Significance
Homeostatic mechanisms maintain stable neuronal and circuit function in the brain, in particular during development and learning, when synapses undergo constant changes. Synaptic scaling is a form of homeostatic synaptic plasticity responsible for maintaining neuronal network activity within a physiological range, mainly through the regulation of AMPA receptors at synaptic sites. However, the intrinsic mechanisms promoting synaptic scaling are still largely unknown. Here, we have uncovered miR-186-5p as an activity-regulated miRNA, which targets the GluA2 AMPA receptor subunit and mediates synaptic scaling triggered by prolonged blockade of synaptic activity.
Keywords: homeostatic plasticity, synaptic scaling, GluA2, microRNAs, miR-186-5p
Abstract
Homeostatic synaptic scaling is a negative feedback response to fluctuations in synaptic strength induced by developmental or learning-related processes, which maintains neuronal activity stable. Although several components of the synaptic scaling apparatus have been characterized, the intrinsic regulatory mechanisms promoting scaling remain largely unknown. MicroRNAs may contribute to posttranscriptional control of mRNAs implicated in different stages of synaptic scaling, but their role in these mechanisms is still undervalued. Here, we report that chronic blockade of glutamate receptors of the AMPA and NMDA types in hippocampal neurons in culture induces changes in the neuronal mRNA and miRNA transcriptomes, leading to synaptic upscaling. Specifically, we show that synaptic activity blockade persistently down-regulates miR-186-5p. Moreover, we describe a conserved miR-186-5p-binding site within the 3′UTR of the mRNA encoding the AMPA receptor GluA2 subunit, and demonstrate that GluA2 is a direct target of miR-186-5p. Overexpression of miR-186 decreased GluA2 surface levels, increased synaptic expression of GluA2-lacking AMPA receptors, and blocked synaptic scaling, whereas inhibition of miR-186-5p increased GluA2 surface levels and the amplitude and frequency of AMPA receptor-mediated currents, and mimicked excitatory synaptic scaling induced by synaptic inactivity. Our findings elucidate an activity-dependent miRNA-mediated mechanism for regulation of AMPA receptor expression.
The dynamic nature of neuronal circuits, constantly changing synapse number and strength during development and learning-related processes, induces powerful destabilization in neuronal networks. These destabilizing forces are counterbalanced by a set of compensatory mechanisms that maintain network stability, known as homeostatic plasticity (1–3). Synaptic scaling, a form of homeostatic plasticity, acts through a negative feedback mechanism, scaling synaptic strength globally by adjusting synaptic AMPA receptor (AMPAR) content (2, 4, 5). Notably, dysfunction of proteins associated with the synaptic scaling machinery has been reported in several neurological disorders, suggesting that defective homeostatic plasticity signaling can contribute to disease pathogenesis (6, 7).
Although many components of the synaptic scaling apparatus have been identified, most associated with synapse stabilization and AMPAR trafficking during development and synaptic plasticity (7–10), full understanding of how transcriptional and translational programs are regulated by these paradigms is still lacking. The time frame of synaptic scaling suggests that transcriptional and posttranscriptional responses may be activated upon chronic changes in neuronal activity, but these alterations are still poorly characterized. Chronic blockade of action potentials with tetrodotoxin (TTX) affects transcription through reduction of CaMKIV activation (11), T voltage-gated calcium channel-dependent Ca2+ signaling, and activity-regulated transcription factors like SRF and ELK1 (12). Proteomic analysis of newly synthesized proteins in day in vitro (DIV) 21 hippocampal neurons subjected to upscaling or downscaling paradigms allowed the identification of bidirectionally regulated proteins, many of which are known regulators of synaptic function (13). Furthermore, adjustments in the postsynaptic density proteome occur during sleep, a process associated with homeostatic downscaling (14). However, little is known about homeostatic plasticity-associated posttranscriptional regulatory mechanisms that control how modifications in the neuronal transcriptome are echoed in the proteome.
MicroRNAs (miRNAs) are small noncoding RNAs that play critical roles in posttranscriptional regulation of gene expression. In neurons, miRNA expression changes rapidly in response to neuronal activity, controlling the translation of multiple transcripts (15–17). Consequently, miRNAs regulate numerous neuronal processes, ranging from differentiation to synaptic plasticity (18–21). However, few miRNAs have been functionally implicated in synaptic upscaling mechanisms: miR-92a levels are decreased in neurons incubated with TTX and the NMDA receptor antagonist APV, allowing the expression of its direct target GluA1 (22), whereas miR-124, increased in neurons treated with TTX and APV, transiently represses GluA2 expression in early stages of synaptic upscaling (23). Therefore, it is likely that the contribution of miRNA-regulated mechanisms to synaptic upscaling is still largely underestimated.
Here, we demonstrate that chronic blockade of glutamate receptors of the AMPA and NMDA (NMDARs) types in hippocampal neurons induces upscaling of AMPAR-mediated miniature excitatory postsynaptic currents (mEPSCs), increases the GluA1 and GluA2 synaptic content, and alters the neuronal transcriptome and miRNA profile. MicroRNA-186-5p (miR-186-5p) was found to be rapidly and persistently down-regulated by synaptic activity suppression, to be a regulator of endogenous GluA2 expression, and of AMPAR-mediated currents. Furthermore, inhibition of basal miR-186-5p expression occluded synaptic upscaling triggered by chronic synaptic inactivity, revealing that miR-186-5p has a functional role in synaptic scaling mechanisms.
Results
Chronic Blockade of AMPARs and NMDARs Promotes Synaptic Upscaling in Hippocampal Neurons.
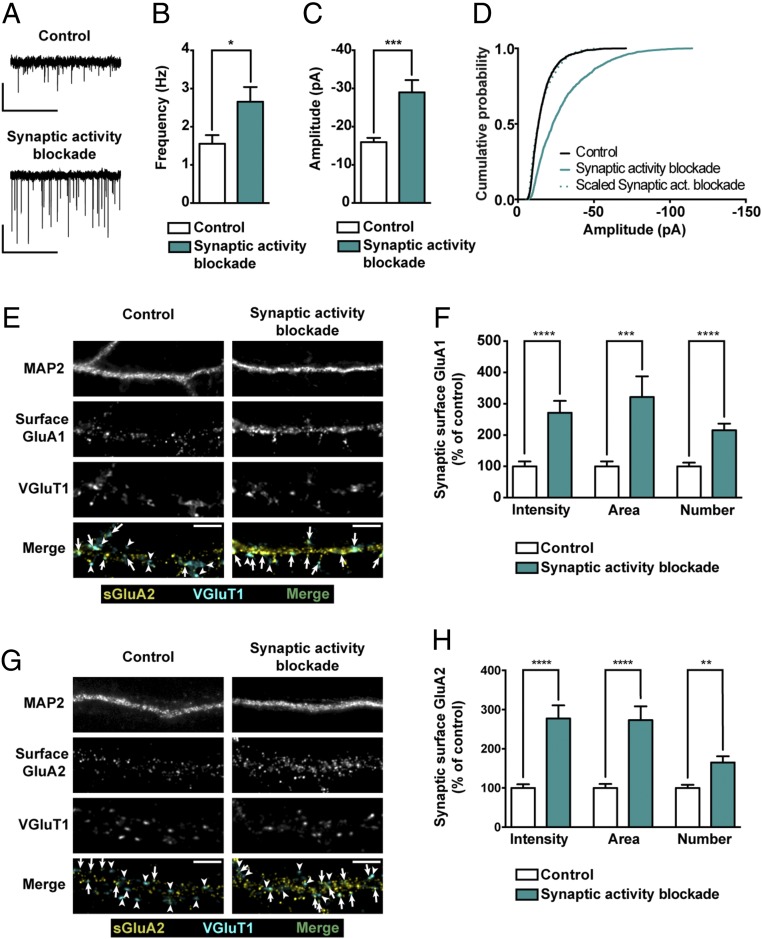
Synaptic upscaling has been modeled using several protocols that result in increased synaptic strength by chronically blocking neuronal and synaptic activity (24–27). Homeostatic regulation of synaptic function can be induced by chronic blockade of action potentials (with TTX), or by prolonged antagonism of AMPARs or NMDARs, or a combination of both (see ref. 28 for a review of different inactivity paradigms used to study homeostatic synaptic adaptations). The responses to decreased action potential firing and silencing of glutamatergic receptors have different underlying mechanisms and probably contribute to different physiological goals, which remain largely unexplored. In this study, we induced synaptic scaling in mature hippocampal neurons with long-term blockade of synaptic activity, through inhibition of AMPA and synaptic NMDA receptors with the noncompetitive antagonists GYKI-52466 (29, 30) and MK-801 (31), respectively. Whole-cell AMPAR-mediated mEPSCs were recorded in hippocampal neurons treated with 50 μM GYKI-52466 and 10 μM MK-801 for 24 h or in control conditions (Fig. 1A). Chronic blockade of AMPARs and NMDARs increased mEPSC frequency and amplitude (Fig. 1 B and C), in agreement with other reports (26, 32, 33). To investigate whether this increase in mEPSC amplitude is multiplicative, a defining feature of synaptic scaling, we ranked control amplitudes of mEPSCs against amplitudes in neurons treated with GYKI-52466 and MK-801 and the data were well fit by a linear function with a slope of 2.22. The cumulative distribution of the mEPSC amplitude data from GYKI-52466– and MK-801–treated hippocampal neurons scaled by the multiplicative factor was almost perfectly superimposable over the distribution of data from control neurons (Fig. 1D), indicating a multiplicative effect. These results show that chronic blockade of AMPARs and NMDARs promotes synaptic upscaling of AMPAR-mediated mEPSC amplitude.
Fig. 1.
Chronic activity blockade of AMPARs and NMDARs upscales AMPARs in hippocampal neurons. (A) Representative whole-cell current traces of AMPAR-mediated mEPSCs from hippocampal neurons in control conditions or treated with 50 μM GYKI-52466 and 10 μM MK-801 for 24 h. (Scale bars: vertical, 20 pA; horizontal, 5 s.) Mean frequency (B) and amplitude (C) of AMPAR-mediated mEPSCs upon chronic synaptic activity blockade (n = 11–13 neurons per condition, three independent preparations; Mann–Whitney test: *P ≤ 0.05, ***P ≤ 0.001). (D) Cumulative probability histograms of mEPSC amplitude. The cumulative probability curve of “synaptic activity blockade” amplitude was scaled by dividing by a factor of 2.22 (n = 1,650–1,950 events recorded from 11 to 13 cells per condition, three independent preparations). (E and G) Representative images of hippocampal neurons in control conditions or treated with GYKI-52466 and MK-801 for 24 h and stained for surface GluA1 (E) or surface GluA2 (G), VGluT1 and MAP2. Arrows indicate colocalization of surface staining (yellow) and VGluT1 puncta (cyan); arrowheads pinpoint VGluT1 puncta (cyan) that do not colocalize with GluA clusters. (Scale bars, 5 μm.) (F and H) Intensity, area and number of synaptic GluA1 (F) or GluA2 (G) clusters (colocalizing with VGluT1 clusters) per dendritic length (n = 29–33 neurons, three independent experiments; Mann–Whitney test: **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
To assess the effects of chronic blockade of AMPARs and NMDARs on surface expression and composition of AMPARs, hippocampal neurons treated with GYKI-52466 and MK-801 for 24 h, or in control conditions, were labeled for surface GluA1 or GluA2, and for the dendritic marker MAP2 and the synaptic marker VGluT1 (Fig. 1 E and G). The intensity, area, and number of VGluT1 clusters were not changed upon synaptic activity blockade (SI Appendix, Fig. S1A), suggesting that the change in the frequency of mEPSCs (Fig. 1 A and B) is not due to a change in synapse number. However, the synaptic surface GluA1 clusters (colocalized with VGluT1 clusters) presented more than twofold increase in their intensity, area, and number upon synaptic activity suppression (Fig. 1F). Moreover, scaled distribution of synaptic GluA1 intensities, in GYKI-52466– and MK-801–treated hippocampal neurons, was nearly superimposed with distribution of intensities from control hippocampal neurons (SI Appendix, Fig. S1B), in agreement with a multiplicative increase on the intensity of synaptic GluA1-containing AMPAR clusters. Similarly, synaptic surface GluA2 clusters (labeled with an anti-GluA2 specific antibody) (SI Appendix, Fig. S2) were increased with synaptic activity suppression for 24 h (Fig. 1H), also in a multiplicative manner (SI Appendix, Fig. S1C). Total cell surface levels of both GluA1 and GluA2 were also increased in hippocampal neurons treated for 24 h with GYKI-52466 and MK-801 (SI Appendix, Fig. S1 D and E). Given previous work suggesting differential roles for calcium-permeable vs. GluA2-containing AMPARs in different synaptic scaling paradigms (25–27, 34–39), we tested the sensitivity of mEPSCs to the calcium-permeable AMPAR inhibitor Naspm, in control conditions and in neurons submitted to synaptic activity blockade for 24 h (SI Appendix, Fig. S3 A and B). We found that synaptic upscaling of AMPAR currents occurred in the presence of Naspm (SI Appendix, Fig. S3C), and that the percentage of inhibition of mEPSCs by Naspm was not significantly changed upon chronic synaptic activity blockade (SI Appendix, Fig. S3D).
In summary, chronic suppression of synaptic activity in hippocampal neurons, through blockade of AMPARs and NMDARs, induces upscaling of synaptic strength, likely by a concurrent regulation of both GluA1 and GluA2 subunits.
Synaptic Scaling Regulates the Neuronal Transcriptome and miRNA Profile in Hippocampal Neurons.
Synaptic scaling has been extensively studied to identify and comprehend the molecular players and signaling pathways behind compensatory homeostatic mechanisms that modulate synaptic strength in response to prolonged changes in neuronal activity (7). In particular, chronic blockade of neuronal and synaptic activity has been shown to promote synaptic scaling through transcription- or translation-dependent mechanisms (11, 12, 35, 36). However, the exact mechanisms and how they impact on the synaptic scaling apparatus are still largely unknown.
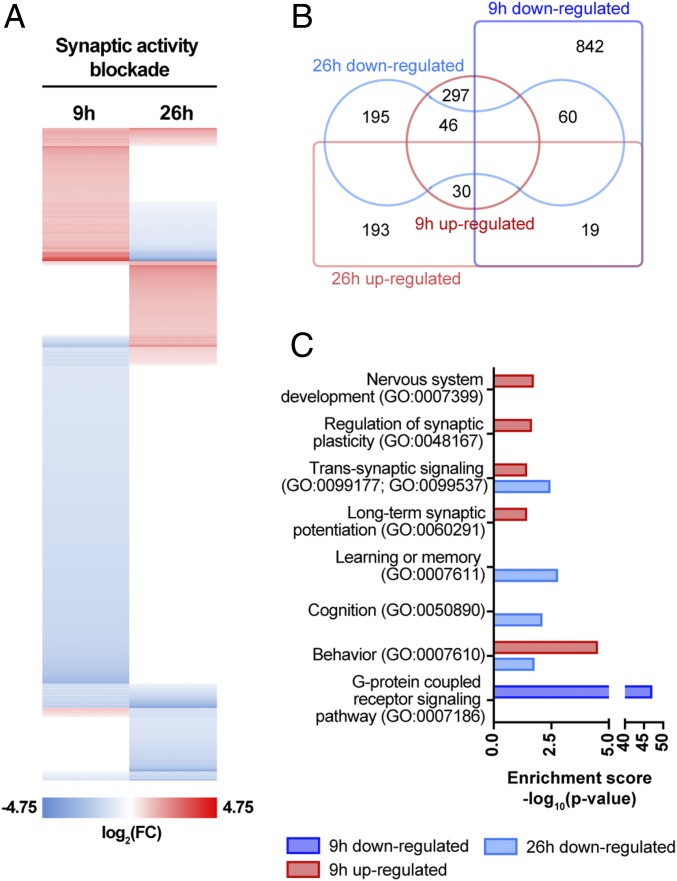
To investigate the effects of synaptic upscaling in the neuronal transcriptome, whole-genome rat gene expression microarray analysis was performed using total RNA from hippocampal neurons in control conditions or treated with GYKI-52466 and MK-801 for 9 or 26 h (Fig. 2A and SI Appendix, Tables S1 and S2) (40). Venn diagram analysis, performed using the jvenn web tool (41), showed that 30 transcripts were up-regulated and 60 transcripts were down-regulated in both experimental conditions. In addition, 46 transcripts were shown to be up-regulated with 9 h of chronic synaptic activity blockade, but their levels were significantly decreased upon 26 h of treatment, whereas 19 transcripts followed the opposite pattern of expression (Fig. 2B). These observations are in agreement with a recent study reporting different sets of newly synthesized proteins in early and late stages of homeostatic responses (42). Subsequently, it was our aim to identify biological processes and signaling pathways involved in synaptic scaling. Sets of up-regulated and down-regulated genes upon chronic blockade of synaptic activity were analyzed with the Gene Ontology Consortium web tool. This analysis aimed to pinpoint gene ontology classes related with neuronal/brain processes and with enriched representation in the altered gene sets, in which a gene can be associated with more than one category. Overall, enrichment of transcripts associated with synaptic signaling, transmission and plasticity, behavior and cognition was detected (Fig. 2C), consistent with many reports linking proteins involved in synaptic plasticity mechanisms with synaptic scaling (7–10). Datasets for down-regulated transcripts upon 9 h of treatment showed enrichment for transcripts associated with G protein-coupled receptor-signaling pathway (Fig. 2C).
Fig. 2.
Chronic synaptic activity blockade alters the transcriptome of hippocampal neurons. (A) Heat-map representation of transcripts significantly changed in whole-genome rat gene-expression microarrays, in hippocampal cultured neurons subjected to synaptic activity blockade with GYKI-52466 and MK-801 for 9 h or 26 h. Transcripts with a fold-change ≥ 2.0 cut-off were considered; each data point refers to averaged fold change for triplicates (n = 3 independent experiments; t test: P ≤ 0.05). (B) Venn diagram representing the overlap of up- and down-regulated gene sets upon activity blockade for 9 or 26 h. (C) Gene ontology analysis exposes enrichment of differently expressed transcripts associated with synaptic transmission and plasticity. Only categories with statistical significance were considered and nonredundant neuroscience related categories were selected for graphical representation (binomial statistical test with Bonferroni correction for multiple testing: P ≤ 0.05).
Among the transcripts regulated by activity suppression, we found several molecules previously associated with synaptic scaling mechanisms, such as Arc, Bdnf, CamKIIβ, Grin1, Plk2, Rps6ka5, and Rara (SI Appendix, Tables S1 and S2) (24, 36, 43–48). Additionally, the levels of Lrrtm2, Mdga1, Nlgn1, Nlgn3, Npas4, and Syn1, associated with synaptogenesis and stability of excitatory and inhibitory synapses (49–59), were also found altered (SI Appendix, Tables S1 and S2), suggesting that homeostatic mechanisms regulating excitatory/inhibitory synaptic balance may also take place with this paradigm of activity blockade (60).
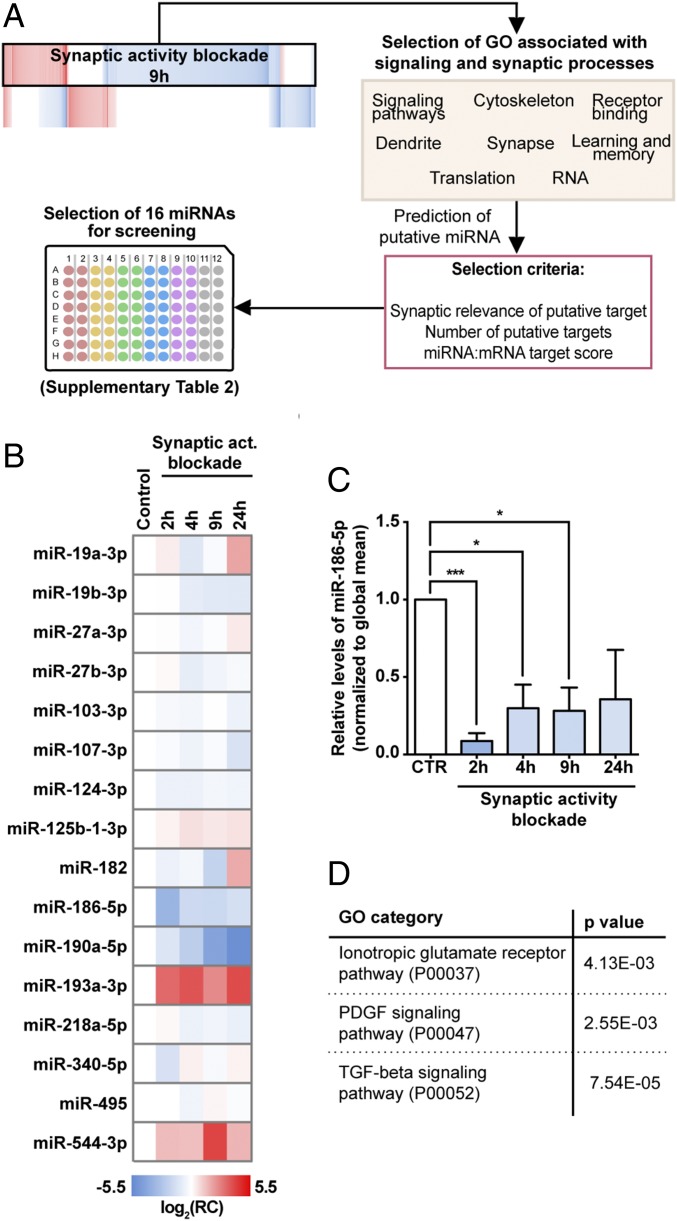
To identify miRNAs regulating synaptic upscaling mechanisms, we built a screening panel of selected miRNAs, based on gene-expression alterations upon 9 h of chronic blockade of activity using GYKI-52466 and MK-801. Briefly, altered transcripts associated with a set of gene ontology categories relevant for synaptic function were chosen to predict possible miRNA regulators, by using three independent prediction algorithms: miRanda, miRTarget2, and TargetScan6.1 (Fig. 3A). We then built a screening panel comprised of selected miRNAs predicted to target a large number of altered transcripts, by at least two different algorithms, and according to synaptic relevance of the putative targets, number of putative targets, and miRNA:mRNA target score (Fig. 3A and SI Appendix, Table S3).
Fig. 3.
Chronic blockade of synaptic activity regulates miRNA levels. (A) Workflow leading to the selection of a miRNA panel. Transcripts differently expressed upon 9 h of synaptic activity blockade, from eight different gene ontology categories, were used to predict putative miRNA regulators (miRanda, miRTarget2, TargetScan 6.1). Sixteen miRNAs were chosen for screening. (B) Heatmap representation of the neuronal miRNA profile in hippocampal cultures in control conditions or upon incubation with GYKI-52466 and MK-801. Data were normalized to global mean and each data point refers relative change. (C) Relative levels of miR-186-5p were down-regulated upon synaptic activity blockade (n = 3–4; one sample t test: *P ≤ 0.05, ***P ≤ 0.001). (D) Prediction of conserved putative targets of miR-186-5p revealed an enrichment of targets associated with pathways of synaptic relevance. Gene ontology analysis was performed for PANTHER pathways using the Gene Ontology Consortium database (binomial statistical test with Bonferroni correction for multiple testing: P ≤ 0.05).
MicroRNA profiling was performed for 16 miRNAs in hippocampal neurons in control conditions or treated for 2, 4, 9, or 24 h with GYKI-52466 and MK-801. We identified several activity-regulated miRNAs, with miR-186-5p, miR-190a-5p, miR-193a-3p, and miR-544-3p exhibiting the most dramatic changes in their expression levels (Fig. 3B). These results reveal that prolonged AMPAR and NMDAR blockade regulates the miRNA profile in neurons, possibly by different parallel mechanisms, because bidirectional alterations in miRNA levels were detected and some miRNAs were more rapidly regulated than others (Fig. 3B). Curiously, with the present activity suppression paradigm, we did not find significant changes in the expression levels of miR-92a-3p or miR-124-3p (SI Appendix, Fig. S4) previously found to be regulated by neuronal activity blockade manipulation (22, 23). To select possible miRNA regulators involved in the synaptic scaling mechanism, three key aspects were taken into consideration: (i) miRNA expression levels in neurons, (ii) how fast the miRNA responded to synaptic activity suppression, and (iii) putative targets with synaptic relevance (besides those identified in the microarray analysis). According to these criteria, miR-186-5p was selected for further scrutiny due to its presence in fair levels in neurons and its rapid and persistent regulation upon synaptic activity blockade (Fig. 3C).
Because a single miRNA has the potential to regulate hundreds of transcripts, we predicted putative targets for miR-186-5p using the TargetScan7.1 algorithm (61) and performed gene ontology enrichment analysis using the Gene Ontology Consortium web tool to assess the possible impact of miR-186-5p in neuronal function. Interestingly, PANTHER pathway analysis showed enrichment in transcripts associated with the ionotropic glutamate receptor pathway, hinting for a function in synaptic plasticity (Fig. 3D and SI Appendix, Fig. S5A). High-throughput strategies have recently been used to understand miRNA targeting and to identify their targets. Moore, et al. (62) performed precipitation and identification of AGO:miRNA-bound RNAs from mouse brain. We used these data to investigate miR-186-5p targets in the mouse brain and found enrichment in transcripts involved in neuronal development, synapse assembly, and synaptic plasticity (SI Appendix, Fig. S5B). Importantly, this analysis also unraveled enrichment in targets related with activation of AMPA receptors, trafficking of GluA2-containing AMPA receptors, and activation of the NMDA receptor upon glutamate binding and postsynaptic events (SI Appendix, Fig. S5B). Together, these results support the hypothesis that miR-186-5p is an important regulator of synaptic processes and synaptic plasticity mechanisms.
miR-186-5p Regulates Endogenous GluA2 Expression.
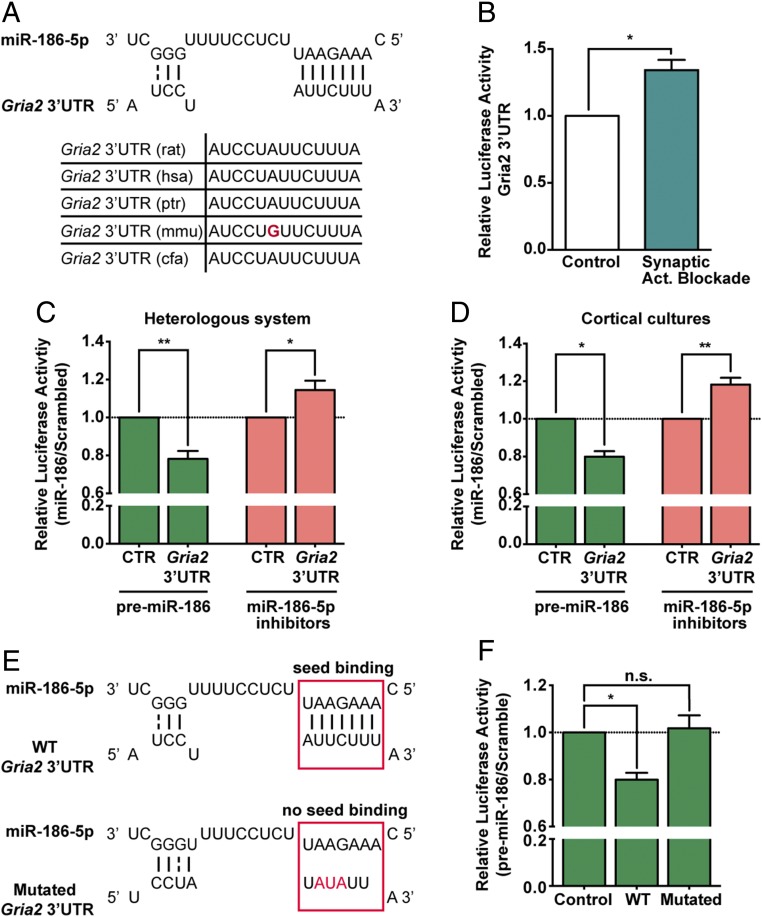
One of miR-186-5p’s possible targets is the GluA2-coding transcript Gria2, which has a highly conserved putative binding site for miR-186-5p in its 3′UTR (Fig. 4A). Moreover, upon chronic blockade of synaptic activity miR-186-5p levels were inversely correlated with GluA2 levels (Figs. 1H and 3C), hinting that miR-186-5p may regulate GluA2. To test this hypothesis we assessed whether synaptic activity deprivation results in translational enhancement dependent on posttranscriptional regulatory mechanisms associated with Gria2 3′ UTR. We used a reporter construct containing the full-length Gria2 3′UTR downstream of the gaussia luciferase coding sequence; secreted alkaline phosphatase (SEAP) expressed from the same vector was used for normalization. Transfected hippocampal neurons were treated for 24 h with GYKI-52466 and MK-801 before luciferase activity was evaluated. Indeed, chronic blockade of synaptic activity increased luciferase activity regulated by the Gria2 3′UTR (Fig. 4B), whereas activity blockade did not significantly affect luciferase expression in neurons expressing the control vector.
Fig. 4.
Gria2 3′UTR is a direct target of miR-186-5p. (A) miR-186-5p:Gria2 3′UTR site interaction is highly conserved in mammals. Putative site prediction performed with TargetScan and RNA hybridization analyzed with RNAhybrid. (B) Hippocampal neurons expressing guassia luciferase under the control of Gria2 3′UTR showed increased expression of guassia luciferase upon chronic blockade of synaptic activity with GYKI-52466 and MK-801 for 24 h (n = 3; one sample t test: *P ≤ 0.05). (C and D) HEK293T cells (C) or cortical neurons (D) expressing luciferase reporter constructs (pGluc-Gria2-3′UTR or pGLuc-Control) and premiR-186 or miR-186-5p inhibitors constructs (or respective scramble sequences) presented an inverse correlation between expression of miR-186-5p and luciferase under the control of Gria2 3′UTR (n = 8 or 3 for premiR-186 expression experiments and n = 6 for miR-186-5p inhibition; one sample t test: *P ≤ 0.05, **P ≤ 0.01). (E) The predicted miR-186-5p seed region targeted site in the Gria2 3′UTR was partially mutated. (F) Cortical neurons expressing luciferase constructs (control pGLuc-Control, pGLuc-GluA2-3′UTR containing the wild-type Gria2 3′UTR or pGLuc-GluA2-3′UTR containing mutated Gria2 3′UTR) and premiR-186 or scramble expressing constructs confirmed this site as a miR-186-5p target site (n = 3; one-sample t test: *P ≤ 0.05; n.s., not significant). (B–D and F) Luciferase expression was normalized for SEAP activity (expressed from the same vector) and with luciferase coexpressed with scramble controls.
To test whether miR-186-5p directly regulates GluA2 expression, we cotransfected HEK293T cells or cortical neuron cultures with the luciferase reporter fused to Gria2 3′UTR and either premiR-186, to overexpress miR-186-5p, or miR-186-5p inhibitors, or the respective control vectors expressing scrambled sequences. In both cell systems, expression of premiR-186 significantly decreased luciferase signal and, conversely, miR-186-5p inhibition increased luciferase expression (Fig. 4 C and D). Therefore, these results suggest that GluA2 is a direct target of miR-186-5p. To determine if direct interaction of miR-186-5p:Gria2 occurs in the predicted target site, we generated a mutant reporter construct containing point mutations in the putative binding site of Gria2 3′UTR (Fig. 4E). Whereas coexpression of premiR-186 and wild-type Gria2 3′UTR decreased luciferase signal, coexpression with the Gria2 3′UTR mutant abolished this effect in cortical neurons (Fig. 4F), demonstrating miR-186-5p:Gria2 3′UTR interaction at this site.
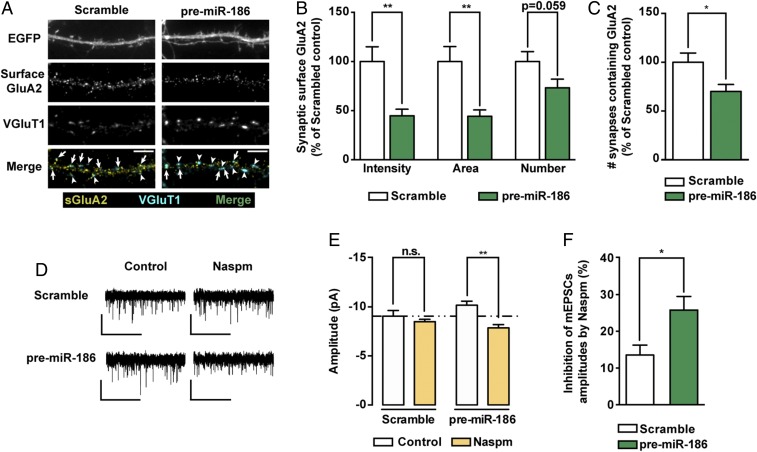
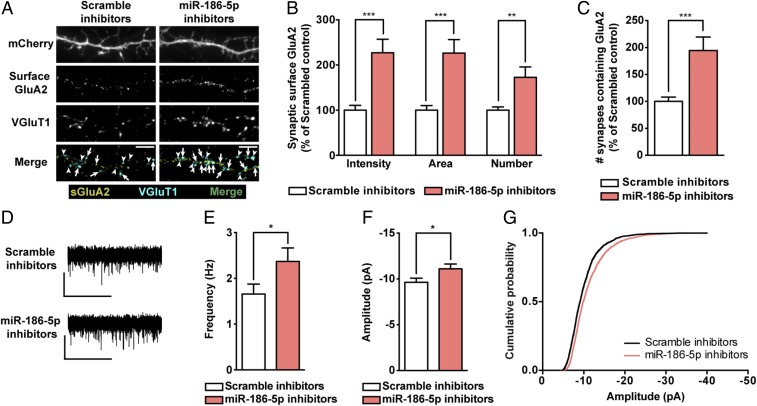
Consequently, we characterized the regulatory role of miR-186-5p on endogenous GluA2 expression in hippocampal neurons by expressing the precursor form of miR-186 or inhibiting miR-186-5p. Expression of premiR-186 decreased the intensity, area, and number of total (SI Appendix, Fig. S6A) and synaptic (Fig. 5 A and B) cell surface GluA2 clusters in hippocampal neurons, and the number of VGluT1+ synapses colocalizing with cell surface GluA2 (Fig. 5C). We then investigated whether AMPAR composition was altered in neurons expressing premiR-186 taking advantage of Naspm. Expression of premiR-186 did not significantly affect the amplitude or frequency of AMPAR-mediated mEPSCs (SI Appendix, Fig. S7), but Naspm produced a higher inhibitory effect in the amplitude of mEPSCs in neurons expressing premiR-186 compared with neurons expressing a scrambled control (Fig. 5 D–F), indicating that miR-186 expression increases the synaptic content of GluA2-lacking AMPARs. Conversely, neurons expressing miR-186-5p inhibitors presented an increase in the intensity, area, and number of total (SI Appendix, Fig. S6B) and synaptic cell surface GluA2 clusters (Fig. 6 A and B), and exhibited more VGluT1+ synapses colocalizing with cell surface GluA2 (Fig. 6C). Inhibiting endogenous miR-186-5p was sufficient to scale up mEPSC frequency and amplitude (Fig. 6 D–G), indicating that endogenous miR-186-5p levels contribute to decrease excitatory transmission. These results show that miR-186-5p regulates the synaptic content of endogenous GluA2-containing AMPARs, and impacts AMPAR-mediated synaptic transmission in hippocampal neurons.
Fig. 5.
Expression of premiR-186 regulates the expression of endogenous GluA2. (A) Representative images of hippocampal neurons expressing premiR-186 or control scramble constructs and stained for surface GluA2, VGluT1, and MAP2. Transfected neurons were identified by expression of EGFP from the premiR-186 expression plasmid. Arrows indicate colocalization of surface GluA2 (yellow) and VGluT1 puncta (cyan); arrowheads pinpoint GluA2-lacking VGluT1 puncta (cyan). (Scale bars, 5 μm.) (B) Intensity, area and number of endogenous synaptic surface GluA2 clusters (colocalizing with VGluT1 clusters) normalized to synapse density (n = 29–32 cells per condition from three independent experiments; Mann–Whitney test: **P ≤ 0.01). (C) Neurons expressing premiR-186 displayed a decreased density of VGluT1+ synapses containing surface GluA2 clusters (n = 29–32 cells per condition, three independent experiments; Mann–Whitney test: *P ≤ 0.05). (D) Representative whole-cell current traces of AMPAR-mediated mEPSCs from hippocampal neurons expressing a scramble control or premiR-186, under control conditions or preincubated with 20 μM Naspm for 30 min. (Scale bars: vertical, 10 pA; horizontal, 5 s.) (E) Naspm decreased the amplitude of AMPAR-mediated mEPSCs of neurons expressing premiR-186 (n = 7–10 cells per condition, five independent experiments; two-way ANOVA with Tukey’s multiple comparison test: **P ≤ 0.01). (F) PremiR-186 expressing neurons displayed a higher percentage of inhibition with Naspm than control neurons (n = 7–9 cells per condition, five independent experiments; Mann–Whitney test: *P ≤ 0.05).
Fig. 6.
Inhibition of miR-186-5p scales up excitatory synaptic strength. (A) Representative images of hippocampal neurons expressing miR-186-5p inhibitors or control scramble constructs and stained for surface GluA2, VGluT1, and MAP2. Transfected neurons were identified by expression of mCherry from the bicistronic miR-186-5p inhibitors expression plasmid. Arrows indicate colocalization of surface GluA2 (yellow) and VGluT1 puncta (cyan); arrowheads pinpoint GluA2-lacking VGluT1 puncta (cyan). (Scale bars, 5 μm.) (B) Intensity, area, and number of endogenous synaptic GluA2 clusters (colocalizing with VGluT1 clusters) normalized to synapse density (n = 49 cells from five independent experiments; Mann–Whitney test: **P ≤ 0.01, ***P ≤ 0.001). (C) Inhibition of miR-186-5p elevated the density of VGluT1+ synapses containing surface GluA2 (n = 49 cells from five independent experiments; Mann–Whitney test: ***P ≤ 0.001). (D) Representative whole-cell current traces of AMPAR-mediated mEPSCs from hippocampal neurons expressing a scramble sequence or miR-186–5p inhibitors. (Scale bars: vertical, 10 pA; horizontal, 5 s.) Expression of miR-186-5p inhibitors increased AMPAR-mediated mEPSC frequency (E) and amplitude (F) (n = 18 cells per condition, 10 independent experiments; Mann–Whitney test: *P ≤ 0.05). (G) Cumulative probability histograms of mEPSC amplitudes display the increase induced by miR-186-5p inhibition (n = 2,400 events recorded from 16 cells per condition, 10 independent experiments).
miR-186-5p Affects Synaptic Upscaling in Hippocampal Neurons.
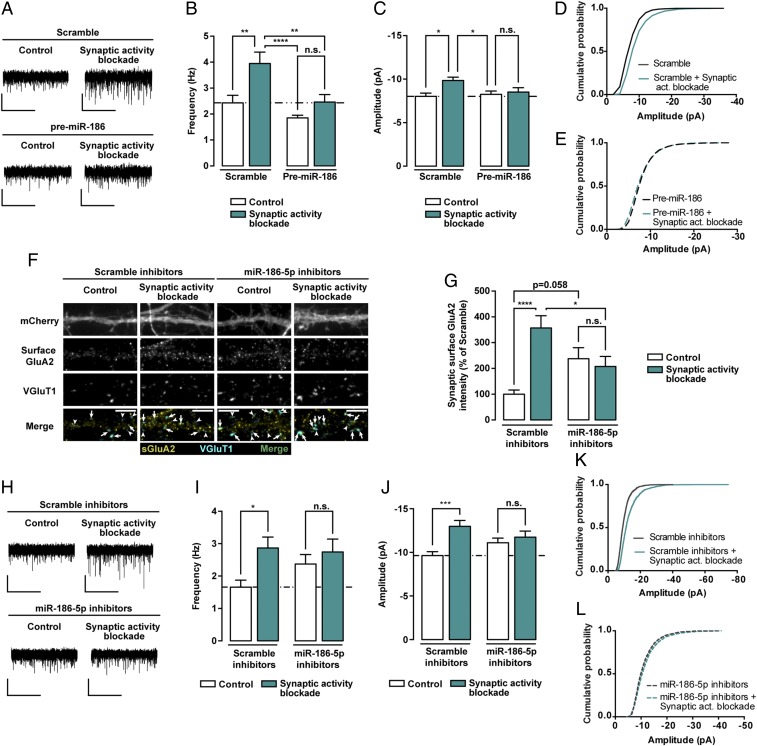
Considering the previous results, we examined whether overexpression or inhibition of miR-186-5p basal expression affects synaptic upscaling triggered by synaptic activity blockade. We expressed a scramble sequence or the precursor form of miR-186 in hippocampal neurons, which were later subjected to synaptic activity suppression for 24 h, before mEPSCs were evaluated (Fig. 7A). Chronic blockade of synaptic activity induced upscaling of mEPSC frequency and amplitude in neurons expressing a scramble sequence (Fig. 7 A–D). However, premiR186 expression blocked the increase in the frequency (Fig. 7B) and amplitude (Fig. 7 C and E) of mEPSCs induced by synaptic activity suppression.
Fig. 7.
Manipulation of basal miR-186-5p levels blocks synaptic scaling-up. (A) Representative whole-cell current traces of AMPAR-mediated mEPSCs from hippocampal neurons expressing a scramble control or premiR-186, and treated with 50 μM GYKI-52466 and MK-801 for 24 h. (Scale bars: vertical, 10 pA; horizontal, 5 s.) (B and C) Expression of premiR-186 hinders the increase of AMPAR-mediated mEPSC frequency (B) and amplitude (C) induced by chronic blockade of synaptic activity (n = 12–14 neurons per condition, six independent preparations). (D and E) Cumulative probability histograms of mEPSC amplitudes display the synaptic scaling associated with chronic blockade of excitatory synaptic activity in neurons expressing scramble control (D), but not in premiR-186–expressing neurons (E) (n = 1,800 events recorded from 16 cells per condition, six independent experiments). (F) Representative images of hippocampal neurons expressing either miR-186-5p inhibitors or control scramble constructs, in control conditions or treated with GYKI-52466 and MK-801 for 24 h, and stained for surface GluA2, VGluT1, and MAP2. Transfected neurons were identified by expression of mCherry from the bicistronic miR-186-5p inhibitors expression plasmid. Arrows indicate colocalization of surface GluA2 (yellow) and VGluT1 puncta (cyan); arrowheads pinpoint GluA2-lacking VGluT1 puncta (cyan). (Scale bars, 5 μm.) (G) Inhibition of basal miR-186-5p expression occluded upscaling of synaptic GluA2 clusters intensity upon blockade of synaptic activity. Synaptic GluA2 clusters (colocalized with VGluT1) were normalized to synapse density (n = 29–30 neurons per condition, three independent experiments). (H) Representative whole-cell current traces of AMPAR-mediated mEPSCs from hippocampal neurons expressing a scrambled sequence or miR-186-5p inhibitors, and exposed to GYKI-52466 and MK-801 for 24 h. (Scale bars: vertical, 10 pA; horizontal, 5 s.) (I and J) Inhibiting the basal levels of miR-186-5p hampered the increase of AMPAR-mediated mEPSC frequency (I) and amplitude (J) induced by blockade of synaptic activity (n = 16–18 neurons per condition, 10 independent preparations). (K and L) Cumulative probability histograms of mEPSC amplitudes display synaptic scaling associated with chronic synaptic activity suppression in neurons expressing scrambled inhibitors (K), but not in miR-186-5p inhibitor-expressing neurons (L) (n = 2,400 events, 16 cells per condition, 10 independent experiments). (B, C, G, I, and J) Statistical analysis was performed using two-way ANOVA with Tukey’s multiple comparison test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
On the other hand, we tested whether inhibition of miR-186-5p expression affected synaptic scaling. Transfected hippocampal neurons expressing miR-186-5p inhibitors, or a scrambled sequence, were subjected to chronic synaptic activity blockade with GYKI-52466 and MK-801 for 24 h or maintained in control conditions. As shown previously (Figs. 1H and 6 A and B), both chronic blockade of synaptic activity and basal inhibition of miR-186-5p increased synaptic cell surface GluA2 levels (Fig. 7 F and G). However, chronic blockade of synaptic activity failed to further up-regulate GluA2 synaptic levels in neurons expressing miR-186-5p inhibitors (Fig. 7G), suggesting that miR-186-5p inhibition up-regulates GluA2 cell surface levels through mechanisms shared with those activated by synaptic activity suppression. Subsequently, the functional role of miR-186-5p in synaptic scaling was evaluated by analyzing AMPAR-mediated mEPSCs. Inhibition of miR-186-5p, which elevated mEPSC amplitude and frequency in basal conditions (Fig. 6 D–G), blocked further scaling-up induced by chronic blockade of AMPARs and NMDARs (Fig. 7 H–J). The combined cumulative probability distribution analysis also evidenced these differences, with synaptic activity blockade inducing a shift toward increased amplitude of AMPAR-mediated mEPSCs in neurons expressing a scrambled sequence (Fig. 7K), but not in neurons expressing miR-186-5p inhibitors (Fig. 7L). In summary, the elevation in mEPSC amplitude and frequency induced by miR-186-5p inhibition occluded further homeostatic adjustment triggered by synaptic activity suppression. Overall, these results support a role for miR-186-5p in synaptic scaling triggered by chronic synaptic activity blockade.
Discussion
Synaptic scaling is an important homeostatic mechanism responsible for maintaining neuronal network activity within a physiological and dynamic range throughout developmental, experience, and environmental changes (2, 63). The mechanisms promoting synaptic scaling can be influenced by neuronal model, developmental age, and activity paradigms (28, 64, 65), revealing the existence of distinct processes underlying scaling (11, 35, 66). Here, we report that chronic blockade of AMPARs and NMDARs for 24 h promotes synaptic scaling of AMPAR-mediated mEPSC amplitude, in accordance with previous studies using this activity-deprivation paradigm in different neuronal models (26, 32, 33) and also reports on neurons subjected to chronic blockade of AMPARs only (24, 27, 66–68). Scaling of AMPAR-mediated currents induced by chronic blockade of AMPARs and NMDARs was accompanied by scaling of synaptic GluA1 and GluA2 content, in agreement with previous observations in spinal cord neurons (26, 69). However, chronic blockade of AMPARs alone enhances postsynaptic contribution of GluA1 homomers in cortical and hippocampal neurons (27, 67), hinting that different mechanisms are responsible for synaptic scaling upon chronic blockade of AMPARs alone or of AMPARs and NMDARs.
The functional relevance of several proteins in synaptic scaling has been investigated, although the underlying mechanisms remain largely unknown, as few high-throughput studies have examined how the neuronal transcriptome is modified at different stages of the scaling process (12, 70). In this work, we analyzed the neuronal transcriptome of hippocampal neurons upon chronic blockade of synaptic activity for 9 and 26 h. Gene ontology enrichment analysis showed altered transcripts associated with synaptic signaling and synaptic plasticity classes. In addition to transcripts previously associated with synaptic scaling—such as Arc, Bdnf, CamKIIβ, Plk2, Rps6ka5, and Rara (24, 36, 43, 44, 47, 48)—we now identify several transcripts associated with synaptogenesis—including Lrrtm2, Mdga1, Nlgn1, Nlgn3, Npas4, and Syn1 (49–59)—suggesting that this paradigm of chronic activity blockade may also elicit adjustments in excitatory/inhibitory synaptic balance (60).
Some paradigms of neuronal activity blockade require de novo protein translation (25, 34, 35, 66), indicating a relevant function for posttranscriptional regulatory mechanisms in this process. The involvement of miRNAs has been reported both in synaptic scaling-down and scaling-up mechanisms: downscaling paradigms regulate miR-485, miR-134, and miR-129-5p levels, which in turn target, respectively, SV2A, Pum2, and ATP2B4 and DCX, important players in downscaling mechanisms (71–73). On the other hand, miR-92a and miR-124 are implicated in upscaling paradigms by targeting GluA1 and GluA2 (22, 23). Here, we performed miRNA profiling of a small set of miRNAs, which are predicted regulators of altered transcripts identified in hippocampal neurons submitted to 9 h of synaptic activity blockade. We observed that synaptic activity deprivation regulates the levels of several miRNAs at different stages of scaling, in agreement with previous studies (17, 22, 23). In addition, our results reveal a dramatic and persistent decrease in miR-186-5p levels after 2 h of treatment with GYKI-52466 and MK-801 and up to 24 h of incubation, suggesting a functional role for this miRNA from early stages of synaptic upscaling induced by synaptic activity blockade.
miR-186-5p is a highly conserved miRNA in mammals, expressed in the spinal cord and across multiple brain subregions, with enriched expression in neurons (74). Furthermore, expression of this miRNA can be regulated as a physiological response to stress in intestinal, prefrontal cortex, and hippocampal tissues (75, 76), and by extrinsic factors, such as methamphetamine or alcohol intake (77, 78). A group of brain-enriched transcripts present a conserved putative binding site for the miR-186-5p seed region (79), and gene ontology analysis of miR-186-5p putative and AGO:miR-186-5p–bound targets (62) revealed an enrichment of neuronal targets associated with neuronal and synaptic development and synaptic plasticity, including several targets related with the ionotropic glutamate receptor pathway. Among this pool of putative and AGO:miR-186-5p–bound targets is Gria2, encoding GluA2. We report that miR-186-5p targets GluA2 and regulates endogenous GluA2 expression under physiological conditions through a direct target site in Gria2 3′UTR. Furthermore, miR-186-5p inhibition elevates the synaptic content of GluA2, and the frequency and amplitude of AMPA receptor-mediated currents, while miR-186 overexpression decreases the synaptic content of GluA2-containing AMPAR, as shown by the increased sensitivity to Naspm, an inhibitor of GluA2-lacking AMPAR. This evidence indicates that miR-186-5p regulates synaptic transmission and the subunit composition of AMPARs. Few miRNAs have been identified as direct regulators of AMPAR expression in a neuronal activity-dependent manner: miR-92a, miR-137, and miR-501-3p were found to regulate GluA1 (22, 80, 81), and miR-124 and miR-181a regulate GluA2 expression (23, 82, 83). We now report miR-186-5p as a regulator of GluA2 expression and AMPAR subunit composition. Interestingly, analysis of Ago:miR-186-5p–precipitated targets (62) shows an enrichment of transcripts associated with the trafficking of GluA2-containing AMPARs—including NSF (84, 85) and PRKCB (86–88)—suggesting that besides regulating GluA2 levels, miR-186-5p may regulate additional mechanisms participating in the GluA2-containing AMPAR trafficking apparatus.
Because miR-186-5p levels are decreased by synaptic activity blockade, and miR-186-5p targets GluA2, the levels of which are up-regulated upon prolonged synaptic activity suppression, we evaluated the role of miR-186-5p on synaptic upscaling. PremiR-186 overexpression blocked synaptic scaling, presumably by down-regulating GluA2 levels. Conversely, miR-186-5p inhibition is sufficient to increase cell surface levels of GluA2 at synapses, and to induce upscaling of AMPAR-mediated mEPSC amplitude. We also report that prolonged synaptic activity blockade fails to further increase cell surface synaptic GluA2 levels and upscaling of mEPSCs in neurons expressing miR-186-5p inhibitors. These results indicate that high levels of miR-186-5p interfere with synaptic scaling, and that miR-186-5p inhibition occludes the synaptic scaling-associated mechanisms that lead to up-regulation of GluA2 levels and increased AMPAR-mediated currents. It is possible that basal miR-186-5p inhibition also negatively interferes with synaptic upscaling mechanisms, since GluA2 synaptic levels detected upon basal miR-186-5p inhibition plus activity blockade do not reach GluA2 levels found upon activity suppression in control neurons. In fact, miR-124 was found to transiently down-regulate GluA2-containing AMPAR upon chronic blockade of action potentials and NMDAR, even though this effect was abolished at later time points, when GluA2-containing receptors were found upscaled (23). However, we did not observe significant changes in the expression of miR-124-3p upon suppression of synaptic activity (SI Appendix, Fig. S4A). Taken together, our results strongly implicate miR-186-5p in synaptic scaling mechanisms associated with suppression of synaptic activity in hippocampal neurons.
Overall, our work demonstrates that chronic blockade of synaptic activity in hippocampal neurons induces upscaling of AMPAR-mediated mEPSC amplitude with an increase in GluA1 and GluA2 synaptic content. The mechanisms underlying synaptic scaling affect the neuronal mRNA transcriptome and miRNA profile, although the signaling pathways triggering these effects remain unclear. Furthermore, suppression of excitatory synaptic activity decreases the levels of miR-186-5p, a negative regulator of GluA2 expression. Expression of the miR-186 precursor abolishes synaptic scaling upon synaptic activity suppression, whereas inhibition of miR-186-5p basal levels enhances excitatory transmission and occludes the increase in cell surface GluA2 content and synaptic scaling-up of AMPAR mediated mEPSCs triggered by synaptic activity blockade. Together, these results indicate an important regulatory function for miR-186-5p in regulating GluA2 and synaptic upscaling mechanisms.
Materials and Methods
mEPSC Recording and Analysis.
Whole-cell voltage-clamp recordings were made at room temperature from 15 DIV hippocampal neurons plated on coverslips. The recording chamber was perfused with extracellular solution supplemented with 100 μM picrotoxin, 500 ηM TTX, and 50 μM APV. For 3 min, AMPAR-mediated mEPSC events were recorded in a gap-free acquisition mode with a sampling rate of 25 kHz after signal filtering at 2.8 kHz. Only events larger than 2× the recording noise and decay τ between 2 and 30 ms were considered. For analysis of AMPARs composition, hippocampal neurons were preincubated with 20 μM of the selective Ca2+-permeable AMPAR antagonist Naspm for 30 min–60 min, and then recorded in the presence of the drug.
Imaging and Quantification.
Fluorescence imaging for puncta analysis was performed using a widefield Zeiss Axio Observer Z1 inverted microscope (Carl Zeiss) equipped with an AxioCam HRm camera and with ZEN blue software (Carl Zeiss). Images were acquired with a 63× Plan-ApoChromat oil objective (numerical aperture 1.4). For each independent experiment, neurons were cultured and stained simultaneously. Dendrites with similar thickness and appearance were randomly selected using MAP2 staining and exposure time was defined to avoid pixel saturation. All experimental conditions within independent preparations were imaged using identical settings.
Images were quantified using image analysis software FIJI (89), taking advantage of a semiautomatic macro designed for this purpose. The area of interest was randomly selected by using MAP2 staining for nontransfected cells and GFP or mCherry staining for transfected cells. Dendritic length was measured in the area of interest, using one of the mentioned channels. To quantify the proteins of interest, images were subjected to a user-defined intensity threshold, to have defined protein clusters, and user-defined background intensity was subtracted in all images. Each protein cluster present in the area of interest was analyzed and measurements of intensity, area, and number were obtained. Synaptic clusters were selected by overlapping clusters of interest with thresholded VGluT1 clusters. All results are normalized to dendritic length or synapse density, as indicated in image captions. This analysis was performed blind to the experimental condition.
Statistical Analysis.
Graphs and statistical analyses were performed using GraphPad Prism 6 software. Outliers were identified using raw experimental data with the ROUT or Grubbs method. Data are presented as mean ± SEM of generally three or more experiments, performed in independent preparations, as indicated in the figure captions. Statistical differences were analyzed as indicated in the figure captions.
Additional details are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was financed by the European Regional Development Fund, through the Centro 2020 Regional Operational Programme under project CENTRO-01-0145-FEDER-000008:BrainHealth 2020, and through the COMPETE 2020, Operational Programme for Competitiveness and Internationalisation and Portuguese national funds via Fundação para a Ciência e a Tecnologia, I.P., under projects POCI-01-0145-FEDER-007440, POCI-01-0145-028541, POCI-01-0145-FEDER-029452, SFRH/BD/51683/2011, and SFRH/BD/131812/2017.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Y.G. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE111384).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900338116/-/DCSupplemental.
References
- 1.Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- 2.Watt AJ, Desai NS. Homeostatic plasticity and STDP: Keeping a neuron’s cool in a fluctuating world. Front Synaptic Neurosci. 2010;2:5. doi: 10.3389/fnsyn.2010.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keck T, et al. Integrating Hebbian and homeostatic plasticity: The current state of the field and future research directions. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160158. doi: 10.1098/rstb.2016.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turrigiano GG. The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turrigiano G. Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wondolowski J, Dickman D. Emerging links between homeostatic synaptic plasticity and neurological disease. Front Cell Neurosci. 2013;7:223. doi: 10.3389/fncel.2013.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes D, Carvalho AL. Mechanisms of homeostatic plasticity in the excitatory synapse. J Neurochem. 2016;139:973–996. doi: 10.1111/jnc.13687. [DOI] [PubMed] [Google Scholar]
- 8.Vitureira N, Goda Y. Cell biology in neuroscience: The interplay between Hebbian and homeostatic synaptic plasticity. J Cell Biol. 2013;203:175–186. doi: 10.1083/jcb.201306030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thalhammer A, Cingolani LA. Cell adhesion and homeostatic synaptic plasticity. Neuropharmacology. 2014;78:23–30. doi: 10.1016/j.neuropharm.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Lau AG, Sarti F. Synaptic retinoic acid signaling and homeostatic synaptic plasticity. Neuropharmacology. 2014;78:3–12. doi: 10.1016/j.neuropharm.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 12.Schaukowitch K, et al. An intrinsic transcriptional program underlying synaptic scaling during activity suppression. Cell Rep. 2017;18:1512–1526. doi: 10.1016/j.celrep.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schanzenbächer CT, Sambandan S, Langer JD, Schuman EM. Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron. 2016;92:358–371. doi: 10.1016/j.neuron.2016.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diering GH, et al. Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science. 2017;355:511–515. doi: 10.1126/science.aai8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krol J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Eacker SM, Keuss MJ, Berezikov E, Dawson VL, Dawson TM. Neuronal activity regulates hippocampal miRNA expression. PLoS One. 2011;6:e25068. doi: 10.1371/journal.pone.0025068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Spronsen M, et al. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS One. 2013;8:e74907. doi: 10.1371/journal.pone.0074907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajman M, Schratt G. MicroRNAs in neural development: From master regulators to fine-tuners. Development. 2017;144:2310–2322. doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- 19.Higa GSV, et al. MicroRNAs in neuronal communication. Mol Neurobiol. 2014;49:1309–1326. doi: 10.1007/s12035-013-8603-7. [DOI] [PubMed] [Google Scholar]
- 20.Ryan B, Joilin G, Williams JM. Plasticity-related microRNA and their potential contribution to the maintenance of long-term potentiation. Front Mol Neurosci. 2015;8:4. doi: 10.3389/fnmol.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y, Xu H, Su X, He X. Role of microRNA in governing synaptic plasticity. Neural Plast. 2016;2016:4959523. doi: 10.1155/2016/4959523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letellier M, et al. miR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat Neurosci. 2014;17:1040–1042. doi: 10.1038/nn.3762. [DOI] [PubMed] [Google Scholar]
- 23.Hou Q, et al. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat Commun. 2015;6:10045. doi: 10.1038/ncomms10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 25.Sutton MA, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 26.O’Brien RJ, et al. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998;21:1067–1078. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- 27.Thiagarajan TC, Lindskog M, Tsien RW. Adaptation to synaptic inactivity in hippocampal neurons. Neuron. 2005;47:725–737. doi: 10.1016/j.neuron.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Queenan BN, Lee KJ, Pak DTS. Wherefore art thou, homeo(stasis)? Functional diversity in homeostatic synaptic plasticity. Neural Plast. 2012;2012:718203. doi: 10.1155/2012/718203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouardouz M, Durand J. GYKI 52466 antagonizes glutamate responses but not NMDA and kainate responses in rat abducens motoneurones. Neurosci Lett. 1991;125:5–8. doi: 10.1016/0304-3940(91)90115-a. [DOI] [PubMed] [Google Scholar]
- 30.Paternain AV, Morales M, Lerma J. Selective antagonism of AMPA receptors unmasks kainate receptor-mediated responses in hippocampal neurons. Neuron. 1995;14:185–189. doi: 10.1016/0896-6273(95)90253-8. [DOI] [PubMed] [Google Scholar]
- 31.Huettner JE, Bean BP. Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: Selective binding to open channels. Proc Natl Acad Sci USA. 1988;85:1307–1311. doi: 10.1073/pnas.85.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-α. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- 33.Lazarevic V, Schöne C, Heine M, Gundelfinger ED, Fejtova A. Extensive remodeling of the presynaptic cytomatrix upon homeostatic adaptation to network activity silencing. J Neurosci. 2011;31:10189–10200. doi: 10.1523/JNEUROSCI.2088-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ju W, et al. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- 35.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maghsoodi B, et al. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2008;105:16015–16020. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierenga CJ, Ibata K, Turrigiano GG. Postsynaptic expression of homeostatic plasticity at neocortical synapses. J Neurosci. 2005;25:2895–2905. doi: 10.1523/JNEUROSCI.5217-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cingolani LA, et al. Activity-dependent regulation of synaptic AMPA receptor composition and abundance by β3 integrins. Neuron. 2008;58:749–762. doi: 10.1016/j.neuron.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anggono V, Clem RL, Huganir RL. PICK1 loss of function occludes homeostatic synaptic scaling. J Neurosci. 2011;31:2188–2196. doi: 10.1523/JNEUROSCI.5633-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva MM, et al. 2019 Synaptic scaling regulates the transcriptome in hippocampal neurons. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111384. Deposited on March 3, 2018.
- 41.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schanzenbächer CT, Langer JD, Schuman EM. Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses. eLife. 2018;7:1–20. doi: 10.7554/eLife.33322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiagarajan TC, Piedras-Renteria ES, Tsien RW. Alpha- and betaCaMKII. Inverse regulation by neuronal activity and opposing effects on synaptic strength. Neuron. 2002;36:1103–1114. doi: 10.1016/s0896-6273(02)01049-8. [DOI] [PubMed] [Google Scholar]
- 45.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 46.Soares C, Lee KFH, Nassrallah W, Béïque J-C. Differential subcellular targeting of glutamate receptor subtypes during homeostatic synaptic plasticity. J Neurosci. 2013;33:13547–13559. doi: 10.1523/JNEUROSCI.1873-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DTS, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrêa SAL, et al. MSK1 regulates homeostatic and experience-dependent synaptic plasticity. J Neurosci. 2012;32:13039–13051. doi: 10.1523/JNEUROSCI.0930-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linhoff MW, et al. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spiegel I, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko J, Soler-Llavina GJ, Fuccillo MV, Malenka RC, Südhof TC. Neuroligins/LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol. 2011;194:323–334. doi: 10.1083/jcb.201101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maćkowiak M, Mordalska P, Wędzony K. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol Rep. 2014;66:830–835. doi: 10.1016/j.pharep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Katzman A, Alberini CM. NLGN1 and NLGN2 in the prefrontal cortex: Their role in memory consolidation and strengthening. Curr Opin Neurobiol. 2018;48:122–130. doi: 10.1016/j.conb.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connor SA, et al. Loss of synapse repressor MDGA1 enhances perisomatic inhibition, confers resistance to network excitation, and impairs cognitive function. Cell Rep. 2017;21:3637–3645. doi: 10.1016/j.celrep.2017.11.109. [DOI] [PubMed] [Google Scholar]
- 57.Fan X, Jin WY, Wang YT. The NMDA receptor complex: A multifunctional machine at the glutamatergic synapse. Front Cell Neurosci. 2014;8:160. doi: 10.3389/fncel.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keck T, Hübener M, Bonhoeffer T. Interactions between synaptic homeostatic mechanisms: An attempt to reconcile BCM theory, synaptic scaling, and changing excitation/inhibition balance. Curr Opin Neurobiol. 2017;43:87–93. doi: 10.1016/j.conb.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 61.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore MJ, et al. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu Rev Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- 64.Wierenga CJ, Walsh MF, Turrigiano GG. Temporal regulation of the expression locus of homeostatic plasticity. J Neurophysiol. 2006;96:2127–2133. doi: 10.1152/jn.00107.2006. [DOI] [PubMed] [Google Scholar]
- 65.Echegoyen J, Neu A, Graber KD, Soltesz I. Homeostatic plasticity studied using in vivo hippocampal activity-blockade: Synaptic scaling, intrinsic plasticity and age-dependence. PLoS One. 2007;2:e700. doi: 10.1371/journal.pone.0000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H-L, Zhang Z, Hintze M, Chen L. Decrease in calcium concentration triggers neuronal retinoic acid synthesis during homeostatic synaptic plasticity. J Neurosci. 2011;31:17764–17771. doi: 10.1523/JNEUROSCI.3964-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gong B, Wang H, Gu S, Heximer SP, Zhuo M. Genetic evidence for the requirement of adenylyl cyclase 1 in synaptic scaling of forebrain cortical neurons. Eur J Neurosci. 2007;26:275–288. doi: 10.1111/j.1460-9568.2007.05669.x. [DOI] [PubMed] [Google Scholar]
- 68.Jakawich SK, et al. Local presynaptic activity gates homeostatic changes in presynaptic function driven by dendritic BDNF synthesis. Neuron. 2010;68:1143–1158. doi: 10.1016/j.neuron.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosen KM, Moghekar A, O’Brien RJ. Activity dependent localization of synaptic NMDA receptors in spinal neurons. Mol Cell Neurosci. 2007;34:578–591. doi: 10.1016/j.mcn.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinmetz CC, et al. Upregulation of μ3A drives homeostatic plasticity by rerouting AMPAR into the recycling endosomal pathway. Cell Rep. 2016;16:2711–2722. doi: 10.1016/j.celrep.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fiore R, et al. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33:2231–2246. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajman M, et al. A microRNA-129-5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J. 2017;36:1770–1787. doi: 10.15252/embj.201695748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J, et al. miR-186 is decreased in aged brain and suppresses BACE1 expression. J Neurochem. 2016;137:436–445. doi: 10.1111/jnc.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nadorp B, Soreq H. Predicted overlapping microRNA regulators of acetylcholine packaging and degradation in neuroinflammation-related disorders. Front Mol Neurosci. 2014;7:9. doi: 10.3389/fnmol.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Babenko O, Golubov A, Ilnytskyy Y, Kovalchuk I, Metz GA. Genomic and epigenomic responses to chronic stress involve miRNA-mediated programming. PLoS One. 2012;7:e29441. doi: 10.1371/journal.pone.0029441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du H-Y, et al. Alterations of prefrontal cortical microRNAs in methamphetamine self-administering rats: From controlled drug intake to escalated drug intake. Neurosci Lett. 2016;611:21–27. doi: 10.1016/j.neulet.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 78.Bekdash RA, Harrison NL. Downregulation of Gabra4 expression during alcohol withdrawal is mediated by specific microRNAs in cultured mouse cortical neurons. Brain Behav. 2015;5:e00355. doi: 10.1002/brb3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olde Loohuis NFM, et al. MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Rep. 2015;11:1876–1884. doi: 10.1016/j.celrep.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 81.Hu Z, et al. miR-501-3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J Cell Biol. 2015;208:949–959. doi: 10.1083/jcb.201404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ho VM, et al. GluA2 mRNA distribution and regulation by miR-124 in hippocampal neurons. Mol Cell Neurosci. 2014;61:1–12. doi: 10.1016/j.mcn.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saba R, et al. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Araki Y, Lin D-T, Huganir RL. Plasma membrane insertion of the AMPA receptor GluA2 subunit is regulated by NSF binding and Q/R editing of the ion pore. Proc Natl Acad Sci USA. 2010;107:11080–11085. doi: 10.1073/pnas.1006584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Evers DM, et al. Plk2 attachment to NSF induces homeostatic removal of GluA2 during chronic overexcitation. Nat Neurosci. 2010;13:1199–1207. doi: 10.1038/nn.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daw MI, et al. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 88.Lin D-T, Huganir RL. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J Neurosci. 2007;27:13903–13908. doi: 10.1523/JNEUROSCI.1750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.