Significance
Primary cilia are antenna-like sensory organelles critical for many signal transduction pathways. One such pathway, called Hedgehog signaling, is essential for normal embryonic development and linked to tissue homeostasis and tumorigenesis. While the tightly regulated spatial and temporal localization of proteins in cilia controls activity, the mechanistic details are not fully understood. To characterize the movements of the Hedgehog receptor Patched1 in the cilium, we used single-molecule fluorescence microscopy, a tool that separates the behavior of individual proteins from the ensemble average, unveiling the underlying hidden physical behaviors. Our study reveals Hedgehog-induced changes in the motion of individual Patched1 molecules, which precede the exodus of Patched1 from cilia. These changes constitute one of the earliest measurable steps of Hedgehog-signal transduction.
Keywords: Hedgehog signaling, primary cilia, Patched, Smoothened, single-molecule tracking
Abstract
The Hedgehog-signaling pathway is an important target in cancer research and regenerative medicine; yet, on the cellular level, many steps are still poorly understood. Extensive studies of the bulk behavior of the key proteins in the pathway established that during signal transduction they dynamically localize in primary cilia, antenna-like solitary organelles present on most cells. The secreted Hedgehog ligand Sonic Hedgehog (SHH) binds to its receptor Patched1 (PTCH1) in primary cilia, causing its inactivation and delocalization from cilia. At the same time, the transmembrane protein Smoothened (SMO) is released of its inhibition by PTCH1 and accumulates in cilia. We used advanced, single molecule-based microscopy to investigate these processes in live cells. As previously observed for SMO, PTCH1 molecules in cilia predominantly move by diffusion and less frequently by directional transport, and spend a fraction of time confined. After treatment with SHH we observed two major changes in the motional dynamics of PTCH1 in cilia. First, PTCH1 molecules spend more time as confined, and less time freely diffusing. This result could be mimicked by a depletion of cholesterol from cells. Second, after treatment with SHH, but not after cholesterol depletion, the molecules that remain in the diffusive state showed a significant increase in the diffusion coefficient. Therefore, PTCH1 inactivation by SHH changes the diffusive motion of PTCH1, possibly by modifying the membrane microenvironment in which PTCH1 resides.
The Hedgehog (Hh)-signaling pathway is critically important in embryonic development and tissue regeneration (1, 2). Defects in the pathway often lead to birth defects and cancer, most commonly basal cell carcinoma and medulloblastoma (3). Pathway-specific inhibitors for treatment of those types of tumors have been approved for use in the clinic (4, 5); however, the frequent appearance of inhibitor resistance has necessitated further investigation into the underlying mechanisms of Hh-signal transduction (6).
The most frequently disrupted step of Hh-signal transduction in cancer is the interaction of two transmembrane proteins: Patched1 (PTCH1), a receptor for extracellular Hh ligands, and Smoothened (SMO), a homolog of G protein coupled receptors (7, 8). PTCH1 inhibits SMO by a still unknown mechanism that possibly involves a small-molecule intermediary (7–9). Several recent publications point to cholesterol as a candidate for an endogenous SMO ligand that would be regulated by PTCH1 (10–13). In addition to the recently described role as an endogenous ligand for SMO, cholesterol is the most abundant lipid in the plasma membrane that organizes membrane microdomains and affects the dynamics and interactions of membrane proteins (14, 15).
At the cellular level, PTCH1 and SMO are found to differentially accumulate in primary cilia: solitary, antenna-like organelles that project from the surface of most cells and are 2–10 μm long and ∼400 nm in diameter (16–18). When the pathway is off, PTCH1 in cilia inhibits SMO and prevents it from accumulating there; when the Hh-ligand Sonic Hedgehog (SHH) binds and inactivates PTCH1, PTCH1 is delocalized from cilia and subsequently degraded (19, 20). This dynamic, reciprocal localization of PTCH1 and SMO in cilia coincides with pathway activation, but the molecular details are still incomplete.
One way to address mechanistic questions about Hh pathway regulation by cilia is to understand the behavior of the pathway components at the single-molecule level in live cells (21–23). Single-molecule tracking and imaging in live cells have been successfully used to further our understanding of signal transduction processes, such as G protein coupled receptor signal transduction (24) and elucidating the protein organization within ciliary structures (25). We recently used single-molecule tracking to demonstrate agonist-induced changes in the movement of single SMO molecules in cilia during pathway activation (26). Here, we use single-molecule tracking and localization-based, superresolution imaging to show that PTCH1, like SMO, undergoes dramatic changes in diffusional dynamics triggered by Hh-pathway agonists, including cholesterol. By extending this approach to simultaneously image PTCH1 and SMO in the same cilium, we determine how these changes relate to SMO movements and localization.
Results
To visualize single PTCH1 molecules in cilia of live cells with high spatial and temporal resolution, we generated a fusion with a small protein-labeling tool, the ACP-tag, which can be rapidly and covalently labeled with a fluorescent small-molecule substrate (27). The ACP-tag was placed in the first extracellular loop of PTCH1, which was also tagged with YFP on its C terminus (19), as diagrammed in Fig. 1A. Full details of reagents and methods are provided in SI Appendix.
Fig. 1.
PTCH1-ACP-YFP protein retains Hh-pathway functional activity. (A) Diagram showing PTCH1-ACP-YFP fusion protein. An ACP-tag was added to the extracellular loop1 of the PTCH1 protein and the YFP-tag is at the C terminus. (B) The PTCH1-ACP-YFP fusion protein was functional in inhibiting the Hh pathway. Levels of Gli1 RNA were assayed by quantitative RT-PCR in Ptch−/− cells infected with an empty retrovirus (vector) only, or with PTCH1-ACP-YFP. (C) PTCH1-ACP-YFP is degraded in response to SHH. PTCH1-ACP-YFP–expressing cells were treated with SHH or with culture media only. Protein extracts from those cells were analyzed by standard immunoblots. PTCH1-ACP-YFP was detected with an anti-GFP antibody, and anti–γ-tubulin was used as a loading control. (D) PTCH1-ACP-YFP is responsive to SHH, as measured by Gli1 target gene expression in cells treated with SHH. Gli1 RNA was measured by quantitative RT-PCR. (E) PTCH1-ACP-YFP localized in cilia of Ptch−/− cells. The protein was detected with an anti-GFP antibody, and cilia were marked with anti-acetylated tubulin antibody. (Scale bar: 1 μm.) (F) PTCH1-ACP-YFP delocalized from cilia in response to SHH. After 24 h of SHH treatment, levels of PTCH1-ACP-YFP were quantified by scanning confocal microscopy after immunostaining. Data are mean ± SEM (*P < 0.05, ***P < 0.001).
The functionality of the PTCH1-ACP-YFP fusion protein was tested in mouse embryonic fibroblast cells (MEFs) lacking endogenous PTCH1 (Ptch1−/− cells), both in a mixed population of cells (Fig. 1B) and in isolated single-cell clones (Fig. 1 C–F). Cells expressing the PTCH1-ACP-YFP construct were able to fully rescue the Ptch1 mutation and block the transcription of the Hh-target gene Gli1, which is otherwise increased in Ptch1−/− cells (Fig. 1B). Treatment with SHH is known to induce degradation of PTCH1 protein (28), and this was also true for the PTCH1-ACP-YFP fusion protein. Immunoblot analysis showed a decrease in PTCH1-ACP-YFP protein levels in lysates from cells treated with SHH, compared with untreated controls (Fig. 1C). Additionally, we found a dose-dependent increase in Gli1 RNA levels in SHH-treated cells, demonstrating the responsiveness to SHH in this cell line (Fig. 1D). We further confirmed that PTCH1-ACP-YFP localized to cilia of cells cultured in control conditions (Fig. 1E) and delocalized from cilia after SHH treatment (representative cilia from scanning confocal images of immunostained cells are shown in SI Appendix, Fig. S1 A and B and average levels of ciliary PTCH1-ACP-YFP are shown in Fig. 1F). Taken together, these results indicate that the PTCH1-ACP-YFP construct retained functional activity. The labeling with fluorescent-ACP substrates was only detected in cells expressing the PTCH1-ACP-YFP construct (SI Appendix, Fig. S1C). Both fluorescent labels overlapped in cilia (SI Appendix, Fig. S1C).
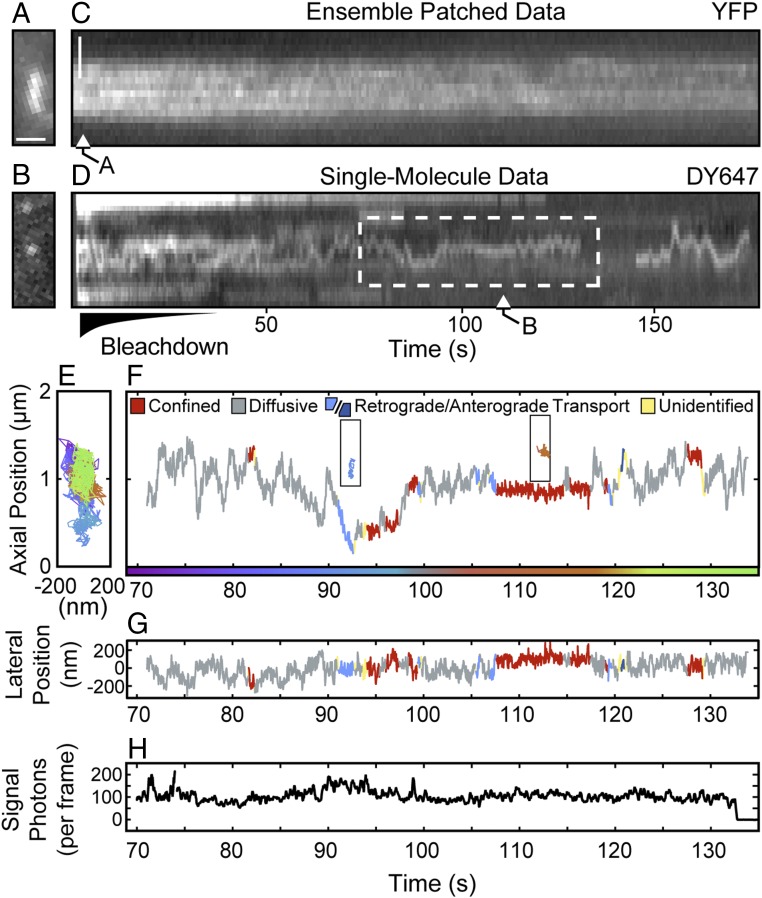
Cells expressing PTCH1-ACP-YFP were imaged by single-molecule localization microscopy using a custom microscope system capable of simultaneously recording two color channels (SI Appendix, Figs. S2 and S3). In those cells, cilia were detected based on the YFP signal derived from the ciliary localization of PTCH1-ACP-YFP (Fig. 2A). Labeling with the ACP-substrate, CoA-DY647 (a derivative of the Dyomics dye DY-647P1), was optimized to obtain low densities of DY647-labeled PTCH1-ACP-YFP molecules in the cilium, suitable for single-molecule tracking after minor photobleaching (Fig. 2B). We recorded movies of individual cilia, for several minutes at a time, profiting from the high photostability of the small-molecule fluorophore label (Movies S1–S3). Rapid alternation between green and red excitation sources (for the YFP and DY647 probes) using acousto-optic modulators (AOMs) enabled near-simultaneous recordings of the two color channels while maintaining low background autofluorescence in the DY647 channel. The sample was exposed to excitation light for 16 ms per color, separated by 4 ms with no illumination during switching, completing each 40-ms cycle. In this way, the contextual information on the distribution of the ensemble of PTCH1 molecules within the cilium was collected from the YFP channel (example kymogram in Fig. 2C), and single-molecule trajectories were collected from the DY647 channel (Fig. 2 B and D shown as a kymogram). The recorded single-molecule trajectories of PTCH1-ACP-YFP often traversed the entire cilium and occasionally lasted longer than a minute (Movies S1–S3). Consistent with the low labeling density, we mostly detected uniform emission brightness for tracked molecules, and single-step bleaching, as expected for single fluorescent molecules (Fig. 2H). However, in several cases out of hundreds, we observed two discrete photon levels in a moving molecule (Movie S2), indicative of in vivo existence of at least a dimer or possibly multimer, consistent with the published biochemical evidence that PTCH1 molecules oligomerize (8, 29).
Fig. 2.
Characterization of PTCH1 motional dynamics in cilia. Representative kymograms and trajectories of PTCH1-ACP-YFP in cilia show three modes of motion: diffusion, confinement, and directional transport. Using two-color imaging, we observe the movement of both the bulk populations of PTCH1 molecules (YFP, in A and C) and single-molecule trajectories (DY647, in B and D). (A) Fluorescent image of YFP in the cilium. Cilia were identified based on the accumulation of YFP. (B) Image of an isolated DY647-labeled PTCH1-ACP-YFP molecule in the cilium shown in A. (C) Kymogram of the YFP channel for the cilium shown in A. (D) Kymogram of the red imaging channel of the same region shown in C. (E) Two-dimensional single-particle trajectory extracted from the period marked with a dashed line in D, color coded for time. (F and G) The same trajectory shown as the long axis (F) or short axis (G) position in time, color coded for the classified movement type. Boxed Insets show the 2D trajectory during an identified period of retrograde transport and confinement, respectively. (H) The signal photons maintain steady levels indicative of a single fluorophore followed by a single-photobleaching event. For clarity, the trace shows the average number of photons from a three-frame window. (Scale bar: 1 μm.)
Two-dimensional single-molecule trajectories obtained from the movies were oriented relative to coordinates defined by the extent of the cilium, as determined by YFP imaging over time; a representative single-molecule trajectory from DY647, color coded for time, is shown in Fig. 2E. The high spatial precision of our single-molecule imaging, ∼50 nm, allowed us to determine both the axial (Fig. 2F) and lateral positions (Fig. 2G) of molecules in each frame (40 ms). This information was analyzed using a statistical algorithm that classified subtrajectories of five or more frames into three modes of motion: confinement, diffusion, or active/directional transport. Briefly, confinement is when the molecule remains at the same position, to the limit of the localization precision; diffusion is when the observed movement is consistent with the random motion of a membrane protein; and active/directional transport is when the trajectory proceeds linearly with a stable velocity (SI Appendix). These types of movement are represented in red, gray, and blue regions in the trajectories of Fig. 2 F and G, respectively. A small fraction of trajectories remained as unclassified and are shown as yellow lines in Fig. 2 F and G. We tested the accuracy of our classification algorithm by analyzing simulated diffusion, binding, and directed motion in confined structures (SI Appendix, Fig. S4).
The predominant mode of motion of PTCH1 in cilia was diffusion, similar to what we previously observed for SMO (26). The diffusion coefficient for periods of clearly diffusive movement was calculated to be 0.1 ± 0.01 μm2/s (mean ± SEM). The analysis of PTCH1 trajectories also revealed intervals of either anterograde (dark blue), or retrograde (light blue) movement, during which molecules moved along the long axis of the cilium without any changes in the lateral position, as if they were directionally moving along the axoneme (Movie S3). Those intervals were usually short and encompassed only a portion of the length of the cilium. The average velocities for the directional transport were 0.35 ± 0.05 µm/s for anterograde and 0.31 ± 0.02 µm/s for retrograde, within the range of previously reported values for other transported proteins in cilia (23, 30–34) and in agreement with kymogram measurements of PTCH1-ACP-YFP clusters and IFT88-YFP (SI Appendix, Fig. S5). Since periods of directional transport were transient and relatively rare events in our experiments, further studies will be necessary to determine whether PTCH1 is a true cargo for active, motor-driven transport in cilia and whether this transport changes for different states of pathway activation.
Both SHH and Cholesterol Depletion Induce an Increase in Confinement of Single PTCH1 Molecules in Cilia.
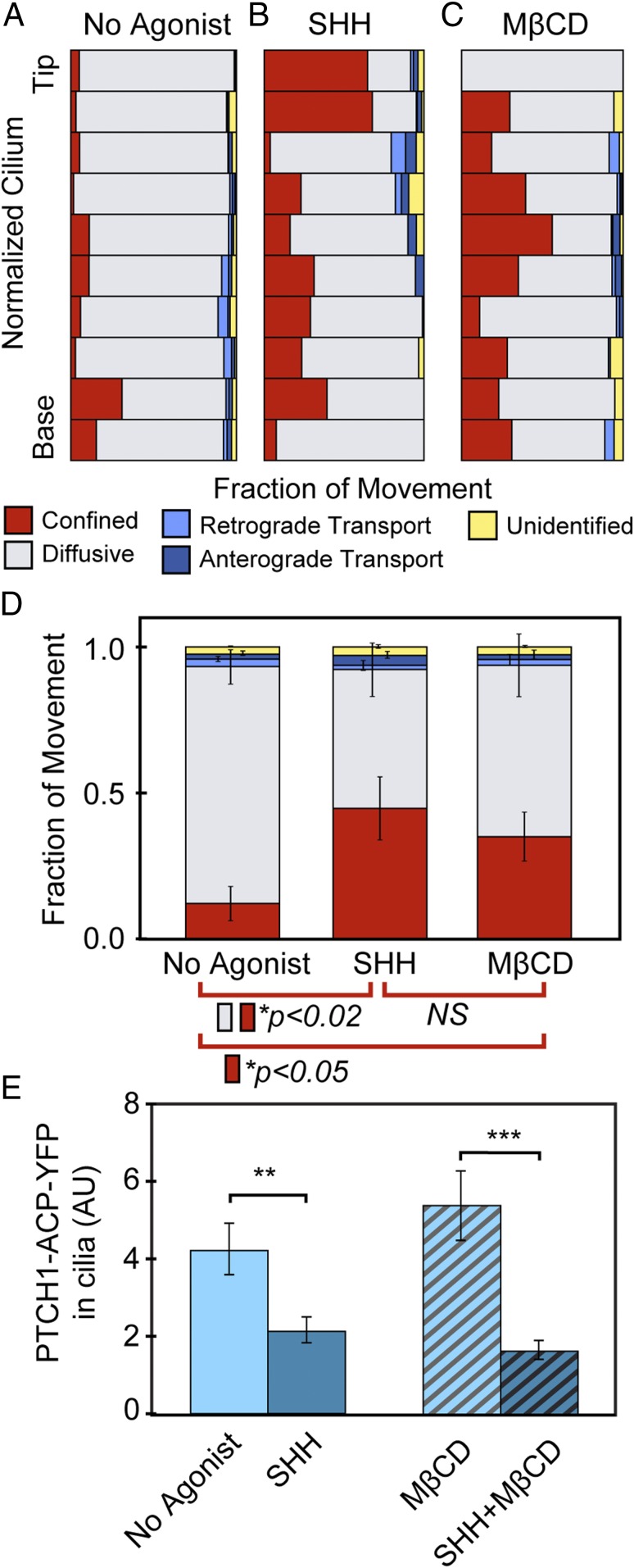
Recorded trajectories show that the mostly diffusional movement of PTCH1 molecules in cilia was frequently interrupted with periods of confinement. Confinement of molecules, defined as motion over five or more frames not exceeding the statistical spatial precision of the measurement (SI Appendix), was observed along the entire cilium length. To analyze data for multiple cilia, we plotted the motion on a normalized cilium long axis, separated into 10 spatial bins, and determined the fraction of different types of motion in each bin (Fig. 3A). When analyzed for the entire length of the cilium, PTCH1 movement in the unstimulated condition consisted of 81% diffusion, 4% directed motion, and 12% confinement; 2% of trajectories were unclassified by our analysis (Fig. 3D).
Fig. 3.
PTCH1 diffusive movement in cilia is reduced in response to SHH and to cholesterol depletion. Single-molecule trajectories were divided into subtrajectories based on the likely mode of motion. (A–C) For each experimental condition, data from multiple cilia were combined by their position along the long axis of a normalized cilium. The relative fraction of each motion type (confined, diffusive, or directed motion) of the total time in each bin was calculated. (D) Pooled motion-type data from all recorded tracks of single PTCH1 molecules in cilia show that both SHH and MβCD treatment induce a significant increase in fraction of time spent in confinement, and a decrease in the fraction of time spent diffusing. No agonist, 85 trajectories from 41 cells; SHH treatment, 86 molecules from 35 cells; MβCD treatment, 17 molecules from 10 cells [not significant (NS), P > 0.05]. (E) Cholesterol depletion does not affect SHH-induced PTCH1 delocalization from cilia. PTCH1-ACP-YFP cells were treated for 4 h with SHH, MβCD, or both, fixed and immunostained, and imaged by confocal microscopy. Images were quantified for cilia localization of PTCH1 (mean ± SEM; **P < 0.01, ***P < 0.001). Treatment with SHH caused delocalization from cilia regardless of whether cells were treated with MβCD or not.
SHH is known to induce removal of PTCH1 from cilia when observed at the bulk protein level (19), but its effect on the dynamics of individual PTCH1 molecules is not known. To address this question PTCH1-ACP-YFP cells were first labeled with the ACP-DY647 substrate and then treated with a saturating concentration of SHH (300 nM), for up to 2 h. During this period, PTCH1 was still present in cilia at levels sufficient for identification and tracking, despite the gradual delocalization from cilia induced by SHH. Treatment with SHH induced a substantial decrease in the fraction of time molecules spent diffusing, to 48% of total recorded time, and an increase in the fraction of time in confinement, to 45% of the time; confinement was especially prominent at the tip of the cilium (Fig. 3 B and D). Directional movement, either anterograde or retrograde, did not substantially change, suggesting that the observed increase in confinement was at the expense of diffusive motion (Fig. 3 B and D).
The diffusive motion of proteins in the membrane could be affected by the cholesterol content in the membrane (35–37). We sought to determine how manipulation of cholesterol levels in cells would affect the motional dynamics of PTCH1 in cilia. Cholesterol was acutely depleted from cells by treatment with methyl-β-cyclodextrin (MβCD) which removes cholesterol and other less abundant sterols from membranes (38–40). The extent of depletion was assessed with the fluorescent probe EGFP-D4H (the domain 4 fragment of bacterial toxin perfingolysin-O fused to GFP) (41, 42). To optimize the conditions for imaging proteins in cilia of cholesterol-depleted cells, SMO agonist (SAG)-treated SNAP-SMO–expressing cells were first treated with MβCD, then fixed and stained with EGFP-D4H (SI Appendix, Fig. S6A). Cilia were marked by antibody staining for SNAP-SMO (SI Appendix, Fig. S6B). The staining with EGFP-D4H in cilia was similar to that in the adjacent membrane (SI Appendix, Fig. S6C). We found that 30 min of treatment with 2 mM MβCD reduced sterol levels in both the ciliary and plasma membranes with no apparent changes in cell morphology (SI Appendix, Fig. S6). Based on this result, live-cell imaging was initiated after 30 min of treatment with MβCD and typically continued for about 1 h.
Depletion of cholesterol from cells had a significant effect on the movement of PTCH1 molecules in cilia. We observed a significant increase in confinement at the expense of diffusion (Fig. 3 C and D): confinement increased to 35% of trajectory time while diffusion decreased to 59% (Fig. 3D), and no increased confinement occurred at the tip. Therefore, cholesterol depletion, similar to SHH treatment, reduces the fraction of time PTCH1 molecules spend freely diffusing in the cilium. Although the effects on single-molecule movements within short timescales were similar between the two treatments, the effects on bulk protein levels in cilia over longer timescales were different. Whereas SHH treatment led to delocalization of PTCH1 from cilia, no change in total PTCH1 levels in cilia was found after 4 h of MβCD treatment (Fig. 3E). Cholesterol depletion did not affect SHH-induced delocalization of PTCH1 from cilia (Fig. 3E). The result is consistent with the idea that only SHH-inactivated PTCH1 is removed from cilia, and that cholesterol depletion does not inactivate PTCH1.
Effect of Cholesterol Depletion on SMO in Cilia.
We hypothesized that cholesterol depletion from cells would also affect the motional dynamics of single SMO molecules in cilia. To test this, we used a cell line in which the Hh pathway is constitutively active due to loss of PTCH1, thus SMO is found at elevated levels in the cilium (Ptch1−/−; SNAP-Smo; PACT-YFP cells). The cells express SNAP-SMO to enable visualization of SMO using an extracellular label, and PACT-YFP to visualize the base of the cilium (26). In agreement with previous publications (11, 12), the addition of 2 mM MβCD to cells resulted in gradual pathway inactivation. Both the bulk SMO protein levels in cilia (Fig. 4 A and B), and Gli1 transcription (Fig. 4C) were reduced by half within 2 h of incubation with MβCD. Based on these results, we established a timeframe for the single-molecule tracking experiments: cells were labeled with the fluorescent SNAP substrate, MβCD was then added, and cells were imaged for the next 30–60 min. During that window of time, SMO was still present in cilia at a level sufficient for recording many single-molecule tracks, which were classified as described above.
Fig. 4.
Cholesterol depletion affects SMO in cilia of Ptch1−/− cells, and of SHH treated cells, but not SAG-treated cells. (A) Cells lacking PTCH1 accumulate SMO in cilia (image on the Left); however, 2-h treatment with MβCD reduces the levels (image on the Right). Fixed cells were stained with anti-SMO and anti-ARL13b to detect cilia and imaged by confocal microscopy. (B) Time-dependent delocalization of SMO from cilia in Ptch1−/− cells after cholesterol depletion [mean ± SEM; not significant (NS), P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001]. (C) Reduction of Gli1 expression after MβCD treatment, quantified by RT-PCR (mean ± SEM). (D) SMO trajectory-motion types in Ptch1−/− cells expressing SNAP-SMO and labeled with Alexa647 fluorescent substrate. Cells were imaged either at baseline, media-only condition, or after 30–90 min of 2-mM MβCD treatment. Trajectories were pooled and organized in bins along the long axis of the cilium. (E) MβCD treatment reduced the bulk SMO protein levels in cilia of Ptch1−/− cells, but did not significantly change the SAG-induced accumulation of SNAP-SMO in cilia. SANT-1 blocked the accumulation of SNAP-SMO in cilia regardless of MβCD treatment. (F) Trajectories of SMO show no increase in binding at the base of cilia after cholesterol depletion in SAG-treated SNAP-SMO; PTCH-YFP cells. (G) When SAG is replaced with SHH, there is a significant increase in SMO binding at the base in cholesterol-depleted cells.
In Ptch1−/− cells not treated with pathway agonists or antagonists, SMO trajectories showed almost entirely diffusive movement (Fig. 4D), consistent with our previous results (26). Treatment with MβCD resulted in a significant increase in SMO confinement at the cilium base (P < 0.01, Fig. 4D), the pattern of motion we previously observed for inactive SMO (26). The exact localization of these confinements relative to ciliary compartments, such as the transition zone and ciliary pocket, was not determined in our experiments, but possibly coincides with the transition zone as recently suggested (25). The observed change in the motional dynamics of SMO in cilia of MβCD-treated Ptch1−/− cells is consistent with SMO inactivation. Based on this result, we propose that after cholesterol depletion from Ptch1−/− cells, SMO molecules are inactivated before exiting cilia.
Treatment of cells with SAG fully restored ciliary accumulation of SMO in MβCD-treated Ptch1−/− cells, while the SMO antagonist SANT-1 completely blocked it, regardless of cholesterol levels (Fig. 4E). Consistent with previously published data, when observed at bulk protein levels, SAG-induced SMO accumulation in cilia and Hh-pathway activation are independent of cholesterol (12). Therefore, the effect of cholesterol depletion on SMO accumulation in cilia is dependent on whether SMO was activated by SHH or by SAG.
To further analyze the effect of cholesterol depletion on SMO dynamics in cilia in the presence of SAG or SHH, and to be able to analyze SMO and PTCH1 at the same time, we constructed cells that express both SNAP-SMO and PTCH1-YFP. Considering SMO first, cells were treated with SAG to induce SMO localization to cilia, labeled with SNAP substrate, and subsequently treated either with SAG, or with SHH, in the presence or absence of 2 mM MβCD. SMO modes of motion in cilia of SAG-treated cells were not significantly affected by MβCD treatment (Fig. 4F). Therefore, when SMO is locked in an active conformation by SAG, cholesterol depletion does not affect SMO dynamics in cilia. After replacing SAG with SHH and after adding MβCD, increased SMO confinement at the cilium base occurred, likely due to a decrease in diffusion (Fig. 4G), consistent with the inactivated SMO motional dynamics (Fig. 4D). We conclude that SMO mobility in cilia is affected by cholesterol depletion only if SMO was activated by inhibiting PTCH1 (i.e., by SHH stimulation or by deletion of Ptch1). Taken together with our analysis of PTCH1 movements, these results suggest that both PTCH1 and SMO alternate between free diffusion and confinement in cilia, and that the fraction of time they spent freely diffusing could be affected by ciliary membrane cholesterol content.
SMO and PTCH1 Segregate to Distinct Domains in the Ciliary Membrane.
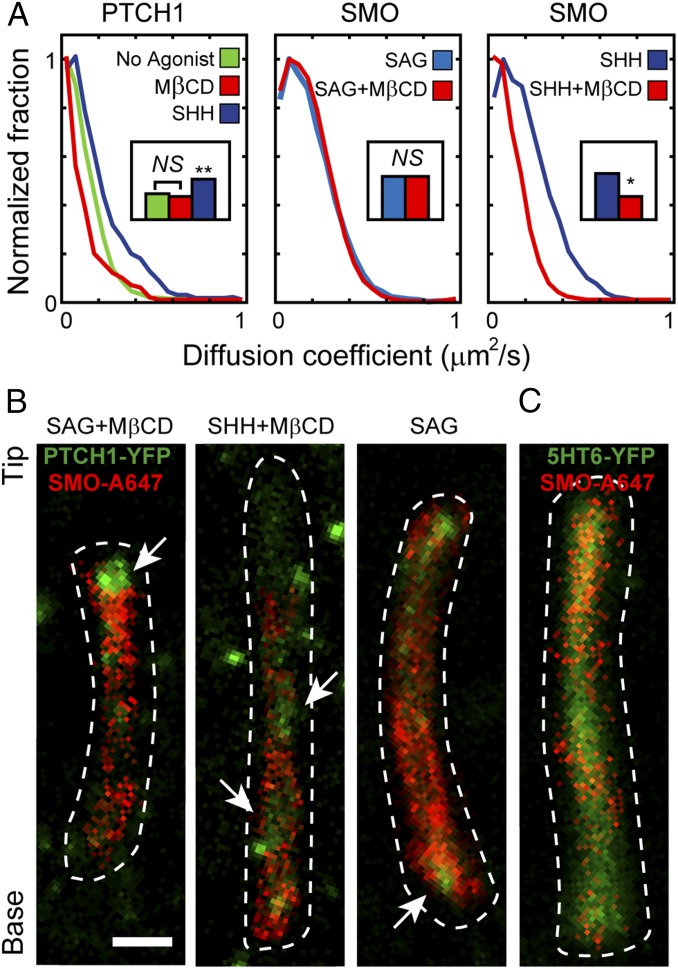
To further characterize and compare the diffusive movements of PTCH1 and SMO in cilia, we calculated the diffusion coefficients for all trajectories determined to be diffusive (distribution curves shown in Fig. 5A). Treatment with SHH induced a significant shift to larger values in the diffusion coefficient for PTCH1 (Fig. 5A, Left). The average value increased from 0.1 ± 0.01 µm2/s at baseline conditions to 0.16 ± 0.01 µm2/s in SHH-treated cells. Therefore, SHH not only reduced the fraction of time PTCH1 molecules spend freely diffusing in the ciliary membrane (Fig. 3), but also increased the diffusion coefficient of the molecules that remain in a diffusive state. Unlike SHH, the depletion of cholesterol from cells did not significantly change the diffusion coefficient for PTCH1 in cilia (Fig. 5A, Left). This result raises the possibility that the observed SHH-induced change in the diffusion coefficient for PTCH1 is a consequence of PTCH1 inactivation by SHH.
Fig. 5.
PTCH1 and SMO are dynamically segregated in the ciliary membrane. (A) Diffusion coefficients for PTCH1 and SMO in cilia are affected by SHH. The diffusion coefficients for PTCH1 and SMO in cilia were determined for clearly diffusing (DY/Alexa) 647-labeled molecules under different experimental conditions, as marked. Graphs show the distribution of measured diffusion coefficients from short subtrajectories determined by mean squared displacement analysis. Insets show the mean diffusion coefficients [not significant (NS), P > 0.05, *P < 0.05, **P < 0.01]. (B and C) Image reconstructions of all SMO-Alexa647 positions in trajectory data (red), superlocalized positions of PTCH1-YFP (green in B), and 5HT6-YFP as a control (green in C). Dotted line outlines cilia. (Scale bar: 500 nm.) Arrows highlight anticorrelation of localizations.
A similar analysis of the diffusion coefficients for SMO showed that the effect of cholesterol depletion was dependent on whether SMO was activated by SHH or by SAG (Fig. 5A, Middle and Right), similar to what we observed for the effect on SMO types of motion (Fig. 4 F and G). The diffusion coefficient for SAG-activated SMO did not change when cells were depleted of cholesterol (Fig. 5A, Middle). Contrary to that, cholesterol depletion significantly reduced the diffusion coefficient of SHH-activated SMO (Fig. 5A, Right).
We hypothesized that the observed differences in diffusive movements of PTCH1 and SMO molecules in cilia reflect their residence in distinct membrane domains. We used the dual-labeled cells to analyze their relative localizations. Many protein positions for PTCH1-YFP over time (43) were determined by making use of the photophysical characteristic of YFP to spontaneously blink on after remaining in long-lived dark states under high illumination intensities (1–5 kW/cm2). This superlocalization approach is analogous to the single-particle tracking photoactivation localization microscopy (spt-PALM) method (22) which records many positions of moving single molecules. We simultaneously captured the trajectories of single SMO-Alexa647 molecules using an interlaced-excitation imaging setup (SI Appendix). The locations found for the blinking PTCH1-YFP molecules are shown along with the many positions observed for the single SMO molecules (Fig. 5B and Movie S4). SMO molecules were rarely seen to enter regions of the cilium with high densities of PTCH1 protein. This anticorrelated behavior was observed in all experimental conditions, although it was most easily observed under cholesterol depletion, perhaps because of the reduced diffusion of PTCH1 (SI Appendix, Fig. S7). As a control, we tracked SMO-Alexa647 in cells transiently transfected with the transmembrane GPCR 5HT6-YFP (Fig. 5C). These two molecules localized independently, and, unlike PTCH1, 5HT6-YFP homogenously distributed in the ciliary membrane (Fig. 5C). We therefore conclude that PTCH1 and SMO can dynamically segregate in distinct domains of the ciliary membrane, possibly related to a different lipid composition or accessibility.
Discussion
Using single-molecule tracking and superlocalization microscopy, we find quantifiable changes in the motional dynamics of single PTCH1 and SMO molecules that may represent some of the earliest measurable events in Hh-signal transduction in cilia. This approach relied on the excellent signal-to-noise ratio achieved by attaching small organic fluorescent probes to the respective proteins compared with fluorescent protein labels. The subsequent time and spatial resolution (∼50 nm, 40 ms per frame) then enabled the parsing of long trajectories of hundreds to thousands of positions by motion type using statistical models for movement.
The predominant mode of motion of PTCH1 is diffusion throughout the entire length of the cilium. This diffusional movement was interspersed with short and transient periods of directional movement and by confinement at different positions along the ciliary axis, like that previously reported for SMO and other transmembrane proteins in primary cilia (23, 26, 44). Upon SHH treatment, the dynamical motion of single PTCH1 molecules undergoes complex changes: an increase in the diffusion coefficient for freely moving molecules, and at the same time an increase in confinement at the expense of free diffusion. A similar decrease in the fraction of time PTCH1 molecules spend diffusing, but not in the diffusion coefficient for freely diffusing molecules, was observed after depletion of cholesterol from cells. Therefore, PTCH1 inactivation by SHH coincides with an increase in its diffusion coefficient.
Both SHH treatment and cholesterol depletion shifted PTCH1 dynamics toward confinement. Changes in cholesterol levels are known to affect the movement of proteins in cholesterol-rich membrane domains (35, 36, 45). PTCH1 has been previously found to be associated with cholesterol-enriched domains often termed lipid rafts, together with Caveolin-1 (28, 46). Our observation that PTCH1 was often found clustered in cilia, and that entire clusters could be seen moving along cilia (SI Appendix, Fig. S5 and Movie S5), raises the possibility that these PTCH1 clusters reside in cholesterol-rich domains in the membrane of cilia. Studies in diverse model organisms indicate that such lipid domains, and cholesterol itself, seem to be enriched in cilia (47–50). In tandem with our results, this raises the possibility that in cilia, PTCH1 preferentially resides in cholesterol-rich domains. Although we were able to detect cholesterol in cilia by using the EGFP-D4H probe and standard confocal microscopy, the available reagents were not suitable for simultaneous superresolution imaging with PTCH1 or SMO. Future studies will be necessary to elucidate the relative localizations of PTCH1, SMO, and membrane domains in cilia.
The unique membrane composition of cilia, enriched in cholesterol, may provide a favorable environment for the efficient regulation of Hh-signal transduction in vertebrate cells. We find that SMO, activated by SHH or by the loss of PTCH1, after additional treatment with MβCD, undergoes changes in motional dynamics in cilia consistent with inactivation. While commonly used to solubilize and deplete cholesterol from membranes, MβCD is known to deplete other, less abundant lipids as well (39, 40). Our observations are consistent with the hypothesis that SMO activation by SHH acts through an intermediary in cilia, possibly cholesterol (10–13, 51) or an endogenous oxysterol (52).
The model for organization of cell membranes, including ciliary membrane, into lateral microdomains and signaling hot spots, and cholesterol-mediated nanodomains, has been supported by many imaging-based observations (24, 44, 53, 54). In addition, membrane proteins are thought to preferentially associate with membrane domains of differing lipid composition to form signaling platforms (14, 45). We find that PTCH1 and SMO dynamically segregate from each other in the cilium membrane, in agreement with what was previously observed by structured illumination microscopy of fixed cells (55). Our observation of PTCH1 and SMO movements in cilia being differently affected by cholesterol depletion is consistent with the hypothesis that they reside in distinct membrane domains with different cholesterol composition or accessibility.
Materials and Methods
To make the construct encoding PTCH1-ACP-YFP, the sequence for ACP-tag (NEB) was inserted into the unique AvrII restriction site of the previously described PTCH1-YFP (19). Newly generated stable cell lines expressing tagged proteins were made using murine stem cell virus-based retroviral vectors. Cells were labeled with SNAP or ACP substrates according to manufacturer’s recommendations (NEB). Single-molecule imaging experiments were performed using an inverted microscope body (IX71, Olympus), outfitted with a motorized stage (M26821LOJ, Physik Instrumente) and a stage-top incubator at 37 °C (INUB-PI-F1, Tokai Hit). Molecular trajectories were analyzed using custom MATLAB scripts described previously (26). In brief, the position of single-molecule emitters was found in each frame by fitting a 2D symmetric Gaussian function (which approximates well the point spread function of the microscope) with a constant offset to a 11 × 11 pixel2 region of the image using the nonlinear least squares fit algorithm lsqnonlin. The algorithm for trajectory analysis classified various periods of motion within a trajectory as confined (remaining static within the precision of the measurement), directed (traveling with a continuous and stable velocity), or diffusive (motion characterized by Brownian diffusion). The parameters used in this analysis were optimized by simulating trajectories in a cylindrical object, to quantitate the relative errors in classification, and by reviewing the output on real data and ensuring rough agreement with manual classification. Measurements were then binned based on their axial position, and the fraction of each type of motion prediction was calculated. Statistical comparisons between the relative abundance of movement types between conditions used all data measured in cilia.
A detailed description of the reagents and methods used in the paper is provided in SI Appendix. All figure data are available through the Stanford Digital Repository, https://purl.stanford.edu/sh175by4999.
Supplementary Material
Acknowledgments
We thank Matthew Scott for early advice, reagents, and support on this project; Rajat Rohatgi and Giovanni Luchetti for reagents and advice; and Takanari Inoue for reagents. This work was supported by grants from the National Institutes of Health (R35GM118067 to W.E.M. and R01GM121424 to T.S.) and by a Stanford Bio-X Interdisciplinary Initiatives Seed Grant (to W.E.M.). L.E.W. was supported by a Bio-X Stanford Interdisciplinary Graduate Fellowship and J.Y. by a National Science Foundation Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: All figure data are available from the Stanford Digital Repository, https://purl.stanford.edu/sh175by4999.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816747116/-/DCSupplemental.
References
- 1.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 2.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 3.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gould SE, et al. Discovery and preclinical development of vismodegib. Expert Opin Drug Discov. 2014;9:969–984. doi: 10.1517/17460441.2014.920816. [DOI] [PubMed] [Google Scholar]
- 5.Pak E, Segal RA. Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 8.Petrov K, Wierbowski BM, Salic A. Sending and receiving hedgehog signals. Annu Rev Cell Dev Biol. 2017;33:145–168. doi: 10.1146/annurev-cellbio-100616-060847. [DOI] [PubMed] [Google Scholar]
- 9.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 10.Byrne EFX, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535:517–522. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luchetti G, et al. Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife. 2016;5:e20304. doi: 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang P, et al. Cellular cholesterol directly activates smoothened in hedgehog signaling. Cell. 2016;166:1176–1187.e14. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci USA. 2017;114:E11141–E11150. doi: 10.1073/pnas.1717891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levental I, Veatch S. The continuing mystery of lipid rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntosh TJ. Overview of membrane rafts. Methods Mol Biol. 2007;398:1–7. doi: 10.1007/978-1-59745-513-8_1. [DOI] [PubMed] [Google Scholar]
- 16.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia G, 3rd, Raleigh DR, Reiter JF. How the ciliary membrane is organized inside-out to communicate outside-in. Curr Biol. 2018;28:R421–R434. doi: 10.1016/j.cub.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 20.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 21.Jacquier V, Prummer M, Segura JM, Pick H, Vogel H. Visualizing odorant receptor trafficking in living cells down to the single-molecule level. Proc Natl Acad Sci USA. 2006;103:14325–14330. doi: 10.1073/pnas.0603942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 23.Ye F, et al. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. eLife. 2013;2:e00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sungkaworn T, et al. Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature. 2017;550:543–547. doi: 10.1038/nature24264. [DOI] [PubMed] [Google Scholar]
- 25.Shi X, et al. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol. 2017;19:1178–1188. doi: 10.1038/ncb3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milenkovic L, et al. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc Natl Acad Sci USA. 2015;112:8320–8325. doi: 10.1073/pnas.1510094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George N, Pick H, Vogel H, Johnsson N, Johnsson K. Specific labeling of cell surface proteins with chemically diverse compounds. J Am Chem Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 28.Yue S, et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. eLife. 2014;3:e02555. doi: 10.7554/eLife.02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Liu S, Kornberg TB. The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes Dev. 2006;20:2539–2551. doi: 10.1101/gad.1461306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa H, Marshall WF. Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol. 2017;9:a021998. doi: 10.1101/cshperspect.a021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besschetnova TY, Roy B, Shah JV. Methods in Cell Biology. Elsevier; Amsterdam: 2009. pp. 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams CL, et al. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun. 2014;5:5813. doi: 10.1038/ncomms6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald F, Prevo B, Acar S, Peterman EJG. Interplay between ciliary ultrastructure and IFT-train dynamics revealed by single-molecule super-resolution imaging. Cell Rep. 2018;25:224–235. doi: 10.1016/j.celrep.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Vrljic M, Nishimura SY, Moerner WE, McConnell HM. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys J. 2005;88:334–347. doi: 10.1529/biophysj.104.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura SY, Vrljic M, Klein LO, McConnell HM, Moerner WE. Cholesterol depletion induces solid-like regions in the plasma membrane. Biophys J. 2006;90:927–938. doi: 10.1529/biophysj.105.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baier CJ, Gallegos CE, Levi V, Barrantes FJ. Cholesterol modulation of nicotinic acetylcholine receptor surface mobility. Eur Biophys J. 2010;39:213–227. doi: 10.1007/s00249-009-0521-2. [DOI] [PubMed] [Google Scholar]
- 38.Radhakrishnan A, McConnell HM. Chemical activity of cholesterol in membranes. Biochemistry. 2000;39:8119–8124. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- 39.Mahammad S, Parmryd I. Cholesterol depletion using methyl-β-cyclodextrin. Methods Mol Biol. 2015;1232:91–102. doi: 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 40.Ohvo H, Slotte JP. Cyclodextrin-mediated removal of sterols from monolayers: Effects of sterol structure and phospholipids on desorption rate. Biochemistry. 1996;35:8018–8024. doi: 10.1021/bi9528816. [DOI] [PubMed] [Google Scholar]
- 41.Maekawa M. Domain 4 (D4) of perfringolysin O to visualize cholesterol in cellular membranes—The update. Sensors (Basel) 2017;17:504. doi: 10.3390/s17030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3:e02882. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biteen JS, et al. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Tan HY, Geneva II, Kruglov A, Calvert PD. Actin filaments partition primary cilia membranes into distinct fluid corrals. J Cell Biol. 2018;217:2831–2849. doi: 10.1083/jcb.201711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 46.Karpen HE, et al. The sonic hedgehog receptor patched associates with caveolin-1 in cholesterol-rich microdomains of the plasma membrane. J Biol Chem. 2001;276:19503–19511. doi: 10.1074/jbc.M010832200. [DOI] [PubMed] [Google Scholar]
- 47.Tyler KM, et al. Flagellar membrane localization via association with lipid rafts. J Cell Sci. 2009;122:859–866. doi: 10.1242/jcs.037721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma AI, et al. Sterol targeting drugs reveal life cycle stage-specific differences in trypanosome lipid rafts. Sci Rep. 2017;7:9105. doi: 10.1038/s41598-017-08770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma AI, Olson CL, Engman DM. The lipid raft proteome of african trypanosomes contains many flagellar proteins. Pathogens. 2017;6:39. doi: 10.3390/pathogens6030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ott C, Lippincott-Schwartz J. Visualization of live primary cilia dynamics using fluorescence microscopy. Curr Protoc Cell Biol. 2012;57:4.26.1–4.26.22. doi: 10.1002/0471143030.cb0426s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang P, et al. Structural basis of smoothened activation in hedgehog signaling. Cell. 2018;174:312–324.e16. doi: 10.1016/j.cell.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raleigh DR, et al. Cilia-associated oxysterols activate smoothened. Mol Cell. 2018;72:316–327.e5. doi: 10.1016/j.molcel.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 54.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 55.Kim J, et al. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal. 2015;8:ra55. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.