Significance
Immediate early genes (IEGs) are a group of genes whose expression is transiently up-regulated upon neural activity. In the present study, we established a method to label neural circuits in an activity-dependent manner, by combining the expression of an insect IEG Hr38 and the powerful genetics of vinegar fly Drosophila melanogaster. With this method, we visualized neurons activated in the male brain when they interacted with a female and identified a neural cluster important for courtship persistency and successful copulation. Furthermore, by activity-dependently expressing light-activatable ion channel channelrhodopsin, we induced male courtship posture by light stimulation in an experience-dependent manner. Activity-dependent labeling can be used as a powerful tool in insects to examine neural circuits regulating innate behavior.
Keywords: immediate early gene, activity-dependent labeling, courtship behavior, Hr38, fruitless
Abstract
Males of Drosophila melanogaster exhibit stereotypic courtship behavior through which they assess potential mates by processing multimodal sensory information. Although previous studies revealed important neural circuits involved in this process, the full picture of circuits that participate in male courtship remains elusive. Here, we established a genetic tool to visualize or optogenetically reactivate neural circuits activated upon specific behavior, exploiting promoter activity of a neural activity-induced gene Hr38. With this approach, we visualized neural circuits activated in the male brain and the ventral nerve cord when a male interacted with a female. The labeling of neural circuits was additively dependent on inputs from antennae and foreleg tarsi. In addition, neural circuits that express the sex-determining gene fruitless or doublesex were extensively labeled by interaction with a female. Furthermore, optogenetic reactivation of the labeled neural circuits induced courtship posture. With this mapping system, we found that a fruitless-positive neural cluster aSP2 was labeled when a male interacted with a female, in addition to previously characterized neurons. Silencing of neurons including aSP2 led to frequent interruption of courtship and significant reduction of mating success rate without affecting latency to start courtship, suggesting that these neurons are required for courtship persistency important for successful copulation. Overall, these results demonstrate that activity-dependent labeling can be used as a powerful tool not only in vertebrates, but also in invertebrates, to identify neural circuits regulating innate behavior.
How the animal brain processes inputs from the environment and controls innate behavior is a fundamental question of neuroscience. Drosophila melanogaster is one of the ideal models to study the neural circuit controlling behavior, because of its small brain size and yet rich behavioral repertoire. Upon encountering a female, Drosophila males exhibit a series of stereotypic courtship behaviors by utilizing multiple types of sensory information (1). The male courtship behavior is regulated by sexually dimorphic neural circuits that express transcription factors fruitless (fru) and doublesex (dsx), involved in sex determination (1, 2). Activation of fru- or dsx-expressing neurons induces courtship behavior of males, whereas silencing of these neurons causes defects in courtship (3–6). To date, anatomical analyses identified candidate fru neural circuits that convey sensory information to the higher center and that descend to flight muscles to control motor outputs (7). Out of the fru neural circuits, a subset (∼20 cells) of male-specific neurons in the posterior brain region which coexpresses fru and dsx, called P1 neurons, plays essential roles in decision making of courtship behavior. P1 neurons are activated upon contact with a female (3, 8), and their artificial activation induces courtship rituals (3, 5). In addition, social experience and multimodal sensory information modulate activity levels of P1 neurons (8–10). Although these extensive studies revealed essential nodes of neural circuits that regulate courtship or copulation, the full picture of neural circuits that participate in these behaviors is still obscure.
Immediate early genes (IEGs) are a class of genes whose expression is rapidly and transiently up-regulated following neural activity. Because of this nature, IEGs such as c-fos and Arc and their promoters have been widely used as powerful tools to visualize and manipulate neural circuits activated by previous experience in vertebrates (11, 12). In insects, however, IEGs have not been reported until recently, and their use in research is limited partly because of the lack of robust and easily applicable systems to detect their expression (13–16). We previously reported Hr38, a transcription factor conserved among insects and up to humans, as an IEG whose expression can be used as a neural activity marker (14). Although detecting Hr38 mRNA by in situ hybridization has been proved useful to analyze neural activity in the insect brain, this approach alone does not allow visualization of the entire neural projections, or targeted transgene expression in the activated neurons, which would be useful for further functional studies.
Here, to achieve activity-dependent targeted transgene expression in the insect brain, we employed the powerful GAL4/upstream-activating sequence (UAS) system widely used in Drosophila (17). Using a newly generated Hr38-GAL4 driver, we show that Hr38-dependent expression of membrane-targeted GFP (mCD8GFP) can label neurons in the male brain that are activated by interaction with a female. In addition, using flies that activity-dependently express channelrhodopsin, we demonstrate that reactivation of neurons labeled during mating experience sufficiently generates male copulation posture. Furthermore, we demonstrate that a cluster of fru-expressing neurons, aSP2, is preferentially activated by interaction with a female. Silencing of neurons including aSP2 caused defects in male courtship and copulation, suggesting the importance of aSP2 activity in sexual behavior.
Results
Labeling of Neurons Activated by Innate Behavior Using a Hr38-GAL4 Driver.
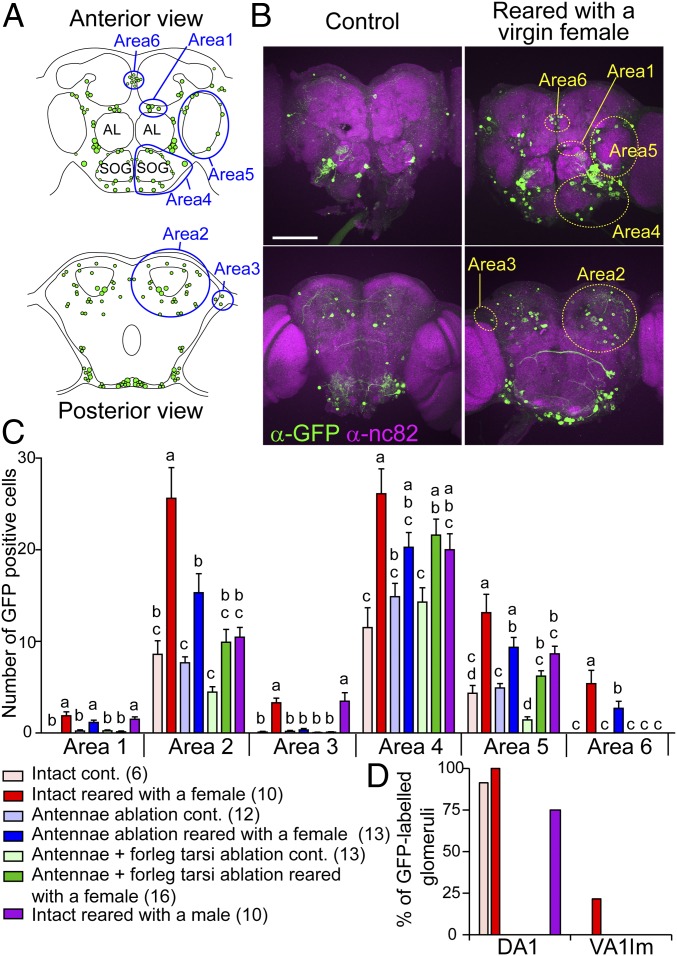
We established a Hr38-GAL4 line and genetic methods so that we can temporally restrict Hr38-GAL4–driven GFP expression to the stimulation period (SI Appendix, Supplementary Text and Fig. S1). To examine whether the Hr38-GAL4 line is useful for activity-dependent labeling of neural circuits, we investigated GFP expression patterns in the brains of males that interacted with a female. In the brains of control males, which were kept alone throughout the experiment, a small number of cells were GFP-positive. When males were placed with a virgin female for 24 h (“reared with a virgin female”), which allowed the males to court and copulate (SI Appendix, Fig. S1H), the number of GFP-expressing cells significantly increased in a wide brain area (Fig. 1 A–C). The expression pattern of GFP was similar to that of Hr38 mRNA previously revealed by in situ hybridization (areas 1–5 in Fig. 1 A and B) (14). In addition to these areas, we noticed a neural cluster, termed area 6, where GFP-positive cells were reproducibly detected. Increase of GFP-posivite cells by interaction with a female was significantly attenuated when sexual behavior was disturbed by surgical removal of antennae and foreleg tarsi of the male, which play important roles in chemical detection of females (Fig. 1C and SI Appendix, Figs. S1H and S2). These results suggest that the GFP expression is related to sensory inputs, or behavior of males, that occur during rearing with a female. We also examined GFP expression induced by rearing with another male (“reared with a male”), and found it strikingly different from that of the male reared with a female (Fig. 1C and SI Appendix, Fig. S2). Interestingly, in area 6, GFP-positive cells were detected only when the male was reared with a female, but not with a male, suggesting that the area 6 neurons are specifically activated by sensory input from the female and/or by regulation of courtship behavior. Increase of GFP-positive cells by rearing with a female was also detected in the ventral nerve cord (VNC) (SI Appendix, Fig. S3), which was partially attenuated by removal of antennae and foreleg tarsi and correlated with activity levels of sexual behavior (SI Appendix, Fig. S1H), indicating that activity-dependent GFP labeling of neural circuits is also applicable to the VNC.
Fig. 1.
Activity-dependent labeling of neural circuits that responded to a female or a male using the Hr38-GAL4 strain. (A) Schematic drawings of the male brain. (Upper) Anterior view. (Lower) Posterior view. Green dots mark the locations of cell bodies with GFP expression, from a representative Hr38 > mCD8GFP male reared with a female. Blue designates areas 1–6. Areas 1–5 symmetrically exist in both left and right hemispheres; only one side is marked in each panel. (B) Pictures of anti-GFP and anti-nc82 (synaptic neuropil marker) staining of male brains. Males were kept alone in a glass vial (control) or reared with a virgin female. (Scale bar: 100 µm.) (C) Number of GFP-positive cells in each area under different conditions. cont., control. Error bars represent SEM. Statistically different groups are indicated by lowercase letters (a–d) [P < 0.05, Tukey–Kramer’s honestly significant difference (HSD) test after ANOVA]. Number of animals analyzed is indicated in parentheses. (D) Proportion of glomeruli labeled by GFP under various conditions. DA1 and VA1lm are male and female pheromone-responsive glomeruli, respectively.
We also analyzed labeling of glomerulus in the antennal lobes (Fig. 1D). DA1, which responds to a male pheromone (18), was frequently labeled by GFP even in control males and the signals disappeared in antennae-ablated males, suggesting that the males smelled their own odor under our experimental condition. In contrast, VA1lm, which responds to a female pheromone (19, 20), was labeled only in males reared with a female. These results indicate that Hr38-GAL4–driven GFP expression is useful for labeling not only cell bodies but also neurites of active neurons.
Activity-Dependent Labeling of Sexually Dimorphic Neural Circuits.
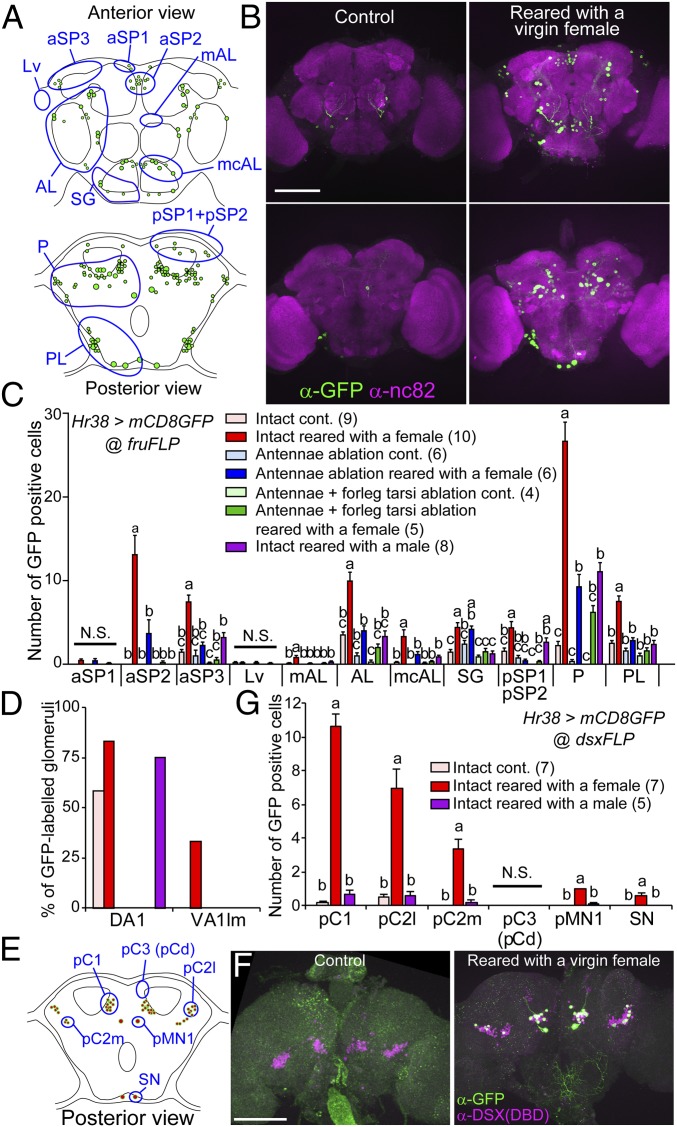
Sexually dimorphic neural circuits that express sex-determining genes, fru or dsx, play critical roles in male courtship behavior (21, 22) and some of the cells respond to female in a contact-dependent manner (3, 8). However, the full picture of these neural circuits that respond to female is not revealed. We found that a part of GFP-positive cells in males reared with a female was Fru-positive (SI Appendix, Fig. S4). Thus, we next took a genetic intersectional approach to restrict the activity-dependent labeling to fru- or dsx-neural circuits using FLP recombinase (Fig. 2 and SI Appendix, Figs. S5–S7). High numbers of GFP-labeled cells were observed in males reared with a female, whereas only a few were labeled in control males. The number of GFP-positive cells was significantly reduced by surgical removal of antennae, and abolished by further removal of foreleg tarsi. Rearing with a male caused a distinct GFP expression pattern compared with rearing with a female in fru-neural circuits (Fig. 2 and SI Appendix, Figs. S5 and S6). Collectively, these results indicate that fru-neural circuits are differentially activated by rearing with a female or a male, and that activity induced by interaction with a female is dependent on chemical sensory inputs. In addition, the labeling pattern of antennal lobes was similar to that observed in the previous experiment (Figs. 1D and 2D), again confirming the usefulness of this labeling system. Interestingly, dsx-neural circuits were selectively activated by rearing with a female (Fig. 2 E–G and SI Appendix, Fig. S7).
Fig. 2.
Activity-dependent labeling of sexually dimorphic neural circuits by an intersectional approach. (A and E) Schematics of fru- or dsx-restricted GFP expression pattern in the brain of an intact male reared with a virgin female. Locations of fru or dsx neural clusters are marked. (B and F) Pictures of anti-GFP immunostaining of male brains. Activity-dependent expression of mCD8GFP was restricted by an intersectional approach using the fruFLP or dsxFLP strain. (Scale bars: 100 µm.) (C and G) Number of GFP-positive cells in each neural cluster under different conditions. Error bars represent SEM. Statistically different groups are indicated by lowercase letters (a–c) (P < 0.05, Tukey–Kramer’s HSD test after ANOVA). N.S., not significant. (D) Proportion of GFP-labeled DA1 and VA1lm glomeruli.
We next focused on neurons that coexpress fru and dsx, which include P1 neurons, since their functions in courtship behavior are intensively characterized (SI Appendix, Fig. S6). These neurons were intensely labeled by GFP by rearing with a female, but not with a male. The GFP labeling of P1 neurons by rearing with a female was not affected by antennal amputation, but attenuated by further amputation of foreleg tarsi. These results are consistent with previous reports showing that P1 neural activity is dependent on contact pheromone, but not on female odor (3, 8). In contrast, the number of GFP-labeled pC2l/m and TN1 neurons decreased by these treatments in a similar manner to those observed in other fru-neural clusters (SI Appendix, Fig. S6C), suggesting that activity of these neurons are regulated by sensory inputs from both antennae and foreleg tarsi.
In addition to these characterized neurons, we identified the newly defined area 6 neurons to be aSP2 cluster, from positions of their soma (Figs. 1B and 2B), Fru expression (SI Appendix, Fig. S4F), and from the characteristics that the labeling of these neurons occurs only when males interacted with a female, and is dependent on inputs from antennae and foreleg tarsi. From previous anatomical studies, aSP2 neurons project their neurites to the lateral protocerebral complex, where multimodal sensory information converges, and show extensive sexual dimorphisms (7), but their function in courtship behavior has not been characterized.
Induction of Male Mating Behavior by Optogenetic Reactivation of Neurons Labeled by Rearing with a Female.
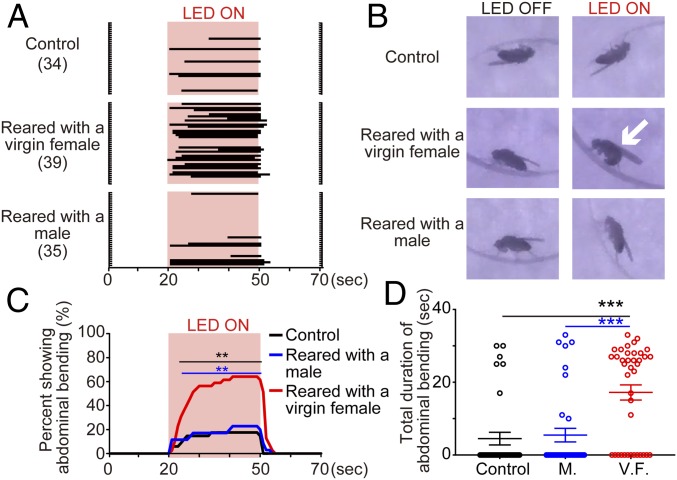
For the analysis of neural regulation of behavior, examination of causal relationships between neural activity and behavior is vital. To evaluate the usefulness of Hr38-GAL4–driven activity-dependent labeling methods, we examined whether optogenetic reactivation of neural circuits labeled by interaction with a female can elicit mating behavior. Here, red light-activatable channelrhodopsin CsChrimson was expressed in fru or dsx neurons in an activity-dependent manner during the interaction period with a female or male (SI Appendix, Fig. S1G), and then the male’s response to red light was examined in absence of another fly (Fig. 3 and SI Appendix, Figs. S8–S10). Without spatial restriction of the expression of CsChrimson, all flies showed knockout phenotype by photostimulation, probably due to excessive neural activity. Upon photostimulation, males reared with a female exhibited abdominal bending (Fig. 3, SI Appendix, Fig. S9, and Movies S1–S7), a characteristic male copulation posture (1). The response was time locked to photostimulation period with a short delay and continued until the end of stimulation once initiated. A fraction of flies showing abdominal bending, and the duration of abdominal bending exhibited, were both significantly greater in males reared with a female than other groups. In males with fru-restricted labeling, unilateral wing extension was mostly not evoked by photostimulation, except for two males reared with a female (Movie S4), but bilateral wing extension was modestly evoked by photostimulation in males reared with a female (SI Appendix, Fig. S8). These behavioral phenotypes are consistent with a previous study which showed that optogenetic stimulation of fru-neural circuits evokes bilateral, rather than unilateral, wing extension (10). A fraction of flies showing either type of wing extension, and duration of the behavior, were significantly higher in males reared with a female than control (SI Appendix, Fig. S8 B and C). In contrast, photostimulation did not evoke wing extension in males with dsx-restricted labeling (SI Appendix, Fig. S9 E–G), although neural circuits for wing extension, such as pC1 and TN1 neurons, were labeled by rearing with a female (Fig. 2 E–G and SI Appendix, Fig. S7). These biased behavioral phenotypes toward abdominal bending than wing extension can be due to intense labeling of abdominal ganglion (SI Appendix, Fig. S7 B and C). Collectively, these results indicate that Hr38-GAL4–driven transgene expression is useful not only for labeling neurons activated by behavior, but also for examining the contribution of labeled neural circuits to behavior, although caution against biased behavioral phenotypes caused by intensely labeled neural circuits is needed.
Fig. 3.
Optogenetic reactivation of fru neurons activity-dependently labeled by previous experience. (A) Raster plots showing the occurrence of abdominal bending in response to red light. Red shades indicate light exposure (30 s). (B) Representative images taken from supplementary movies (Movies S1–S3). White arrow indicates a male showing abdominal bending. (C) Percentage of flies showing abdominal bending, based on A. **P < 0.01, Fisher’s exact test performed for each second. Black and blue lines indicate the time range where significant differences were observed when males reared with a virgin female were compared with control, and to males reared with a male, respectively. (D) Total duration of abdominal bending during the observation period. Each data point is indicated as a circle. Error bars represent SEM. M., reared with a male; V.F., reared with a virgin female. ***P < 0.001, Mann–Whitney U test.
To evaluate whether prior behavioral differences can be detected as differences in behavioral phenotypes caused by optogenetic reactivation, we compared photostimulation-induced phenotypes between males reared with a virgin or mated female (SI Appendix, Fig. S10). Virgin females are receptive to male courtship and copulate soon upon encounter with males (SI Appendix, Fig. S1H), but mated females reject copulation attempts of males. Thus, it is expected that males reared with a virgin female spend longer periods on copulation than males reared with a mated female. In both cases of fru- or dsx-restricted labeling, the difference in fraction of flies exhibiting abdominal bending compared with control reached statistical significance earlier in males reared with a virgin female than males reared with a mated female, although there was no obvious difference in wing extension (SI Appendix, Fig. S10). These results suggest that Hr38-GAL4–driven transgene expression can also be useful for detecting differences, at least in part, of prior behavior and/or experience by post hoc photostimulation.
OK371fru Neural Activity Is Required for Proper Courtship and Copulation.
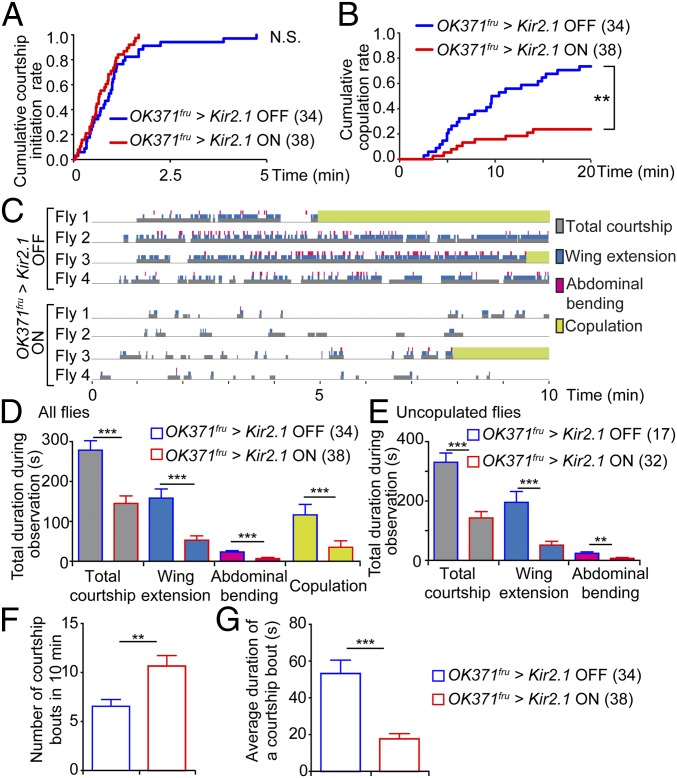
Since our approach can label neural circuits relevant to behavior, we hypothesized that aSP2 neurons, which were labeled by rearing with a female, have functions in mating behavior of male flies. In the present study, we took an intersectional approach using a combination of OK371-GAL4 and fruFLP to label aSP2 neurons according to a previous report (7) (SI Appendix, Fig. S11A). Since this combination also labels cells in other areas such as suboesophageal ganglion and abdominal ganglion, we hereafter refer to this combination as OK371fru to avoid confusion. We investigated the effects of silencing OK371fru neurons by expression of inward rectifying potassium channel Kir2.1 (Fig. 4). With GAL80ts, expression of Kir2.1 was limited to 24 h preceding the behavioral test (SI Appendix, Fig. S11A). There was no difference in time required for the male to initiate courtship with this treatment, suggesting that silencing of OK371fru neurons does not significantly affect detection of the target female, initial decision to court, or motor ability (Fig. 4A and SI Appendix, Fig. S11 B and D). On the other hand, silencing of OK371fru neurons caused a significantly lower copulation success rate, suggesting that activity of OK371fru neurons is required for normal copulation (Fig. 4B and SI Appendix, Fig. S11 C and E).
Fig. 4.
OK371fru neural activity is required for proper courtship and copulation. (A) Cumulative rate of flies that initiated courtship. (N.S., not significant, log-rank test). (B) Cumulative copulation rate plotted against time. (**P < 0.01, log-rank test). (C) Raster plots of courtship behavior for the first 10 min, from representative flies. Courtship bouts (durations of flies showing any of the following courtship behavior: orientation, chasing, wing extension, and abdominal bending) are shown in gray. Abdominal bending and wing extension are represented in pink and blue, respectively. Copulations are shown in green. Plots from all flies are shown in SI Appendix, Fig. S11. (D and E) Durations of courtship components within the 10-min observation period. Results from all observed flies (D) and flies which did not copulate within the first 10 min (E). (F) Number and (G) average duration of courtship bouts. (D–G) **P < 0.01, ***P < 0.001, Mann–Whitney U test. Error bars represent SEM.
To further characterize the roles of OK371fru neurons in mating behavior, we performed detailed examinations of male behavior during the first 10 min of the test period (Fig. 4C and SI Appendix, Fig. S11F). From our video analysis, we noticed that males with silenced OK371fru neurons spent less time courting; durations of total courtship, wing extension, and abdominal bending were significantly decreased (Fig. 4D). Comparing only the males which did not copulate during the first 10 min also gave similar results (Fig. 4E), indicating low courtship vigor in OK371fru neuron-silenced male flies. Furthermore, silencing of OK371fru neurons caused frequent interruption of courtship behavior. Here, we defined “courtship bout” as a period when the male fly was showing any of the following behavior: orientation, chasing, wing extension, and abdominal bending. The end of a bout was scored when the male stopped courting and did not restart for longer than 1 s. OK371fru neuron-silenced males showed a significantly higher number of courtship bouts within the observation period, whereas their average duration of one courtship bout was much shorter compared with control (Fig. 4 F and G). These quantifications indicate that OK371fru neuron-silenced males can initiate courtship but are not able to sustain courtship activity, resulting in significantly less time spent courting.
We also addressed effects of activation of OK371fru neurons on mating behavior. Artificial activation of OK371fru neurons by warmth-sensor channel dTrpA1 did not affect courtship or copulation latency (SI Appendix, Fig. S12 A and B). Moreover, this treatment did not reverse the reduced courtship activity of antennae-ablated males (SI Appendix, Fig. S12 C and D). These results indicate that OK371fru neurons do not promote mating behavior, at least by themselves.
Discussion
In the present study, we demonstrate that an IEG Hr38-dependent transgene expression can be used as a powerful tool to visualize and map neurons activated in the central nervous system of D. melanogaster. With this approach, we identified aSP2 as a neural cluster activated upon interaction with a female, and further showed that activity of OK371fru neurons is required for proper courtship and copulation. These results collectively demonstrate that Hr38-dependent labeling enables identification of neurons functionally important for certain behavior.
To date, resources for labeling active neurons in the insect nervous system remain scarce. Previous studies have reported two techniques which take advantage of the increase of intracellular calcium upon cell activity to label neurons in Drosophila: calcium-dependent nuclear import of LexA (CaLexA) and transcriptional reporter of intracellular Ca2+ (TRIC) (20, 23). Although the purpose of labeling neural circuits in an activity-dependent manner is conceptually similar among CaLexA, TRIC, and Hr38-dependent labeling, their underlying mechanisms and properties are different. CaLexA and TRIC utilize intracellular calcium-dependent nuclear translocation of nuclear factor of activated T cells (NFAT) and calcium-dependent binding activity of calmodulin (CaM) to its target peptide, respectively. Since intracellular signaling pathways reflected to reporter expression by these methods are clear, labeling intensity is supposed to correlate with levels of neural activity (24–26). In contrast, since the intracellular signaling network that regulates activity-dependent Hr38 expression is still unclear, what and how neural activity is reflected in neural labeling is obscure in Hr38-dependent labeling. Thus, cautious interpretation for correlation of labeling intensity and level of neural activity is needed. A drawback related to this is that there is a bias in Hr38-dependent labeling among cell types and brain regions, since Hr38 mRNA is expressed only in 55–75% of neurons and almost not in optic lobe neurons (14). These biases could cause biased behavioral response by optogenetic reactivation of neural circuits labeled by courtship behavior (Fig. 3 and SI Appendix, Figs. S8–S10). We speculate that differences in abundance and/or type of intracellular signaling components among cell types and brain regions cause the biases. Future studies that clarify the intracellular signaling network that regulates Hr38 expression will help us improve and/or optimize the Hr38-dependent labeling method. With Hr38-dependent labeling, identification of individual neurons was challenging except for some cases in the antennal lobe neurons, since the majority of labeling was detected strongly in the cell bodies and the labeling intensity was weak in neural processes. This is also a current limitation of the Hr38-dependent labeling method that needs future improvements. Despite these limitations, the Hr38-dependent labeling method has merits compared with other methods in that it reflects endogenous and behaviorally relevant gene expression with a high signal-to-noise ratio in response to neural activity. In addition, using the Hr38-dependent labeling method, here we demonstrate that reactivation of neurons labeled by interaction with a female is sufficient to recapitulate courtship. Thus, combinatorial or complemental use of our method and calcium-dependent labeling techniques will be useful for identifying neural circuits underlying many behaviors. In addition, in combination with the powerful genetic tools of D. melanogaster, our system will enable activity-dependent silencing of neurons, which will contribute to the understanding of neural circuit mechanisms. Furthermore, since Hr38 is conserved as a neural activity marker between two insect species of different orders (14), it is highly likely that our activity-dependent labeling approach is applicable to species other than D. melanogaster. Recent studies using other Drosophila species have started to reveal the neural mechanisms regulating species-specific sexual behaviors (27, 28). Activity-dependent labeling will provide additional means to analyze similarities and differences of neural circuits among different species and will support elucidation of the neural and evolutionary mechanisms underlying their various behaviors.
Recent advancements of optogenetic techniques have enabled researchers to analyze the functions of specific neurons by altering neural activity in a noninvasive and reversible manner. By combining activity-dependent labeling of neurons with IEGs and optogenetic control of neural activity, one can now examine the effects of reactivation of neurons that were once activated under certain conditions. For example, in mice, optogenetic reactivation of hippocampal neurons activated under fear learning induces fear response in a different context (29), and reactivation of amygdala neurons activated during reward learning causes appetitive response (30). Such approach, however, has not been applied to invertebrates so far. Here, using the Hr38-GAL4 driver, we demonstrate that reactivation of fru-positive neurons activated by rearing with a female can induce male courtship behavior in flies. This suggests that the transcriptional activity of Hr38 is sufficient to label neurons required for initiation of specific behavior and provide support for potential application of these methods to identification and functional analyses of cell assemblies associated with different behaviors.
A recent study reported that a subpopulation of aSP2 neurons expressing octopamine receptor Oamb promotes male aggression behavior (31). Interestingly, it was also shown that silencing these neurons by expression of Kir2.1 did not significantly affect copulation latency or the duration of courtship behavior (unilateral wing extension) toward a female. The apparent difference with our results where we found defects in copulation and courtship of OK371fru neuron-silenced males is likely due to the differences in the cell populations being analyzed. Watanabe et al. used a cis-regulatory motif of Oamb, R47A04, whereas we used OK371fru driver to specify aSP2 neurons. Observations that R47A4-labeled neurons include glutamatergic and GABAergic neurons (31), whereas OK371fru driver labels glutamatergic neurons (32), also support the idea that the two neural populations are not identical, although they could be partially overlapping. Otherwise, we cannot exclude the possibility that OK371fru-labeled cells other than aSP2 neurons contribute the behavioral phenotype observed in the present study. Future work using strains specific to aSP2 neurons or their subclasses is necessary to pin down responsible neural circuits. In addition, since we were unable to establish useful Hr38-LexA or Hr-38-QF strains (SI Appendix, Supplemental Text), we could not confirm that area 6 cells that we assumed to be aSP2 neurons overlap with OK371fru cells by a double-labeling system. Establishment of useful Hr38-dependent labeling methods independent of the GAL4/UAS system will expand the usefulness and accuracy of cell population analyses in future study.
Specifically, what role do aSP2 neurons play in the neural circuit regulating courtship and copulation? Anatomically, they reside in a higher order brain region which receives multiple sensory inputs, presumably important for integration of sensory information and regulation of premotor neurons (7, 33). For a successful copulation, male flies are required not only to perform courtship for a short period, but to sustain courtship behaviors for a certain amount of time. Our results suggest that activity of OK371fru neurons is important for continuation of courtship behavior. One possible underlying mechanism is that, upon receiving sensory information from females, aSP2 neurons affect the excitability of the courtship-controlling neural circuit by neuromodulatory effects, and thereby contribute to sustained courtship behavior. Identifying neurotransmitters involved and analyzing connections of aSP2 neurons will be of interest for future studies to further characterize the role of aSP2 neurons in the neural circuit regulating fly courtship behavior.
Materials and Methods
Fly Strains.
Flies (D. melanogaster) were maintained on standard fly medium, under normal 12-h light/12-h dark conditions. Temperature was kept at 25 °C unless otherwise noted. For stimulation or behavioral experiments, eclosed male flies were maintained individually in small glass tubes with food (10 mm in diameter and 75 mm in height, and ∼1 mL of standard fly medium) until use. In all experiments, Canton-S (CS) or white-eyed CS (2022u) flies 3–7 d after eclosion were used for stimulation. In experiments using males with foreleg and/or antennae amputation, the foreleg tarsi on both sides and/or antennae were amputated on the day of eclosion. Complete genotypes of flies used in this study are listed in SI Appendix, Table S1.
Generation of the Hr38-GAL4 Strain.
A previously characterized Hr38-LacZ line was genetically converted to the Hr38-GAL4 line using Δ2–3 (34). The transgenes used were as follows: target LacZ transgene, P{PZ}Hr3802306 (35) and donor GAL4 transgene, P{IT.GAL4} (36). See SI Appendix, Supplementary Text for details.
Immunohistochemistry.
The brains and VNCs of male flies were dissected and fixed in 4% formaldehyde (Nisshin EM 3152)/PBS for 30 min at room temperature. The brains were washed three times with PBS containing 0.3% Triton X-100 (PBT), blocked in 7% normal donkey or goat serum in PBT for 30 min at room temperature, and incubated with primary antibodies for 3 d at 4 °C with continuous shaking. After three washes in PBT, secondary antibodies were added and incubated for 2 d at 4 °C. The brains were washed three times with PBT, once with PBS, mounted with Vectashield (Vector Labs) onto glass coverslips, and subjected to imaging. Antibodies used in the present study are as follows: rabbit anti-GFP antibody (1:1,000; Invitrogen A6455), rabbit anti-Fru antibody (1:200; gift from Daisuke Yamamoto, National Institute of Information and Communications Technology, Kobe, Japan), mouse anti-DSXDBD monoclonal antibody [1:20; gift from Bruce S. Baker, Janelia Research Campus, Ashburn, VA, now available from Developmental Studies Hybridoma Bank (DSHB)] (37), mouse anti-nc82 antibody (1:50; DSHB), mouse anti–β-GAL (1:200, Z3783; Promega), goat anti-rabbit Alexa 488 (1:200; Jackson ImmunoResearch Laboratories), and goat anti-mouse Cy3 (1:200; Invitrogen). For detection of DSX, antibody reaction was conducted using Can Get Signal Immunostain solution B according to manufacturer’s instructions (Toyobo). Images were taken using confocal microscope LSM5 (Carl Zeiss) or FV10i (Olympus). The cell whose GFP signal was visible over counterstaining was counted as a positive cell.
Behavioral Assay.
Male flies were reared at 18 °C, collected within a few hours after eclosion, and single reared in 10 × 75 mm borosilicate glass culture tubes (Fisher Scientific) with standard medium until subjected to experiments (SI Appendix, Fig. S11A). One day before the assay, males of the experimental group were transferred to 30 °C to allow expression of Kir2.1 (shown as “ON” in Fig. 4 and SI Appendix, Fig. S11). Control males were kept in 18 °C (shown as “OFF”). After 24 h, both males were brought to room temperature (25 °C) and allowed to rest for 1 h. For examination of male courtship behavior, a wild-type virgin female (Canton-S, 3–5 d after eclosion) was transferred into the culture tube as a mating target. Male behavior was videorecorded for 20 min. Courtship latency and copulation latency were scored manually. Initiation of courtship was scored when the male exhibited wing extension toward the female for the first time during the observation period. For Fig. 4C, the first 10 min of the videos were manually analyzed and each component of courtship behavior was scored.
Optogenetic Assays.
Flies were raised at 18 °C throughout development. Within a few hours after eclosion, male flies were transferred to 10 × 75 mm borosilicate glass culture tubes (Fisher Scientific) with food containing 400 µM all-trans-retinal and single reared in the dark until use. Males 5–10 d after eclosion were used for the experiment. On the day of the experiment, a virgin white (2022u) female or a male (3–5 d posteclosion) was transferred into each tube. Fly tubes were immediately moved to 30 °C and kept at that temperature for 24 h. Control males were also moved to 30 °C and experienced the temperature shift. After the temperature-shift treatment, the flies were brought to room temperature (25 °C), white flies were removed, and the flies were allowed to rest for 30 min. Exposure to light was performed essentially as previously described (10) with modifications. Briefly, each fly was transferred into a circular arena (15.0-mm diameter, 5.0-mm depth) in a custom-made acrylic chamber, and exposed to red light (627 nm LED, 9.9 and 28.2 µW/mm2 for fru and dsx restricted, respectively) for 30 s. Behavior of each fly was videorecorded, and the durations of wing extension and abdominal bending were scored manually.
Supplementary Material
Acknowledgments
We thank Makoto Sato, Takaomi Sakai, Barry J. Dickson, Stephen F. Goodwin, Gerald M. Rubin, Julie H. Simpson, Anne C. von Philipsborn, and Kenta Asahina, and the Bloomington Stock Center and the Drosophila Genetic Resource Center for fly strains; and Bruce S. Baker, Daisuke Yamamoto, and the Developmental Studies Hybridoma Bank for antibodies. This work was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) Grants 26850218 and 16H05053, the Sasakawa Scientific Research Grant from the Japan Science Society, the Naito Foundation, the Yamada Science Foundation, the Hokuriku Bank, the Takeda Science Foundation, the Asahi Glass Foundation, and the Uehara Memorial Foundation (to T.K.), and JSPS KAKENHI Grant 18K14813, JSPS Research Fellow 201800091, the Sasakawa Scientific Research Grant from the Japan Science Society, and the Japan Prize Foundation (to S.T.-K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814628116/-/DCSupplemental.
References
- 1.Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci. 2013;14:681–692. doi: 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- 2.Pavlou HJ, Goodwin SF. Courtship behavior in Drosophila melanogaster: Towards a ‘courtship connectome’. Curr Opin Neurobiol. 2013;23:76–83. doi: 10.1016/j.conb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohatsu S, Koganezawa M, Yamamoto D. Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron. 2011;69:498–508. doi: 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Robinett CC, Baker BS. Turning males on: Activation of male courtship behavior in Drosophila melanogaster. PLoS One. 2011;6:e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Philipsborn AC, et al. Neuronal control of Drosophila courtship song. Neuron. 2011;69:509–522. doi: 10.1016/j.neuron.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu JY, Kanai MI, Demir E, Jefferis GSXE, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Clowney EJ, Iguchi S, Bussell JJ, Scheer E, Ruta V. Multimodal chemosensory circuits controlling male courtship in Drosophila. Neuron. 2015;87:1036–1049. doi: 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koganezawa M, Kimura K, Yamamoto D. The neural circuitry that functions as a switch for courtship versus aggression in Drosophila males. Curr Biol. 2016;26:1395–1403. doi: 10.1016/j.cub.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki HK, et al. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat Methods. 2014;11:325–332. doi: 10.1038/nmeth.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr Opin Neurobiol. 2007;17:567–571. doi: 10.1016/j.conb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawashima T, Okuno H, Bito H. A new era for functional labeling of neurons: Activity-dependent promoters have come of age. Front Neural Circuits. 2014;8:37. doi: 10.3389/fncir.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Rahman R, Guo F, Rosbash M. Genome-wide identification of neuronal activity-regulated genes in Drosophila. eLife. 2016;5:e19942. doi: 10.7554/eLife.19942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita N, et al. Visualization of neural activity in insect brains using a conserved immediate early gene, Hr38. Curr Biol. 2013;23:2063–2070. doi: 10.1016/j.cub.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 15.Kiya T, Kunieda T, Kubo T. Increased neural activity of a mushroom body neuron subtype in the brains of forager honeybees. PLoS One. 2007;2:e371. doi: 10.1371/journal.pone.0000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ugajin A, Kunieda T, Kubo T. Identification and characterization of an Egr ortholog as a neural immediate early gene in the European honeybee (Apis mellifera L.) FEBS Lett. 2013;587:3224–3230. doi: 10.1016/j.febslet.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 19.Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 20.Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura K, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 22.Siwicki KK, Kravitz EA. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr Opin Neurobiol. 2009;19:200–206. doi: 10.1016/j.conb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao XJ, et al. A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat Neurosci. 2015;18:917–925. doi: 10.1038/nn.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo F, Chen X, Rosbash M. Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc Natl Acad Sci USA. 2017;114:E8780–E8787. doi: 10.1073/pnas.1706608114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayser MS, Yue Z, Sehgal A. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science. 2014;344:269–274. doi: 10.1126/science.1250553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Liu Q, Tabuchi M, Wu MN. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell. 2016;165:1347–1360. doi: 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seeholzer LF, Seppo M, Stern DL, Ruta V. Evolution of a central neural circuit underlies Drosophila mate preferences. Nature. 2018;559:564–569. doi: 10.1038/s41586-018-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka R, Higuchi T, Kohatsu S, Sato K, Yamamoto D. Optogenetic activation of the fruitless-labeled circuitry in Drosophila subobscura males induces mating motor acts. J Neurosci. 2017;37:11662–11674. doi: 10.1523/JNEUROSCI.1943-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redondo RL, et al. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–430. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, et al. A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron. 2017;95:1112–1128.e7. doi: 10.1016/j.neuron.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: A marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Lee G, et al. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 34.Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlova T, et al. Drosophila hormone receptor 38 functions in metamorphosis: A role in adult cuticle formation. Genetics. 1998;149:1465–1475. doi: 10.1093/genetics/149.3.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gohl DM, et al. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellert DJ, Robinett CC, Baker BS. Doublesex functions early and late in gustatory sense organ development. PLoS One. 2012;7:e51489. doi: 10.1371/journal.pone.0051489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.