Abstract
Human CD8+ cytotoxic T lymphocytes (CTLs) contribute to antimicrobial defense against intracellular pathogens through secretion of cytotoxic granule proteins granzyme B, perforin, and granulysin. However, CTLs are heterogeneous in the expression of these proteins, and the subset(s) responsible for antimicrobial activity is unclear. Studying human leprosy, we found that the subset of CTLs coexpressing all three cytotoxic molecules is increased in the resistant form of the disease, can be expanded by interleukin-15 (IL-15), and is differentiated from naïve CD8+ T cells by Langerhans cells. RNA sequencing analysis identified that these CTLs express a gene signature that includes an array of surface receptors typically expressed by natural killer (NK) cells. We determined that CD8+ CTLs expressing granzyme B, perforin, and granulysin, as well as the activating NK receptor NKG2C, represent a population of “antimicrobial CTLs” (amCTLs) capable of T cell receptor (TCR)–dependent and TCR-independent release of cytotoxic granule proteins that mediate antimicrobial activity.
INTRODUCTION
CD8+ cytotoxic T lymphocytes (CTLs) are known to contribute to host defense against intracellular pathogens through production of interferon-γ (IFN-γ) and by killing of infected target cells. In animal studies, both conventional and nonconventional T cells appear to contribute to protection against Mycobacterium tuberculosis (1). Human CD8+ T cells have been shown not only to lyse macrophages infected with intracellular mycobacteria (2) but also to have the capacity to exert antimicrobial activity independent of their ability to secrete IFN-γ, mediated by a secretory granule-dependent mechanism (3). A number of potential mediators of antimicrobial activity have been delineated, including granzyme B (GZMB), perforin (PRF), and granulysin (GNLY) (4, 5). PRF is largely responsible for lysing infected cells recognized by CD8+ T cells, GZMB can kill intracellular parasites by degrading their defenses against oxygen radicals, and GNLY is important for intracellular killing of bacteria and pathogens (6, 7). Multiple lines of evidence indicate the importance of CD8+ CTLs in host defense against one such intracellular pathogen, M. tuberculosis. Depletion of CD8+ T cells in mice (8) and nonhuman primates results in marked increases in growth of the pathogen. Furthermore, the clinical use of infliximab has established the importance of CD8+ T cells in human resistance to tuberculosis. This drug binds to cell surface tumor necrosis factor (TNF) expressed by cytotoxic CD8+ T effector memory RA (TEMRA) cells, which express GNLY, resulting in their depletion, and this is associated with reactivation of tuberculosis (9). In addition, GNLY is up-regulated along with PRF after Bacille Calmette-Guérin (BCG) vaccination (10).
CD8+ T cells are heterogeneous in their expression of cytotoxic granule proteins, with some cells expressing only one cytotoxic protein and others expressing multiple cytotoxic proteins (11). Therefore, the specific CTL granule components that are necessary and sufficient for intracellular killing remain unclear. The heterogeneous expression of cytotoxic molecules across the CTL compartment creates a situation in which some CTL populations may have the capacity to lyse infected cells, but not kill the intracellular bacteria contained within. In this case, CTLs with the capacity only to lyse infected cells may release viable bacilli and contribute to the spread of infection. However, if some CTLs have the capacity to not only lyse the infected macrophages but also deliver antimicrobial molecules, the pathogens could then be killed intracellularly, limiting dissemination. Consistent with this hypothesis, it has recently been established that the frequency of T cells expressing these three cytotoxic proteins [GZMB, PRF, and GNLY; i.e., tricytotoxic Tlymphocytes (T-CTLs)] correlates with protection against M. tuberculosis (11). Two reasons limit exploration of which CTL subsets have the functional antimicrobial activity. First, GNLY is not naturally expressed in mice (12), and therefore, studies on the role of GNLY are limited to either human models of infection that prohibit deletion of specific immune populations or mice rendered transgenic for human GNLY. Second, the CTL compartment is heterogeneous in the expression of cytotoxic granule proteins such that identification of CTL subsets expressing GNLY or other granule proteins requires permeabilization and chemical fixation, thus precluding functional studies.
To characterize the human CTL subsets responsible for host defense against intracellular pathogens, we took advantage of the human disease leprosy, caused by infection with the intracellular bacterium Mycobacterium leprae. Leprosy has provided an extraordinary model to investigate the human immune system’s response to a microbial infection because it presents as a clinical and immunologic spectrum (13, 14), providing an opportunity to study resistance versus susceptibility to widespread infection. Patients with the resistant tuberculoid form (T-lep) manifest strong cell-mediated immunity (CMI) to the pathogen, skin lesions are few, and bacilli are rare. However, CMI is absent or diminished in the progressive lepromatous form (L-lep) (14, 15), skin lesions are numerous, and growth of the pathogen in macrophages is unabated. Clinical presentations of leprosy correlate with the innate and adaptive cytokine profiles at the site of disease (15, 16). Of relevance, expression of the antimicrobial protein GNLY in leprosy patient lesions has been shown to correlate with host defense against M. leprae (5). Here, we addressed whether distinct CTL subsets differentially contribute to the host antimicrobial responses against human intracellular pathogens, including M. leprae.
RESULTS
CTLs exist in heterogeneous populations that differ in a susceptible versus protected state of infection
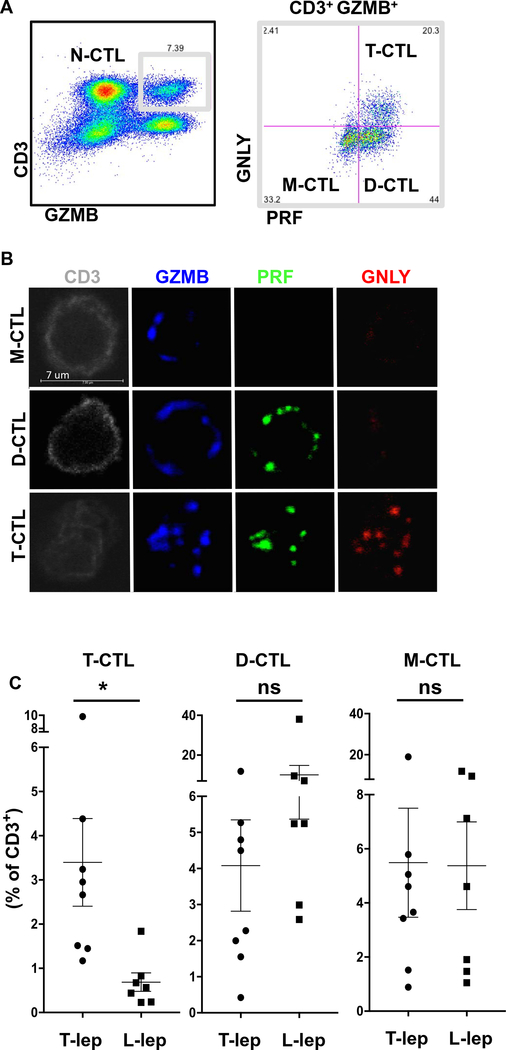
To elucidate which cells may be responsible for delivering the necessary microbicidal payload to control intracellular infection, we examined CTL subsets in healthy donors by flow cytometry (Fig. 1A) and confocal microscopy (Fig. 1B). In general, we found three major subpopulations of CTLs: monocytotoxic T lymphocytes (M-CTLs) expressed only GZMB; dicytotoxic T lymphocytes (D-CTLs) expressed both PRF and GZMB; and T-CTLs expressed all three proteins GZMB, PRF, and GNLY. The defined M-CTL, D-CTL, and T-CTL populations comprised 95 to 99% of CD8+ CTLs in the nine donors studied, with the remainder composed of other combinations of cytotoxic proteins including GZMB and GNLY, or PRF and GNLY.
Fig. 1. T-CTLs are defined by expression of GZMB, PRF, and GNLY and enriched in T-lep versus L-lep.
(A) PBMCs were stained with antibodies to CD3, GZMB, PRF, and GNLY. T-CTL, D-CTL, M-CTL, and N-CTL cells were delineated by flow cytometry. (B) T cells from a healthy donor were sorted from PBMCs and stained for GNLY, PRF, and GZMB. Cells were examined by confocal microscopy, and representative images of the types of cells seen are shown. (C) PBMCs from T-lep (n = 8) or L-lep (n = 7) donors were examined and compared for the percentage of CD3+ T cells that coexpress GZMB, PRF, and GNLY (T-CTLs). *P < 0.05. ns, not significant.
We examined the percentage of T-CTLs in peripheral blood of patients across the spectrum of leprosy to learn which populations were expanded to a greater extent in resistant T-lep versus progressive L-lep states of infection. Our results indicated that the frequency of T-CTLs in leprosy is greatest in the group of patients able to restrict the infection (Fig. 1C and fig. S1).
Cytokines control the T-CTL compartment
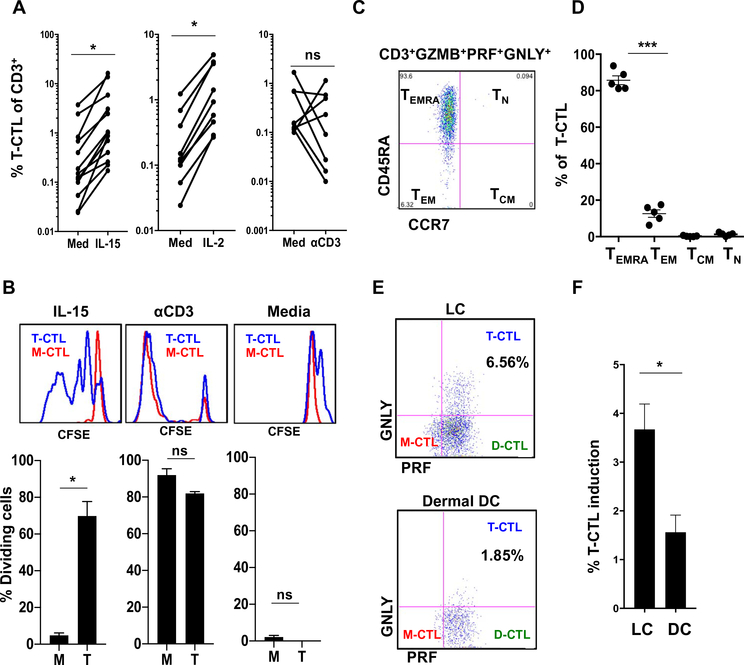
Because the clinical presentation of leprosy correlates with distinct cytokine profiles (15), we reasoned that the difference in percentage of T-CTLs found in T-lep, as opposed to L-lep, patients might be explained by the ability of distinct cytokines to trigger T-CTL differentiation and/or expansion. Interleukin-15 (IL-15) is preferentially expressed in T-lep lesions (17) and has been previously shown in separate experiments to induce GNLY (18), PRF (19–21), and GZMB (20) in T cells. Therefore, we examined CTL subsets for expression of the high-affinity IL-15 receptor subunit IL2Rβ. We found that IL2Rβ is preferentially expressed on T-CTLs, as compared with M-CTLs and N-CTLs (noncytotoxic T lymphocytes; fig. S2A). The expression of IL2Rβ was variable on D-CTLs such that we subsequently compared the response of T-CTLs and M-CTLs with cytokines. Treatment of healthy donor peripheral blood mononuclear cells (PBMCs) with IL-15 or IL-2 induced an about fivefold expansion of the T-CTL subset, as measured by flow cytometry (Fig. 2A). There was also an increase in the M-CTL subset (fig. S2B). However, stimulation through the T cell receptor (TCR) did not cause consistent expansion of T-CTLs (Fig. 2A), indicating that the effect of IL-15 was a specific function of its receptor.
Fig. 2. T-CTLs are CD8+ TEMRA cells that can be expanded by IL-15 and IL-2 through selective proliferation and can be induced by LCs.
(A) PBMCs from healthy donors were treated with IL-15 (n = 13), IL-2 (n = 9), or anti-CD3 plus anti-CD28 (n = 8) for 5 days and compared with medium treatment. (B) PBMCs were labeled with CFSE and treated with medium, IL-15, or aCD3 stimulation for 5 days. Proliferation of T-CTL and M-CTL populations was compared by CFSE dilution; one representative experiment is shown above, and average proliferation is shown below (n = 3). (C) One example shows that the T-CTL compartment was examined and found to consist primarily of CCR7−, CD45RA+ (TEMRA) cells. (D) The T-CTL subset was examined for memory markers (n = 5). (E) Naïve CD8+ T cells were cocultured with allogeneic dermal CD14+ DCs or LCs isolated from human skin. CTL subset formation was interrogated after 7 days of culture. One representative experiment is shown. (F) Composite bar graph demonstrating the significantly greater ability of LCs versus DCs to induce T-CTLs (LCs, n = 9; DCs, n = 6). *P < 0.05, ***P < 0.001.
IL-15 causes selective proliferation of T-CTLs
To determine whether IL-15 triggers the specific expansion of the M-CTL and/or T-CTL subsets, we used carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling to interrogate cell division. We found that IL-15 induced cell division within the T-CTL but not the M-CTL compartment (Fig. 2B and fig. S2C). IL-15 also up-regulated GZMB expression, leading to the induction of M-CTLs (fig. S2, B and C). Thus, IL-15 leads to the induction of M-CTLs and expansion due, at least in part, to the proliferation of T-CTLs.
Characterization of T-CTLs as CD8+ TEMRA cells
Previous findings indicated that CD8+ TEMRA cells (T effector memory cells defined as CCR7−, CD45RA+) contain the CTLs that express GNLY (9). Because both IL-15 and IL-2 have been shown to influence the memory CD8+ T cell compartment (22), the ability of T-CTLs to respond to IL-15 suggested a link to CD8+ TEMRA cells (22), which we investigated by flow cytometry. Interrogation of the T-CTL population for CD8, CCR7, and CD45RA expression revealed that a mean of 75% (47 to 93%) of T-CTLs were composed of CD8+ T cells in seven donors analyzed (fig. S3A) and a mean of 86% (81 to 94%) of T-CTLs were composed of CCR7−, CD45RA+ TEMRA cells in five donors analyzed (Fig. 2, C and D).
Epidermal Langerhans cells efficiently differentiate naïve CD8 T cells into T-CTLs
Further characterization of the memory T cell compartments revealed that although T-CTLs were composed primarily of CD8+ TEMRA cells, naïve CD8+ T cells were found to be devoid of any cytotoxic protein expression (fig. S3B). The addition of individual or combinations of cytokines—including IL-2, IL-15, IL-21, IL-18, IL-12, and IFN-γ, known to induce cytotoxic granule proteins as well as influence the memory T cell compartments—failed to induce naïve CD8+ T cells to differentiate into T-CTLs. Because dendritic cells (DCs) are characterized by their ability to differentiate CD8+ naïve T (TN) cells, we examined the capacity of different DC subsets in skin to induce T-CTLs. Langerhans cells (LCs), but not dermal CD14+ DCs, better facilitated the differentiation of naïve CD8+ T cells into T-CTLs (Fig. 2, E and F).
Identification of specific T-CTL surface markers receptors by RNA sequencing
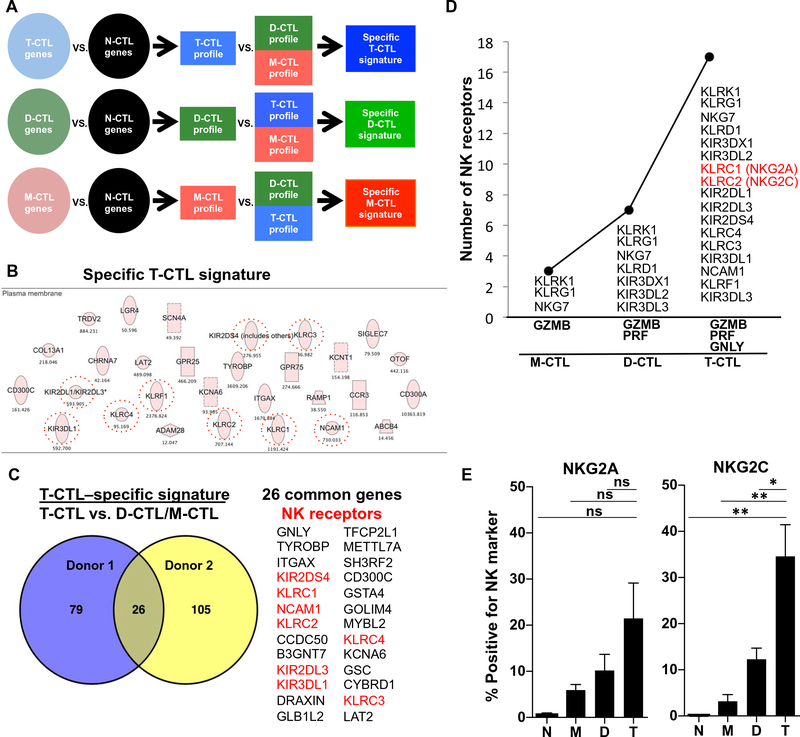
The identification of T-CTL subsets currently requires intracellular staining of cytotoxic granule proteins, which necessitates fixation and permeabilization and thereby precludes functional studies. To identify specific T-CTL surface markers to permit sorting of live cells, we performed RNA sequencing (RNA-seq) on fixed and sorted CTL subsets. Using this transcriptome sequencing data, we subsequently identified gene profiles for the T-CTL, D-CTL, and M-CTL populations relative to the N-CTL population in detail in two donors, as defined by all genes expressed twofold greater in each CTL population over the N-CTL population. A common signature of 520 genes in T-CTLs, D-CTLs, and M-CTLs, as compared with N-CTLs, was up-regulated in both donors (fig. S4A). Congruent with flow cytometry data, these common genes included the cell surface marker CD8 as well as the cytotoxic granule protein genes encoding GZMB, PRF, and GNLY and the IL-2 and IL-15 common cytokine receptor gene IL2RB. In addition, CTL transcription factors EOMES and RUNX3 were enriched in this common signature, known to drive memory CTL differentiation (23). Consistent with flow cytometric data, further analysis of RNA-seq data demonstrated that although GNLY, PRF1, and GZMB mRNA were up-regulated in the joint CTL signature as compared with N-CTLs, expression was greatest in the T-CTLs > D-CTLs > M-CTLs (fig. S4B). In addition, T-CTLs (and D-CTLs) did not express CCR7 mRNA transcripts (fig. S4B). Analysis of the genes differentially expressed in the T-CTLs versus N-CTLs showed an ~70% overlap between donors (fig. S4C).
A T-CTL–specific signature was derived by comparing the T-CTL gene profile with the D-CTL and M-CTL gene profiles, as outlined (Fig. 3A). There were 105 and 131 genes identified in the T-CTL–specific signatures in each donor, respectively. Bioinformatics analysis unexpectedly identified “natural killer cell signaling” and “cross-talk between DCs and natural killer cells” as the top two canonical pathways in the specific T-CTL signature of both donors. Subsequent analysis of the specific T-CTL signature from the first donor revealed 23 genes that encode cell membrane proteins. We noticed that nine of these genes belong to the family of natural killer (NK) receptors plus TYROBP (DAP12), a known signaling component of this receptor family (Fig. 3B and fig. S5), and similar results were observed with the specific T-CTL signature in the second donor (fig. S6). Overlap of the specific T-CTL signatures between the donors revealed 26 common genes, 8 of which belonged to the family of NK receptors (Fig. 3C). Analysis of surface NK receptor gene expression revealed that the number of NK receptors correlated with cytotoxic molecule expression. Specifically, we observed that as the number of cytotoxic molecular species increased from M-CTLs to D-CTLs to T-CTLs, the level of expression of NK receptor genes concomitantly increased (Fig. 3D and fig. S7). In total, there were 15 NK receptor genes expressed by T-CTLs that were common in the donors studied (fig. S6). Eight genes were the previously identified NK genes specific to T-CTLs and included KLRC1, KLRC2, KIR2DL3, KIR2DS4, KLRC4, KLRC3, KIR3DL1, and NCAM1. Seven NK genes were expressed by one or more of the other subsets of CTLs in addition to T-CTLs, and there were two additional NK receptor genes expressed by T-CTLs in donor 1 (fig. S6). Analysis of TCR genes in the T-CTL RNA-seq data revealed that T-CTLs likely originate from oligoclonal or diverse αβ T cells in the different donors (fig. S8).
Fig. 3. RNA-seq of cytotoxic cell populations identifies a specific T-CTL gene signature composed of NK receptors that correlate with the number of cytotoxic molecules expressed and that are enriched on T-CTLs.
(A) RNA-seq was performed on sorted populations of T-CTL, D-CTL, M-CTL, and N-CTL cells. Gene profiles of T-CTL, D-CTL, and M-CTL cells were created by selecting genes increased twofold or more in each population over the N-CTL population. Specific gene signatures were then created by selecting genes more highly expressed in one profile when compared with the others. (B) Genes comprising the T-CTL specific signature were analyzed by Ingenuity (Qiagen) and sorted for surface expression. NK receptors are circled. (C) The “specific T-CTL signatures” between two donors were overlapped using VENNY (69), and common genes are listed, with NK receptors highlighted in red. (D) The number of surface NK receptors expressed in a population (y axis) is graphed as a function of the number of cytotoxic molecules expressed by that population (x axis). NK receptor expression by a population is defined by greater than twofold expression over each preceding population (or 1.5-fold for T-CTLs versus D-CTLs). (E) CTL subsets from 10 donors were analyzed for the denoted NK marker expression by flow cytometry. The average percentage of T-CTL, D-CTL, M-CTL, and N-CTL cells expressing these markers in each respective donor is shown. *P < 0.05, **P < 0.01.
NK cell surface markers are expressed on T-CTLs and can be used to purify viable T-CTLs
RNA-seq identified eight-candidate NK surface markers specific to T-CTLs: KLRC1, KLRC2, KIR2DL3, KIR2DS4, KLRC4, KLRC3, KIR3DL1, and NCAM1 (Fig. 3, C and D). To select genes for further study, we interrogated these surface markers to determine which had the highest differential expression on T-CTLs versus the next most closely related subset, D-CTLs. From the RNA-seq data, three markers—KLRC2 (NKG2C), KLRC1 (NKG2A), and NCAM1 (CD56)—best distinguished T-CTLs from D-CTLs (fig. S9). Therefore, we measured expression of NKG2C, NKG2A, and CD56 by flow cytometry on T-CTLs from healthy donors.
We found that T-CTLs expressed the greatest percentage of NKG2C+, NKG2A+, and CD56+ cells, as compared with the D-CTL, M-CTL, and N-CTL populations (Fig. 3E and fig. S10), validating the RNA-seq data. Across 10 donors, ~30% of all T-CTLs were NKG2C+, albeit with donor-to-donor variability congruent with the known genetic variation in NK receptor usage (24). We confirmed that the NK receptor–expressing T-CTLs were in the CD8+ compartment (fig. S11A). Furthermore, we found that NKG2C selected for T-CTLs such that, generally, NKG2C+ cells were more highly enriched for T-CTLs than NKG2A+ cells and there were few T-CTLs in the NKG2A−NKG2C− population (figs. S11B and S12, A and B). Hence, NKG2C and NKG2A can be used as molecular markers to isolate enriched populations of T-CTLs and investigate their functional capacity, which was previously precluded by the need for intracellular flow cytometry to identify these cells.
Modulatory NK receptors function to augment and inhibit cytotoxic granule release on CD8+ CTLs
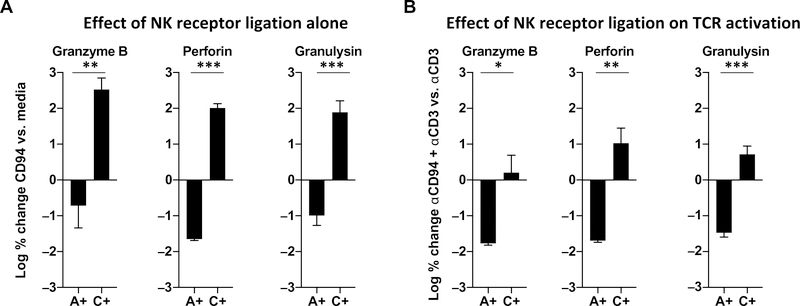
NKG2C and NKG2A form heterodimers with CD94, which, upon activation, triggers a stimulatory versus an inhibitory response, respectively (25–27). We sorted CD8high cells, confirmed to be all CD3+, which were either NKG2C+, or NKG2A+ or double-negative, given that NKG2C and NKG2A expression was mutually exclusive in the donors tested. We investigated the function of these NK receptors on cytotoxic granule protein release by triggering sorted single-positive NKG2C and NKG2A populations using plate-bound anti-CD94 antibodies (25), given that both populations contain T-CTLs. Stimulation of sorted CD8+ NKG2C+ T cells with anti-CD94 resulted in the release of all three cytotoxic granule proteins (GZMB, PRF, and GNLY). In contrast, stimulation of sorted CD8+ NKG2A+ T cells via anti-CD94 had little effect on release of cytotoxic granule proteins and was significantly different than the activation induced in NKG2C+ cells (Fig. 4A). Stimulation with isotype-matched control antibody had no effect on cytotoxic granule release in either NKG2A+ or NKG2C+ T-CTLs.
Fig. 4. NK receptors on T-CTLs are functional and may modulate TCR-induced release of cytotoxic granules.
T-CTLs were enriched from other subsets by cell sorting for either NKG2A (A+) or NKG2C (C+). (A) Each population was stimulated with anti-CD94 or left in medium for 20 hours. GZMB, PRF, and GNLY release was measured in the culture supernatants by ELISA. The log % change of the anti-CD94 stimulation relative to medium is shown. (B) Each population was stimulated with anti-CD3 or anti-CD3 and anti-CD94 for 20 hours, and cytotoxic granule proteins were measured by ELISA. The log % change of the anti-CD94 plus anti-CD3 relative to anti-CD3 alone is shown. *P < 0.05, **P < 0.01, ***P < 0.001.
We further explored whether NKG2A or NKG2C might modulate release of cytotoxic granule proteins from CD8+ T cells in the context of TCR activation by comparing stimulation with anti-CD3 plus anti-CD94 versus anti-CD3 alone. Activation of both NKG2A+ and NKG2C+ T cells with anti-CD3 alone induced the release of GZMB, PRF, and GNLY. Treatment of NKG2A+ T cells with anti-CD3 plus anti-CD94 resulted in a reduction of all three cytotoxic granule proteins, as compared with anti-CD3 stimulation alone. On the other hand, stimulation of NKG2C+ T cells with anti-CD3 plus anti-CD94 trended toward an increase in GZMB, PRF, and GNLY release, and was significantly different than the inhibition induced in NKG2A+ cells (Fig. 4B). These findings reveal that triggering of NKG2C activates T-CTLs to release cytotoxic granule proteins in a TCR-independent and TCR-dependent manner relative to triggering of NKG2A, which inhibits cytotoxic granule release.
T-CTLs reduce the viability of intracellular mycobacteria more efficiently than other CD8+ subsets
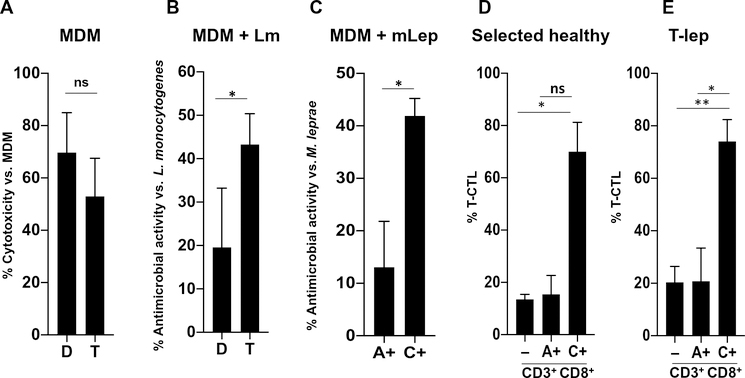
To compare directly the antimicrobial and cytotoxic capacity of the T-CTL subset with that of the D-CTL subset in homogeneous T cell populations, we generated stable CD8+ CTL clones derived from peripheral blood and screened them for cytotoxic molecule expression. We screened these T cell clones by flow cytometry and cytotoxic protein expression by enzyme-linked immunosorbent assay (ELISA) after anti-CD3 activation, selecting one T-CTL clone (clone “T”) and one D-CTL clone (clone “D”) for further study (fig. S13). Both clone T and clone D displayed equal levels of cytotoxicity toward anti-CD3–coated monocyte-derived macrophages (MDMs; Fig. 5A). We tested these clones for their antimicrobial activity against Listeria monocytogenes given the role of CD8+ CTLs in protective immunity against this intracellular bacterium (28, 29). The T-CTL clone mediated an antimicrobial activity of about double that of the D-CTL clone against L. monocytogenes in infected MDMs (Fig. 5B). This indicates that although both the T-CTL and D-CTL clones can lyse target cells to the same degree, the T-CTL clone has greater antimicrobial activity.
Fig. 5. T-CTLs can be purified using NK receptors and display greater antimicrobial activity as compared with other CD8+ T cells.
(A) MDMs coated with anti-CD3 were admixed with the T-CTL and D-CTL clones in an E:T ratio of 2:1, as indicated. LDH was measured in culture supernatants after 4.5 hours. The percent cytotoxicity was calculated by comparing the amount of LDH released in each condition to the amount of LDH released by MDMs alone (n = 4). (B) L. monocytogenes (Lm)–infected MDMs coated with anti-CD3 were admixed with the T-CTL and D-CTL clones at an E:T ratio of 2:1, as indicated. After 20 hours, MDMs were lysed and the amount of L. monocytogenes present was assessed by CFU. Percent antimicrobial activity was determined by normalizing the amount of bacterial growth in each condition to the amount of bacterial growth found in infected MDMs without admixed T cells (n = 4). (C) M. leprae (mLep)–infected MDMs coated with anti-CD3 were admixed with sorted populations of CD8+ T cells based on NKG2A and NKG2C expression in an E:T ratio of 2:1, as indicated. The T-CTL, D-CTL, M-CTL, and N-CTL composition of each sorted population was delineated at the time of sorting. After 24 hours, RNA and DNA were isolated and the ratio of bacterial RNA to DNA was calculated. Antimicrobial activity was determined by normalization of each condition to M. leprae–infected MDMs admixed with NKG2A− and NKG2C− cells (n = 4). (D) The percent of T-CTLs in total CD3+CD8+ (−), CD3+CD8+NKG2A+ (A+), and CD3+CD8+NKG2C+ (C+) cells was determined in four healthy donors by flow cytometry and is compared. (E) The percent of T-CTLs in total CD3+CD8+ (−), CD3+CD8+NKG2A+, and CD3+CD8+NKG2C+ cells was determined in T-lep donors by flow cytometry and is compared (n = 4). *P < 0.05, **P < 0.01.
We also tested the antimicrobial capacity of the different CD8+ CTL subpopulations against M. leprae, studying primary cells enriched from a group of four donors in which the CD8+ NKG2C+ T cells contained, on average, ~70% T-CTLs (Fig. 5D and fig. S12A, donors marked by an asterisk). From these donors, we cocultured NKG2C+ or NKG2A+ T cells with M. leprae–infected MDMs. After 24 hours, we measured bacterial viability by assessing the bacterial RNA–to–DNA ratio in the entire sample, because it is not possible to culture M. leprae, and determined the antimicrobial activity (30, 31). After coculture with M. leprae–infected MDMs, the NKG2C+ T cells (average, ~70% T-CTLs) exhibited a significantly higher level of antimicrobial activity than NKG2A+ cells (average, ~15% T-CTLs) or the NKG2C−NKG2A− cells (average, ~2% T-CTLs; Fig. 5C). In the CD8+ NKG2C+ effector T cell population, the absence of anti-CD3 reduced antimicrobial activity (fig. S14). There was a significant positive correlation of antimicrobial activity with T-CTL enrichment but not with D-CTLs or M-CTLs; reciprocally, there was a significant negative correlation of antimicrobial activity with N-CTL enrichment (fig. S15, A to D). These results suggest that T-CTLs have greater antimicrobial activity than the other CD8+ CTL subsets.
NKG2C labels CD8+ T-CTLs in T-lep donors
Last, given that GZMB+PRF+GNLY+ CTLs were greater in the PBMCs from T-lep versus L-lep patients, we investigated whether in T-lep patients NKG2C was a molecular marker for CD8+ T-CTLs. In four donors, CD3+CD8+NKG2C+ marked a population that contained ~75% T-CTLs, whereas NKG2A+ and total CD3+CD8+ CTLs contained ~20% T-CTLs (Fig. 5E and fig. S16). Together, these results define antimicrobial CTLs (amCTLs) as T-CTLs expressing NKG2C, capable of TCR-dependent and TCR-independent release of cytotoxic granule proteins that mediate antimicrobial activity.
DISCUSSION
A key function of CTLs is their ability to kill infected target cells, which, in viral infection, aborts maturation into infectious particles but, for bacterial infection, can lead to the release of viable organisms with the potential to disseminate. We identified at least one subset of amCTLs, defined by the functional capacity to potently kill intracellular bacteria, coexpression of the three cytotoxic granule proteins (T-CTLs), as well as the enrichment of the activating receptor NKG2C. These studies identify at least one unique functional T cell subset in host defense against some intracellular pathogens.
Because it has previously been possible to identify the cells expressing different granule components only on fixed and permeabilized cells, our result using RNA-seq that T-CTLs expressed NK receptors permitted the isolation and functional study of enriched T-CTLs, allowing the identification of amCTLs. Antibodies directed against two NK modulatory receptors, NKG2A and NKG2C, were used to directly isolate CTL subsets for functional experiments, revealing that T-CTLs had the greatest antimicrobial activity against M. leprae in infected macrophages. Furthermore, direct comparison of T-CTL and D-CTL subsets by generation of stable clones confirmed that T-CTLs displayed higher antimicrobial activity, whereas T-CTLs and D-CTLs displayed equal levels of cytotoxic activity toward host cells. These results suggest that the three cytotoxic proteins—GZMB, PRF, and GNLY—act together to equip amCTLs with the lethal weapons required to kill intracellular bacteria such as M. leprae and L. monocytogenes. The most comprehensive model would suggest that PRF disrupts the membrane of the targeted eukaryotic cell, allowing GZMB and GNLY to gain access to the compartment in which the bacteria reside. Next, GNLY binds to the intracellular bacterium damaging its membrane. In the case of intracellular parasitic infection, GZMB triggers host cell apoptosis and enhances killing of the bacterium via free radical generation by GZMB cleavage of oxidative stress defense proteins (6, 7). Oxidative radicals, which are known to be microbicidal for many pathogens, may not be sufficient in the amounts produced to kill mycobacteria because they are highly resistant to killing by reactive oxygen species (32). Conversely, several mycobacterial proteins that have important roles in metabolic and biosynthetic pathways have been shown to be GZMB substrates (6). Thus, GNLY may play a more direct role in antimicrobial activity directly through damage to the bacterial cell wall (3, 4) and/or indirectly through permeabilization of the bacterial cell wall, allowing GZMB to enter and cleave bacterial substrates (6).
Through examination of the gene expression signature of CTL subsets, we unexpectedly found that the number of cytotoxic granule proteins expressed by the CTL subsets correlated with the frequency of NK receptors, with the most NK receptors found in T-CTLs, followed by D-CTLs, M-CTLs, and N-CTLs, respectively. Previous studies have implicated that several surface NK receptors have modulatory functions (activating or inhibitory) of cytolytic activity (25–27, 33). Of the NK receptors we identified on T-CTLs, five are known to be activating, including NKG2C, and four have been defined as inhibitory, including NKG2A. Here, we show that these two receptors both marked the T-CTL subset but were, in general, mutually exclusively expressed at the individual cell level. We found that NKG2C and NKG2A had opposing effects on CTL function as defined by cytotoxic granule release. Stimulation through NKG2C alone is sufficient to trigger cytotoxic granule release and had little effect on TCR function, whereas stimulation through NKG2A does not induce cytotoxic granule release and inhibits TCR function. Therefore, the expression of NKG2A on some T-CTLs may serve to negatively regulate their function. In addition, we show that the frequency of NK receptors increased as CTLs expressed more granule proteins. Together, our data suggest that CTL antimicrobial activity may be controlled by both TCR ligation and engagement of activating and inhibitory NK receptors such that these receptors may act as checkpoints or costimulators to modulate CTL function. The identification of other activating receptors expressed on terminally differentiated T-CTLs, including KLRC3, KLRC4, KIR2DS4, and KLRF1, raises the possibility that there may be a spectrum of other subsets of amCTLs yet to be defined.
The result that NKG2C activation of CD8+ T-CTLs induced cytotoxic granule release in the absence of anti-CD3 stimulation revealed a TCR- and hence antigen-independent mechanism of T-CTL activation. This finding elucidates an innate-like pathway for activation of this antimicrobial T cell subpopulation. Activation of CD8+ T cells via NKG2D, independent of TCR ligation, is thought to contribute to liver injury in hepatitis A infection (34). NKG2A and NKG2C, when complexed with CD94, both bind human leukocyte antigen E (HLA-E) loaded with major histocompatibility complex (MHC) class I leader peptides(35).HLA-E–restricted CD8+ T cells that recognize M. tuberculosis have been reported to vary in their cytotoxic versus immunoregulatory capacity (36–42) such that examination of the antimicrobial capacity of these T cells is now warranted.
In mouse models of bacterial and viral infection, CD8+ T cells bearing NK receptors have been observed and are thought to represent clones of antigen-specific T cells (33). In humans, NK receptor–bearing CD8+ αβ T cells have been found to comprise up to 5% of circulating CD8+ T cells (43) and are typically thought to represent subsets of terminally differentiated CD8+ memory or effector T cells (44). Furthermore, NK receptor–expressing CD8+ T cells have been shown to have cytotoxic activity (45, 46). Our result that amCTLs are a subset of TEMRA cells is consistent with the reported finding that some TEMRA cells also express NK receptors (47), consistent with the ordered expression of cytotoxic granule proteins during CD8 T cell differentiation (9, 48, 49). In addition, NK receptors are expressed on a subpopulation of CD4+ T cells (50), which are present at sites of viral infection (51).
Several lines of translational evidence point to a key role for amCTLs in human host defense against intracellular mycobacteria. We reported previously that PRF and GNLY were more strongly expressed in T-lep versus L-lep lesions (5). Here, we found that the frequency of T-CTLs is greater in PBMCs from patients with the self-limited T-lep form of leprosy compared with the progressive L-lep form and the T-CTLs in T-lep express the amCTL marker NKG2C.Similarly, the frequency of T-CTLs in peripheral blood correlates with protective immunity in tuberculosis (11). Finally, reports showing that the anti–TNF-α biologic infliximab causes reactivation of latent tuberculosis and depletes CD8+ TEMRA cells, a cell population that we show contains the T-CTL subset, in vitro and in vivo (9), are compelling examples of the importance of these cells in resistance to intracellular pathogens in humans.
Identification of the signals that govern induction and expansion of the amCTL compartment is pivotal to the development of strategies to modulate this response in the prevention and/or treatment of clinical infection caused by intracellular pathogens. The finding that T-CTLs constitutively express high levels of IL2Rβ led to the realization that these cells are a subset of effector memory T cells that can be expanded by either IL-15 or IL-2. In addition, we found that epidermal LCs, but not dermal CD14+ DCs, induce naïve CD8+ T cells to differentiate into T-CTLs, consistent with the known ability of LC to facilitate the differentiation of naïve CD8+ T cells into CTLs (52). Together, these results reveal a distinct functional capacity for skin LCs and suggest that the tissue microenvironment in which the immune system in the skin encounters pathogens dictates differential immune responses. The stronger expression of IL-15 in T-lep versus L-lep patients may contribute to the induction and expansion of T-CTLs in T-lep patients.
Our characterization of amCTLs, a subset of CD8+ CTLs that can exert antimicrobial activity, as well as establishing the methods to induce them with LCs and expand them with IL-15 or IL-2, offers new possibilities for translational application and potential therapy. It appears that production of IFN-γ by CD4+ T cells within granulomas is not sufficient to assure protection against tuberculosis. CD8+ T cells are localized to the periphery of granulomas in both leprosy (53) and tuberculosis (54, 55). One possible function of these cells, we would suggest, may be to serve as guardians of the granuloma, killing macrophages and their intracellular pathogens in the process of escaping the granuloma, thus reducing dissemination of the infection.
Unlike T-CTLs, D-CTLs do not express GNLY, indicating that the antimicrobial protein GNLY is essential for human host defense against intracellular pathogens. GNLY is not present in mice (12, 56) such that the critical function of this antimicrobial protein by CTLs in human host defense cannot be directly studied in standard mouse models of infection. Consistent with our data, transgenic mice engineered to express human GNLY indicated a synergy of these granule proteins against Escherichia coli and intracellular parasites (6, 7). However, it remains difficult to draw conclusions about the differentiation and function of transgenic CTL subsets and how they may compare with the true function of a human immune response. In addition, the effect of NK modulatory receptor function on antigen-specific TCR activation remains to be fully determined because we were obliged to rely upon anti-CD3 redirected TCR activation given that antigen-specific CD8+ CTL T cell clones from leprosy patients have been difficult to isolate and maintain. Last, our results included two intracellular bacteria (M. leprae and L. monocytogenes) such that it remains to be determined the extent to which the present findings will extend to other bacterial species or intracellular parasites.
In terms of translational application, the association of T-CTLs with host defense in leprosy and tuberculosis (11) suggests that a combination of the identified T-CTL cell surface receptors described here could be used as biomarkers to monitor the role of CD8+ T cells in protective immunity in ongoing vaccine trials. In addition, regulating amCTL function by small molecules or cytokines may enhance immunotherapy. Last, the finding that CTLs contain activating and inhibitory surface NK receptors suggests that the license to kill a target may depend not only on cognate TCR-MHC recognition but also on the matrix of inhibitory or activating receptors and their ligands expressed by the CTLs and the target cell. In summary, the present studies have defined and characterized a functional subset of human CD8+ T cells that has antimicrobial activity against intracellular pathogens, the conditions for their differentiation from TN cells, and a complex framework that modulates their activity.
MATERIALS AND METHODS
Study design
This study was aimed at identifying the human CD8+ cytotoxic T cell subsets that may be important for controlling intracellular bacterial infection. Human peripheral blood was collected at two locations, either at University of California, Los Angeles (UCLA) or through collaborations with University of Southern California, as outlined below. Researchers were not blinded as to the source (healthy donors versus leprosy patients) of the blood samples. All blood collections were carried out with informed consent according to institutional review board protocols.
Patients and healthy donors
Leprosy blood specimens were obtained through collaborations with M.T.O. at the Los Angeles County/University of Southern California Medical Center. The diagnosis of leprosy was established by means of clinical criteria according to Ridley and Jopling (13). Healthy donors served as controls and were used for baseline examination. We were blinded as to the race of the leprosy patients, but on the basis of epidemiology of the leprosy patients in Los Angeles, most patients are of Hispanic or Asian descent; a large proportion of healthy donor blood comes from donors in these ethnic/gender categories to best match the population of leprosy donors.
Isolation and expansion of CTL subsets from PBMCs and human T cells using cytokines
PBMCs were isolated from the peripheral blood of healthy donors, or patients with a diagnosis of leprosy, using Ficoll-Paque gradients (Amersham Biosciences). T cells were negatively selected by subjecting PBMCs to magnetic bead separation using immunomagnetic negative selection (EasySep, STEMCELL Technologies) and then cultured in RPMI 1640 with 10% fetal calf serum (FCS; HyClone) with or without cytokines for 5, 7, or 12 days. The following cytokines were used: IL-15 (15 ng/ml; R&D Systems and a gift from T. Waldman at the National Institutes of Health), 50 nM IL-2 (Chiron), and anti-CD3/CD28 microbeads (Dynabeads, Gibco).
Confocal microscopy
CD3+ T cells were adhered to poly-L-lysine–coated slides (1 mg/ml; Sigma). The cells were surface-stained with anti-human CD3–Pacific Blue (clone UCHT1, BioLegend) or isotype-matched control antibody. After fixation and permeabilization (BD Cytofix/Cytoperm, BD Biosciences), the cells were stained intracellularly with anti-human GNLY (clone DH4), anti-human PRF (clone δG9, BD Biosciences), and anti-human GZMB (clone GB11, Thermo Fisher Scientific) or isotype-matched control antibodies. Isotype-specific fluorochrome-conjugated (Alexa Fluor 488, 568, and 647) secondary antibodies were incubated with the cells, and slides were mounted with ProLong gold antifade reagent and a coverslip. Immunofluorescence images were taken using a Leica SP5 microscope at the Advanced Light Microscopy/Spectroscopy Core Facility Laboratory, California NanoSystems Institute, UCLA.
Calculating the percentage of CTL subsets
T-CTLs were defined as CD3+ cells coexpressing GZMB, PRF, and GNLY. The %T-CTL of CD3+ T cells was calculated using multicolor flow cytometry by dividing the number of T-CTL events by the total number of CD3+ events. FlowJo software (FlowJo LLC, Ashland, OR) was used to analyze flow cytometry data.
Analysis of memory subpopulations
Fluorescence-activated cell sorting (FACS) was used to analyze memory subpopulations of T-CTLs. Cells were labeled with combinations of CD3, CD8, CCR7, and CD45RA to distinguish between TN, TCM (central memory T cells), TEM (effector memory T cells), and TEMRA cells.
T cell proliferation
PBMCs were isolated from donors as described above. Cells were labeled ex vivo with 0.5 μM CFSE (CellTrace, Invitrogen) and cultured with IL-15 (15 ng/ml), medium, or αCD3/αCD28 microbeads. Flow cytometry was used with CD3, CD8, CD4, GZMB, PRF, GNLY, and CFSE staining to interrogate proliferation within the T-CTL, D-CTL, M-CTL, and N-CTL compartments by examining CFSE dilution. FlowJo software (FlowJo LLC) was used to analyze flow cytometry data.
Priming CTL subsets using skin DCs
LCs or dermal CD14+ DCs were isolated from cells that migrated from skin specimens, as described (52, 57). DCs (live Lin−HLA-DR+) were sorted on the basis of CD1a, CD1c, and CD14 expression. Naïve T cells were enriched using the Pan-Naïve T Cell Isolation Kit (STEMCELL Technologies) according to the manufacturer’s protocol and sorted as CCR7+CD45RA+CD8+ cells. T cells were then CFSE-labeled and cultured with allogeneic sorted skin DCs at a DC:T cell ratio of 1:40 for 7 days in the presence of IL-7 (10 ng/ml) and IL-2 (10 IU/ml; added on day 2).CD40L (100 ng/ml)was used to activate the DCs, and GZMB, PRF, and GNLY expression was assessed by flow cytometry.
Cell sorting of CTL populations and RNA isolation from fixed and sorted cells
RNA was isolated from fixed sorted cells based on the MARIS (method for analyzing RNA following intracellular sorting) protocol, as described by Hrvatin et al. (58). Briefly, we used FACS to obtain highly purified populations of T-CTLs, D-CTLs, M-CTLs, and N-CTLs from donors based on staining with CD3, GZMB, PRF, and GNLY, as described above. Before sorting, cells were fixed in 2% electron microscopy–grade paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 0.5% deoxyribonuclease (DNase)/ribonuclease (RNase)–free saponin (Sigma) to permit intracellular staining. All staining and sorting took place in DNase/RNase-free phosphate-buffered saline (PBS) supplemented with microbiology-grade bovine serum albumin (Gemini Bio-Products) in the constant presence of RNasin plus RNase inhibitor (Promega) 1:25 to 1:100 (1:100 for washes and 1:25 for staining and sorting). After sorting, RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion), as per the manufacturer’s instructions, with the same modification to the protocol used as described by Hrvatin et al. (58).
RNA-seq of cytotoxic cell populations
Sequencing libraries were constructed from RNA using Illumina TruSeq Stranded Total RNA Sample Prep and sequenced at the Clinical Microarray Core at UCLA by single-end sequencing on an Illumina HiSeq3000.
Analysis of RNA-seq data
RNA-seq analysis was performed as described (59). Briefly, sequenced reads were demultiplexed and aligned to the human reference genome hg19 (University of California, Santa Cruz) using TopHat (version 2.0.6). The HTSeq package was then used to assign uniquely mapped reads to exons and genes using the gene annotation file for build hg19 from Ensembl to generate raw count data. Data normalization and differential expression analysis using a negative binomial model was performed in the R statistical programming environment using the DESeq2 Bioconductor package.
Analysis of genes expressed across CTL populations and generation of the CTL-specific signatures
To determine the M-CTL, D-CTL, and T-CTL gene profiles, we selected genes that were expressed twofold or greater in each subset compared with N-CTLs. To determine the M-CTL–, D-CTL–, and T-CTL–specific signatures, we performed three-way comparisons of the gene profiles. For example, to generate the “T-CTL–specific signature,” we compared the “T-CTL gene profile” with the “D-CTL gene profile” and “M-CTL gene profile,” selecting genes that were more strongly expressed in the T-CTL profile versus both the D-CTL and M-CTL profiles. For the comparisons of M-CTLs with either T-CTLs or D-CTLs, we used a twofold cutoff. For comparison of T-CTLs with D-CTLs, a cutoff of 1.5-fold was used because of the similarities of these signatures. This specific signature was then analyzed by Ingenuity (Qiagen) to sort genes expressed on the cell membrane as candidate T-CTL markers to test for validation.
Validation of T-CTL markers by flow cytometry
Once identified (as outlined above), surface markers were validated. PBMCs or T cells isolated as described above were interrogated by flow cytometry for expression of CD56, NKG2A, and NKG2C across T-CTL, D-CTL, M-CTL, and N-CTL populations.
Assessment of NK receptor function
High-expressing CD8+ cells exclusively expressing either NKG2C or NKG2A were sorted from PBMCs. Concomitant portions of PBMCs were stained with CD3, CD8, NKG2C, NKG2A, GZMB, PRF, and GNLY, allowing confirmation that the CD8high population was entirely CD3+ and allowing us to delineate the percentage of T-CTLs, D-CTLs, M-CTLs, and N-CTLs in each sorted population. Sorted CD8+NKG2C+ or CD8+NKG2A+ cells were incubated overnight in RPMI 1640 (Gibco) supplemented with 10% AB serum (Sigma), penicillin, streptomycin, and L-glutamine (Gibco), sodium pyruvate (Gibco), 55 μM 2-mercaptoethanol (Gibco), 12.5 mM Hepes (Gibco), 0.5× nonessential amino acids (Gibco), 0.25× essential amino acids (Gibco), gentamicin (10 μg/ml; Gibco), 1 nM IL-2 (Chiron), IL-7 (5 ng/ml; BioLegend), and IL-15 (5 ng/ml; R&D Systems). After overnight incubation, cells were stimulated with medium alone, plate-bound anti-CD94 (100 μg/ml), plate-bound anti-CD20 isotype-matched control (100 μg/ml), anti-CD3 microbeads, or anti-CD3 microbeads in the presence of plate-bound anti-CD94 (100 μg/ml). For these experiments, we used a suboptimal dose of anti-CD3 so that it would be possible to augment and inhibit responses—10% of the manufacturer’s recommended concentration was used as determined by titration experiments. After 24 hours of stimulation, GZMB (Abcam), PRF (Abcam), and GNLY (R&D Systems) release was measured by ELISA as per the manufacturer’s instructions. The effect of anti-CD94 versus medium was calculated as log {[(αCD94 − medium)/medium] × 100}. The effect of anti-CD94 plus anti-CD3 versus anti-CD3 was calculated as log {[(αCD94&αCD3 − αCD3)/medium] × 100}.
Generation of clones
T cell clones were generated as previously described (60, 61). Briefly, PBMCs were isolated from whole blood of T-lep donors, and DCs were generated by CD14+ negative selection without CD16 depletion (STEMCELL Technologies), followed by culture in RPMI 1640 with 10% FCS, granulocyte-macrophage colony-stimulating factor (GM-CSF; 800 IU/ml), and IL-4 (1000 IU/ml). On day 5, DCs were harvested and cultured with RPMI 1640 medium supplemented with 12.5 mM Hepes, 4 mM L-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), 50 μM β-mercaptoethanol, and 10% AB human serum and pulsed with ML0049 peptide pool (10 μg/ml) for 2 hours. These cells were then mixed with CD8+ T cells isolated by negative selection (STEMCELL Technologies) from the same donor in 96-well U-bottom plates to create CD8+ T cell lines. T cell lines were refed or split 1 to 2 every 3 to 4 days depending on confluence, using the same medium with IL-7 (5 ng/ml; BioLegend), IL-15 (5 ng/ml; R&D Systems), and 1 nM IL-2 (Chiron). After 2 weeks, all cells were collected and positively selected CD8+ T cells were then cloned by limiting dilution in 96-well U-bottom plates in the presence of 200,000 irradiated (75 Gy) allogeneic PBMCs, 40,000 lymphoblastoid cell line (provided by D. Lewinsohn), anti-CD3 (10 ng/ml; clone OKT3, eBioscience), IL-7 (5 ng/ml), IL-15 (5 ng/ml), and 1 nM IL-2. Clones were either split 1 to 2 or refed with fresh medium and cytokines every 3 to 4 days. After 2 to 4 weeks, clonal populations were tested for expression of GZMB, PRF, and GNLY by flow cytometry.
Generation of target cells
To assess the antimicrobial activity directed toward M. leprae– or L. monocytogenes–infected cells, we used infected MDMs as targets. MDMs are known to have a high capacity to engulf and become infected by mycobacteria but a low intrinsic ability to kill without the presence of IFN-γ (62), making them ideal targets for our assays. Briefly, 5 to 7 days before infection with bacteria, monocytes were purified from peripheral blood using Ficoll-Paque gradients (Amersham Biosciences) followed by negative selection of CD14+ CD16+ cells using the EasySep Human Monocyte Enrichment Kit without CD16 Depletion (STEMCELL Technologies). Negatively selected cells were cultured for 5 to 7 days in RPMI 1640 with 10% superlow immunoglobulin FCS without antibiotics in the presence of M-CSF to differentiate into MDMs, as previously described (62–64). After 5 to 7 days, MDMs were infected with M. leprae at a multiplicity of infection (MOI) of 10 or with L. monocytogenes at an MOI of 1.
Coating MDMs with αCD3
Because isolated effector CTL subsets were of varying TCR specificities, we coated MDM target cells with αCD3 (20 ng/ml) by incubating with this antibody for 15 min before admixing effector cells, as previously described (7, 26). Briefly, 24 hours after infection with bacteria, MDMs were washed and then admixed with αCD3 (20 ng/ml) in complete medium. Cells were subsequently washed and were then ready to be mixed with effector cells.
Cell sorting of viable cytotoxic cell populations
FACS was used to purify CD8+ T-CTLs from other populations of CTLs by labeling cells with αCD3, αCD8, and combinations of the identified surface markers that were previously validated. To sort, staining was performed in sterile PBS with 10% FCS, and sorting was performed in complete medium.
Bacteria
M. leprae was provided by R. Lahiri (National Hansen’s Disease Programs, 6 Health Resources Service Administration, Baton Rouge, LA). M. leprae was grown in the footpad of nu/nu mice, as described (65). The Lm-ActA–attenuated 10403S—strain of L. monocytogenes—was provided by G. Chen (UCLA) (66).
Antimicrobial assays
CTL subsets or clones were admixed with differentiated, infected, and anti-CD3–coated (or not) MDM cells, as described above in effector-to-target (E:T) ratios of 2:1. After 24 hours of incubation, all cells were lysed and antimicrobial activity was quantified as described below.
Determination of cellular cytotoxicity
Eukaryotic cytotoxicity of T cell subsets was determined by lactate dehydrogenase (LDH) release using the CytoTox 96 NonRadioactive Cytotoxicity Assay Kit (Promega), as per the manufacturer’s instructions.
Determination of L. monocytogenes viability
L. monocytogenes viability was determined by colony-forming unit (CFU) assays after plating total lysates and supernatant from experimental wells (67).
Determination of M. leprae viability and quantification of antimicrobial activity
Because it is not possible to culture M. leprae, traditional methods of assessing viability that rely on using CFU are precluded. Therefore, M. leprae viability was determined using the bacterial RNA–to–DNA ratio, as previously described (31, 59). Because RNA from lysed cells degrades more rapidly than RNA from intact cells and DNA is more stable than RNA, the mycobacterial RNA–to–mycobacterial DNA ratio, determined through quantitative polymerase chain reaction (qPCR), can be used as a surrogate indicating viability. RNA and DNA were isolated using TRIzol reagent by using the phenol-chloroform and back-extraction method, respectively (manufacturer’s suggested protocol). Isolated RNA was DNase (Qiagen)–treated, and complementary DNA (cDNA) was subsequently synthesized using the iScript cDNA Synthesis Kit (Bio-Rad). Bacterial viability was determined by a qPCR-based method, as previously described (30, 31). Briefly, the levels of bacterial 16S ribosomal RNA (rRNA) and a genomic DNA element, RLEP for M. leprae, were measured by qPCR. The 16S rRNA and DNA element values were determined by using the ΔΔCT analysis, with the DNA value serving as a housekeeping gene. The ratio of RNA to DNA was calculated from averages, and the percent of bacterial viability was calculated relative to MDMs admixed with predominantly noncytotoxic T cells (CD8+NKG2C−NKG2A− population). Primers for M. leprae 16S and RLEP were previously reported (30, 31, 68). Antimicrobial activity was determined by normalizing the RNA-to-DNA ratio from conditions with admixed NKG2C+ or NKG2A+ T cells to the RNA-to-DNA ratio calculated from conditions with NKG2A−NKG2C− T cell subset (control) and calculated according to the equation below.
Flow cytometry antibodies
The following antibodies were used: αCD3-PerCP (peridinin chlorophyll protein) (clone SK7, BD Biosciences), αCD3–Pacific Blue (clone UCHT1, BioLegend), αCD3-UV395 (clone SK7, BD Biosciences), αCD8-BV605 (clone RPA-T8, BioLegend), αCD4–PE (phycoerythrin)–Cy7 (clone OKT4, BioLegend), αGZMB-APC (allophycocyanin) (clone GRB05, Invitrogen), αGZMB–Pacific Blue (clone GB11, BioLegend), αPRF-FITC (fluorescein isothiocyanate) (BD Biosciences), αPRF–PE-Cy7 (clone dG9, eBioscience), αGNLY-PE (clone DH2, eBioscience), αCCR7-BV605 (clone GO43H7, BioLegend), αCCR7–APC-Cy7 (clone G043H7, BioLegend), αCD45RA–PE-Cy7 (clone Hl100, eBioscience), αCD45RO-BV421 (clone UCHL1, BioLegend), αCD56-PerCP (clone HCD56, BioLegend), αCD56-APC (clone CMSSB, eBioscience), αNKG2A-FITC (Miltenyi), and αNKG2C-APC (Miltenyi).
Statistical analysis
Data sets were analyzed for normality using Prism. If normal, a Student’s two-tailed t test was used for comparisons and carried out in Microsoft Excel. If data sets were not normally distributed, a nonparametric test was used to determine significance as indicated in the supplement and tests were carried out in Prism. If more than two data sets were compared, one-way analysis of variance (ANOVA) with Newman-Keuls multiple comparison posttest was used in Prism. Error bars in figures indicate SEM.
Supplementary Material
Table S1. Raw data used to generate figures.
Fig. S1. T-CTLs are expanded in tuberculoid leprosy as compared with lepromatous leprosy.
Fig. S2. T-CTLs express the IL-15 receptor, and IL-15 causes an increased expression of GZMB and causes selective proliferation of T-CTLs over M-CTLs.
Fig. S3. T-CTLs are CD8+ TEMRA cells.
Fig. S4. Generation and examination of CTL profiles.
Fig. S5. Analysis of the specific T-CTL signature reveals an enrichment of NK surface modulatory receptors.
Fig. S6. NK receptor expression on CTL subsets correlates with the number of cytotoxic molecules expressed.
Fig. S7. Comparison of NK receptors between cytotoxic cell populations between two donors.
Fig. S8. T-CTLs express unique αβ TCRs.
Fig. S9. NCAM1, KLRC1, and KLRC2 are the most differentially expressed NK receptors between T-CTL and D-CTL subsets across two donors.
Fig. S10. CD56, NKG2C, and NKG2A are enriched in the T-CTL subset.
Fig. S11. NKG2C and NKG2A mark CD8+ T-CTLs.
Fig. S12. Across healthy donors, NKG2C marks T-CTLs.
Fig. S13. Generation and confirmation of a CD3+CD8+ D-CTL and T-CTL clone.
Fig. S14. Antimicrobial activity is increased by aCD3 coating.
Fig. S15. Antimicrobial activity correlated with CTL subset composition.
Fig. S16. NKG2C marks T-CTLs within T-lep donors.
Acknowledgments:
We would like to thank G. Chen, S. Aliyari, and M. Chapon for sharing resources; P. Liu and R. Teles for assistance with statistical analysis and helpful discussion; A. Legaspi for tissue culture assistance; D. Korenfield for technical assistance; M. Schibler and the UCLA, California NanoSystems Institute, Advanced Light Microscopy Core Facility for assistance with the confocal studies; and I. Schmid and the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility for assistance with flow cytometry and cell sorting. Funding: This work was supported by Dermatology Foundation grants 20151604 and 20141510 (S.J.B.); NIH grants R01HL119068, R01AI022553, R01HL129887, R01AR040312, and P50AR063020 (R.L.M.); grant AI068129; and the Parker Institute for Cancer Immunotherapy (L.L.L.).
Footnotes
Competing interests: S.J.B., R.L.M., and M.P. are authors on a pending patent application PCT/US17/25842 pertaining to the use of T-CTLs in health and disease. The other authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data are in the process of being uploaded to the Gene Expression Omnibus database.
REFERENCES AND NOTES
- 1.Lin PL, Flynn JL, CD8 T cells and Mycobacterium tuberculosis infection. Semin. Immunopathol 37, 239–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan JS, Canaday DH, Boom WH, Balaji KN, Schwander SK, Rich EA, Human alveolar T lymphocyte responses to Mycobacterium tuberculosis antigens: Role for CD4+ and CD8+ cytotoxic T cells and relative resistance of alveolar macrophages to lysis. J. Immunol 159, 290–297 (1997). [PubMed] [Google Scholar]
- 3.Stenger S, Mazzaccaro RJ, Uyemura K, Cho S, Barnes PF, Rosat J-P, Sette A, Brenner MB, Porcelli SA, Bloom BR, Modlin RL, Differential effects of cytolytic T cell subsets on intracellular infection. Science 276, 1684–1687 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Stenger S, Hanson DA, Teitelbaum R, Dewan P, Niazi KR, Froelich CJ, Ganz T, Thoma-Uszynski S, Melián A, Bogdan C, Porcelli SA, Bloom BR, Krensky AM, Modlin RL, An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282, 121–125 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Ochoa M-T, Stenger S, Sieling PA, Thoma-Uszynski S, Sabet S, Cho S, Krensky AM, Rollinghoff M, Sarno EN, Burdick AE, Rea TH, Modlin RL, T-cell release of granulysin contributes to host defense in leprosy. Nat. Med 7, 174–179 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Walch M, Dotiwala F, Mulik S, Thiery J, Kirchhausen T, Clayberger C, Krensky AM, Martinvalet D, Lieberman J, Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 157, 1309–1323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dotiwala F, Mulik S, Polidoro RB, Ansara JA, Burleigh BA, Walch M, Gazzinelli RT, Lieberman J, Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat. Med 22, 210–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn JL, Goldstein MM, Triebold KJ, Koller B, Bloom BR, Major histocompatibility complex class I-restricted T cells are required for resistance to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. U.S.A 89, 12013–12017 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruns H, Meinken C, Schauenberg P, Härter G, Kern P, Modlin RL, Antoni C, Stenger S, Anti-TNF immunotherapy reduces CD8+ T cell–mediated antimicrobial activity against Mycobacterium tuberculosis in humans. J. Clin. Invest 119, 1167–1177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semple PL, Watkins M, Davids V, Krensky AM, Hanekom WA, Kaplan G, Ress S, Induction of granulysin and perforin cytolytic mediator expression in 10-week-old infants vaccinated with BCG at birth. Clin. Dev. Immunol 2011, 438463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busch M, Herzmann C, Kallert S, Zimmermann A, Höfer C, Mayer D, Zenk SF, Muche R, Lange C, Bloom BR, Modlin RL, Stenger S; TBornotTB Network, Lipoarabinomannan-responsive polycytotoxic T cells are associated with protection in human tuberculosis. Am. J. Respir. Crit. Care Med 194, 345–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krensky AM, Clayberger C, Granulysin: A novel host defense molecule. Am. J. Transplant 5, 1789–1792 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Ridley DS, Jopling WH, Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis 34, 255–273 (1966). [PubMed] [Google Scholar]
- 14.Bloom BR, Learning from leprosy: A perspective on immunology and the third world. J. Immunol 137, i–x (1986). [PubMed] [Google Scholar]
- 15.Yamamura M, Uyemura K, Deans RJ, Weinberg K, Rea TH, Bloom BR, Modlin RL, Defining protective responses to pathogens: Cytokine profiles in leprosy lesions. Science 254, 277–279 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Montoya D, Cruz D, Teles RM, Lee DJ, Ochoa MT, Krutzik SR, Chun R, Schenk M, Zhang X, Ferguson BG, Burdick AE, Sarno EN, Rea TH, Hewison M, Adams JS, Cheng G, Modlin RL, Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe 6, 343–353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jullien D, Sieling PA, Uyemura K, Mar ND, Rea TH, Modlin RL, IL-15, an immunomodulator of T cell responses in intracellular infection. J. Immunol 158, 800–806 (1997). [PubMed] [Google Scholar]
- 18.Hogg AE, Bowick GC, Herzog NK, Cloyd MW, Endsley JJ, Induction of granulysin in CD8+ T cells by IL-21 and IL-15 is suppressed by human immunodeficiency virus-1. J. Leukoc. Biol 86, 1191–1203 (2009). [DOI] [PubMed] [Google Scholar]
- 19.White L, Krishnan S, Strbo N, Liu H, Kolber MA, Lichtenheld MG, Pahwa RN, Pahwa S, Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV). Blood 109, 3873–3880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Catalfamo M, Li Y, Henkart PA, N.-p. Weng, IL-15 mimics T cell receptor crosslinking in the induction of cellular proliferation, gene expression, and cytotoxicity in CD8+ memory T cells. Proc. Natl. Acad. Sci. U.S.A 99, 6192–6197 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banchereau J, Thompson-Snipes L, Zurawski S, Blanck JP, Cao Y, Clayton S, Gorvel JP, Zurawski G, Klechevsky E, The differential production of cytokines by human Langerhans cells and dermal CD14+ DCs controls CTL priming. Blood 119, 5742–5749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schluns KS, Lefrancois L, Cytokine control of memory T-cell development and survival. Nat. Rev. Immunol 3, 269–279 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, Pipkin ME, The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilches C, Parham P, KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol 20, 217–251 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Angelini DF, Zambello R, Galandrini R, Diamantini A, Placido R, Micucci F, Poccia F, Semenzato G, Borsellino G, Santoni A, Battistini L, NKG2A inhibits NKG2C effector functions of γδ T cells: Implications in health and disease. J. Leukoc. Biol 89, 75–84 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Arlettaz L, Villard J, de Rham C, Degermann S, Chapuis B, Huard B, Roosnek E, Activating CD94:NKG2C and inhibitory CD94:NKG2A receptors are expressed by distinct subsets of committed CD8+ TCR αβ lymphocytes. Eur. J. Immunol 34, 3456–3464 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Gumá M, Busch LK, Salazar-Fontana LI, Bellosillo B, Morte C, Garcia P, López-Botet M, The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur. J. Immunol 35, 2071–2080 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Kaufmann SH, Effective antibacterial protection induced by a Listeria monocytogenes-specific T cell clone and its lymphokines. Infect. Immun 39, 1265–1270 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufmann SH, Hug E, de Libero G, Listeria monocytogenes-reactive T-lymphocyte clones with cytolytic activity against infected target cells. J. Exp. Med 164, 363–368 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, Moraes MO, Williams DL, Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J. Clin. Microbiol 47, 2124–2130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, Graeber TG, Modlin RL, MicroRNA-21 targets the vitamin D–dependent antimicrobial pathway in leprosy. Nat. Med 18, 267–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan J, Xing Y, Magliozzo RS, Bloom BR, Killing of virulent Mycobacterium tuberculosis by the reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med 175, 1111–1122 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braud VM, Aldemir H, Breart B, Ferlin WG, Expression of CD94–NKG2A inhibitory receptor is restricted to a subset of CD8+ T cells. Trends Immunol 24, 162–164 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Chang DY, Lee HW, Lee H, Kim JH, Sung PS, Kim KH, Hong SH, Kang W, Lee J, Shin SY, Yu HT, You S, Choi YS, Oh I, Lee DH, Lee DH, Jung MK, Suh KS, Hwang S, Kim W, Park SH, Kim HJ, Shin EC, Innate-like cytotoxic function of bystander-activated CD8+ T cells is associated with liver injury in acute hepatitis A. Immunity 48, 161–173.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Braud VM, Allan DS, O’Callaghan CA, Söderström K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ, HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Heinzel AS, Grotzke JE, Lines RA, Lewinsohn DA, McNabb AL, Streblow DN, Braud VM, Grieser HJ, Belisle JT, Lewinsohn DM, HLA-E–dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med 196, 1473–1481 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joosten SA, van Meijgaarden KE, van Weeren PC, Kazi F, Geluk A, Savage NDL, Drijfhout JW, Flower DR, Hanekom WA, Klein MR, Ottenhoff THM, Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8+ T-cells with cytotoxic as well as regulatory activity. PLOS Pathog. 6, e1000782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caccamo N, Pietra G, Sullivan LC, Brooks AG, Prezzemolo T, La Manna MP, Di Liberto D, Joosten SA, van Meijgaarden KE, Di Carlo P, Titone L, Moretta L, Mingari MC, Ottenhoff THM, Dieli F, Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA-E during active tuberculosis and express type 2 cytokines. Eur. J. Immunol 45, 1069–1081 (2015). [DOI] [PubMed] [Google Scholar]
- 39.van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH, Joosten SA, Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLOS Pathog. 11, e1004671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harriff MJ, Wolfe LM, Swarbrick G, Null M, Cansler ME, Canfield ET, Vogt T, Toren KG, Li W, Jackson M, Lewinsohn DA, Dobos KM, Lewinsohn DM, HLA-E presents glycopeptides from the Mycobacterium tuberculosis protein MPT32 to human CD8+ T cells. Eur. J. Immunol 7, 4622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurtrey C, Harriff MJ, T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLOS ONE 12, e0188288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prezzemolo T, van Meijgaarden KE, Franken KLMC, Caccamo N, Dieli F, Ottenhoff THM, Joosten SA, Detailed characterization of human Mycobacterium tuberculosis specific HLA-E restricted CD8+ T cells. PLOS ONE 48, 293–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon CW, Raulet DH, Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol 13, 465–470 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D, Pitard V, Merville P, Moreau JF, Delfraissy JF, Dechanet-Merville J, Bonneville M, Venet A, Vivier E, Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J. Immunol 173, 7223–7229 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Correia MP, Costa AV, Uhrberg M, Cardoso EM, Arosa FA, IL-15 induces CD8+ T cells to acquire functional NK receptors capable of modulating cytotoxicity and cytokine secretion. Immunobiology 216, 604–612 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Jabri B, Selby JM, Negulescu H, Lee L, Roberts AI, Beavis A, Lopez-Botet M, Ebert EC, Winchester RJ, TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity 17, 487–499 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez RM, Suarez-Alvarez B, Lavín JL, Mosén-Ansorena D, Baragaño Raneros A, Márquez-Kisinousky L, Aransay AM, Lopez-Larrea C, Epigenetic networks regulate the transcriptional program in memory and terminally differentiated CD8+ T Cells. J. Immunol 198, 937–949 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Bratke K, Kuepper M, Bade B, Virchow JC Jr., W. Luttmann, Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol 35, 2608–2616 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Azuma M, Phillips JH, Lanier LL, CD28- T lymphocytes. Antigenic and functional properties. J. Immunol 150, 1147–1159 (1993). [PubMed] [Google Scholar]
- 50.Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D, Sette A, Vijayanand P, Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol 3, eaan8664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marshall NB, Vong AM, Devarajan P, Brauner MD, Kuang Y, Nayar R, Schutten EA, Castonguay CH, Berg LJ, Nutt SL, Swain SL, NKG2C/E marks the unique cytotoxic CD4 T cell subset, ThCTL, generated by influenza infection. J. Immunol 198, 1142–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, Briere F, Chaussabel D, Zurawski G, Palucka AK, Reiter Y, Banchereau J, Ueno H, Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity 29, 497–510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Modlin RL, Hofman FM, Kempf RA, Taylor CR, Conant MA, Rea TH, Kaposi’s sarcoma in homosexual men: An immunohistochemical study. J. Am. Acad. Dermatol 8, 620–627 (1983). [DOI] [PubMed] [Google Scholar]
- 54.van den Oprd JJ, de Wolf-Peeters C, Facchetti F, Desmet VJ, Cellular composition of hypersensitivity-type granulomas: Immunohistochemical analysis of tuberculous and sarcoidal lymphadenitis. Hum. Pathol 15, 559–565 (1984). [DOI] [PubMed] [Google Scholar]
- 55.Ulrichs T, Kosmiadi GA, Trusov V, Jörg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH, Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. J. Pathol 204, 217–228 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Jongstra J, Schall TJ, Dyer BJ, Clayberger C, Jorgensen J, Davis MM, Krensky AM, The isolation and sequence of a novel gene from a human functional T cell line. J. Exp. Med 165, 601–614 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Artyomov MN, Munk A, Gorvel L, Korenfeld D, Cella M, Tung T, Klechevsky E, Modular expression analysis reveals functional conservation between human Langerhans cells and mouse cross-priming dendritic cells. J. Exp. Med 212, 743–757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hrvatin S, Deng F, O’Donnell CW, Gifford DK, Melton DA, MARIS: Method for analyzing RNA following intracellular sorting. PLOS ONE 9, e89459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Realegeno S, Kelly-Scumpia KM, Dang AT, Lu J, Teles R, Liu PT, Schenk M, Lee EY, Schmidt NW, Wong GC, Sarno EN, Rea TH, Ochoa MT, Pellegrini M, Modlin RL, S100A12 is part of the antimicrobial network against Mycobacterium leprae in human macrophages. PLOS Pathog. 12, e1005705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, Greenberg PD, Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood 110, 201–210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewinsohn DM, Alderson MR, Briden AL, Riddell SR, Reed SG, Grabstein KH, Characterization of human CD8+ T cells reactive with Mycobacterium tuberculosis–infected antigen-presenting cells. J. Exp. Med 187, 1633–1640 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabri M, Stenger S, Shin D-M, Yuk J-M, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, Teles R, Montoya D, Iyer SS, Bruns H, Lewinsohn DM, Hollis BW, Hewison M, Adams JS, Steinmeyer A, Zugel U, Cheng G, Jo EK, Bloom BR, Modlin RL, Vitamin D is required for IFN-γ–mediated antimicrobial activity of human macrophages. Sci. Transl. Med 3, 104ra102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, Lee ZW, Lee SH, Kim JM, Jo EK, Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 6, 231–243 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Yang C-S, Shin D-M, Kim K-H, Lee Z-W, Lee C-H, Park SG, Bae YS, Jo E-K, NADPH oxidase 2 interaction with TLR2 is required for efficient innate immune responses to mycobacteria via cathelicidin expression. J. Immunol 182, 3696–3705 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Lahiri R, Randhawa B, Krahenbuhl J, Application of a viability-staining method for Mycobacterium leprae derived from the athymic (nu/nu) mouse foot pad. J. Med. Microbiol 54, 235–242 (2005). [DOI] [PubMed] [Google Scholar]
- 66.O’Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng G, Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TANK-binding kinase 1. J. Immunol 174, 1602–1607 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G, Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416, 190–194 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Wheelwright M, Kim EW, Inkeles MS, De LA, Pellegrini M, Krutzik SR, Liu PT, All-trans retinoic acid-triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2. J. Immunol 192, 2280–2290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliveros JC, VENNY. An interactive tool for comparing lists with Venn diagrams (2007–2015); http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw data used to generate figures.
Fig. S1. T-CTLs are expanded in tuberculoid leprosy as compared with lepromatous leprosy.
Fig. S2. T-CTLs express the IL-15 receptor, and IL-15 causes an increased expression of GZMB and causes selective proliferation of T-CTLs over M-CTLs.
Fig. S3. T-CTLs are CD8+ TEMRA cells.
Fig. S4. Generation and examination of CTL profiles.
Fig. S5. Analysis of the specific T-CTL signature reveals an enrichment of NK surface modulatory receptors.
Fig. S6. NK receptor expression on CTL subsets correlates with the number of cytotoxic molecules expressed.
Fig. S7. Comparison of NK receptors between cytotoxic cell populations between two donors.
Fig. S8. T-CTLs express unique αβ TCRs.
Fig. S9. NCAM1, KLRC1, and KLRC2 are the most differentially expressed NK receptors between T-CTL and D-CTL subsets across two donors.
Fig. S10. CD56, NKG2C, and NKG2A are enriched in the T-CTL subset.
Fig. S11. NKG2C and NKG2A mark CD8+ T-CTLs.
Fig. S12. Across healthy donors, NKG2C marks T-CTLs.
Fig. S13. Generation and confirmation of a CD3+CD8+ D-CTL and T-CTL clone.
Fig. S14. Antimicrobial activity is increased by aCD3 coating.
Fig. S15. Antimicrobial activity correlated with CTL subset composition.
Fig. S16. NKG2C marks T-CTLs within T-lep donors.