SUMMARY
Cell survival is a critical and ubiquitous endpoint in biology. The broadly accepted colony formation assay (CFA) directly measures a cell’s ability to divide; however, it takes weeks to perform and is incompatible with high-throughput screening (HTS) technologies. Here, we describe the MicroColonyChip, which exploits microwell array technology to create an array of colonies. Unlike the CFA, where visible colonies are counted by eye, using fluorescence microscopy, microcolonies can be analyzed in days rather than weeks. Using automated analysis of microcolony size distributions, the MicroColonyChip achieves comparable sensitivity to the CFA (and greater sensitivity than the 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide [XTT] assay). Compared to CellTiter-Glo, the MicroColonyChip is as sensitive and also robust to artifacts caused by differences in initial cell seeding density. We demonstrate efficacy via studies of radiosensitivity and chemosensitivity and show that the approach is amenable to multiplexing. We conclude that the MicroColonyChip is a rapid and automated alternative for cell survival quantitation.
Graphical Abstract:
In Brief
The gold standard for cytotoxicity testing is the colony formation assay (CFA), which requires visible colonies in large dishes. Ngo et al. describe the MicroColonyChip, which directly measures the ability of cells to divide. This automated miniaturized assay retains the sensitivity of the CFA and takes days instead of weeks.
INTRODUCTION
Cell fate, whether to die, divide, or senesce, is an underlying driver of cancer and disease. Therefore, cell survival is a broadly used metric in a number of contexts in the life sciences. For example, cell viability assays enable basic research studies of molecular pathways and also applied studies of chemical safety. Viability assays are also a mainstay in the pharmaceutical industry, where they are used to predict adverse effects, as well as for establishing efficacy of compounds designed to target cancer cells. Accurate cell survival testing thus plays a significant role in health. For example, in terms of public health, a false negative result for toxicity could lead to people being exposed to a hazardous chemical. For the pharmaceutical industry, a false positive result could mean that an effective drug does not make it to the market, while a false negative result could mean that patients get exposed to toxic pharmaceuticals.
The broadly accepted method for cell survival quantitation is the colony formation assay (CFA) (Cook and Mitchell, 1989), wherein cells are exposed to an agent and the ability of single cells to form colonies is quantified by eye (Franken et al., 2006). While the assay has an impressive dynamic range (over several orders of magnitude), it is inconvenient and relatively low throughput due to the need for 2 to 3 weeks of incubation time. In addition, to prevent colony overlap, cells are plated in large dishes, which require large amounts of media and thus high amounts of test compounds, which can be problematic when studying small molecule libraries, which generally have limited quantities. Further, manual colony counting is time consuming, varies from person to person and lab to lab, and can be prone to bias (Cook and Mitchell, 1989).
Due to the significant limitations of the CFA, faster viability assays have become popular alternatives. A popular approach is to measure mitochondrial function via metabolism-based endpoints. For example, tetrazolium assays (e.g., 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide [MTT] and 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide [XTT]) are based upon the underlying principle that a live cell can reduce tetrazolium salts to formazan derivatives, causing a change in color that can be measured by absorbance (Berridge et al., 2005). Because of their relative ease of use and affordability, tetrazolium assays are commonly used despite their low sensitivity and propensity to artifacts. For example, artifacts can arise from spontaneous reduction of the tetrazolium salts by reducing agents in cell media (Cook and Mitchell, 1989) or changes in the absorbance induced by pH changes Plumb et al., 1989). An analogous approach is to quantify intracellular ATP levels (Crouch et al., 1993). The most popular assay for this approach is the CellTiter-Glo (CTG) assay, wherein luciferin-luciferase luminescence is used to estimate the levels of ATP. Although CTG is exquisitely sensitive, ATP assays are affected by viability-independent perturbations, such as nutrient depletion and pH changes (Galluzzi et al., 2009; Kepp et al., 2011), and can underestimate toxicity for short incubation periods Sumantran, 2011). Importantly, as described below, we have also found that CTG can yield highly variable results depending on the initial cell density used in the assay.
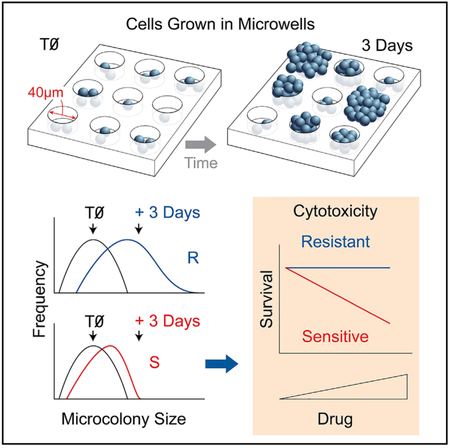
Here, we set out to create an alternative approach to cell proliferation analysis that combines several key advantages of existing microtiter plate-based assays with a direct measure of cell division. The result is the “MicroColonyChip” (μCC). For the CFA, the number of cells able to form colonies is used as a metric for cell survival, which is accomplished by counting colonies above a threshold size (estimated by eye). In contrast, for the μCC, the total number of cell divisions is used as a metric of cell survival, which is estimated based on colony sizes (where the number of cells per colony is quantified using a fluorescent DNA stain). Specifically, the μCC approach depends on an alternative metric for cell growth, namely the change in the distribution of microcolony sizes (which reflects the total number of cell divisions). Since colonies are assessed using a microscope, instead of counting by eye, the assay takes 3–5 days instead of 2–3 weeks (note that for both assays, longer times may be required for slower-growing cell types). Furthermore, growing cells in a microarray enables close packing of colonies, while suppressing colony overlap, making it possible to move from large dishes to a 96-well plate format (requiring ~250× less surface area compared to the CFA). Remarkably, the sensitivity of the μCC is nearly identical to the CFA. Furthermore, we find that the μCC is more sensitive than the XTT assay and has comparable sensitivity to CTG, while also being more robust against artifacts caused by culture conditions. Finally, we show that the μCC can be applied to studies of chemical toxicity in metabolically relevant conditions, an important factor for predicting liver toxicity. Taken together, the μCC provides a robust, rapid, automated, and sensitive platform for cell survival quantitation.
RESULTS
Patterning of Cells Using a Microwell Array Platform
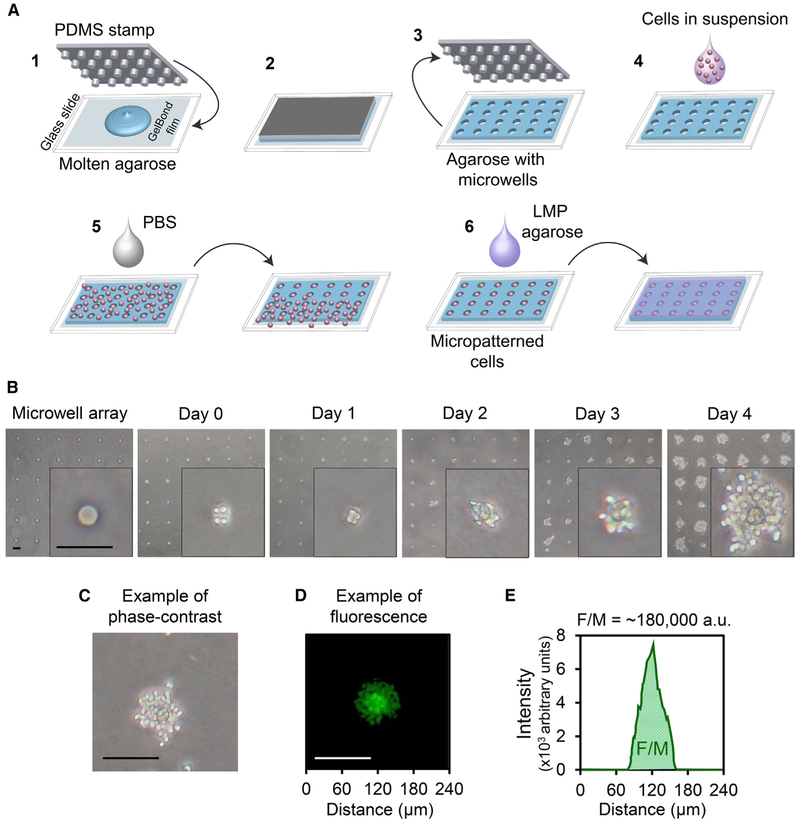
The μCC is based upon the ability to grow cells in a microarray. Here, we use a cell microarray approach that was previously been described by our laboratory in collaboration with S. Bhatia (Wood et al., 2010). Briefly, a polydimethylsiloxane (PDMS) mold (created by photolithography and soft lithography) is pressed into molten agarose. The agarose is allowed to gel, and the mold is removed to create an array of microwells. For the experiments described here, each microwell is ~40 μm in both diameter and depth, spaced 240 μm apart from one another; however, the microwell array platform provides a tunable physical distance between microcolonies and tunable well sizes to accommodate different cell types. A bottomless 96-well plate is then compressed on top of the microwell array to create macrowells, each containing ~300 microwells. Cells are then loaded by gravity and excess cells are removed by sheer force (Figure 1A). Since excess cells can be washed away, the assay is robust across a remarkably broad range of initial cell numbers (2,000–200,000 cells/macrowell). Cells are then trapped by layering low melting point agarose above the cells.
Figure 1. Construction of μCC and Quantification of Total DNA Content.
(A) Micropatterning cells in μCC. (1) APDMS stamp with microposts is pressed into molten agarose; (2) agarose is allowed to cool and gelate; (3) stamp is lifted to reveal patterned microwells on agarose; (4) cell suspension is added; (5) cells settle into microwells via gravity, and excess cells are washed off via sheer force; and (6) cells are trapped by an overlay of 0.3% low melting point agarose.
(B) Phase-contrast images of patterned TK6 microcolonies (40x magnification). Far left: empty agarose microwell array. Day 0: micropatterned cells after loading. Day 1 to day 4: images of microcolonies after growth for the indicated time.
(C) Phase-contrast image of a TK6 microcolony after 4 days in culture.
(D) A fluorescent image of a different TK6 microcolony stained with Vybrant DyeCycle Green.
(E) A plot of the average fluorescence intensity of each pixel column from the left to the right of the colony shown in (D), after background correction (binary mask using Otsu thresholding method). F/M is the total area under the curve.
All scale bars, 100 μm.
TK6 lymphoblastoid cells are ~15 μm in diameter and can be readily micropatterned with one to seven cells per well at ~40 μm in diameter (Figure 1B). Suspension cells that naturally grow without attachment have previously been shown to form colonies in soft agar (Imamura and Moore, 1968; Valiathan et al., 2012). Consistent with these studies, TK6 microcolonies grow on the microwell array platform. The appearance of cells growing out of the microwell boundary was noted as soon as 2 days in culture (Figure 1B, day 2). By day 4, an average microwell harbored a microcolony with more than ~60 cells, which is consistent with a doubling time of ~20 h (Furth et al., 1981) and an average of approximately 3 cells/well on day 0 (Figure 1B, day 4).
High-Throughput Quantification of Microcolony Size via Nucleic Acid Fluorescence Staining
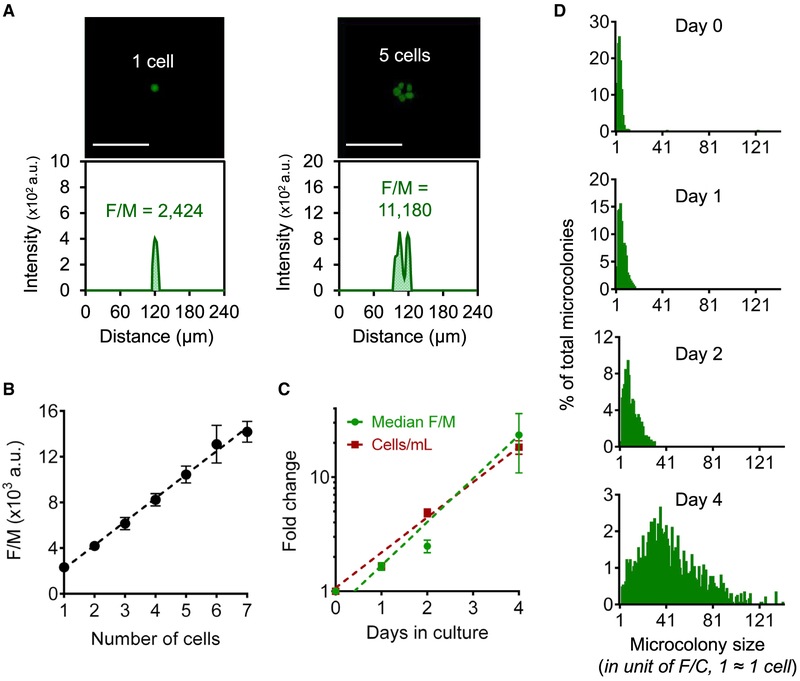
We estimated the number of cells per microcolony based on the total DNA content, using a fluorescence DNA stain that is membrane permeable (Vybrant DyeCycle Green; Figure 1C) (Blaheta et al., 1991). In non-synchronous cultures, the average DNA content per cell stays relatively constant (Jones et al., 2001). We imaged microcolonies (Figure 1D) and then used an in-house MATLAB program “uCCanalyzer” (Data S1) for microcolony analysis. The “uCCanalyzer” was adapted from previous software (Wood et al., 2010) to specifically detect the locations of the microcolonies, generate images of individual microcolonies, and calculate the integrated fluorescence intensity per microcolony (F/M) (Figure 1E).
To explore the efficacy of the F/M parameter, the number of distinct fluorescent nuclei in microcolonies was counted manually as an estimate of the total cell number (examples in Figure 2A). As shown in Figure 2B, the number of cells per microcolony increases linearly with the microcolony’s F/M value (R2 = 0.92, p < 0.0001; note that the fluorescence level for seven cells is not significantly different from expected, given a linear increase in signal). We found that F/M for a single TK6 cell, or fluorescence intensity per cell (F/C), is 2,300 ± 500 (arbitrary fluorescence unit). To analyze efficacy for larger colonies, we monitored the change in the median F/M value over 4 days in culture. We found that TK6 have a doubling time of ~20 h, which is consistent with what was observed for TK6 cells grown in liquid culture (Figure 2C). We conclude the doubling time of TK6 cells is not significantly affected by culturing in the microarray (as opposed to liquid culture) when analyzed 4 days post plating and that F/M is a sensitive and robust measure of microcolony size (since there is no apparent saturation of signal when analyzed 4 days post plating).
Figure 2. Total DNA Content as a Measure of Microcolony Size.
(A) Calculation of F/M for microwells with one cell (left) and five cells (right). Top: fluorescent images of TK6 cells stained with Vybrant DyeCycle Green. Bottom: fluorescence intensity plots of the corresponding fluorescent images. Scale bars, 100 μm.
(B) Average F/M values for one to seven TK6 cells. Dotted black line = linear fit of data (R2 = 0.92, p < 0.0001).
(C) Fold change of median F/M for TK6 microcolonies in μCC (green) and TK6 cell density in liquid culture (red). Dotted lines = exponential fits of data (R2 = 0.99 for both).
(D) TK6 microcolony size distributions. F/M values of >700 microcolonies were analyzed for each distribution and converted to cell numbers by dividing by the value of one F/C. The y axis for each plot is individually scaled.
In (B) and (C), all data are means of at least three independent experiments (n ≥ 3). Error bars are SEM.
See also Figure S1.
Construction of Microcolony Size Distribution Using F/M Values
We monitored the growth of the TK6 microcolonies over the course of 4 days. On each day, a μCC was removed and microcolonies were analyzed for DNA content (Figure 2D). Consistent with the traditional CFA, some microcolonies remained very small, while others had grown extensively. After 4 days, microcolony F/M values ranged from 1 F/C to 150 F/C, corresponding to ~1 cell up to ~150 cells. It is important to note that a colony that initially had 7 cells could readily double to form a colony of more than 150 cells over the course of 4 days (approximately 4.5 doubling times). Figure 2D shows that the F/M distribution of TK6 microcolonies is very tight on day 0 and that as the microcolonies grow, the distribution both shifts to the right and broadens. One can readily see the extent to which the populations become broader when all plots have the same scale for the y axis (Figure S1A). We postulate that the broadening of F/M distributions is attributable to the difference in starting microcolony sizes, as well as the rates of cell division and cytotoxicity.
Toxicity Measurement with μCC
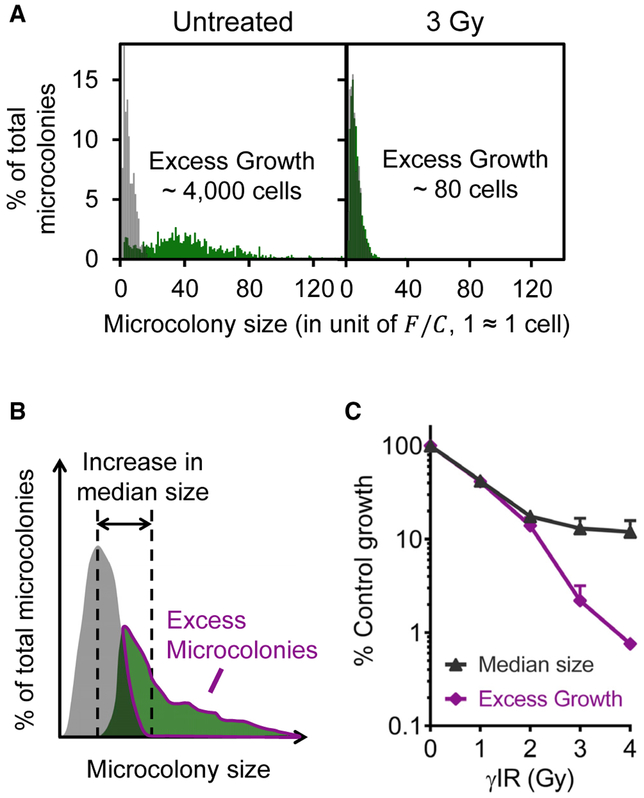
To perform a toxicity assay, cells are exposed to a toxic agent for a fixed time and analyzed after at least three cell divisions. As shown in Figure 2D, microcolony growth leads to a shift of the microcolony size distribution toward higher frequencies of larger microcolonies compared to the initial population. Figure 3A illustrates that a toxic treatment (e.g., γ-ionizing radiation [γIR]) leads to a much smaller shift, indicating the presence of far fewer surviving cells. Based on this observation, we aimed to measure toxicity by estimating the total number of surviving cells. Specifically, for each microcolony size, we subtracted the relative frequency in the initial population from the relative frequency in the final population. We defined the positive values of the subtractions to be an approximation of the excess microcolonies. For each microcolony size, we multiplied excess microcolonies by the number of cells per microcolony to obtain an estimation of the total number of cells in the excess microcolonies (the total number of surviving cells). A summation of the total number of cells in the excess microcolonies across all available microcolony sizes constitutes excess growth (EG). Thus, EG reflects the extent to which colonies have grown beyond the starting population in size (see hand-drawn illustration in Figure 3B). EG values for each treatment condition are then calculated and compared to a negative control EG (unexposed cells) to obtain percent control growth. In practice, data are analyzed in an automated fashion using “uCCHistogram,” a package of in-house Python scripts (Data S2).
Figure 3. Comparison of Microcolony Size Distributions for Toxicity Assessment.
(A) TK6 microcolony size distributions. Left: distribution before γIR (starting population, gray) overlaying with untreated distribution after 3 days in culture (untreated, green). Right: distribution before γIR (starting population, gray) overlaying with γ-irradiated distribution after 3 days in culture (3 Gy, green).
(B) Illustration of microcolony size distributions before (starting population, gray) and after growth (final population, green). EG is the total number of cells in excess microcolonies (magenta outlined).
(C) μIR-induced toxicity (% control growth) derived from normalization of γ-irradiated microcoionies by “untreated” distribution using median microcolony sizes or EG. n ≥ 3. Error bars are SEM.
To explore the sensitivity of EG as a metric for cell survival, we treated micropatterned TK6 cells with γIR. Previous studies using the CFA have shown that TK6 cells are highly sensitive to γIR (Amundson et al., 1993; Wenz et al., 1998). We examined the microcolony size distributions for TK6 microcolonies 3 or 4 days after γ-radiation. As shown in Figure 3A, after 3 days, non-irradiated cells gave rise to a broad distribution of microcolony sizes, with approximately a 9-fold increase in median colony size and EG of ~4,000 cells. In contrast, microcolonies that were exposed to 3 Gy of γIR were growth inhibited, due to either cell death or inability to divide, leading to overall smaller microcolonies after 3 days and EG of ~80 cells (note that the final population is nearly overlapping with the starting population).
Having established EG as a parameter for cell survival, we compared it to a more established metric, the median microcolony size. We first used the median colony size to estimate toxicity and found that we could detect about one-log difference in cell survival. Under the exact same conditions, we calculated EG. We found that more than a two-log change in cell survival is detected using EG (Figure 3C). Thus, the intuitive approach of simply comparing median values is relatively ineffective compared to the EG values.
To explore the efficacy of the EG approach for cells with different amounts of total DNA (e.g., different mammalian cell types), we explored whether derivation of F/C is required. Specifically, because F/C is used to estimate EG values, we tested the effect of varying F/C in order to learn about the potential impact of variation in total DNA content per cell. Remarkably, EG is robust against a 25-fold range of F/C values (Figure S1B; note that F/C = 2,300 was derived from the standard curve for TK6 in Figure 2B), obviating the need for re-deriving F/C for different mammalian cell types. Therefore, for all subsequent studies, all the microcolony size distributions are analyzed for EG values using F/C = 2,300, and the method will simply be referred to as “μCC.”
Radiosensitivty of Cancer Cells: Comparison of μCC, XTT, CTG, and CFA
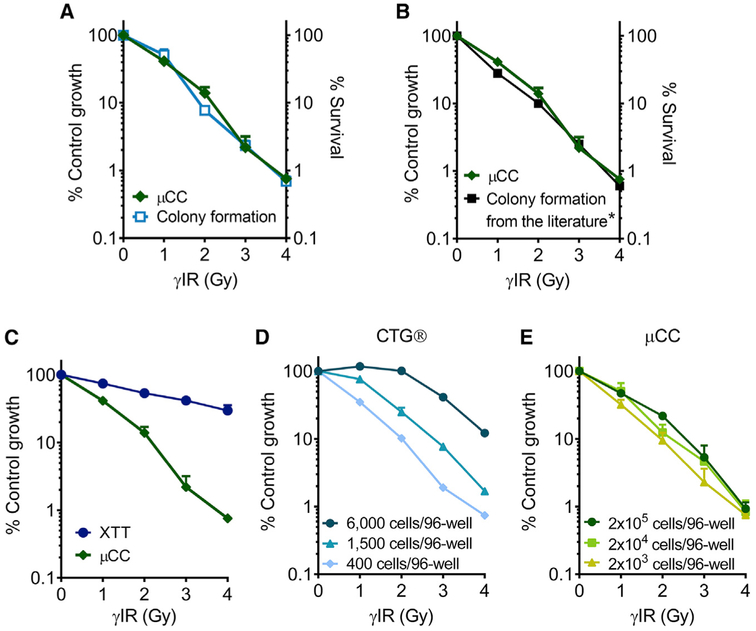
To investigate the sensitivity of the μCC for measuring γIR-induced toxicity, we compared its performance to that of several other assays. Specifically, we compared μCC with the CFA (Furth et al., 1981; Kraemer et al., 1980) and two commercially available methods, XTT and CTG. For the CFA, we tracked the appearance of colonies in 96-well plates over 18 days and observed that some colonies were not obvious until the last day (Figure S2). Therefore, we decided on a timescale of 3 weeks before counting the colonies in order to maximize the assay’s sensitivity. In contrast, the μCC data were obtained 3 days after γIR. Remarkably, we observed that the μCC yields an exponential toxicity curve that is virtually undistinguishable from the survival curve obtained from the CFA (Figure 4A). We then specifically compared the μCC results to a study performed in 1998 (Wenz et al., 1998) using the same cell type but analyzed using the traditional CFA. Interestingly, the μCC assay also yielded nearly identical results to the historic data (Figure 4B). We conclude that the μCC assay is as sensitive as the CFA, while being far faster (3 days versus 3 weeks).
Figure 4. Comparisons among Toxicity Assays for TK6 Cells Exposed to γIR.
(A) Data from CFA (light blue) and μCC (green).
(B) CFA data obtained from Wenz et al. 1998 (black) were reproduced with permission from Radiation Research.
(C) Data from XTT (dark blue).
(B and C) The μCC data are the same as in (A).
(D) CTG data were obtained from different cell seeding densities.
(E) The μCC data were obtained from different cell loading densities.
n ≥ 3. Error bars are SEM.
See also Figure S3.
Many microtiter-plate methods for measuring cell viability have been developed as faster and more convenient alternatives to the CFA. We therefore sought to compare the μCC with two of the most popular assays, XTT and CTG. The XTT method is a widely used colorimetric assay that estimates the number of viable cells by measuring the ability of cells to reduce the faint yellow salt (XTT) to a bright orange water-soluble formazan dye (Scudiero et al., 1988). Remarkably, the μCC assay is orders of magnitude more sensitive than the popular XTT assay (Figure 4C).
The CTG assay (from Promega) is based on luminescent quantification of cellular ATP, which is proportional to the number of metabolically active cells (Crouch et al., 1993). We found that when using 400 cells per well on a 96-well plate, the CTG assay and the μCC assay yield comparable results (compare Figure 4D and Figure 4E). Flowever, unexpectedly, the CTG results are highly dependent on the initial cell number (Figure 4D; Figure S3A). Thus, results can potentially vary considerably from one experiment to the next if the concentration of cells is not optimized. Unlike CTG, the μCC is highly consistent across a two-log range of cell loading densities (Figure 4E; Figure S3B). Importantly, the number of days in culture also changes the results of CTG, while having a minimal effect on the μCC (Figure S3).
Use of μCC as a Tool for Study of DNA Repair in Chemotherapeutic Sensitivity
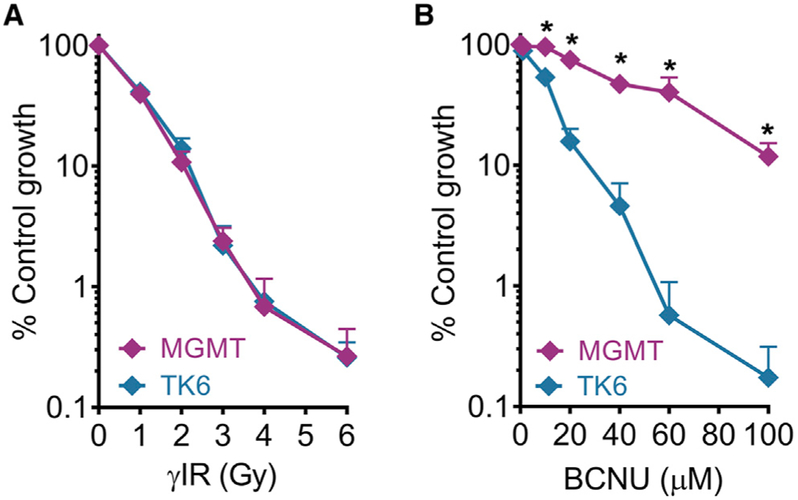
There is a great interest in DNA damaging agents, because they can both cause cancer and be used to treat cancer. Here, we explored the utility of the μCC as a tool for studies of the role of DNA repair in preventing cytotoxicity caused by two DNA damaging agents used for cancer chemotherapy, namely γIR and N,N’-bis (2-chloroethyl)-N-nitrosourea (BCNU). BCNU, a chemotherapeutic alkylating agent used to treat brain cancers (Durando et al., 2003; Hochberg et al., 1981; Valiathan et al., 2012), induces highly cytotoxic DNA interstrand crosslinks (Gonzaga et al., 1992). DNA crosslinks are formed when O6-chloroe-thylguanine lesions form an ethano ring that can then react with cytosine to form an interstrand crosslink (Tong et al., 1982). The O6-methylguanine DNA methyltransferase (MGMT) protein removes the O6-chloroethyl adducts and thus prevents ethano lesions from forming crosslinks (Gerson, 2004; Kaina et al., 1991a).
TK6 cells are deficient in MGMT and are very sensitive to BCNU toxicity, while TK6 cells stably transfected with cDNA expressing the MGMT protein (designated as MGMT in Figures 5A and 5B) are resistant to BCNU (Gerson, 2004; Kaina et al., 1991b; Valiathan et al., 2012). As a control, we studied the effects of γIR, for which MGMT is not expected to play a role. Results show that TK6 and TK6+MGMT are similarly sensitive to γIR (Figure 5A), consistent with the fact that MGMT is not involved in the repair of γIR-induced DNA damage. In contrast, TK6+MGMT displays vastly increased resistance to BCNU compared to MGMT-deficient TK6 cells, as expected (Figure 5B). These results again show that the μCC accurately recapitulates results obtained using the gold standard CFA (Valiathan et al., 2012).
Figure 5. Application of μCC to Study the Role of DNA Repair Protein in Mediating Toxicity Induced by DNA Damaging Agents.
(A) γIR treatment for TK6 and TK6+MGMT (labeled MGMT).
(B) BCNU treatment for TK6 and TK6+MGMT. *p < 0.05, Student’s t test, two-tailed, unequal variance. n ≥ 3. Error bars are SEM.
Application of μCC to Test for Chemical Toxicity under Metabolically Relevant Conditions
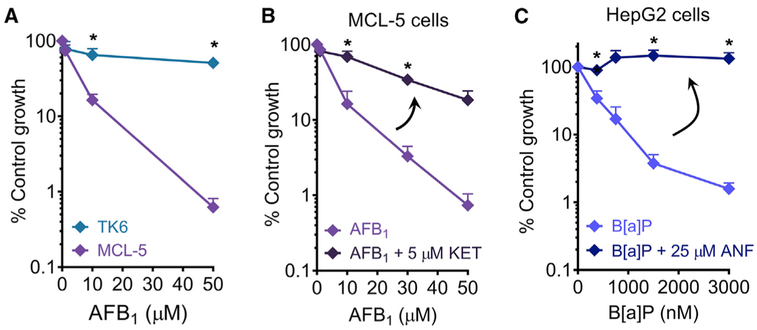
In the human body, foreign chemicals are extensively metabolized, mainly by hepatocytes in the liver (Golan and Tashjian, 2012). This process can result in reactive intermediates that form mutagenic and cytotoxic DNA adducts (Nebert and Dalton, 2006). While viability assays are widely used to monitor the potential health impact of industrial and pharmaceutical chemicals, a major drawback is the use of cells that lack metabolic capacity. The cytochrome P450s (CYP450s) are a superfamily of metabolizing enzymes, responsible for ~70%–80% of phase I metabolism in the liver (Evans and Relling, 1999). Here, we employed metabolically competent cell lines, MCL-5 and HepG2. MCL-5 is a human B-lymphoblastoid cell line engineered to stably express human cytochrome P450 CYP1A1, CYP1A2, CYP2A6, CYP2E1, and CYP3A4 (Crespi et al., 1991). Together, these metabolic enzymes are responsible for approximately 50% of P450 activity in phase I metabolism (Evans and Relling, 1999). HepG2 is a human hepatocyte cell line derived from hepatocellular carcinoma. HepG2 cells retain partial liver-specific functions, including relatively high expressions of the CYP1 family (CYP1A1, CYP1A2, and CYP1B1) and are a classic cell model for metabolism studies (Castell et al., 2006).
Millions of people worldwide are exposed to aflatoxin B1 (AFB1), a procarcinogen that can be metabolized in the liver to form a potent liver carcinogen. AFB1 is present in the molds Aspergillus flavus and A. parasiticus, which can be found on grains. Genotoxic metabolites are generated via oxidation of AFB1 mainly by CYP3A4 and CYP1A2 (Gallagher et al., 1994). We applied the μCC to measure the toxicity of AFB1 in MCL-5 cells. To control for the effects of metabolism, we included TK6 as a negative control cell line, since it lacks expression of phase I enzymes. As expected, MCL-5 cells are significantly more sensitive than TK6 (Figure 6A). Further, we confirmed that MCL-5’s sensitivity to AFB1 is due to metabolic activation of AFB1 by using ketoconazole (KET), a potent inhibitor of CYP3A4 activity (Greenblatt et al., 2011). Whereas MCL-5 cells are extremely sensitive to AFB1, inhibition of metabolism by KET efficiently suppresses this sensitivity (Figure 6B).
Figure 6. Application of μCC to Study Toxicity Induced by Metabolic Activation in Metabolically Competent Cell Lines.
(A) AFB1 exposure for TK6 and MCL-5. *p < 0.05, Student’s t test, two-tailed, unequal variance.
(B) Treatment of MCL-5 cells with AFB1 alone or in the presence of 5 μM KET.
(C) Treatment of HepG2 cells with B[a]P alone or in the presence of 25 μM ANF.
In (B) and (C), *p < 0.05, Student’s t test, two-tailed, paired. n ≥ 3. Error bars are SEM.
In metabolism studies, HepG2 is one of the most commonly used cell lines. Therefore, to further demonstrate the utility of the μCC, we applied the assay in HepG2 cells to measure the toxicity of benzo[a]pyrene (B[a]P), another human procarcinogen. Most people are exposed to B[a]P daily via breathing in fuel exhaust, cigarette smoke, or consuming charred food (International Agency for Research on Cancer, 2012). B[a]P is metabolized in the liver mainly by the CYP1 family to form highly carcinogenic metabolites (International Agency for Research on Cancer, 2012). Because HepG2 is an adherent cell type, we incorporated a layer of collagen type 1 into the μCC to enable cell attachment. The HepG2 microcolonies were cultured in the presence of B[a]P for 5 days and then assessed for toxicity. In order to control for the effect of B[a]P metabolism, we used α-naphthoflavone (ANF), a potent inhibitor of CYP1A2 (Cho et al., 2003). As expected, while HepG2 is highly sensitive to B[a]P, the suppression of B[a]P metabolism via ANF makes the cells completely resistant (Figure 6C). Taken together, the incorporation of metabolically competent cells, such as MCL-5 and HepG2, into the μCC platform yields a rapid, sensitive, and physiologically relevant method for chemical toxicity testing.
DISCUSSION
Despite the fact that a large number of cytotoxicity tests are performed every year worldwide, there have been relatively few fundamental advances in cytotoxicity assays in recent decades. Although having been developed decades ago, the CFA is still the most broadly accepted method for cell survival. Major drawbacks of the assay include its low throughput and laborious procedure as well as incompatibility with high-throughput screening. Alternative faster approaches have been developed, but they rely on indirect measures of cell viability. With nearly identical sensitivity, we show here that the μCC rivals the CFA with its speed and automation and thus presents a valuable alternative. The results presented in this work highlight a few important advantages of the μCC assay, as follows: (1) analysis of microcolony size distribution is a direct measure of cells’ ability to divide, enabling quantification of cell survival in ~3–5 days (instead of ~2–3 weeks for the CFA); (2) by arraying cells in microwells, the μCC shrinks the area needed for cell growth to ~1/250 of the area needed for the CFA, enabling compatibility with the microtiter 96-well plate format; (3) the μCC is capable of measuring cell survival on a multi-log scale, enabling sensitive quantification of toxicity; and (4) the μCC is robust against experimental noise introduced by differences in initial cell numbers, unlike CTG. Perhaps the most important observations are that results from the μCC are nearly identical to data from the traditional CFA performed in our laboratory as well as to data collected using the same approach ~20 years ago by leading toxicology laboratories (Hickman and Samson, 1999; Valiathan et al., 2012; Wenz et al., 1998).
Today, common approaches for measuring cell viability are the tetrazolium and ATP assays. The underlying principle for these assays is that the number of viable cells is estimated by the cells’ mitochondrial functions, either to reduce a tetrazolium salt or to maintain intracellular ATP level. Although tetrazolium assays have enjoyed popularity due to the convenient microtiter plate format, they suffer from limited sensitivity (Carmichael et al., 1987a, 1987b; Cook and Mitchell, 1989). In contrast, as demonstrated in this work, the μCC is capable of measuring multi-log survival levels in the same amount of time and is more sensitive than the XTT assay, one of the most common tetrazolium assays. Importantly, the μCC relies solely on DNA content, which is a reliable and robust measure of cell number (Blaheta et al., 1991; Jones et al., 2001; Quent et al., 2010; Rago et al., 1990). In contrast, tetrazolium assays depend on mitochondrial activity, which can vary due to factors unrelated to cell division (Cook and Mitchell, 1989; Petty et al., 1995; Plumb et al., 1989).
Similar to tetrazolium assays, ATP assays are also a staple in cell viability testing. The CTG assay is amendable to microtiter plate format and is highly sensitive, and the luminescence output is very convenient to measure. In this work, we observed that CTG results are highly variable depending on both initial cell number and the number of days in culture. This is in sharp contrast to the μCC assay, which is consistent across more than two orders of magnitude variation in initial cell number and number of days in culture. The observed dependence of the CTG assay on initial cell number is consistent with the fact that metabolic functions are highly influenced by non-lethal changes in culture conditions (Galluzzi et al., 2009; Kepp et al., 2011; Ng et al., 2005; Petty et al., 1995; Sumantran, 2011). In addition, the relationship between intracellular ATP level and cell viability is not always linear (Huang et al., 2010). Finally, ATP-based assays are not appropriate to assess toxicity of chemicals that might interfere with ATP biosynthesis (e.g., atractyloside inhibits ADP/ATP translocases, inhibiting ATP biosynthesis) (Schütt et al., 2012).
The μCC has several important advantages compared to the traditional CFA. In particular, the CFA requires that colonies above 50 cells/colony be counted by eye. Counting colonies is prone to bias, vulnerable to inter-laboratory differences (since it is necessary to decide if a colony is countable or not), and laborious. By using image analysis software, bias is eliminated and inter-laboratory differences are suppressed. In terms of labor, with the exceptions of casting the gel (~10 min of labor) and that colony counting is replaced by imaging and image analysis, the remaining steps are similar. Depending on the density of colonies, counting colonies for a single experiment can take many hours. In contrast, imaging takes minutes, and analysis can be performed rapidly using in-house software. Thus, the μCC has advantages beyond the difference of incubation time (weeks versus days), since the μCC reduces bias, inter-laboratory variability, and labor.
For the μCC approach, microcolony size reflects the ability of cells to divide between one and four times post exposure. Thus, quantitation of early cell division events is an effective predictor of colony forming ability, as revealed by the CFA. Interestingly, both approaches are impacted by cell cycle arrest and delayed cell death, which can give rise to smaller colonies. For the μCC, fewer cells would be present, whereas for the CFA, colonies may be too small to count (for the CFA, colonies with fewer than 50 cells are not counted). Thus, both assays are sensitive to arrest and delayed cell death. Importantly, using an independent approach, it was previously shown in the Samson laboratory that estimates of the number of cells that divide once, twice, or three times post exposure yield results comparable to the CFA (Valiathan et al., 2012). Thus, quantification of early cell division events post exposure is an effective predictor of toxicity when estimated by the CFA, even though the basis for the assays is somewhat different (ability of cells to divide for the μCC and ability of a cell to form a colony for the CFA).
The advantages of the μCC over commonly used cell viability assays can be leveraged to provide an efficient and sensitive screening platform in drug discovery. Quantification of cell proliferation provides a critical endpoint for discovery of effective cancer treatments. For example, in cancer therapy research, large chemical libraries are screened for their ability to induce toxicity in cancer cells (Brown and Attardi, 2005). On the other hand, compounds are also selected for their ability to stimulate cell growth, which is relevant for tissue repair and certain medical procedures, such as hematopoietic regeneration after bone marrow transplantation (Zhang et al., 2015). In these cases, the μCC can provide an effective screening platform to select candidate drugs that stimulate cell division.
In drug development, it is also important to understand the underlying cell death mechanism induced by a potential candidate. Specifically, cell death modalities, such as apoptosis and necrosis, can be exploited as therapeutic targets. For example, necrosis inducers can be used to target apoptosis-resistant tumors (Galluzzi et al., 2009; Guidicelli et al., 2009). Further, the μCC can potentially be used in a multiplex fashion to both assess cytotoxicity and study cell death mechanism. For example, in addition to EG, we have shown that established viability biomarkers, such as plasma membrane integrity (ethidium homodimer staining), esterase activity (calcein acetoxymethyl [AM] staining), and apoptosis (Annexin V staining) can be probed simultaneously to provide information about cytotoxic levels as well as cell death modalities (Figure S4).
In the broad context of chemical toxicity testing, a major limitation of cell-based assays is the lack of metabolic activation (Coecke et al., 2006). Therefore, in vitro assays need to account for the toxic effects of both the parent chemicals and their metabolites (Coecke et al., 2006). By employing MCL-5 and HepG2 cells, which have stable expression of critical metabolic enzymes, the μCC can provide a rapid and sensitive platform for compounds that require metabolic activation.
The μCC’s compatibility with the microtiter plate format is potentially adaptable for high-throughput screening (HTS) for chemical safety testing in the context of environmental health. Due to the large number of chemicals that come in contact with humans and the environment, governmental agencies, such as the U.S. National Toxicology Program and the EU’s Registration, Evaluation, Authorisation, and Restriction of Chemicals, have heavily invested in large-scale chemical safety studies. Cell sensitivity is a critical endpoint in these studies, and microtiter plate viability assays are often the top choice due to their compatibility with HTS. With its many advantages, the μCC could provide more accurate assessment of cytotoxicity in these contexts.
In this work, we mainly focused on lymphoblastoid cells that can grow in suspension. Many different cell types can similarly be grown in a non-adherent fashion, making them compatible with the agarose-based μCC platform. However, for some cell types, a charged surface or a biological ligand is needed for adherent growth in culture (Giancotti and Ruoslahti, 1999; Gumbiner, 1996). We have explored the potential utility of additives to make the μCC effective for adherent cells. For example, cells loaded into the agarose microwells can be overlaid with collagen type 1 followed by low melting point agarose. We found that HeLa cells and HepG2 cells appear to attach to the collagen ligands and form adherent microcolonies (Figure S5, top and middle). Further, we showed that HepG2 cultured on the collagen-modified μCC are highly sensitive to B[a]P (Figure 6C), demonstrating that μCC can be used to study toxicity cells that are normally cultured on tissue culture plastic. Other extracellular matrix component proteins including laminin 1, fibronectin, vitronectin, and various collagens (Geiger et al., 2001) can also potentially be added to the μCC. Additionally, microwell diameters and distances between the microwells can be varied to accommodate different cell types. For example, for larger cells, a microwell with a larger diameter may be desirable, and for cells that migrate away from the microcolony (such as fibroblasts), an increase in the inter-microwell distance may be advantageous.
Because of its high-throughput capacity and its multi-log sensitivity, we also anticipate that the μCC platform can be applied to personalized medicine, as well as to studies of environmental chemicals. For personalized medicine, it is possible that for many cancers, tumor cells could be cultured on the μCC, since anchorage-independent growth is a characteristic of many cancer cells. As such, it may be possible to culture cells from a biopsy on the μCC in order to predict responses of primary cancer cells to anticancer drugs (Rotem et al., 2015). In terms of differences in sensitivity for normal cells, many studies have revealed interindividual differences in radiosensitivity among mitogen-stimulated T lymphocytes obtained from different individuals (Geara et al., 1992). We expect that the μCC is well suited for studies of toxicity in normal and cancer cells, in part because of its ability to yield sensitive measurements after only a few days in culture.
In conclusion, we have developed the μCC, a rapid and sensitive cell survival assay that combines key advantages of commonly used viability assays with an alternative metric for cell division, namely colony size distribution. With the short assay time and large dynamic range, the μCC platform provides a more rapid alternative to the CFA, which continues to be the most broadly accepted method in cell survival testing. The companion publicly available software enables automated image analysis of thousands of microcolonies in a few minutes, making it easy for a new user to learn and use the assay. Finally, the high-throughput and potential multiplexing capacity make the μCC a powerful tool for many applications, including screens for drug development, epidemiological studies, and chemical safety studies.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bevin P. Engelward (bevin@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
TK6 (Liber and Thilly, 1982; Skopek et al., 1978), TK6+MGMT (Hickman and Samson, 1999), and MCL-5 (Crespi et al., 1991) human B-lymphoblastoid cell lines were cultured in 1X RPMI 1640 medium with GlutaMAX™ supplemented with 10% FBS and 100 U/ml Pen-Strep. Human liver carcinoma-derived HepG2 (Aden et al., 1979) (ATCC® HB-8065) cell line was obtained from the American Type Culture Collection (Manassas, VA). HepG2 cells were cultured in high-glucose DMEM supplemented with 10% FBS, 1X GlutaMAX, and 100 U/ml Pen-Strep. Human skin fibroblast (Ellison et al., 1998; Staresincic et al., 2009) cell line was cultured in high-glucose DMEM supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/ml Pen-Strep. HeLa (Gey et al., 1952) cells were cultured in high-glucose DMEM supplemented with 10% FBS and 100 U/ml Pen-Strep. TK6, TK6+MGMT, MCL-5, HepG2, and human skin fibroblast cells were derived from male patients, and HeLa cells were derived from a female patient. The cell lines have not been authenticated. All cells were cultured at 37°C with 5% atmospheric CO2.
METHOD DETAILS
μCC Fabrication
Microwells were fabricated as described previously (Ge et al., 2013, 2015; Weingeist et al., 2013; Wood et al., 2010). Briefly, 1% w/v agarose solution in phosphate-buffered saline (PBS) was prepared. When the molten agarose solution cooled down to ~65°C, Pen-Strep was aseptically added to a final concentration of 200 U/ml. A polydimethylsiloxane (PDMS) mold with an array of micropegs was pressed into the molten agarose solution on top of the hydrophilic side of a sheet of GelBond® Film (Lonza, Hopkinton, MA). The agarose was allowed to gelate at room temperature for ~15 minutes. After the mold was removed, a bottomless 96-well plate was pressed on top of the agarose chip to create 96 wells, each with ~300 microwells at its base.
A 0.6% w/v low melting point agarose stock solution in PBS was kept molten at 43°C. A culture medium stock (RPMI 1640 with GlutaMAX™ supplemented with 20% FBS and 200 U/ml Pen-Strep) was kept at 37°C. To prepare the 0.3% w/v overlay agarose solution, one volume of the agarose stock solution was combined with one volume of the culture medium stock. Cells were loaded into the microwells by gravity, and the gel was then covered with 0.3% w/v overlay agarose, prepared as described above. After 15 min at room temperature (RT), another layer of the overlay agarose was added. After another 15 min at RT, the gel was transferred to 4°C for 15 min to ensure maximal gelation of the overlayed agarose.
The RatCol® Collagen Solution kit (5153-1KIT) was purchased from Advanced BioMatrix, Inc., San Diego, CA. A solution of collagen type 1 was prepared on ice according to the manufacturer’s 3-D gel preparation procedure using the supplied neutralization solution and was diluted in culture medium to a final concentration of 1 mg/ml collagen. For HepG2, HeLa, and human skin fibroblasts, after the cells were loaded into the microwells, the agarose gel was covered with a thin layer of the 1 mg/ml collagen solution. After 15 min at RT and 15 min at 37°C (for maximal collagen gel formation), a thin layer of 0.3% w/v overlay agarose was added on top of the collagen gel layer. The gel was further incubated for 15 min at RT, followed by 15 min at 4°C.
Microcolony Culture and Treatment with DNA Damaging Agents
The μCC was incubated in fresh culture medium at 37°C and 5% atmospheric CO2 overnight. The μCC was then either exposed to γIR or submerged in treatment solutions (Note that an effective alternative is to treat cells prior to loading the μCC.).
Cell Fixation
The μCC was submerged in 10% formalin and kept at 4°C for at least 2 hours.
Treatment with γIR
The μCC was exposed to γ-rays from a 137Cesium source at 1 Gy/min (Gammacell 40 Exactor, Best Theratronics Model C-440). Following irradiation, the μCC was submerged in culture medium and incubated at 37°C and 5% atmospheric CO2 for three or four days before fixation.
BCNU Treatment
The μCCs were removed from culture medium and rinsed by submerging in PBS for 5 minutes. PBS was aspirated and replaced with BCNU diluted in warm RPMI 1640 (+ GlutaMAX™) supplemented with 100 U/ml Pen-Strep. The maximum final concentration of ethanol was 0.1%. BCNU exposure was for one hour at 37°C with 5% atmospheric CO2. BCNU solution was aspirated and residual BCNU was rinsed off twice by submerging the μCC in PBS for 5 minutes each time. Following exposure, the μCC was submerged in culture medium and incubated at 37°C and 5% atmospheric CO2 for three days before fixation.
AFB1 Treatment
The μCC was incubated in AFB1 solution for 24 hours at 37°C and 5% atmospheric CO2. The maximum final concentration of DMSO was 0.5%. After 24 hours, the AFB1-supplemented medium was removed, and the μCC was washed three times by submerging in PBS for 5 minutes each time. Following exposure, the μCC was submerged in culture medium and incubated at 37°C and 5% atmospheric CO2 for three days before fixation.
Inhibition of AFB1 Metabolism with KET
To inhibit AFB1 metabolism, 5 μM KET was added to culture medium at the start of AFB1 treatment. The maximum final concentration of DMSO was 0.5%.
B[a]P Treatment
The μCC was incubated in B[a]P solution for five days at 37°C and 5% atmospheric CO2. The maximum final concentration of DMSO was 0.003%. The μCC was fixed immediately following exposure.
Inhibition of B[a]P Metabolism with ANF
To inhibit B[a]P metabolism, 25 μM ANF was added to culture medium at the start of B[a]P treatment. The maximum final concentration of DMSO was 0.1%.
Imaging and Analysis
After cell fixation, the μCC was stained with Vybrant® DyeCycle™ Green (dissolved in PBS to a final concentration of 1.5 μM) at 4°C overnight, protected from light. Fluorescent images were captured using an epifluorescence microscope (Nikon Eclipse 80i, Nikon Instruments, Inc., Melville, NY) with a 480 nm excitation filter and a fixed aperture time. Images were analyzed using “uCCanalyzer” (Data S1), a freely available custom software written in MATLAB (The MathWorks Inc., Natick, MA), as previously described (Wood et al., 2010), except with modifications to enable quantification of F/M. Distributions of F/M values were generated and exported into spreadsheet files using “uCCHistogram” (Data S2), a package of custom scripts written in Python (Python Software Foundation, Python version 2.7.10). Analyses of F/M distributions were performed in Microsoft Excel (Microsoft Office Suite 2016).
Correlation of F/M Values with Cell Numbers per Microwell
TK6 cells were loaded onto a μCC and stained with Vybrant® DyeCycle Green. The number of distinct fluorescent nuclei was counted manually and compared to the F/M value of the corresponding microwell to determine fluorescence/cell (F/C).
Calculation of Percent Control Growth
F/M is used to approximate microcolony size, which is then used to construct microcolony size distributions. For each microcolony size, we subtracted the relative frequency in the initial population from the relative frequency in the final population. We defined the positive values of the subtractions to be an approximation of the excess microcolonies. For each microcolony size, we multiplied the approximate excess microcolonies by the number of cells per microcolony to obtain an estimation of the total number of cells in the excess microcolonies. EG is calculated by adding the total number of cells in the excess microcolonies across all available microcolony sizes. The EG value for each treatment condition was calculated and compared to a negative control EG (unexposed cells) to obtain percent control growth. In practice, data is analyzed in an automated fashion, using uCCHistogram.
CFA in Microtiter Plates
The CFA was performed as previously described (Furth et al., 1981; Kraemer et al., 1980). Briefly, TK6 cells growing in suspension were exposed to γIR and then distributed over U-bottom 96-well plates. After three weeks, wells were scored for the absence of colony growth, and the surviving fraction was then calculated (Furth et al., 1981; Kraemer et al., 1980). Phase-contrast pictures were taken with an Olympus TG-860 Tough Stylus Digital Camera (Olympus America, Inc., Center Valley, PA) at 5X magnification.
XTT Assay
The XTT kit was purchased from Cell Signaling Technology® (Danvers, MA). Three days after μIR treatment, the viability of TK6 cells was measured according to the manufacturer’s instructions.
CTG® Assay
The CTG® kit was purchased from Promega (Madison, WI). Three or four days after μIR treatment, the viability of TK6 cells was measured according to the manufacturer’s instructions.
Live/Dead Staining
The LIVE/DEAD® Viability/Cytotoxicity kit for mammalian cells was obtained from Thermo Fisher Scientific (Waltham, MA). The μCC was rinsed three times by submergence in fresh 1X PBS for 5 minutes. A solution of calcein-AM and ethidium homodimer-1 (EthD-1) was prepared according to the manufacturer’s instruction such that the final concentrations for calcein-AM and EthD-1 were 2 μM and 4 μM, respectively. The μCC was then submerged in the calcein-AM/EthD-1 solution and incubated at 37°C for 1 hour, protected from light. Microcolonies were imaged with an epifluorescence microscope using 480 nm and 560 nm excitation filters for calcein and EthD-1, respectively.
Apoptosis Staining
A Tali® Apoptosis kit was purchased from Thermo Fish Scientific (Waltham, MA). The μCC was rinsed three times via submergence in 1X Annexin binding buffer for 5 minutes. Annexin V Alexa Fluor® 488 stock solution was diluted 1:20 in 1X Annexin binding buffer. The final solution was used to completely cover the μCC. After 30 minutes of incubation at 37°C (protected from light), EthD-1 stock solution from the LIVE/DEAD® Viability/Cytotoxicity kit was added to a final concentration of 4 μM. After incubation at 37°C for 30 minutes, microcolonies were imaged with 480nm and 560nm excitation filters for Annexin V and EthD-1, respectively.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
All statistical details of experiments can be found in the figure legends, where n represents the number of independent experiments. All experiments were repeated at least three times. At least 700 microcolonies were analyzed per condition. The Student’s t test was used to evaluate significance (p < 0.05). Error bars (SEM) and curve fitting were performed using GraphPad Prism version 6.00 for Windows, GraphPad Software, La Jolla California USA, https://www.graphpad.com.
DATA AND SOFTWARE AVAILABILITY
MATLAB software “uCCanalyzer” for quantification of F/M: Data S1.
Python script package “uCCHistogram” for generating F/M distributions and importing to Microsoft Excel: Data S2.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Fetal bovine serum | Atlanta Biologicals | Cat#S11150 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,3-Bis(2-chloroethyl)-1-nitrosourea | Sigma-Aldrich | Cat#C0400 |
| aflatoxin B1 | Sigma-Aldrich | Cat#A6636 |
| ketoconazole | Sigma-Aldrich | Cat#K1003 |
| benzo[a]pyrene | Sigma-Aldrich | Cat#B1760 |
| dimethyl sulfoxide | Sigma-Aldrich | Cat#D45040 |
| alpha-naphthoflavone | Sigma-Aldrich | Cat#N5757 |
| Vybrant DyeCycle Green | Thermo Fisher Scientific | Cat#V35004 |
| RPMI 1640 medium | Thermo Fisher Scientific | Cat#61870-036 |
| DMEM medium | Thermo Fisher Scientific | Cat#11965092 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050061 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140-122 |
| 10% formalin solution | Thermo Fisher Scientific | Cat#5755 |
| Critical Commercial Assays | ||
| XTT cell viability kit | Cell Signaling Technology | Cat#9095 |
| CellTiter-Glo | Promega | Cat#G7570 |
| LIVE/DEAD Viability/Cytotoxicity kit | Thermo Fisher Scientific | Cat#L3224 |
| Tali Apoptosis kit | Thermo Fisher Scientific | Cat#A10788 |
| Experimental Models: Cell Lines | ||
| Human: TK6 cells | W. G. Thilly (Liber and Thilly, 1982; Skopek et al., 1978) | N/A |
| Human: MCL-5 cells | G. J. Jenkins (Crespi et al., 1991) | N/A |
| Human: TK6+MGMT cells | L. D. Samson (Hickman and Samson, 1999) | N/A |
| Human: HepG2 cells | ATCC | HB-8065 |
| Human: HeLa cells | ATCC | CCL-2 |
| Human skin fibroblasts | O. D. Scharer (Ellison et al., 1998; Staresincic et al., 2009) | N/A |
| Software and Algorithms | ||
| uCCAnalyzer | This paper, modified from guicometanalyzer (Wood et al., 2010) | N/A |
| uCCHistogram | This paper | N/A |
| Prism 6.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Microsoft Excel | Microsoft | Microsoft Office Suite 2016 |
| Other | ||
| RatCol Collagen Solution kit | Advanced BioMatrix | Cat#5153-1KIT |
| 137Cesium γ-rays | Best Theratronics | Gammacell 40 Exactor |
| GelBond Film | Lonza | Cat#53761 |
| UltraPure Agarose | Thermo Fisher Scientific | Cat#16500500 |
| UltraPure Low Melting Point Agarose | Thermo Fisher Scientific | Cat#16520050 |
Highlights.
The MicroColonyChip (μCC) is a rapid cell survival quantitation platform
Colony micropatterning enables 250× miniaturization of the colony formation assay
Analysis of microcolony sizes enables rapid automated analysis of cell survival
μCC is more sensitive and robust than commonly used cytotoxicity assays
ACKNOWLEDGMENTS
This work was primarily supported by the National Institute of Environmental Health Sciences (NIEHS) (R44ES024698) with partial support from the NIEHS Superfund Basic Research Program, NIH (P42 ES027707). L.P.N. was also partly supported by NIH grant R01 ES022872 and NIEHS DP1 ES022576. T.K.C. was supported by the Singapore National Research Foundation, which is administered by the Singapore-MIT Alliance for Research and Technology. J.G. was supported by the Siebel Scholars Program. We also thank the Center for Environmental Health Sciences (P30-ES002109). We would like to thank W.G. Thilly (Massachusetts Institute of Technology, Cambridge, MA, USA) for TK6 cells, G.J. Jenkins (Swansea University, Wales, UK) for MCL-5 cells, and O.D. Scharer (Harvard University, Cambridge, MA, USA) for the human skin fibroblast cell line.
Footnotes
DECLARATION OF INTERESTS
L.P.N., T.K.C., J.G., and B.P.E. applied for a patent related to the approach of using cell-micropatterning technology and microcolony size distribution analysis for toxicity quantification.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and two data files and can be found with this article online at https://doi.org/10.1016/j.celrep.2019.01.053.
REFERENCES
- Aden DP, Fogel A, Plotkin S, Damjanov I, and Knowles BB (1979). Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 282, 615–616. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Xia F, Wolfson K, and Liber HL (1993). Different cytotoxic and mutagenic responses induced by X-rays in two human lymphoblastoid cell lines derived from a single donor. Mutat. Res. 286, 233–241. [DOI] [PubMed] [Google Scholar]
- Berridge MV, Herst PM, and Tan AS (2005). Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11, 127–152. [DOI] [PubMed] [Google Scholar]
- Blaheta RA, Franz M, Auth MK, Wenisch HJ, and Markus BH (1991). A rapid non-radioactive fluorescence assay for the measurement of both cell number and proliferation. J. Immunol. Methods 142, 199–206. [DOI] [PubMed] [Google Scholar]
- Brown JM, and Attardi LD (2005). The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer 5, 231–237. [DOI] [PubMed] [Google Scholar]
- Carmichael J, DeGraff WG, Gazdar AF, Minna JD, and Mitchell JB (1987a). Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 47, 936–942. [PubMed] [Google Scholar]
- Carmichael J, DeGraff WG, Gazdar AF, Minna JD, and Mitchell JB (1987b). Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of radiosensitivity. Cancer Res. 47, 943–946. [PubMed] [Google Scholar]
- Castell JV, Jover R, Martínez-Jiménez CP, and Gómez-Lechón MJ (2006). Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opin. Drug Metab. Toxicol. 2, 183–212. [DOI] [PubMed] [Google Scholar]
- Cho US, Park EY, Dong MS, Park BS, Kim K, and Kim KH (2003). Tight-binding inhibition by alpha-naphthoflavone of human cytochrome P450 1A2. Biochim. Biophys. Acta 1648, 195–202. [DOI] [PubMed] [Google Scholar]
- Coecke S, Ahr H, Blaauboer BJ, Bremer S, Casati S, Castell J, Combes R, Corvi R, Crespi CL, Cunningham ML, et al. (2006). Metabolism: a bottleneck in in vitro toxicological test development. The report and recommendations of ECVAM workshop 54. Altern. Lab. Anim. 34, 49–84. [DOI] [PubMed] [Google Scholar]
- Cook JA, and Mitchell JB (1989). Viability measurements in mammalian cell systems. Anal. Biochem. 179, 1–7. [DOI] [PubMed] [Google Scholar]
- Crespi CL, Gonzalez FJ, Steimel DT, Turner TR, Gelboin HV, Penman BW, and Langenbach R (1991). A metabolically competent human cell line expressing five cDNAs encoding procarcinogen-activating enzymes: application to mutagenicity testing. Chem. Res. Toxicol. 4, 566–572. [DOI] [PubMed] [Google Scholar]
- Crouch SP, Kozlowski R, Slater KJ, and Fletcher J (1993). The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J. Immunol. Methods 160, 81–88. [DOI] [PubMed] [Google Scholar]
- Durando X, Lemaire JJ, Tortochaux J, Van-Praagh I, Kwiatkowski F, Vincent C, Bailly C, Verrelle P, Irthum B, Chazal J, and Bay JO (2003). High-dose BCNU followed by autologous hematopoietic stem cell transplantation in supratentorial high-grade malignantgliomas: a retrospective analysis of 114 patients. Bone Marrow Transplant. 31, 559–564. [DOI] [PubMed] [Google Scholar]
- Ellison AR, Nouspikel T, Jaspers NG, Clarkson SG, and Gruenert DC (1998). Complementation of transformed fibroblasts from patients with combined xeroderma pigmentosum-Cockayne syndrome. Exp. Cell Res. 243, 22–28. [DOI] [PubMed] [Google Scholar]
- Evans WE, and Relling MV (1999). Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286, 487–491. [DOI] [PubMed] [Google Scholar]
- Franken NAP, Rodermond HM, Stap J, Haveman J, and van Bree C (2006). Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319. [DOI] [PubMed] [Google Scholar]
- Furth EE, Thilly WG, Penman BW, Liber HL, and Rand WM (1981). Quantitative assay for mutation in diploid human lymphoblasts using microtiter plates. Anal. Biochem. 110, 1–8. [DOI] [PubMed] [Google Scholar]
- Gallagher EP, Wienkers LC, Stapleton PL, Kunze KL, and Eaton DL (1994). Role of human microsomal and human complementary DNA-expressed cytochromes P4501A2 and P4503A4 in the bioactivation of aflatoxin B1. Cancer Res. 54, 101–108. [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, Bazan NG, Blagosklonny MV, Blomgren K, Borner C, et al. (2009). Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Wood DK, Weingeist DM, Prasongtanakij S, Navasumrit P, Ruchirawat M, and Engelward BP (2013). Standard fluorescent imaging of live cells is highly genotoxic. Cytometry A 83, 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Chow DN, Fessler JL, Weingeist DM, Wood DK, and Engelward BP (2015). Micropatterned comet assay enables high throughput and sensitive DNA damage quantification. Mutagenesis 30, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geara FB, Peters LJ, Ang KK, Wike JL, Sivon SS, Guttenberger R, Callender DL, Malaise EP, and Brock WA (1992). Intrinsic radiosensitivity of normal human fibroblasts and lymphocytes after high- and low-dose-rate irradiation. Cancer Res. 52, 6348–6352. [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, and Yamada KM (2001). Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2, 793–805. [DOI] [PubMed] [Google Scholar]
- Gerson SL (2004). MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 4, 296–307. [DOI] [PubMed] [Google Scholar]
- Gey GO, Coffman WD, and Kubicek MT (1952). Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 12, 264–265. [Google Scholar]
- Giancotti FG, and Ruoslahti E (1999). Integrin signaling. Science 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- Golan DE, and Tashjian AH (2012). Principles of pharmacology: the pathophysiologic basis of drug therapy, Third Edition (Wolters Kluwer Health/Lippincott Williams & Wilkins; ). [Google Scholar]
- Gonzaga PE, Potter PM, Niu TQ, Yu D, Ludlum DB, Rafferty JA, Margison GP, and Brent TP (1992). Identification of the cross-link between human O6-methylguanine-DNA methyltransferase and chloroethylnitro-sourea-treated DNA. Cancer Res. 52, 6052–6058. [PubMed] [Google Scholar]
- Greenblatt DJ, Zhao Y, Venkatakrishnan K, Duan SX, Harmatz JS, Parent SJ, Court MH, and von Moltke LL (2011). Mechanism of cytochrome P450-3A inhibition by ketoconazole. J. Pharm. Pharmacol. 63, 214–221. [DOI] [PubMed] [Google Scholar]
- Guidicelli G, Chaigne-Delalande B, Dilhuydy MS, Pinson B, Mahfouf W, Pasquet JM, Mahon FX, Pourquier P, Moreau JF, and Legembre P (2009). The necrotic signal induced by mycophenolic acid overcomes apoptosis-resistance in tumor cells. PLoS ONE 4, e5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, and Samson LD (1999). Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. USA 96, 10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg FH, Parker LM, Takvorian T, Canellos GP, and Zervas NT (1981). High-dose BCNU with autologous bone marrow rescue for recurrent glioblastoma multiforme. J. Neurosurg. 54, 455–460. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang X, Li S, Liu N, Lian W, McDowell E, Zhou P, Zhao C, Guo H, Zhang C, et al. (2010). Physiological levels of ATP negatively regulate proteasome function. Cell Res. 20, 1372–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, and Moore GE (1968). Ability of Human Hematopoietic Cell Lines to Form Colonies in Soft Agar. Proc. Soc. Exp. Biol. Med. 128, 1179–1183. [Google Scholar]
- International Agency for Research on Cancer (2012). Chemical Agents and Related Occupations. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Volume 100F (IARC). [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Gray M, Yue ST, Haugland RP, and Singer VL (2001). Sensitive determination ofcell number using the CyQUANT cell proliferation assay. J. Immunol. Methods 254, 85–98. [DOI] [PubMed] [Google Scholar]
- Kaina B, Fritz G, and Coquerelle T (1991a). Identification of human genes involved in repair and tolerance of DNA damage. Radiat. Environ. Biophys. 30, 1–19. [DOI] [PubMed] [Google Scholar]
- Kaina B, Fritz G, Mitra S, and Coquerelle T (1991b). Transfection and expression of human O6-methylguanine-DNA methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of MGMT in protection against the genotoxic effects of alkylating agents. Carcinogenesis 12, 1857–1867. [DOI] [PubMed] [Google Scholar]
- Kepp O, Galluzzi L, Lipinski M, Yuan J, and Kroemer G (2011). Cell death assays for drug discovery. Nat. Rev. Drug Discov. 10, 221–237. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Waters HL, and Buchanan JK (1980). Survival of human lymphoblastoid cells after DNA damage measured by growth in microtiter wells. Mutat. Res. 72, 285–294. [DOI] [PubMed] [Google Scholar]
- Liber HL, and Thilly WG (1982). Mutation assay at the thymidine kinase locus in diploid human lymphoblasts. Mutat. Res. 94, 467–485. [DOI] [PubMed] [Google Scholar]
- Nebert DW, and Dalton TP (2006). The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960. [DOI] [PubMed] [Google Scholar]
- Ng KW, Leong DT, and Hutmacher DW (2005). The challenge to measure cell proliferation in two and three dimensions. Tissue Eng. 11, 182–191. [DOI] [PubMed] [Google Scholar]
- Petty RD, Sutherland LA, Hunter EM, and Cree IA (1995). Comparison of MTT and ATP-based assays for the measurement of viable cell number. J. Biolumin. Chemilumin. 10, 29–34. [DOI] [PubMed] [Google Scholar]
- Plumb JA, Milroy R, and Kaye SB (1989). Effects of the pH dependence of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide-formazan absorption on chemosensitivity determined by a novel tetrazolium-based assay. Cancer Res. 49, 4435–4440. [PubMed] [Google Scholar]
- Quent VM, Loessner D, Friis T, Reichert JC, and Hutmacher DW (2010). Discrepancies between metabolic activity and DNA content as tool to assess cell proliferation in cancer research. J. Cell. Mol. Med. 14, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago R, Mitchen J, and Wilding G (1990). DNA fluorometric assay in 96-well tissue culture plates using Hoechst 33258 after cell lysis by freezing in distilled water. Anal. Biochem. 191, 31–34. [DOI] [PubMed] [Google Scholar]
- Rotem A, Janzer A, Izar B, Ji Z, Doench JG, Garraway LA, and Struhl K (2015). Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. USA 112, 5708–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütt F, Aretz S, Auffarth GU, and Kopitz J (2012). Moderately reduced ATP levels promote oxidative stress and debilitate autophagic and phagocytic capacities in human RPE cells. Invest. Ophthalmol. Vis. Sci. 53, 5354–5361. [DOI] [PubMed] [Google Scholar]
- Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, Currens MJ, Seniff D, and Boyd MR (1988). Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 48, 4827–4833. [PubMed] [Google Scholar]
- Skopek TR, Liber HL, Penman BW, and Thilly WG (1978). Isolation of a human lymphoblastoid line heterozygous at the thymidine kinase locus: possibility for a rapid human cell mutation assay. Biochem. Biophys. Res. Commun. 84, 411–416. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, and Schärer OD (2009). Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumantran VN (2011). Cellular chemosensitivity assays: an overview. Methods Mol. Biol. 731, 219–236. [DOI] [PubMed] [Google Scholar]
- Tong WP, Kirk MC, and Ludlum DB (1982). Formation of the cross-link 1-[N3-deoxycytidyl),2-[N1-deoxyguanosinyl]ethane in DNA treated with N,N′-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 42, 3102–3105. [PubMed] [Google Scholar]
- Valiathan C, McFaline JL, and Samson LD (2012). A rapid survival assay to measure drug-induced cytotoxicity and cell cycle effects. DNA Repair (Amst.) 11, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingeist DM, Ge J, Wood DK, Mutamba JT, Huang Q, Rowland EA, Yaffe MB, Floyd S, and Engelward BP (2013). Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors. Cell Cycle 12, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz F, Azzam EI, and Little JB (1998). The response of proliferating cell nuclear antigen to ionizing radiation in human lymphoblastoid cell lines is dependent on p53. Radiat. Res. 149, 32–40. [PubMed] [Google Scholar]
- Wood DK, Weingeist DM, Bhatia SN, and Engelward BP (2010). Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl. Acad. Sci. USA 107, 10008–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Desai A, Yang SY, Bae KB, Antczak MI, Fink SP, Tiwari S, Willis JE, Williams NS, Dawson DM, et al. (2015). TISSUE REGENERATION. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science 348, aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MATLAB software “uCCanalyzer” for quantification of F/M: Data S1.
Python script package “uCCHistogram” for generating F/M distributions and importing to Microsoft Excel: Data S2.