Abstract
Globally, Streptococcus pyogenes poses a continuous burden on human health, causing both self-limiting and life-threatening diseases. Therefore, studying the profile of virulence genes and their combinations is essential to monitor the epidemiology and pathogenic potential of this important species. Thus, the aim of this study was to analyze related genetic features of clinical strains collected in Italy in 2012 in order to obtain a valid picture of their virulence profile that could be compared to similar studies made in other countries approximately in the same period. We conducted emm typing and fibronectin-collagen-T antigen (FCT) region typing in 122 Streptococcus pyogenes strains. Furthermore, several additional virulence genes were screened by polymerase chain reaction. We found correlations between emm types and FCT region profiles. emm1 strains were mainly associated with FCT2 and FCT6, while emm89 and emm12 strains were associated with FCT4. FCT5 was mainly represented in emm4, emm6, and emm75 strains. Significantly, we defined subtypes for each FCT type based on the differences in single and double loci compared to the reference scheme used for the classification of the FCT region. In addition, new FCT-region variants with differences in multiple loci were also recorded. Cluster analysis based on virulence gene profiling showed a non-random distribution within each emm type. This study added new data to existing studies conducted worldwide and revealed new variability scores in circulating Streptococcus pyogenes strains and new assortments in well-established virulence gene signatures.
Keywords: FCT, Virulence, GAS, emm type
Introduction
Streptococcus pyogenes (group A Streptococcus, GAS) is an important human pathogen that colonizes the pharynx and the skin, causing an array of diseases ranging from mild sore throat and impetigo to invasive and life-threatening infections (Cunningham, 2000).
Like most pathogens, GAS produces many virulence factors (Cunningham, 2000). Currently, eleven virulence factors have been identified as superantigens: three chromosomally encoded genes (speG, speJ, and smeZ) and eight genes harbored by temperate phages (speA, speC, speH, speI, speK, speL, speM, and ssa) (Friães et al., 2013). Superantigens are effective exotoxins produced, in addition to GAS, by group C/G Streptococcus dysgalactiae ssp. equisimilis, Staphylococcus aureus, Yersinia pseudotuberculosis, and Mycoplasma arthritidis (Barnett et al., 2015). Their pathogenic mechanism involves the ability to bypass the typical antigen presentation process generating an overstimulation of the immune inflammatory cascade of events, eventually leading to a toxic shock syndrome (STSS). STSS and invasive diseases have been frequently found in strains carrying speA and ssa, together with speK, and smeZ (Commons et al., 2014). Scarlet fever has been correlated with the carriage or acquisition of ssa, speA, and speC in different studies (Barnett et al., 2015). A robust correlation has been seen between M18 isolates with speL and speM and acute rheumatic fever (Barnett et al., 2015). Also speK-positive M89 isolates have been associated with the last disease. Two additional virulence factors are encoded by genes located on phages: a phospholipase A2 (Sla) and a DNAse (Sdn) (Friães et al., 2013).
SpeB and SpeF, whose genes map instead on the bacterial chromosome and were originally described as exotoxins, are actually a cysteine protease and a DNAse, respectively (Friães et al., 2013). SpeB, is a secreted enzyme possessing a relatively indiscriminant substrate specificity, degrading, on the host side, several substrates such as the extracellular matrix, cytokines, chemokines, complement components, immunoglobulins, and serum protease inhibitors. On the pathogen side, SpeB proteolytic cleavage regulates other streptococcal proteins (Nelson, Garbe & Collin, 2011).
However, the primary virulence factor for GAS remains the major surface M protein that is encoded by the emm gene (Metzgar & Zampolli, 2011). It is also the basis for the major typing scheme for GAS, thanks to the considerable sequence variability of the 5′-end of the emm genes, for which more than 220 variants have been identified. As such, a large number of functions, and interactions with host molecules, have been ascribed to different M protein variants. Among others, this protein has antiphagocytic properties and elicits a substantial host specific immune response.
emm types are associated with specific superantigen profiles, and these associations differ in GAS populations collected from different geographical areas (Commons et al., 2008). It is still unknown whether environmental or biological factors may play a role in these associations.
Additionally, genes encoding several virulence factors involved in GAS pathogenesis are located in the fibronectin-collagen-T antigen (FCT) region (Bessen & Kalia, 2002). The FCT region is highly variable in genetic content and may comprise five to 10 Open Reading Frames (ORFs). In addition, all regions contains either rofA or nra regulatory genes at one end and genes that encode the major pilin backbone protein (BP, often referred to as FctA), one or two accessory pilin proteins (for example, AP2 or FctB), and at least one sortase.
Different FCT types are associated with different subsets of emm types, and a genetic linkage between emm type and FCT region profile is associated with GAS tissue specificity (Kratovac et al., 2007).
At last, it is worth mentioning the association between emm types and the different recognized arrangements of emm and emm-like genes within the GAS chromosomal region comprising the emm gene itself, named emm patterns. Five emm patterns, named A–E, have been recognized so far (McGregor et al., 2004). As the first three share basic structural properties and are grouped together, one may consider three main emm patterns only, namely A–C, D, and E. Notably, also the structure of the emm gene associated to each emm pattern group is characteristic (McMillan et al., 2012). GAS strains with different emm patterns show a different tissue tropism. Those having patterns A–C are throat specialists; those with pattern D are skin specialists, while pattern-E-positive strains have no obvious tissue preference (Bessen, 2009).
Our study described 122 GAS isolated in 2012 in three regions of central Italy (Marche, Lazio, and Umbria). Our investigation focused on the distribution of virulence factors and their correlation with emm types as well as with FCT-region types. The set of genes considered for profiling the population under investigation was modulated on the basis of previous investigations to allow inter-studies comparisons and to increment the knowledge on the epidemiology of GAS populations.
Methods
Sample collection and identification
In 2012, 122 GAS isolates were collected from symptomatic patients with pharyngotonsillitis from hospital clinical microbiology laboratories in Roma (Ospedale Pediatrico Bambino Gesù), Perugia (Ospedale S. Maria della Misericordia), and Macerata (Laboratorio di Analisi ASL9). Ethical approval and patient informed consent were not required because the work was on isolates only (no specific sensible data on patients were collected) and entirely conducted between 2011 and 2015 when approval for this kind of studies was not mandatory. Isolates were from throat (n = 107), rectal (n = 1), and vaginal swabs (n = 2), skin and wound (n = 6), bronchoalveolar lavage (n = 1), and normally sterile fluids (blood and pleural fluid, n = 5). Identification of clinical GAS was confirmed in our laboratory by standard procedures such as colony morphology, β-hemolysis on blood agar, serogrouping using a latex agglutination test (Streptococcal Grouping kit; Oxoid, Basingstoke, UK), bacitracin susceptibility test (Oxoid, Basingstoke, UK), and, additionally, by polymerase chain reaction (PCR) detection of dnaB (Slinger et al., 2011).
DNA extraction and emm typing
DNA extraction was performed using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA). emm typing was performed according to the Center for Disease Control and Prevention (CDC) guidelines (https://www.cdc.gov/streplab/protocol-emm-type.html). To determine the emm type, each sequence was compared to the reference sequences available in the CDC database. The Streptococcus pyogenes SF370 was used as the reference strain to check for the general quality of DNA extraction/purification and for PCR conditions.
Virulence gene profiling
The presence of speA, speB, speC, speH, speI, speK, speL, speM, smeZ, ssa, sdn, and sla virulence genes was tested by PCR using specific primer sequences (Table S1). This set of genes is referred to as non-FCT throughout the text. Briefly, each 25 μL test tube contained one μg of chromosomal DNA, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleotide triphosphates (dNTPs), one μM oligonucleotide primers, and 0.5 U Taq polymerase (AmpliTaq Gold; Applied Biosystems, Foster City, CA, USA). The cycling program included an initial denaturation at 95 °C for 2 min, followed by 30 cycles at 95 °C for 40 s, at primer-specific annealing temperature for 40 s, at 72 °C for 1 min, and one cycle at 72 °C for 5 min. Various quality controls were applied to uncover potential problems with false positive or false negative results. Controls for the template DNA and components of the PCR mixture included amplification of the dnaB housekeeping gene (Slinger et al., 2011). Two different operators repeated each PCR independently in different days. Whenever results were discordant, genomic DNA was prepared again from freshly cultured cells and PCR repeated. For the specific case of speB, initial screening gave a high proportion of negative PCR (about 25%). As speB is known to belong to the core genome and found present in almost all surveys of GAS populations so far, an additional set of primers was used to validate the first result (speB-2, Table S1).
As a positive control for each superantigen gene PCR, we used the DNA of an isolate positive to that specific gene. The Streptococcus pyogenes SF370 was used as the general reference strain to check for quality of DNA preparations to be used in PCR. In particular, it was used as a positive control for amplification of speB, emm, and dnaB.
FCT typing
PCR amplification using chromosomal DNA and the general design described in the paragraph “Virulence gene profiling” was performed using 13 different primer pairs targeting various virulence genes within the FCT region (Table S2). The sorted profiles allowed the identification of “FCT-region variants,” based upon but not equivalent to the general scheme by Kratovac et al. (2007) (Table 1). At first, the strains whose profiles were matching one Kratovac’s scheme FCT type were assigned the corresponding type number (one to eight, Table 1), referred to as “reference FCT-region variant,” and designated with the capital letter A after the type number (e.g., 1A, 2A, 3A, and so on). Those strains showing an FCT region pattern matching a Kratovac’s scheme FCT type but for a single PCR product identified a “single FCT-region variant” that was named with the capital letter B after the respective reference FCT type number (e.g., 1B, 2B, 3B, and so on). The same rationale was used to classify double FCT-region variants indicated by the letter C after the corresponding reference FCT type number (e.g., 1C, 2C, 3C, and so on). An example is given by FCT-region variant 5. A total of 21 strains had an FCT-region profile matching that of the Kratovac’s scheme (prtF1+rofA+) and are designated as 5A. Three strains had the profile prtF1+rofA+ and were also positive to the bridge region. This last difference made them a “single FCT-region variant” of type 5, indicated as 5B.
Table 1. Patterns of genes or locus defining the FCT-region typing scheme according to Kratovac et al. (2007).
| FCT-type§ | Gene or locus within the FCT region | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| prtF1 | cpa | prtF2 | sipA2 | Bridge | fctA | srtC2 | fctB | srtB | rofA | nra | |
| 1 | + | − | − | − | − | − | − | − | + | + | − |
| 2 | − | − | − | − | − | − | − | − | + | + | − |
| 3 | − | + | + | + | + | + | + | + | − | − | + |
| 4 | + | + | + | + | + | + | + | + | + | + | − |
| 5 | + | − | − | − | − | − | − | − | − | + | − |
| 6 | − | − | − | − | − | − | − | − | − | + | − |
| 7 | + | − | + | + | + | + | + | + | + | + | − |
| 8 | + | + | + | + | + | + | + | + | + | – | + |
Note:
FCT-type 9 is not listed since it is not discriminated using the Kratovac et al. (2007) PCR typing scheme.
In conclusion, the introduced classification is less stringent and depicts general variation within the classical FCT-type scheme by Kratovac and coll. (Kratovac et al., 2007). For convenience and reference, we have provided the whole raw data set (see the raw data in the Supplemental File).
Data analysis and statistics
Cluster analysis was done in MEGA v6 (Tamura et al., 2013) and confirmed using Statgraphics Centurion v17.1.08 (Statpoint Technologies Inc., Warrenton, VA, USA). The latter software was used to perform the principal component analysis (PCA) as well as the Chi-square tests when indicated (p < 0.05 was considered significant for the rejection of the null hypothesis). Data for PCA were input as a binary matrix of 0 (absence of the gene) and 1 (presence of the gene). Display and annotation of the phylogenetic tree were obtained with Interactive Tree Of Life (iTOL) v3 online tools (Letunic & Bork, 2016).
Results
Correlation between non-FCT virulence gene profiles and emm types
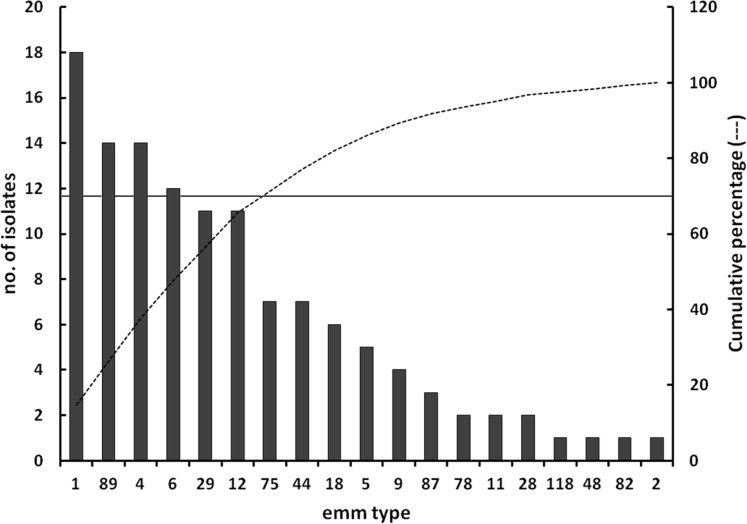
The distribution of emm types within the collected strains is reported in Fig. 1. Six emm types (1, 4, 89, 6, 12, and 29) accounted for nearly 70% of the population.
Figure 1. Distribution of emm types.
Distribution of emm types within the Streptococcus pyogenes population under investigation. The principal Y-axis reports the absolute number of isolates distributing into each emm type group, while the secondary Y-axis refers to the cumulative function (dotted line) computed from the distribution of emm types sorted in a descending order of isolates relative abundance. The continuous horizontal line, referring to the secondary Y-axis, indicates the 70% of the study population.
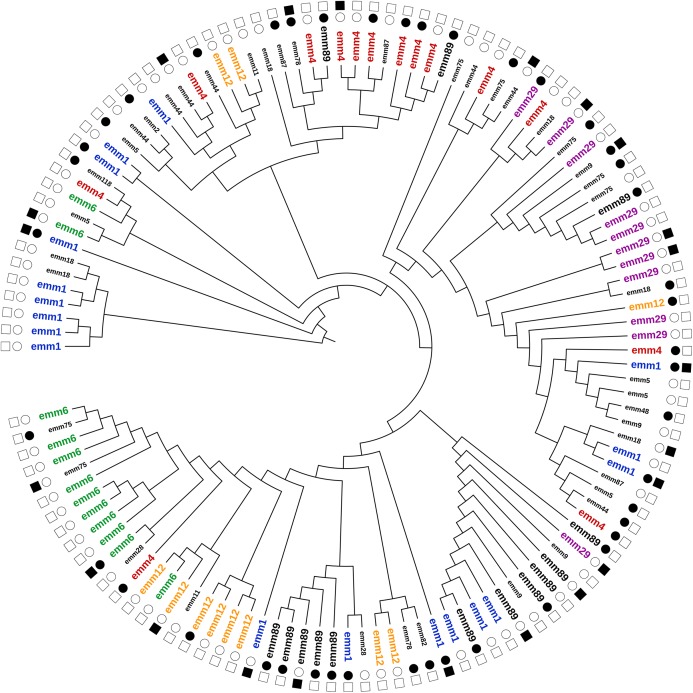
Although we detected a consistent heterogeneity of non-FCT virulence genes across all emm types (Fig. 2), we considered their distribution in the most prevalent emm types, namely emm1, emm4, emm89, emm6, emm12, and emm29 (Table 2; Fig. 2—labels with a larger font size). speB was detected in all strains. The distribution of the other virulence genes in each emm type was variable (Fig. 2). For instance, emm1 strains were present in five distant clusters as well as emm89 strains (four different clusters). Only six strains were negative to the whole set of virulence genes (excluding speB), whereas speK and sla were the least common (15% and 14%, respectively).
Figure 2. Cluster analysis of S. pyogenes strains based on the profile of the entire set of screened virulence genes.
For each strain, only the emm type (inner circle) and the presence/absence of the genes encoding the two major superantigens, speA and speC (outer circle) are shown. Filled or empty symbols indicate the presence or the absence of the screened gene, respectively. The emm types names are in different font size to reflect their relative abundance within the study population.
Table 2. Distribution of virulence genes within each emm type group.
| Total | emm1 | emm4 | emm89 | emm6 | emm12 | emm29 | |
|---|---|---|---|---|---|---|---|
| n = 80 | n = 18 | n = 14 | n = 14 | n = 12 | n = 11 | n = 11 | |
| speC | 30 | 3 | 10 | 7 | 7 | 3 | 0 |
| speA | 15 | 9 | 3 | 0 | 3 | 0 | 0 |
| ssa | 16 | 2 | 10 | 2 | 0 | 1 | 1 |
| sdn | 18 | 2 | 8 | 0 | 4 | 1 | 3 |
| sla | 11 | 0 | 1 | 0 | 10 | 0 | 0 |
| speK | 12 | 1 | 0 | 0 | 7 | 4 | 0 |
| speH | 22 | 2 | 2 | 0 | 11 | 7 | 0 |
| speI | 14 | 3 | 1 | 0 | 4 | 6 | 0 |
| speL | 14 | 0 | 2 | 1 | 0 | 2 | 9 |
| speM | 10 | 2 | 2 | 0 | 0 | 2 | 4 |
| speB | 80 | 18 | 14 | 14 | 12 | 11 | 11 |
| smeZ | 22 | 8 | 9 | 1 | 1 | 0 | 3 |
speC was identified in 71% of the emm4, 58% of the emm6, 50% of emm89, 27% of emm12, and 17% of the emm1 strains. Strains positive to speA belonged to emm1 (50%), emm6 (25%), and emm4 (21%). The emm29 strains were the sole type uniformly negative to both speC and speA.
sdn was found in the emm4 (57%), emm6 (33%), and emm1 (11%) strains. smeZ was detected in the emm4 (64%), emm1 (44%), emm89 (7%), and emm6 (8%) strains. We did not observe any emm89 strain positive to speA, sdn, sla, speK, speH, speI, and speM. We did not detect ssa, speL, and speM in the emm6 strains as well as sla and speL in the emm1 strains, whereas in the emm4 strains only speK was not detected (Table 2).
Within emm1, emm4, emm6, and emm89, the number of different non-FCT virulence gene profiles was limited (Table S3). The analysis identified 40 genotypes, and the two most common genotypes, G38 and G39, were exclusively found in emm89 strains (five and six strains, respectively). Similarly, the four G7 strains belonged to emm1. Only genotypes G5 and G26 contained three strains. G5 was associated with emm1, whereas G26 was associated with emm4 and emm89 strains. The rest of genotypes were characterized by one or at least two strains and were not considered for further comparative analysis and discussion.
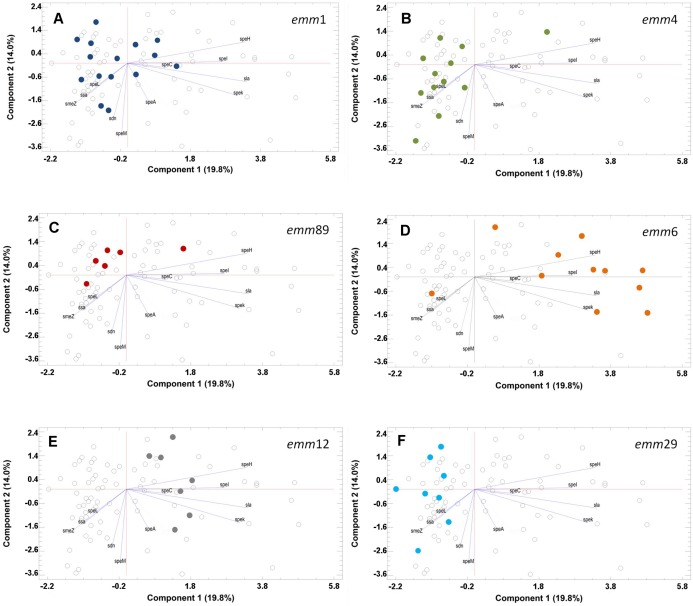
Variability in non-FCT virulence genes among emm1 strains unmasked a substantial heterogeneity wherein none of the sorted principal components accounted for most of the variability (Fig. 3). There were strains containing no virulence genes and groups having speC or sdn as the major contributors to variability. Almost the same trend was found in emm4, emm29, and emm89 strains. The latter group, however, was composed by strains mostly devoid of virulence genes. The emm6 strains were characterized by the presence of speH/speI and speK/sla. Similarly, the emm12 strains were diversified by the presence of speC.
Figure 3. Principal component analysis (PCA) of the contribution of virulence genes to diversity.
Only plots referring to the six more prevalent emm types in the study population are reported. Each panel shows, as colored dots (different for each emm type), the strains sharing at least the same emm type (A: emm1–dark blue; B: emm4–green; C: emm89–red; D: emm6–orange; E: emm12–grey; F: emm29–light blue). Strains sharing similar virulence genes’ profiles tend to cluster in the same area of the 2D plot and those having the very same profile spot on the same point within the 2D plot. Hence, the number of spots does not necessarily reflect the number of strains belonging to a definite emm type group as per the distribution given in Fig. 1.
FCT typing
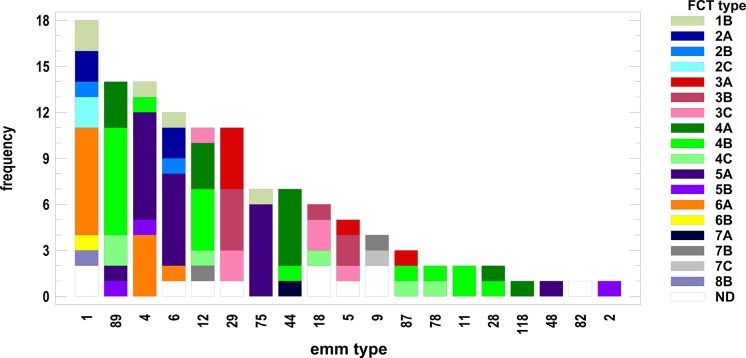
A total of 41 (34%) GAS strains showed reference FCT-region variants as previously described (Table 1; Kratovac et al., 2007). The two most frequent were the FCT5 (16%) and FCT4 (10%), which are referred to as FCT5A and FCT4A, respectively. FCT4B was the most prevalent single FCT-region variant (n = 18). Compared to the reference FCT-region variant FCT4A, FCT4B lacked fctA (n = 12) or the bridge region (n = 6). Overall, 73 out of 122 strains (60%) showed a single FCT-region variant or a double FCT-region variant of the reference variant. The association of 11 strains (9%) to a specific reference FCT-region variant or a single/double variant was not possible. Their FCT region genetic profile showed extensive differences compared to the reference.
In order to establish a possible association, we cross-tabulated emm types and FCT-region variants. We run the test to determine whether or not to reject the hypothesis that the emm type and FCT-region variants classifications were independent (Chi-square test). Since the p-value was less than 0.001, we rejected the hypothesis that emm types and FCT variants were independent at the 99.9% confidence level. Thus, we looked for the associations between FCT-region variants and the most represented emm types. As it is shown in Fig. 4, emm1 strains were mainly associated with FCT regions 2 and 6, while in emm89 and emm12 strains the most represented region was FCT4. A good proportion of both emm4 and emm6 strains were positive to FCT5. At last, emm29 strains were almost completely characterized by the FCT3 group of variants.
Figure 4. Distribution of fibronectin-collagen-T antigen (FCT) region types variants within each emm type group of strains.
Frequency on the Y-axis refers to the number of strains.
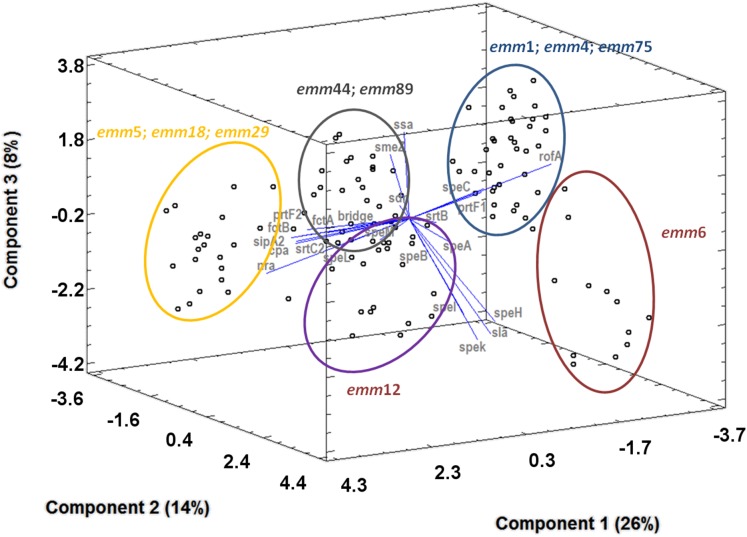
The PCA based on the emm type, non-FCT virulence genes, and FCT-region pattern showed an association between emm typing, FCT-region variants typing, and virulence gene profiling (Fig. 5). In terms of variability/associations, component 1 was mostly and equally contributed by fctB and prtF2, followed by fctA, nra, srtB, and speL. rofA, prtF1, srtB, and speH/I contributed to variability within component 2, followed by speK/sla and speC. At last, component 3 included ssa, rofA, prtF1, smeZ, and speC, followed by srtB. Based on this variability analysis, same emm type strains clustered together. Five major groups were identifiable. The group circled in black (Fig. 5) was mainly composed by emm44 and emm89 strains in which there was not a great extent of variability in presence/absence of many genes (proximity to the center of the plot). For instance prtF1, prtF2, fctA, fctB, cpa, srtC2 together with srtB were overall present, while ssa with speC contributed to variability within the group as seen by the spread along component 3 axis). The group circled in yellow had emm type 5, 18, and 29 strains. They provided a strong contribution to the variability of the component 1 and were positively associated to the presence of prtF2, fctB, cpa, srtC2, sipA2, nra. The opposite could be seen in the case of the cluster circled in blue (emm1, emm4, emm75). Here, the associated genes were mainly rofA, speA, smeZ, and prtF1. The two remaining clusters, in violet and red, were made up by strains belonging to emm12 and emm6, respectively. The latter was shifted toward the positive values along the component 2, with a dominant positivity to srtB, speH/I, speK/sla, and speC. Additionally, emm types associable to the same emm pattern composed the same cluster. For instance, emm44 and emm89 (Fig. 5, black circle) are associated to the emm pattern E. emm5, emm18, and emm29, belonging to the emm pattern A–C, clustered together (Fig. 5, yellow circle). The cluster made by emm1, emm4, and emm75 was an exception (Fig. 5, blue circle), in that emm1 is usually associated to emm pattern A–C, while emm4 and emm75 belong to the emm pattern E (McMillan et al., 2012).
Figure 5. Principal component analysis (3D plot) showing the relative contribution of each virulence gene to the diversity among strains.
Clusters of strains (spots) according to their emm type is indicated by circles of different colors.
Overall, when the complete set of virulence genes were considered (i.e., emm, non-FCT, and FCT), a total of 83 different profiles were discriminated (Supplemental File), supporting the extremely high genetic heterogeneity of the strains.
Geographical distribution of emm and FCT-region variants
The strains were isolated from three different areas of Central Italy, namely Roma (RM; n = 48), Perugia (PG; n = 50) and Macerata (MC; n = 26). We investigated if there was any association between the isolation area and specific virulence traits. Cross-tabulation of emm types by areas and the Chi-square test showed that the hypothesis of independence could be rejected (p < 0.001). The measurement of the degree of association between emm types and areas showed that there was a 43.1% reduction in error when the emm type was used to predict the geographical area where the strain came from. A focus on the six most common emm types (1, 4, 89, 6, 12, and 29) showed that the proportions of emm1 and emm89 were almost equally represented within the RM, PG, and MC groups. Actually, the RM group had slightly more emm1 strains than the other two groups, whereas emm4 strains were scarcely represented in the PG cohort. Conversely, the PG area consistently contributed to the total number of emm6 and emm29 strains.
The same analysis was performed to investigate the association between the area of isolation and FCT-region variants typing. The hypothesis of independence for these two variables could be rejected (p < 0.01). Given the overall association between the major emm types and the FCT-region variants and between emm types and geographical area of isolation, a general association between FCT types and geographical area of isolation was expected. In fact, we observed an association between FCT3 variants and the PG area as well as between FCT2/FCT6 variants and the RM area.
Discussion
Our investigation revealed a high variability of emm types and virulence genes among GAS strains from three different areas of Central Italy. Some virulence traits are mostly represented in one area over the other two. This result was not biased by the possible occurrence of major clones circulating in a single area, for instance as a result of epidemics. In fact, the large number of virulence traits considered had a high discriminatory power sorting out 83 different profiles. When compared to the total number of strains (n = 122), this number excluded clonality from being a factor influencing clustering and interpretation of variable associations.
Our data confirmed a trend observed in other countries in which emm1, emm4, emm89, and emm6 (listed in descendent order) were among the prevalent circulating types (Zampaloni et al., 2003; Commons et al., 2008; Steer et al., 2009; Shea et al., 2011), followed by emm12 and emm29. The association between emm type and source of isolation was not investigated because the number of strains isolated from throat was highly predominant and would have undermined the significance of the correspondence analysis. Genetic changes in emm89 strains have recently generated clonal lineages responsible for invasive infections in different parts of the globe (Steer, Danchin & Carapetis, 2007; Beres et al., 2016). The high prevalence of non-invasive emm89 type strains in our collection confirmed the increasing expansion of this emm type that, together with its propensity to switch to more virulent variants, is of concern and claims the continuous monitoring of its diffusion into the population (Latronico et al., 2016). The analysis of the recorded patterns of virulence genes revealed the presence of major clones as already described in previous studies (Vlaminckx et al., 2003; Commons et al., 2008; Friães et al., 2012). We found emm4 strains with the G25 pattern of virulence genes. They had a wide distribution and were recorded in invasive and non-invasive infections over a long period of time (Commons et al., 2008; Friães et al., 2012). Among the emm6 strains, the FCT5 and G35 pattern of virulence genes were identified. A similar pattern was present in invasive GAS isolated in The Netherlands from 1992 to 1996 (Vlaminckx et al., 2003). In addition, we described emm1/G16 pattern strains that differed for the presence of smeZ from emm1 strains isolated in Portugal (Friães et al., 2012).
Analysis of the results obtained considering the entire set of data and leading to the general picture of Fig. 5 showed different clusters of profiles identified by emm types. This observation reinforces the evidence on how emm typing continues to represent an exceptional marker of the genetic diversity within a GAS population. The same clustering has been additionally considered in relation to the emm patterns, whose general correlation with emm types and strains’ source of isolation is well known (Commons et al., 2008; Metzgar & Zampolli, 2011; Friães et al., 2012). Our collection is almost completely made of isolates from cases of pharyngotonsillitis. As a consequence, it is expected to be composed of emm types corresponding to patterns A–C (emm1, 5, 6, 12, 18) and E (emm4, 44, 75, 89). Clusters identified by our total analysis confirmed the emm type/emm pattern association in all cases but for the one composed by emm1, emm4, and emm75 (Fig. 5, blue circle). For instance, emm1 is usually associated to pattern A–C while the latters to pattern E. This observation clearly indicates that the emm type/emm pattern association may hinder dissimilarities in important subset of virulence genes, namely superantigens and FCT-region associated ones. As a matter of facts, emm types 5, 18, and 29, grouping into one defined cluster by PCA (Fig. 5), were mostly FCT3 and as such positive to the Nra transcriptional regulator gene (Table 1). However, nra-positive strains have usually been found associated to emm pattern D (skin specialist), while emm types 5, 18, and 29 are well known to be associated to emm pattern A–C (throat specialist) (Bessen, 2009; McMillan et al., 2012).
Conclusions
The distribution of emm types revealed a complex epidemiological scenario of GAS strains. The detection of 12 virulence genes showed a great degree of clonal heterogeneity within the GAS collection. There was a great structural variability within the FCT region, higher than that previously described (Kratovac et al., 2007), with new FCT-region variants recorded, indicating overall an extremely high degree of genetic shuffling among different parts of the FCT region in GAS. This is most probably related to the evolutionary pressure of the immune system on the different elements exposed on the surface of GAS and encoded by genes mapping within the FCT region such as fibronectin-binding proteins and pili (Mora et al., 2005). Further and continuous molecular epidemiologic studies are needed to increase our understanding of possible associations of virulence determinants and their variants that facilitate host-pathogen interactions. This understanding may help in guiding the design of vaccines against GAS infections.
Supplemental Information
Funding Statement
This work was supported by funds from the Italian Ministry of Education, University, and Research (MIUR, grant FUTURO IN RICERCA No. RBFR10X4YN_001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Daniela Bencardino performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Maria Chiara Di Luca performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Dezemona Petrelli conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Manuela Prenna contributed reagents/materials/analysis tools, approved the final draft.
Luca Agostino Vitali conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data are available in a Supplemental File.
References
- Barnett et al. (2015).Barnett TC, Cole JN, Rivera-Hernandez T, Henningham A, Paton JC, Nizet V, Walker MJ. Streptococcal toxins: role in pathogenesis and disease. Cellular Microbiology. 2015;17(12):1721–1741. doi: 10.1111/cmi.12531. [DOI] [PubMed] [Google Scholar]
- Beres et al. (2016).Beres SB, Kachroo P, Nasser W, Olsen RJ, Zhu L, Flores AR, De la Riva I, Paez-Mayorga J, Jimenez FE, Cantu C, Vuopio J, Jalava J, Kristinsson KG, Gottfredsson M, Corander J, Fittipaldi N, Di Luca MC, Petrelli D, Vitali LA, Raiford A, Jenkins L, Musser JM. Transcriptome remodeling contributes to epidemic disease caused by the human pathogen Streptococcus pyogenes. mBio. 2016;7(3):e00403-16. doi: 10.1128/mBio.00403-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen (2009).Bessen DE. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infection, Genetics and Evolution. 2009;9(4):581–593. doi: 10.1016/j.meegid.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen & Kalia (2002).Bessen DE, Kalia A. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infection and Immunity. 2002;70(3):1159–1167. doi: 10.1128/IAI.70.3.1159-1167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commons et al. (2008).Commons R, Rogers S, Gooding T, Danchin M, Carapetis J, Robins-Browne R, Curtis N. Superantigen genes in group A streptococcal isolates and their relationship with emm types. Journal of Medical Microbiology. 2008;57(10):1238–1246. doi: 10.1099/jmm.0.2008/001156-0. [DOI] [PubMed] [Google Scholar]
- Commons et al. (2014).Commons RJ, Smeesters PR, Proft T, Fraser JD, Robins-Browne R, Curtis N. Streptococcal superantigens: categorization and clinical associations. Trends in Molecular Medicine. 2014;20(1):48–62. doi: 10.1016/j.molmed.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Cunningham (2000).Cunningham MW. Pathogenesis of group A streptococcal infections. Clinical Microbiology Reviews. 2000;13(3):470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friães et al. (2012).Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J. Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. BMC Microbiology. 2012;12(1):280. doi: 10.1186/1471-2180-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friães et al. (2013).Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J. Superantigen gene complement of Streptococcus pyogenes—relationship with other typing methods and short-term stability. European Journal of Clinical Microbiology & Infectious Diseases. 2013;32(1):115–125. doi: 10.1007/s10096-012-1726-3. [DOI] [PubMed] [Google Scholar]
- Kratovac et al. (2007).Kratovac Z, Manoharan A, Luo F, Lizano S, Bessen DE. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. Journal of Bacteriology. 2007;189(4):1299–1310. doi: 10.1128/JB.01301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latronico et al. (2016).Latronico F, Nasser W, Puhakainen K, Ollgren J, Hyyryläinen H-L, Beres SB, Lyytikäinen O, Jalava J, Musser JM, Vuopio J. Genomic characteristics behind the spread of bacteremic group A Streptococcus type emm 89 in Finland, 2004–2014. Journal of Infectious Diseases. 2016;214(12):1987–1995. doi: 10.1093/infdis/jiw468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic & Bork (2016).Letunic I, Bork PC. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor et al. (2004).McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, Beall B, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. Journal of Bacteriology. 2004;186(13):4285–4294. doi: 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan et al. (2012).McMillan DJ, Sanderson-Smith ML, Smeesters PR, Sriprakash KS. Host-Pathogen Interactions in Streptococcal Diseases. Berlin, Heidelberg: Springer; 2012. Molecular markers for the study of streptococcal epidemiology; pp. 29–48. [DOI] [PubMed] [Google Scholar]
- Metzgar & Zampolli (2011).Metzgar D, Zampolli A. The M protein of group A Streptococcus is a key virulence factor and a clinically relevant strain identification marker. Virulence. 2011;2(5):402–412. doi: 10.4161/viru.2.5.16342. [DOI] [PubMed] [Google Scholar]
- Mora et al. (2005).Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AGO, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and lancefield T antigens. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, Garbe & Collin (2011).Nelson DC, Garbe J, Collin M. Cysteine proteinase SpeB from Streptococcus pyogenes-a potent modifier of immunologically important host and bacterial proteins. Biological Chemistry. 2011;392:1077–1088. doi: 10.1515/BC-2011-208. [DOI] [PubMed] [Google Scholar]
- Shea et al. (2011).Shea PR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Beres SB, Flores AR, Low DE, Willey BM, Musser JM. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerging Infectious Diseases. 2011;17(11):2010–2017. doi: 10.3201/eid1711.110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger et al. (2011).Slinger R, Goldfarb D, Rajakumar D, Moldovan I, Barrowman N, Tam R, Chan F. Rapid PCR detection of group A Streptococcus from flocked throat swabs: a retrospective clinical study. Annals of Clinical Microbiology and Antimicrobials. 2011;10(1):33. doi: 10.1186/1476-0711-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer, Danchin & Carapetis (2007).Steer AC, Danchin MH, Carapetis JR. Group A streptococcal infections in children. Journal of Paediatrics and Child Health. 2007;43(4):203–213. doi: 10.1111/j.1440-1754.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- Steer et al. (2009).Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infectious Diseases. 2009;9(10):611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaminckx et al. (2003).Vlaminckx BJM, Mascini EM, Schellekens J, Schouls LM, Paauw A, Fluit AC, Novak R, Verhoef J, Schmitz FJ. Site-specific manifestations of invasive group a streptococcal disease: type distribution and corresponding patterns of virulence determinants. Journal of Clinical Microbiology. 2003;41(11):4941–4949. doi: 10.1128/JCM.41.11.4941-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampaloni et al. (2003).Zampaloni C, Cappelletti P, Prenna M, Vitali LA, Ripa S. emm gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenes. Journal of Clinical Microbiology. 2003;41(3):1307–1310. doi: 10.1128/JCM.41.3.1307-1310.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in a Supplemental File.