-coordinate Tris(thiolate) Cu(I) complex upon inner-sphere substrate binding, before oxygen binding and activation. Finally, in the reaction of the preformed enzyme–substrate complex with O2, a transient intermediate is observed by stopped-flow absorbance spectroscopy. Together, these results represent the structural characterization of Cu-bound FGE in both redox states, and definitively identify the O2 activating site.
Results
Discovery and Characterization of a Transient Cu(II)–FGE Complex.
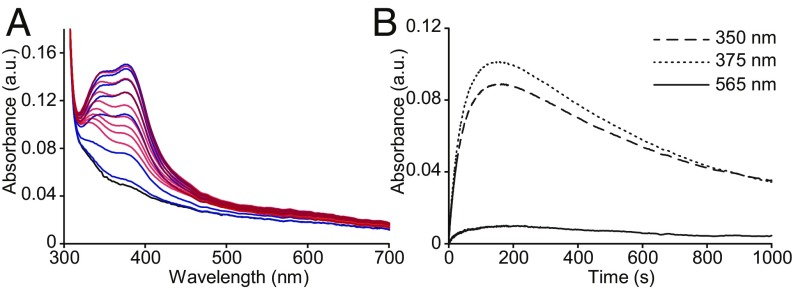
Initial interrogation of the S. coelicolor FGE (scFGE) copper center was pursued using UV-vis absorption and EPR (Figs. 2 and 3), revealing characteristic features of an explicit Cu(II)–FGE complex. Aerobic mixing of Cu(II) solution with apo-scFGE resulted in a vivid blue solution. This color change is associated with at least two absorbance features with maxima at 348 and 375 nm that saturate in intensity at ∼1:1 Cu(II):FGE (ε375 = 4,940 M−1⋅cm−1; SI Appendix, Fig. S1). Curiously, these solutions turned colorless after ∼10 min. Stopped-flow UV-vis absorption spectroscopy was employed to measure the kinetics of the chromophoric species formed by mixing 1:1 apo-scFGE and Cu(NO3)2 solution anaerobically. Absorbance features in the 320- to 450-nm region form immediately and maximize in intensity within the first 175 s, then decay to baseline over 17 min (Fig. 2A). The observed rate constants for formation and decay of the 375-nm species at 100 µM (apparent) Cu(II)–scFGE were k1 = 2.45 × 10−2 s−1, and k2 = 8 × 10−4 s−1, respectively. These features are absent in the inactive C272A/C277A double mutant (SI Appendix, Fig. S2), providing evidence for the formation of a transient Cu(II)–Cys(272,277) complex. However, the presence of additional ligands from solvent or protein is not discernible from these data.
Fig. 2.
Addition of Cu(II) to FGE forms a copper complex distinguished by transient UV-vis absorbance features. (A) Stopped-flow UV-vis spectra of apo scFGE following mixing with one equivalent of Cu(NO3)2. Features of the Cu(II)–scFGE complex grow from 1 s (black) to a maximum at 175 s (blue), and decay from 175 to 1,000 s (red). (B) The baseline-subtracted kinetic traces of the 350-, 375-, and 565-nm absorbance features. The kinetic trace of the 375-nm feature was fit to a double-exponential equation, Y = A(1 − e−k1t) + Ae−k2t + C, using nonlinear regression, yielding rate constants k1 = 2.45 × 10−2 ± 1 × 10−4 s−1 for growth and k2 = 8 × 10−4 ± 2.4 × 10−6 s−1 for decay. Errors are the SEs of the fit.
Fig. 3.
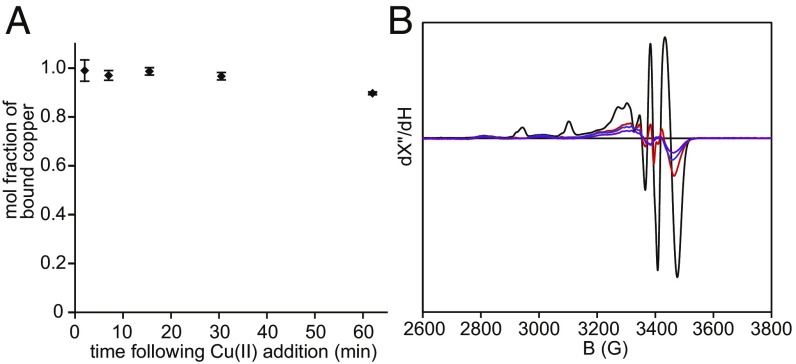
The Cu(II)–FGE complex undergoes autoreduction to Cu(I)–FGE. (A) The fraction of FGE-bound Cu remaining after anaerobic addition of one equivalent of Cu(NO3)2 to apo FGE and incubation at 25 °C, as determined by biquinoline assay of buffer-exchanged aliquots at each time point. Error bars represent the SD of three replicates. (B) 77 K, X-band EPR quantification of total Cu(II) following addition of Cu(NO3)2 to apo scFGE in a parallel experiment to A, but without buffer exchange following incubation. Each spectrum represents a single time point of an anaerobic solution of Cu(II)–scFGE: 90 s, black; 490 s, red; 1,260 s, blue; 3,470 s, purple.
The instability of the Cu(II)–Cys(272,277) complex can result from copper dissociation or 1e− reduction to form a transparent Cu(I)–FGE complex, among other mechanisms. To evaluate the dissociation pathway, the fraction of FGE-bound copper following anaerobic reconstitution was monitored as a function of time (Fig. 3A). Compared with the complete loss of absorption bands (Fig. 2A), the Cu:FGE ratio only declined from 0.99 to 0.90. Therefore, the disappearance of the chromophoric Cu(II)–Cys(272,277) complex cannot be attributed to copper release. To assess whether a reductive pathway is instead responsible, X-band EPR spectra were collected in a parallel time course. The EPR spectra (Fig. 3B) contain a mixture of at least three species. Two species having similar g values and hyperfine coupling constants are already formed by 86-s post-Cu(II) addition (black), and nearly disappear over 1 h (purple). A third intense feature near g = 2.00 may be tentatively assigned as an organic radical. The loss of EPR-active copper occurs with comparable kinetics to the decay of Cu(II)–scFGE absorbance features and is consistent with the autoreduction of Cu(II)–scFGE to Cu(I)–scFGE. Nonreducing SDS-PAGE demonstrates concomitant disulfide-mediated oligomerization of scFGE following Cu(II) treatment (SI Appendix, Fig. S4). Thus, reducing equivalents for this process ultimately originate from FGE cysteine thiols. Binding of Cu(I) to scFGE was also observed; anaerobic addition of one equivalent of Cu(MeCN)4+PF6− yielded stable holoenzyme with >0.98:1 Cu:FGE after buffer exchange.
Single-Turnover Activity of Cu(I)– and Cu(II)–FGE.
The catalytic properties of FGE in each copper oxidation state have not been addressed directly in prior studies. Although the activity of Cu(I)-loaded FGE has been demonstrated under steady-state conditions (9, 10), our data prove that apo-FGE binds both Cu(I) and Cu(II). The observed rate constant for the autoreductive decay of Cu(II)–scFGE (8 × 10−4 s−1) is more than two orders of magnitude lower than that of kcat for scFGE (0.3 s−1) (8). Therefore, it is kinetically feasible that Cu(II)–scFGE could be catalytically competent in the presence of substrate and O2. As the oxidation state of the resting enzyme places specific constraints on the mechanistic pathway available for substrate oxidation, explicitly testing the catalytic ability of each Cu–FGE redox state is of fundamental importance.
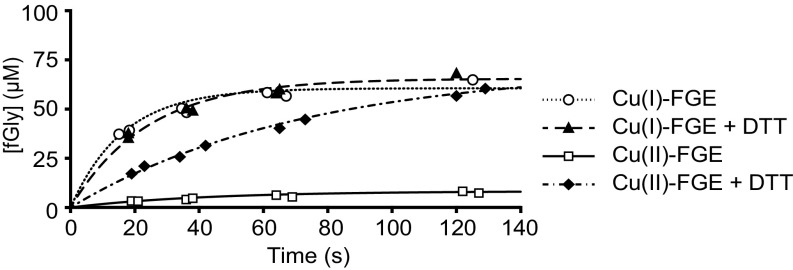
Single-turnover reactions containing 1:1 scFGE:substrate were employed to compare the activity of Cu(I)– and Cu(II)–scFGE during the first turnover (Fig. 4). These assays used a 15-mer peptide substrate (DNP-scP15) containing a 13-residue recognition motif from an S. coelicolor sulfatase and an N-terminal chromophore. To account for the inherent instability of the Cu(II)–scFGE complex, one equivalent of Cu(NO3)2 was added to apo-scFGE immediately before each reaction time course. Within 2 min, Cu(I)–scFGE achieves nearly 70% conversion of substrate to fGly, compared with 10% conversion by Cu(II)–scFGE. When the reaction was preceded by a 5-min treatment with excess DTT, Cu(II)–scFGE was activated approximately eightfold. Unlike during multiturnover conditions (9), DTT does not further activate Cu(I)–scFGE for a single turnover, suggesting that the input of external electrons is not required to resolve an intermediate and instead occurs after product formation. These data conclusively identify Cu(I)–scFGE as the catalytic resting enzyme.
Fig. 4.
Single-turnover activity assays of Cu(I)- and Cu(II)-loaded scFGE. Reactions contained 100 µM FGE and 100 µM DNP-scP15 peptide substrate at 4 °C, in the presence or absence of 2 mM DTT. The progress curves were fit to a single-exponential equation, Y = Ao(1 − e−kobst), using nonlinear regression (solid and dashed lines).
X-Ray Crystal Structure of the Cu–scFGE Holoenzyme.
With the benefit of an apo X-ray crystal structure (15), scFGE was likewise chosen for structural studies of the reconstituted holoenzyme. Using the copper activation procedure described previously (8), a crystallization screen was performed around the conditions used to generate the apo enzyme crystals (15). Initial screens with premetallated enzyme and with supplemented copper salts failed to yield copper-containing crystals. This suggested that copper may be released during crystallization, or that a disulfide active-site conformation that excluded copper entirely was a prerequisite for crystallization. Successful crystallization of the scFGE C277A mutant excludes the latter model. Instead, an irreversible copper dissociation mechanism likely accounts for the absence of bound copper among crystallization trials. Following copper binding, the extended period of crystal growth may allow copper to dissociate via oxidation of the putative cysteine ligands, yielding apo crystals. The apparent instability of Cu–FGE may be related to the autoreduction reaction pathway of Cu(II)–scFGE and is consistent with previous biochemical findings (10).
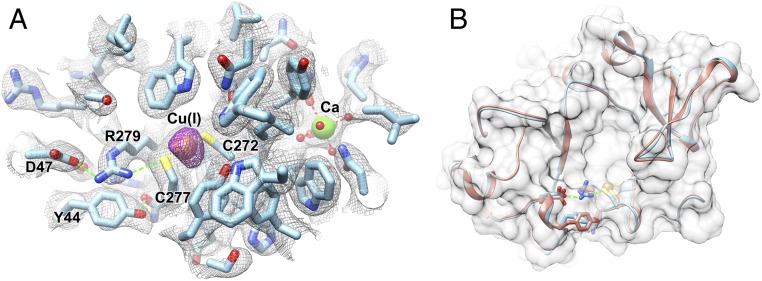
With this in mind, a multisolution crystal soaking procedure was employed. In brief, crystals were harvested, reduced with DTT, soaked with CuCl2, and last, soaked in cryoprotectant for storage in liquid N2. This sequence resulted in a reconstituted holoenzyme crystal structure of FGE with copper, its native and sole functional metal cofactor (Fig. 5). Holoenzyme crystals diffracted to a resolution of 2.2 Å, and phases were determined by molecular replacement using apo-scFGE (PDB ID 2Q17). The resulting structure contains five molecules per asymmetric unit with RMSD ∼0.21–0.27 Å between molecules. X-ray data collection and refinement statistics support the accuracy of the refined structure (Table 1). The well-conserved structure in all crystallographically independent molecules improves confidence in the active-site structure and geometry for providing mechanistic understanding, as seen for the dioxygen reactive sites in superoxide dismutases (16–18). From the crystal packing, a dimeric form (chain AB or DE or C2) of Cu(I)–scFGE was implicated as a stable assembly by PISA analysis (19) (surface area, 21,390 Å2; buried area, 5,160 Å2; ΔGassembly = −115.2 kcal/mol; ΔGdiss = 5.1 kcal/mol). However, size-exclusion chromatography coupled multiangle light scattering demonstrates that both apo-scFGE, and Cu(I)–scFGE with peptide substrate, are monomers in solution (SI Appendix, Fig. S5A).
Fig. 5.
X-ray crystal structure of copper-bound scFGE. (A) Cu bound at the FGE active site, coordinated by C272, C277, shown with adjacent active-site residues. The electron density map is overlaid as gray mesh (2Fo-Fc, 1 σ), and the anomalous difference map as purple mesh (4 σ). Cu and Ca ions are displayed as brown and green spheres, respectively. (B) Alignment of the Cu–scFGE holoenzyme crystal structure (cyan) to the apo enzyme (orange; PDB ID 2Q17) shown for the active-site region, with RMSD ∼0.38 Å.
Table 1.
Crystallography data collection and refinement statistics
| Parameters | Cu–scFGE |
| Data collection | |
| Space group | P3121 |
| Cell dimensions | |
| a = b, c, Å | 140.0, 217.1 |
| α = β, γ (°) | 90, 120 |
| Resolution, Å | 37.89–2.25 (2.33–2.25) |
| Redundancy | 10.8 (8.8) |
| Completeness (%) | 93.95 (83.18) |
| Mean I/sigma (I) | 12.72 (0.65) |
| CC1/2 | 0.998 (0.538) |
| Refinement | |
| No. reflections | 110,236 |
| Rwork/Rfree | 0.2024/0.2456 |
| No. atoms | |
| Protein | 11,267 |
| Ligand/ion | 80 |
| Water | 653 |
| Average B-factor | 48.58 |
| Protein | 48.54 |
| Ligand/ion | 64.87 |
| Water | 47.19 |
| RMSDs | |
| Bond lengths, Å | 0.008 |
| Bond angles, ° | 0.95 |
| PDB code | 6MUJ |
Statistics for the highest-resolution shell are shown in parentheses.
Each molecule contains a Cu ion, identified by a strong anomalous peak in the experimental electron density map, that is bound to two cysteine residues (C272, C277) and a Ca atom bound to D208 and N197 (Fig. 5A). The coordination environment of the Cu center features a linear, two-coordinate metal-binding site composed of both active-site cysteines, is isostructural to that of the inactive Ag(I)–FGE (PDB ID 5NXL; RMSD, 0.61 Å), and is superficially similar to active sites of cuprous trafficking proteins (Fig. 5A) (12, 20). The S1–[Cu]–S2 angle is 171°, and S–Cu bond distances are between 2.1 and 2.2 Å for C272 and C277 among five molecules. The apo and holoenzyme structures align closely with RMSD ∼0.38 Å (Fig. 5B), suggesting a largely preformed Cu atom binding site in the apo enzyme.
Notably, a hydrogen bond between the side chains of the conserved (SI Appendix, Fig. S5B) D47 and R279 residues is specific to the holoenzyme and appears to organize an interaction network that may be important for positioning of the thiolate ligand of C277 (Fig. 5A). Although the redox state of copper in crystallo was not explicitly known, Cu(I) is inferred from the two-coordinate linear ligand geometry, and the propensity for copper photoreduction during X-ray diffraction data collection at 77 K (21). This crystal structure is particularly revelatory because this binding motif is not present in previously described copper oxidases/oxygenases (11), and must confer reactivity that is uniquely suited to catalysis of O2-dependent fGly formation.
XAS and Extended X-ray Absorption Fine Structure of Cu(I)–scFGE.
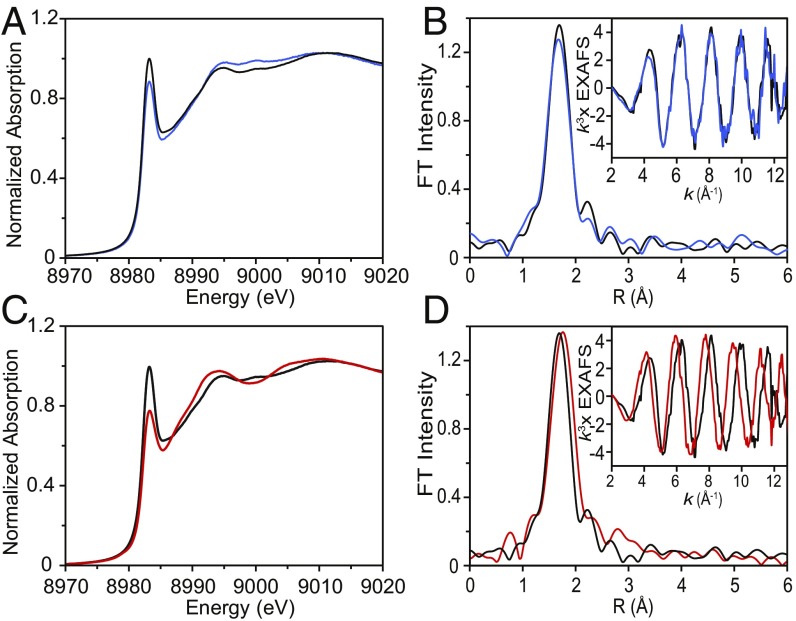
Our biochemical data and the detailed atomic structure provided by X-ray crystallography define the importance of the Cu(I)–scFGE complex. XAS (22) was used to determine the coordination number, ligand identity, and bond distances of Cu(I)–scFGE in solution. The Cu(I)–scFGE holoenzyme was prepared anaerobically and found to be stable at a concentration of 1 mM in 50 mM Tris, pH 9.0, 0.5 M NaCl, and 10% glycerol. The Cu K-edge XAS spectrum of a frozen solution of Cu(I)–scFGE (Fig. 6A, black) revealed an intense absorption feature in the 8,984-eV region, with a normalized intensity near 1.0, corresponding to the Cu(I) (1s → 4p) transition for a linear two-coordinate complex (22, 23).
Fig. 6.
XAS of the Cu(I)–scFGE, Cu(II)–scFGE → Cu(I)–scFGE (autoreduced), and Cu(I)–scFGE:scP14 complexes. (A) Cu K-edge XAS spectra, and (B) EXAFS data (Inset) and non–phase-shift-corrected Fourier transforms for Cu(I)–FGE (black) and Cu(II)–scFGE → Cu(I)–scFGE (autoreduced) (blue). (C) Cu K-edge XAS spectra, and (D) EXAFS data (Inset) and non–phase-shift-corrected Fourier transforms for Cu(I)–FGE (black) and Cu(I)–FGE:scP14 (red). These data are representative of at least three replicates.
Extended X-ray absorption fine structure (EXAFS) spectra were collected to identify the ligand environment at copper (24). EXAFS data and its Fourier transform are shown in Fig. 6B (black). Using density functional theory (DFT)-optimized parameters as a starting point, these EXAFS data were used to extract information about bond lengths and ligand identities. The best-fit model of the Cu(I)–scFGE EXAFS data included two sulfur ligands at an average Cu–S bond distance of 2.14 Å, in good agreement with the crystal structure (Table 2). Full EXAFS fitting results are shown in SI Appendix, Fig. S6. In addition to collecting and analyzing XAS and EXAFS spectra of Cu(I)–scFGE, we also characterized the fully autoreduced enzyme. Samples of autoreduced scFGE were prepared anaerobically by allowing Cu(II)–scFGE to decay to the colorless Cu(I)–scFGE complex (∼1 h). Comparison of the Cu K-edge XAS data for both complexes revealed that autoreduction results in an identical cuprous complex to that formed via direct Cu(I) binding to scFGE (Fig. 6 A and B, blue).
Table 2.
EXAFS fitting results
| Complex | Fit | CN/path* | R, ņ | σ2, Å2‡ | ΔE0, eV | Error F§ |
| Cu(I)–FGE | 1A | 1 Cu-S | 2.14 | 67 | −13.68 | 0.41 |
| 1B | 2 Cu-S | 2.14 | 390 | −13.79 | 0.29 | |
| 1C | 3 Cu-S | 2.14 | 641 | −14.14 | 0.34 | |
| Cu(I)–scFGE | 2A | 1 Cu-S | 2.13 | 97 | −15.75 | 0.43 |
| (autoreduced) | 2B | 2 Cu-S | 2.13 | 420 | −15.60 | 0.30 |
| 2C | 3 Cu-S | 2.13 | 669 | −15.82 | 0.34 | |
| Cu(I)–FGE:scP14 | 3A | 2 Cu-S | 2.23 | 365 | −9.79 | 0.36 |
| 3B | 3 Cu-S | 2.22 | 583 | −10.61 | 0.30 | |
| 3C | 2 Cu-S | 2.17 | 265 | −13.01 | 0.28 | |
| 1 Cu-S | 2.28 | −64 | ||||
| 3D | 2 Cu-S | 2.22 | 365 | −10.78 | 0.35 | |
| 1 Cu-O | 2.57 | 1,284 | ||||
| 3E | 4 Cu-S | 2.22 | 775 | −10.96 | 0.33 |
Errors in coordination numbers are ±25%, and those in the identity of the scatterer Z are ±1. The best acceptable fit for each complex is shown in bold font.
CN is coordination number.
The estimated SDs in R are ±0.02 Å.
The σ2 values are multiplied by 105.
The error F is given by [∑k6(χexptl − χcalcd) (2)/∑k6χexptl2]1/2.
XAS and EXAFS of Substrate-Bound Cu(I)–FGE.
Previous experimental and computational studies using Cu-free apo-FGE have identified a shallow binding pocket for peptide substrates that may orient the substrate cysteine residue toward C272 and C277 (25). However, the interaction of substrate with the copper center has remained ambiguous. XAS was utilized to test whether the substrate thiolate binds merely in the vicinity of the copper center or ligates it directly (12). Two factors were of concern in evaluating substrate binding. First, that Cu(I)–scFGE and its peptide substrate will form an inert complex under anaerobic conditions and, second, that substrate binds with sufficient affinity to the Cu(I)–scFGE complex to achieve sample concentrations required for XAS studies.
A 14-mer substrate (scP14) composed of 13 residues from an S. coelicolor sulfatase sequence (National Center for Biotechnology Information: WP_011031740.1), and a C-terminal tyrosine was selected for these studies. The KM of scP14 was found to be >10-fold lower (6.0 ± 0.8 µM; SI Appendix, Fig. S7) than that used in a prior study (8). Therefore, we expected that this feature may increase the equilibrium fraction of the Cu(I)–scFGE:scP14 complex for high-quality XAS samples.
The Cu(I)–scFGE:scP14 complex was isolated anaerobically by the addition of one equivalent peptide to enzyme at a final concentration of 1 mM each. The Cu K-edge XAS spectrum of the Cu(I)–scFGE:scP14 complex revealed a decrease in the intensity of the 8,983- to 8,985-eV feature compared with that of Cu(I)–scFGE (0.77 vs. 1.0). An intensity decrease of this magnitude is indicative of a change from two- to three-coordinate, for a sizable fraction of the enzyme (Fig. 6C) (23). The increase in the coordination number is also supported by the corresponding EXAFS data, which were best fit with three Cu–S bonds at 2.22 Å (Table 2, Fit 3B). The fit with two short and one longer Cu–S was attempted but excluded because of the negative σ2 value (Table 2, Fit 3C). Replacement of one Cu–S path by a Cu–O path was also tested but led to a larger error and very high σ2 value for the Cu–O path (Table 2, Fit 3D). These data provide experimental verification of substrate binding directly to the Cu(I)-scFGE active site.
O2 Activation at the FGE Copper Center Reveals a Catalytic Intermediate.
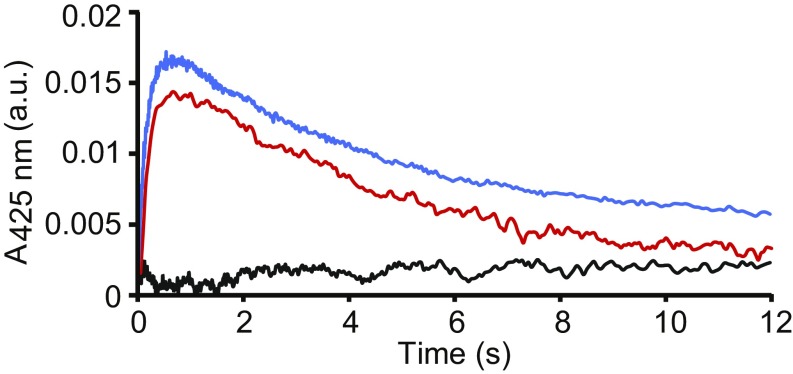
To interrogate Cu–FGE during catalysis, stopped-flow UV-vis absorption spectroscopy was used to monitor changes in absorption spectra recorded from 5 ms to 20 min following mixing of Cu(I)–scFGE:scP14 (anaerobic) with O2-saturated buffer. The time-dependent absorption spectra of this reaction revealed a transient feature at 425 nm (Fig. 7, blue).
Fig. 7.
Observation of an intermediate during FGE catalysis. Stopped-flow UV-vis spectra of a transient absorbance feature at 425 nm that is dependent on the presence of Cu holoenzyme, substrate, and O2. Kinetic traces at 425 nm are shown for the following: anaerobic Cu(I)–scFGE mixed with O2-saturated buffer (black), anaerobic preformed Cu(I)–scFGE:scP14 complex mixed with O2-saturated buffer (blue), and anaerobic autoreduced Cu(I)–scFGE:scP14 with O2-saturated buffer (red).
However, mixing Cu(I)–scFGE (anaerobic) alone with O2-saturated buffer did not produce this feature (Fig. 7, black). Interestingly, the reaction of autoreduced Cu(II)–scFGE:scP14 (anaerobic) with O2-saturated buffer yielded an identical chromophoric species that accumulated to approximately the same amounts with the same time dependence as those observed for the Cu(I)-loaded scFGE:scP14 (Fig. 7, red). This result confirms that when Cu(II)–scFGE is allowed to completely autoreduce, it converts to a form that is also catalytically competent. These observations identify the 425-nm species as the key intermediate observed during FGE catalysis, and its characterization is currently underway in our laboratories.
Discussion
Here, we present the X-ray crystal structure of the FGE holoenzyme containing its native copper cofactor, revealing a Cu binding site that shows a surprising linear coordination with two cysteine ligands, and is validated in solution by XAS/EXAFS. Our Cu(I)–scFGE active site is in good agreement with the Ag(I)-bound crystal structure (PDB ID 5NXL) (12). Single-turnover activity of Cu(I)–FGE supports its assignment as the catalytic redox form and corroborates the relevance of our structural data. We also report the discovery and characterization of a noncatalytic, or perhaps precatalytic, Cu(II)–scFGE complex. The kinetics of Cu(II)-dependent UV-vis and EPR features demonstrate the instability of this complex, which decays to a species that is spectroscopically and catalytically equivalent to Cu(I)–scFGE. These data provide a coherent model for an autoreduction process that maintains bound metal, but converts Cu(II)–scFGE to a stable Cu(I) conformation capable of catalysis. It is unclear whether the ligand environment of transient Cu(II)–scFGE is analogous to that of the inactive Cd(II)–FGE structure (PDB ID 5NYY) (12).
FGE is functionally distinct from, and phylogenetically unrelated to, known Cu(I) binding proteins (14, 20, 26). For example, the Cu-responsive transcriptional activator (CueR; PDB ID 1Q05) also binds copper in a linear bis(thiolate) geometry, but with 104-fold greater affinity (Kd ∼10−21 M) than FGE (27) and with a disparate Cβ1–S1–[Cu]–S2–Cβ2 torsion angle (φ) of φ = 19° (FGE) vs. φ = 108° (CueR). The unique deviation of φ in FGE compared with CueR and other Cu(I) binding proteins (20) may be viewed as a structural indication that its copper site has evolved to perform the dynamic requirements of substrate binding and oxidase/oxygenase catalysis. The functional divergence of these two copper sites is likely related to structural differences. The FGE Cu center is surrounded by polar residues and is accessible to substrate, solvent, and O2 (SI Appendix, Fig. S8A). CueR forms a buried Cu(I) site: It is closed by a shift at the dimer interface of three helices (28) and is protected from solvent and O2 binding by hydrophobic residues (SI Appendix, Fig. S8B). Although the O2 reactivity of CueR has not been tested directly, the role of CueR under aerobic copper stress in Escherichia coli suggests it is inert (29).
The prediction that the substrate thiolate binds directly at copper in the Michaelis complex is verified experimentally by a decrease in intensity of the 8,984-eV XAS peak and a markedly different EXAFS beat pattern relative to Cu(I)–scFGE (Fig. 6D). The change in coordination number upon substrate binding is correlated with an elongation of the Cu–S bonds from ∼2.14 Å in Cu(I)–scFGE to ∼2.22 Å in Cu(I)–scFGE:scP14. The Cu–scFGE crystal structure, combined with enhanced information from XAS, forms the basis of more detailed electronic structure calculations of the Cu(I)–FGE and Cu(I)–scFGE:scP14 copper centers. DFT geometry optimization of crude scFGE:substrate docking models show that bond elongation is accompanied by a change in S–Cu–S bond angle from 172° in resting Cu(I)–scFGE to 134° in Cu(I)–scFGE:substrate. Importantly, these structures can be used to provide valuable insight regarding changes to the electronic structure of the copper active site, which in turn alter the O2 reactivity of the scFGE complex.
Stopped-flow experiments show Cu(I)–scFGE is relatively stable under both aerobic and anaerobic conditions. However, upon substrate binding, an O2-activation reaction is available to generate an intermediate. To our knowledge, no other mononuclear copper enzyme is known to bind substrate and a reactive O2 intermediate simultaneously at the metal center. For comparison, galactose oxidase catalyzes the 2e– oxidation of d-galactose to d-galacto-hexodialdose directly from a Cu(II)/cofactor radical pair through an inner-sphere complex with the substrate hydroxyl, and only then binds and reacts with O2 to regenerate the oxidized resting state (30). Thus, the unique susceptibility of FGE’s thiol substrate to nonproductive oxidation appears to necessitate a mechanism that tightly controls the location of both activated oxygen species and the substrate sulfur atom during turnover.
A mechanistic proposal consistent with the results of this study (Fig. 8) supports and extends predictions made by Seebeck and coworkers (10, 12), with evidence for (1) and (2) presented here. Substrate binds to the two-coordinate Cu(I) active site via the cysteine sulfur to yield a three-coordinate Cu(I)–scFGE:substrate complex that is primed for reaction with O2. H-atom abstraction (HAA) of the pro-(R)-β-hydrogen from the substrate cysteine is the rate-determining step (kH/kD = 3.0–3.7) (12) and involves an activated oxygen species, putatively a Cu(II)–OO·– (3). HAA by a superoxo intermediate would generate a Cu(II)–OOH species with a carbon-centered radical on substrate (4a) or, depending of the rate of product release relative to that of C=S bond formation, species (4b). In the latter, HAA from the substrate results in the rapid reduction of Cu(II) to Cu(I) and the concomitant formation of a thioaldehyde weakly bound to the metal via the substrate sulfur lone pair. Indeed, this process resembles that proposed for the nonheme iron(II)-dependent oxidase isopenicillin N synthase (IPNS). Calculations for IPNS predict that an analogous thioaldehyde is formed at cysteine via two 1e– transfer steps from an inner-sphere ligated Fe-thiolate complex (31–33). From (4a) or (4b), dissociation and rapid hydrolysis (34) of thioaldehyde (5) gives the final fGly product. At this stage, FGE would undergo a two-electron reduction to return to its Cu(I) resting state (1).
Fig. 8.
Proposed catalytic cycle of FGE inspired by this study.
In the mechanism presented in Fig. 8, the requirement of external reducing agents would occur following product formation, but this process remains speculative. Stoichiometric DTT oxidation during multiple turnovers of FGE (9) requires that 4e– ultimately reduce O2 to H2O. It has been proposed that the Cu(I)–OOH (5) intermediate is resolved by transfer of an oxidative equivalent to one of the enzyme’s sulfur ligands; the formation of an S–O intermediate would accompany a change in the ligand occupancy at Cu(I), and reduction from an external thiol source would follow (12). This proposal may be verified experimentally by chemical trapping, or spectroscopically using the EXAFS approach in this report. The direct 2e– reduction of Cu(I)–OOH offers an alternative pathway but would rely on a sufficiently high Cu reduction potential to forestall detrimental formation of Cu(II)/·OH.
A mechanistic understanding of FGE catalysis has direct significance to the understanding of human sulfatase biology and diseases (35), and can be exploited to facilitate the production of therapeutics and biotechnology tools (6). More broadly, the detailed findings of this investigation provide foundational knowledge of monocopper enzyme chemistry and the diversity of biological reaction pathways for O2 activation.
Materials and Methods
Detailed procedures for protein purification, reagent synthesis, mass spectrometry, Cu reconstitution, stopped-flow UV/vis absorption spectroscopy, enzyme activity assays, X-ray crystallography, size exclusion chromatography–multiangle light scattering analysis, EPR spectroscopy, molecular docking and DFT computations, and XAS/EXAFS are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank George Q. Fei for providing scFGE C272A/C277A, and Dr. Stacy Malaker for assistance with mass spectrometry. This study was supported by National Institutes of Health (NIH) Grant R01DK031450 (to E.I.S.), Ruth L. Kirschstein National Research Service Award F32GM116240 (to K.K.M.), and NIH Grant CA227942 (to C.R.B.). H.L. was supported by an Abbott Laboratories Stanford Graduate Fellowship. J.A.T. is supported by NIH Grant R35CA22043, a Robert A. Welch Chemistry Chair, and the Cancer Prevention and Research Institute of Texas. See SI Appendix, SI Acknowledgements.
Footnotes
Conflict of interest statement: C.R.B. is a cofounder and member of the Scientific Advisory Board of Redwood Bioscience (a subsidiary of Catalent, Inc.), which has exclusive rights to the SMARTag technology based on protein modification by FGE. C.R.B. is also a cofounder of Palleon Pharmaceuticals, Enable Biosciences, and InterVenn Biosciences, and a member of the Board of Directors of Eli Lilly & Co.
This article is a PNAS Direct Submission. J.E.P.-H. is a guest editor invited by the Editorial Board.
Data deposition: The X-ray crystallography data have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6MUJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818274116/-/DCSupplemental.
References
- 1.Appel MJ, Bertozzi CR. Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem Biol. 2015;10:72–84. doi: 10.1021/cb500897w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosma MP, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 3.Dierks T, et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 4.Rabuka D, Rush JS, deHart GW, Wu P, Bertozzi CR. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc. 2012;7:1052–1067. doi: 10.1038/nprot.2012.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drake PM, et al. Aldehyde tag coupled with HIPS chemistry enables the production of ADCs conjugated site-specifically to different antibody regions with distinct in vivo efficacy and PK outcomes. Bioconjug Chem. 2014;25:1331–1341. doi: 10.1021/bc500189z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.York D, et al. Generating aldehyde-tagged antibodies with high titers and high formylglycine yields by supplementing culture media with copper(II) BMC Biotechnol. 2016;16:23. doi: 10.1186/s12896-016-0254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fetzner S, Steiner RA. Cofactor-independent oxidases and oxygenases. Appl Microbiol Biotechnol. 2010;86:791–804. doi: 10.1007/s00253-010-2455-0. [DOI] [PubMed] [Google Scholar]
- 8.Holder PG, et al. Reconstitution of formylglycine-generating enzyme with copper(II) for aldehyde tag conversion. J Biol Chem. 2015;290:15730–15745. doi: 10.1074/jbc.M115.652669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knop M, Engi P, Lemnaru R, Seebeck FP. In vitro reconstitution of formylglycine-generating enzymes requires copper(I) ChemBioChem. 2015;16:2147–2150. doi: 10.1002/cbic.201500322. [DOI] [PubMed] [Google Scholar]
- 10.Knop M, Dang TQ, Jeschke G, Seebeck FP. Copper is a cofactor of the formylglycine-generating enzyme. ChemBioChem. 2017;18:161–165. doi: 10.1002/cbic.201600359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon EI, et al. Copper active sites in biology. Chem Rev. 2014;114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meury M, Knop M, Seebeck FP. Structural basis for copper-oxygen mediated C–H bond activation by the formylglycine-generating enzyme. Angew Chem Int Ed Engl. 2017;56:8115–8119. doi: 10.1002/anie.201702901. [DOI] [PubMed] [Google Scholar]
- 13.Wernimont AK, Huffman DL, Lamb AL, O’Halloran TV, Rosenzweig AC. Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat Struct Biol. 2000;7:766–771. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 14.Rubino JT, Franz KJ. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J Inorg Biochem. 2012;107:129–143. doi: 10.1016/j.jinorgbio.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Carlson BL, et al. Function and structure of a prokaryotic formylglycine-generating enzyme. J Biol Chem. 2008;283:20117–20125. doi: 10.1074/jbc.M800217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tainer JA, Getzoff ED, Richardson JS, Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983;306:284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 17.Borgstahl GE, et al. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 18.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel superoxide dismutase structure and mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudmundsson M, et al. Structural and electronic snapshots during the transition from a Cu(II) to Cu(I) metal center of a lytic polysaccharide monooxygenase by X-ray photoreduction. J Biol Chem. 2014;289:18782–18792. doi: 10.1074/jbc.M114.563494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon EI, et al. Electronic absorption spectroscopy of copper proteins. Methods Enzymol. 1993;226:1–33. doi: 10.1016/0076-6879(93)26003-r. [DOI] [PubMed] [Google Scholar]
- 23.Kau LS, et al. X-ray absorption edge determination of the oxidation state and coordination number of copper. Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 24.Penner-Hahn JE. Characterization of “spectroscopically quiet” metals in biology. Coord Chem Rev. 2005;249:161–177. [Google Scholar]
- 25.Roeser D, et al. A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme. Proc Natl Acad Sci USA. 2006;103:81–86. doi: 10.1073/pnas.0507592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huffman DL, O’Halloran TV. Function, structure, and mechanism of intracellular copper trafficking proteins. Annu Rev Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 27.Changela A, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 28.Philips SJ, et al. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015;349:877–881. doi: 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 30.Whittaker JW. The radical chemistry of galactose oxidase. Arch Biochem Biophys. 2005;433:227–239. doi: 10.1016/j.abb.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Brown CD, Neidig ML, Neibergall MB, Lipscomb JD, Solomon EI. VTVH-MCD and DFT studies of thiolate bonding to [FeNO]7/[FeO2]8 complexes of isopenicillin N synthase: Substrate determination of oxidase versus oxygenase activity in nonheme Fe enzymes. J Am Chem Soc. 2007;129:7427–7438. doi: 10.1021/ja071364v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown-Marshall CD, Diebold AR, Solomon EI. Reaction coordinate of isopenicillin N synthase: Oxidase versus oxygenase activity. Biochemistry. 2010;49:1176–1182. doi: 10.1021/bi901772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundberg M, Siegbahn PEM, Morokuma K. The mechanism for isopenicillin N synthase from density-functional modeling highlights the similarities with other enzymes in the 2-His-1-carboxylate family. Biochemistry. 2008;47:1031–1042. doi: 10.1021/bi701577q. [DOI] [PubMed] [Google Scholar]
- 34.Kida Y, Class CA, Concepcion AJ, Timko MT, Green WH. Combining experiment and theory to elucidate the role of supercritical water in sulfide decomposition. Phys Chem Chem Phys. 2014;16:9220–9228. doi: 10.1039/c4cp00711e. [DOI] [PubMed] [Google Scholar]
- 35.Dierks T, et al. Molecular basis of multiple sulfatase deficiency, mucolipidosis II/III and Niemann-Pick C1 disease - Lysosomal storage disorders caused by defects of non-lysosomal proteins. Biochim Biophys Acta. 2009;1793:710–725. doi: 10.1016/j.bbamcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.