Abstract
White grain disorder is a recently emerged wheat disease in Australia, caused by Eutiarosporella darliae, E. pseudodarliae, and E. tritici-australis. The disease cycle of these pathogens and the molecular basis of their interaction with wheat are poorly understood. To address this knowledge gap, we undertook a comparative genomics analysis focused on the secondary metabolite gene repertoire among these three species. This analysis revealed a diverse array of secondary metabolite gene clusters in these pathogens, including modular polyketide synthase genes. These genes have only been previously associated with bacteria and this is the first report of such genes in fungi. Subsequent phylogenetic analyses provided strong evidence that the modular PKS genes were horizontally acquired from a bacterial or a protist species. We also uncovered a secondary metabolite gene cluster with three polyketide/nonribosomal peptide synthase genes (Hybrid-1, -2, and -3) in E. darliae and E. pseudodarliae. In contrast, only remnant and partial genes homologous to this cluster were identified in E. tritici-australis, suggesting loss of this cluster. Homologues of Hybrid-2 in other fungi have been proposed to facilitate disease in woody plants, suggesting a possible alternative host range for E. darliae and E. pseudodarliae. Subsequent assays confirmed that E. darliae and E. pseudodarliae were both pathogenic on woody plants, but E. tritici-australis was not, implicating woody plants as potential host reservoirs for the fungi. Combined, these data have advanced our understanding of the lifestyle and potential host-range of these recently emerged wheat pathogens and shed new light on fungal secondary metabolism.
Keywords: phytopathogen, horizontal gene transfer, gene loss, modular polyketide synthase, Eutiarosporella
Introduction
Fungi occupy a wide range of trophic niches across the world. While many blend into the environment unseen, a small but significant number are damaging phytopathogens, responsible for substantial losses to agricultural, horticultural, and forestry industries (Agrios 2005). Understanding how fungal species evolved to persist in the hostile environment of a host plant, whether as an endophyte or a pathogen, is a major focus of mycological research.
Small molecules, known as secondary metabolites (SMs) are often used by microbes to enable unique trophic lifestyles (Walton 1996; Friesen et al. 2008; Stergiopoulos et al. 2013). These small molecules can promote growth in environments where there is strong competition from other microbes or environmental stress (Stergiopoulos et al. 2013; Shabuer et al. 2015). Historically, it has been challenging to study the clusters of genes involved in secondary metabolite synthesis as they can span large regions of the genome. However, whole genome sequencing has led to a rapid expansion of our knowledge of the diversity and number of microbial SM gene clusters (Chooi and Solomon 2014). Similarly, synthetic biology approaches that manipulate or isolate these clusters has led to the identification of many novel compounds providing a glimpse of their structural and functional diversity (Chooi and Tang 2012).
The Dothideomycetes are a prominent class of fungi encompassing many economically significant phytopathogens. In recent years, a large number of Dothideomycete species’ genomes have been sequenced and analyzed (Ohm et al. 2012). These genomes have enabled the discovery and characterization of the genes and small molecules that these fungi use to infect plants (Oliver and Solomon 2010). Dothideomycetes also produce a wide range of SMs (Muria-Gonzalez et al. 2015), some of which have been shown to facilitate disease on specific host genotypes (Walton 1996; Friesen et al. 2008; Stergiopoulos et al. 2013). A selection of these host specific Dothideomycete SMs include: HC-toxin, produced by a nonribosomal peptide synthase (NRPS) in Cochliobolus carbonum (Walton 2006); T-toxin, produced by a polyketide synthase (PKS) in Cochliobolus heterostrophus (Yang et al. 1996; Baker et al. 2006); and ACT toxin and AK toxin, produced by two PKSs in tangerine and Japanese pear pathovars of Alternaria alternata, respectively (Nakashima et al. 1982; Kohmoto et al. 1993; Tanaka et al. 1999; Miyamoto et al. 2010).
Some SMs produced by phytopathogenic fungi are also a significant threat to human health due to their extremely toxic activity in mammals (Solomon 2011). This is particularly important with phytopathogens that contaminate food supplies. A prominant example is Fusarium graminearum, the causal agent of the wheat grain disease, fusarium head blight. Wheat grain infected with F. graminearum is often tainted with mycotoxins, including deoxynivalenol (Solomon 2011). Similarly, wheat-infecting Dothideomycetes are known to produce the mycotoxin, alternariol, during infection (Tan et al. 2009). As such, it is important to learn whether new and emerging pathogens are capable of producing mycotoxins.
White grain disorder (WGD) is a disease of wheat first observed in Queensland, Australia during the late 1990s, which causes the shrivelling and discoloration of grain heads (Wildermuth et al. 2001; Platz 2011). In the past decade, the prevalence of WGD has increased within Australia and the disease has now been observed in wheat-growing regions in Western Australia, South Australia, and Queensland (Evans 2013; Thomas and Jayasena 2015). The causal agents of WGD are three closely related species: Eutiarosporella darliae, E. pseudodarliae, and E. tritici-australis (Thynne et al. 2015), from the Botryosphaeriaceae family within the Dothideomycetes. It is unknown how these three species emerged as wheat pathogens, where they originated, and if they interact with any nongrass hosts. Similarly, it is not known whether these pathogens are capable of producing any of the known fungal mycotoxins, which would contaminate the infected grain.
In this study, we performed a comparative genomics analysis of the WGD Eutiarosporella spp. with the objective of determining the prevalence and importance of SMs in these fungi and to further our understanding of the lifestyle and potential host range of these recently emerged pathogens.
Materials and Methods
WGD Fungi Sequencing, Assembly, and Annotation
Genome assemblies are described in Thynne et al. (2015) for each of E. tritici-australis, E. darliae, and E. pseudodarliae and are available at the European Nucleotide Archive (E. darliae accession: GFXH01000000; E. pseudodarliae accession: GFXI01000000, E. tritici-australis accession: GFXG01000000). Illumina paired-end sequencing was performed using a HiSeq 2000 (Illumina) at the John Curtain School of Medical Research, at The Australian National University (ANU). The resulting sequenced reads were quality trimmed using Trimmomatic v0.27 (Lohse et al. 2012) and assembled de novo with SPAdes v2.5.0, with –k mer values 21, 33, 55, and 77 (Bankevich et al. 2012). Genome size was estimated from the trimmed Illumina reads using BBmap v38.34 and the kmercountexact script with default settings (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbmap-guide/). Gene sets for each species were predicted and annotated with the MAKER genome annotation pipeline with the gene sets from Botryosphaeria dothidea and Parastagonospora nodorum used as training genes (Cantarel et al. 2007). Protein sets used for this analysis, in a format compatible with OrthoMCL analysis (Li et al. 2003) are available at doi: 10.6084/m9.figshare.7448402. Where required, genes were reannotated with FGNESH+ on the online server at softberry.com (Solovyev 2001). Annotations were visualized within Geneious v7.1.8 (Biomatters, New Zealand). Secondary metabolite genes were predicted with the online antiSMASH (v.2) server (Weber et al. 2015). Protein domains were predicted with Interproscan v.1.0.6. (Quevillon et al. 2005). OrthoMCL (Li et al. 2003) was used to cluster orthologous protein sequence groups among the three Eutiarosporella spp. and four relatives, B. dothidea, Neofusicoccum parvum, Macrophomina phaseolina, and Diplodia seriata (Islam et al. 2012; Blanco-Ulate et al. 2013; van der Nest et al. 2014; Morales-Cruz et al. 2015; Marsberg et al. 2017). OrthoMCL was run using the OrthoMCL pipeline, with default settings (all-vs-all BLASTp e-value: 1e-5). All of the sequences predicted as unique clusters within the WGD species, but not the other Botryosphaeriaceae species, were manually submitted as BLASTp queries against the NCBI nr database (e-value cut-off of 1e-4) to confirm that Botryosphaeriaceae sequences were not best-BLAST hits. The curated data, raw data output and protein sets with fasta headings compliant with orthoMCL are also listed in Supplementary Material online (doi: 10.6084/m9.figshare.7448402). Each protein set from E. darliae, E. pseudodarliae, B. dothidea, M. phaseolina, N. parvum, and D. seriata, were submitted to the online Carbohydrate active enzyme (CAZyme) online server, dbCAN (Yi et al. 2012). The CAZyme sequences were extracted, and an additional OrthoMCL clustering analysis was performed to identify CAZyme orthologues not present among the WGD species, or unique among these Botryosphaeriaceae species to the WGD species.
Genome Analysis of Eutiarosporella and Macrophomina Modular PKSs and the Hybrid PKS–NRPS Cluster
The relative genome positions of the modular PKS genes (mPKS), hybrid PKS–NRPSs, and surrounding genes were determined in Geneious v7.1.8, through manual analysis of the assembled scaffolds. Annotated genes were used as BLASTn and tBLASTx queries to search for these genes and compare synteny on the genome-scaffolds of the alternative WGD species.
Phylogenetic Analysis and Alternative Topology Testing of Eutiarosporella KS Domains
All alignments used to build the trees presented in this manuscript can be downloaded from figshare.com (doi: 10.6084/m9.figshare.7448402). The ketoacyl synthase (KS) domains for the predicted polyketide synthase (PKS) amino-acid sequences from each of the WGD Eutiarosporella spp. were used as BLASTp (Altschul et al. 1990) queries to search the NCBI nonredundant (NR) database for close homologues. Similarly, the KS domain of Penicillium aethiopicum VrtA amino acid sequence and the Alternaria alternata PKSj amino acid sequence were used to search for additional fungal PKS homologues. Selections of these PKS homologues were downloaded from NCBI and the protein domains for each predicted with Interproscan v.1.0.6 (Quevillon et al. 2005). The predicted KS domains from all of the sequences were extracted and aligned with MUSCLE v3.5 (Edgar 2004) and the internal Geneious aligner (Geneious v7.1.8; Biomatters, New Zealand) (doi: 10.6084/m9.figshare.7448402). Ep-mPKSa KS1, 2, and 3, and Ed-mPKSa KS1 and 2, were removed from this tree due to decreased amino acid sequence length (these were included in a secondary tree comparing the Botryosphaeriaceae mPKS–KS domains). A maximum likelihood tree with 500 bootstrap replications was created using RaxML v8 (Stamatakis 2014) with the rapid bootstrapping command “-f a,” the random number seed and starting tree commands of “-p 1234” and “-x 1234,” respectively.
A second tree was generated with the addition of the RaxML multifurcating tree command “-g” with a kingdom segregated constraint file. File used for Kingdom segregation can be downloaded from figshare.com (doi: 10.6084/m9.figshare.7448402). The “best-tree” output from both the constrained and unconstrained trees were concatenated and used as the input file to generate a site-log likelihood file from RaxML using the “-f g” command. The output file in treepuzzle format was fed into Consel (Shimodaira and Hasegawa 2001) to perform Kishino–Hasegawa (KH) and Shimodaira–Hasegawa (SH) tests to compare the constrained and unconstrained trees.
Phylogenetic Analyses of KS Domains from the Hybrid PKS–NRPS Genes
The KS domains from the encoded amino acid sequences of the hybrid PKS–NRPS genes were used as BLASTp queries to search the NCBI nonredundant (NR) database and the JGI MycoCosm portal for close homologues. Some of the predicted protein sequences were shorter than expected (EKG15258.1, KKY13504.1) and were reannotated using FGNESH+ on the online server at softberry.com. The domains for each homologue were predicted with Interproscan v.1.0.6 (Quevillon et al. 2005) and the online antiSMASH (v.2) server (Weber et al. 2015). Images used were generated in Geneious v7.1.8 (Biomatters, New Zealand) and the online antiSMASH (v.2) server (Weber et al. 2015). A maximum likelihood tree with 500 bootstrap replications was created using RaxML 7.2.8 (Stamatakis 2014) in Geneious v7.1.8 (Biomatters, New Zealand) with the “rapid bootstrapping and search for best-scoring ML tree” command selected. The tree was visualized and annotated in FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
qRT-PCR Analysis of Eta-mPKSa and b, and of Ed-Hybrid1 and 2, and FSTF Genes
For qRT-PCR analysis of Eta-mPKSa and b: E. tritici-australis isolate 153 was grown on potato dextrose agar (PDA, 10 g/l Difco Potato dextrose, 15 g/l Agar). After 3-day postinoculation, plugs of healthy growing mycelia from the PDA plates were used to inoculate the three different growth substrates, potato dextrose broth (PDB, 10g/l Difco Potato dextrose), wheat leaves, and stems (roughly cut, and placed within a petri dish), and fries medium (FM, 5 g/l of ammonium tartrate, 1.0 g/l of ammonium nitrate, 0.5 g/l of magnesium sulfate, 1.3 g/l of potassium phosphate [dibasic], 2.6 g/l of potassium phosphate [monobasic], 30.0 g/l of sucrose, 1.0 g/l of yeast extract). After 3-day postinoculation, mycelial samples were harvested, flash frozen in liquid nitrogen, and ground in a mortar and pestle. Liquid cultures were grown at 22°C, 12 h light/dark and shaken at 130 rpm. The mycelium was harvested by centrifugation to remove the liquid. Mycelial pellets were flash frozen in liquid nitrogen and ground with a mortar and pestle for RNA extraction.
For qRT-PCR analysis of EdHybrid1, 2, and FSTF E. darliae isolate 2E2 was grown on potato dextrose agar (PDA). At 3-day postinoculation, plugs of healthy growing mycelia from the PDA plates were used to inoculate the two different growth substrates, PDA and wood from a Hakea branch (cut, and placed within a petri dish). At 3-day postinoculation, mycelial samples were harvested and ground in a mortar and pestle with liquid nitrogen.
All RNA was extracted with the ZR plant RNA MiniPrep kit (Zymo Research). cDNA was synthesized with the SuperScript IV reverse transcriptase kit according to the manufacturers protocol (ThermoFisher Scientific). Quantitative PCR (qPCR) was performed with the Fast SYBR Green Master Mix (ThermoFisher Scientific) in a 10-μl reaction. Each sample had four technical replicates and a no-template control was included in each qPCR reaction. The cycling conditions used were 95 °C for 20 s, followed by 40 cycles with a 1-s denaturation at 95 °C, 20 s annealing and extension at 60 °C. A final melting curve was measured for each sample to control for unspecific amplification. The primers used for each assay (normalized in each to β-tubulin) are listed in supplementary table 1, Supplementary Material online.
Infection Assays
Leaves of a several unknown Eucalyptus spp. and Hakea salicifolia were obtained from The Australian National University campus and surrounding suburbs. Hakea salicifolia was mainly used for infection assays in the study due to the pronounced disease symptoms. Leaves were wounded in an “X” pattern (∼1–2 cm) with a scalpel. Wound sites were inoculated with agar plugs covered in growing mycelia and incubated in high humidity. Disease symptom development was observed over the following week.
Both seedlings and leaves from mature plants were used. To grow the seedlings, H. salicifolia seeds were obtained from seedpods from a mature plant. To access the seed, pods were heated at 60°Celsius for 6 h (or until open). Seeds were germinated in seed-raising mix (The Scotts Miracle-Gro Company). When established shoots appeared, the seedlings were further fertilized with 5 g Osmocote (The Scotts Miracle-Gro Company).
Results
The WGD Eutiarosporella spp. Assembled Genomes Are Smaller than Their Close Relatives, and Have Reduced Secondary Metabolite Potential
The de novo genome assemblies of E. darliae, E. pseudodarliae, and E. tritici-australis used in this analysis were ∼27 Mb and predicted to encode between 8,500 and 8,750 genes (table 1). In addition, a kmer based estimate of genome size using unassembled Illumina reads showed these genomes were similar in size to the assembled scaffolds (table 1). Both estimates showed that the genomes of the three WGD species are smaller than other sequenced members of the Botryosphaeriaceae. Comparisons against the genome assemblies of the Botryosphaeriaceae species: Botryosphaeria dothidea, Diplodia seriata, Neofusicoccum parvum, and Macrophomina phaseolina are displayed in table 1 (Islam et al. 2012; Blanco-Ulate et al. 2013; Morales-Cruz et al. 2015). For additional comparison with species outside of the Botryosphaeriaceae, we also include three other Dothideomycete species: Zymoseptoria tritici, Mycosphaerella populorum, and M. populicola (Ohm et al. 2012; Dhillon et al. 2015). The Core Eukaryotic Gene Mapping Approach (CEGMA) scores for estimating genome assembly completeness are >96% for each of the Eutiarosporella spp. genomes (table 1). This suggests that, although the genome assemblies are smaller than those of the other Dothideomycete species listed, they are relatively complete (Parra et al. 2007).
Table 1.
Basic Genome and Secondary Metabolite Cluster Statistics for the Three WGD Species in Comparison to Selected Other Dothideomycetes
| Dothideomycete Species | Estimated Genome Size (Mb)a | Assembled Genome Size (Mb)b | Number of Predicted Proteins | CEGMAc Score | References |
|---|---|---|---|---|---|
| Botryopshaeriaceae | |||||
| Eutiarosporella tritici-australis | 26.6 | 27.1 | 8571 | 96.77 | This work |
| Eutiarosporella darliae | 27.9 | 27.8 | 8735 | 97.18 | This work |
| Eutiarosporella pseudodarliae | 28.0 | 27.3 | 8678 | 97.58 | This work |
| Macrophomina phaseolina | 49.3 | 14249 | 98.79 | Islam et al. (2012) | |
| Botryosphaeria dothidea | 34 | 14998 | N/A | Marsberg et al. (2017) | |
| Neofusicoccum parvum | 43.7 | 13124 | 97.2 | Blanco-Ulate et al. (2013) | |
| Diplodia seriata | 37.1 | 9398 | 96 | Morales-Cruz et al. (2015) | |
| Mycosphaerellaceae | |||||
| Zymoseptoria tritici | 39.7 | 10933 | N/A | Goodwin et al. (2011) | |
| Mycosphaerella populicola | 33.2 | 9739 | 94.4 | Dhillon et al. (2015) | |
| Mycosphaerella populorum | 29.3 | 10233 | 96 | Dhillon et al. (2015) | |
| Predicted secondary metabolite clusters | |||||
| Polyketide synthase (PKS) | Non ribosomal peptide synthase (NRPS) | Hybrid PKS/NRPS | Terpene synthase | ||
| Eutiarosporella tritici-australis | 4 | 3 | 0 | 7 | |
| Eutiarosporella darliae | 5 | 5 | 1 | 7 | |
| Eutiarosporella pseudodarliae | 5 | 6 | 1 | 7 | |
| Macrophomina phaseolina | 35 | 28 | 12 | N/A | Islam et al. (2012) |
Genome size estimated from quality trimmed, unassembled Illumina reads with a kmer size of 31.
Sum of the total length of bases assembled into contigs.
Core Eukaryotic Gene Mapping Approach (CEGMA), used to estimate genome assembly completeness by identifying conserved genes in the assembled contigs.
To investigate which genes or gene families were potentially unique to the WGD species, we performed an OrthoMCL orthologous protein clustering on all annotated WGD genes and other sequenced Botryosphaeriacae species. The raw orthoMCL data output predicted 203 orthologous protein groups that were present in the WGD species but absent from other Botryosphaeriaceae species (doi: 10.6084/m9.figshare.7448402). To assess the robustness of this prediction, we performed a manual BLASTp of these 203 WGD “unique” groups against the NCBI NR database, using the protein sequences from E. darliae as a query. This analysis reduced the number of unique WGD orthologous groups from 203 to 123. Among these unique WGD orthologs was a small protein (98 AA) with a predicted secretion signal that has best-BLAST hits among cereal pathogens and endophytes, including: Epicoccum nigrum (accession: OSS49457, e-value 4e-44, 75% shared sequence identity), Parastagonospora nodorum (accession: XP_001806258, e-value 5e-38, 67% shared sequence identity), and Rhynchosporium commune (accession: CZTO5762, e-value 5e-32, 60% shared sequence identity) (Relative to E. darliae sequence EDA00091). This analysis gave us a broad overview of orthologous sequence groups across the Botryosphaeriaceae species.
We also compared the carbohydrate-active enzymes (CAZymes) content of each of the WGD species. These analyses revealed that each have a similar number of predicted CAZyme sequences (405 sequences for E. darliae, 402 sequences for E. pseudodarliae, and 401 sequences for E. tritici-australis), but, as with the overall predicted gene numbers, they have a reduced complement in comparison with the other Botryosphaeriaceae species (592 sequences for N. parvum, 698 sequences for M. phaseolina, 682 sequences for B. dothidea, and 506 sequences for D. seriata). We clustered each of the predicted carbohydrate active enzymes into 571 orthologous groups, using orthoMCL (as above). Among these, 267 orthologous sequence groups were putatively shared among all of the Botryosphaeriaceae species (including the WGD species) and 66 orthologue groups were putatively shared among the Botryosphaeriaceae species, but not found in the WGD pathogens. Only five orthologue groups were predicted as unique to the WGD species. Three of these had best-BLAST hits on the NCBI NR database with Botryosphaeriaceae sequences, leaving only two unique WGD groups. As above, one of these unique sequences has close BLAST hits to other wheat infecting species (e.g., Gaeumannomyces tritici, NCBI accession XP_009226784, e-value 2e-141, 65% shared sequence identity), and Magnaporthiopsis poae, NCBI accession KLU91681.1, e-value 4e-141, 67% shared sequence identity) (relative to E. darliae sequence EDA07266). This CAZyme gene has a glycosyl hydrolase family 43 domain (cd08983).
To further dissect the genomes of the WGD pathogens, we used antiSMASH to identify genes within predicted SM clusters. For comparative purposes, we have included M. phaseolina’s predicted SM gene counts (Islam et al. 2012) (table 1). Similar to overall predicted gene numbers, the WGD species have a reduced SM complement to M. phaseolina. BLASTp was used to compare the SM sequences against the NCBI and JGI databases to determine the distribution of similar genes in other organisms (table 2). No SM gene clusters with any similarity to known mycotoxin-related SM genes were identified suggesting that these pathogens are unlikely to produce harmful toxins. The only SM gene with high similarity to a characterized gene whose metabolite is known is mellein synthase (Chooi et al. 2015). This gene is shared by all sequenced Botryosphaeriaceae species, and mellein been isolated from liquid cultures of fungi from this family (Venkatsubbaiah et al. 1991). In addition to the putative mellein synthase, most of the secondary metabolite amino acid sequences have best-BLAST hits to sequences from other members of the Botryosphaeriaceae. However, among the NRPSs identified, three had best-BLAST hits from Sordariomycetes: NRPS2 had a best-BLAST hit to Helicocarpus griseus; NRPS3, which is absent in E. tritici-australis, had a best BLAST hit from Trichophyton benhamiae; NRPS5, which is only present in E. pseudodarliae, had a best-BLAST hit to an NRPS from Colletotrichum gloeosporoides. However, each of these NRPSs have only a low sequence identity with these putative homologues (<50% shared identity). The Terpene synthases (TSs) and iterative PKSs (iPKSs) are largely conserved among the three Eutiarosporella species. Among the three WGD pathogens, the PKS–NRPS hybrid genes appeared to be absent in E. tritici-australis. Our SM analyses also unexpectedly revealed the presence of multiple modular PKSs, discussed in detail below (tables 1 and 2).
Table 2.
Summary of Secondary Metabolite Genes in the Three WGD Species and Their Closest BLAST Hits on NCBI
| Individual SM Synthase Genes | Presence/Absence (+/−) | Best-BLAST Species | Accession | Percent Identity | Predicted SM Type | ||
|---|---|---|---|---|---|---|---|
| TSs | E. tritici-australis | E. darliae | E. pseudodarliae | ||||
| 1 | + | + | + | Diplodia seriata | KKY22841.1 | 86 | TS |
| 2 | + | + | + | D. corticola | OJD35063.1 | 70 | TS |
| 3 | + | + | + | Neofusicoccum parvum | XP_007581866.1 | 86 | TS |
| 4 | + | + | + | D. seriata | OMP85614.1 | 85 | TS |
| 5 | + | + | + | D. corticola | XP_020126315.1 | 78 | TS |
| 6 | + | + | + | D. seriata | KKY14050.1 | 64 | TS |
| 7 | + | + | + | N. parvum | EOD50659.1 | 86 | TS |
| NRPSs | |||||||
| 1 | + | + | + | D. corticola | XP_020135006.1 | 73 | NRPS |
| 2 | + | + | + | Helicocarpus griseus | PGH17632.1 | 37 | NRPS |
| 3 | − | + | + | Trichophyton benhamiae | DAA73007.1 | 48 | NRPS |
| 4 | − | + | + | N. parvum | EOD50138.1 | 66 | PKS–NRPS |
| 5 | − | − | + | C. gloeosporioides | EQB52787.1 | 44 | NRPS |
| PKS–NRPSs | |||||||
| 1 | − | + | + | Macrophomina phaseolina | EKG15176.1 | 90 | PKS–NRPS |
| 2 | − | + | + | M. phaseolina | EKG15173.1 | 85 | PKS–NRPS |
| PKSs | |||||||
| 1 | + | + | + | D. corticola | XP_020132262.1 | 74 | PKS |
| 2 | + | + | + | D. seriata | KKY20157.1 | 90 | PKS |
| 3 | + | + | + | N. parvum | EOD47055.1 | 80 | PKS |
| mPKSs | |||||||
| a | + | + | + | M. phaseolina | EKG12504.1 | 68 | mPKS |
| b | + | + | + | M. phaseolina | EKG12506.1 | 73 | mPKS |
| c | − | + | + | M. phaseolina | EKG12504.1 | 74 | mPKS |
| d | − | − | + | M. phaseolina | EKG12506.1 | 67 | mPKS |
The Modular PKS Genes in the Wheat-Infecting Eutiarosporella spp. Exist in Pairs with Missing Acyltransferase Domain in Some Modules
Based on antiSMASH secondary metabolite cluster predictions, four modular PKS genes (mPKSs) were identified within the WGD Eutiarosporella genomes (named a, b, c, and d) (fig. 1A–C). The longest of the mPKS genes in each species is mPKSa or mPKSc, which encode proteins that are ∼10,000 amino acids long. These are the longest predicted proteins in the genomes of each WGD species. mPKSs were characterized in bacteria and protist species, and differ from fungal iterative PKSs (iPKSs), in that each enzymatic domain can be represented many times in the protein, forming “modules” of enzymatic domains. In these large proteins each module is used only once during the synthesis of the metabolite product and the growing intermediate is passed to the next module in an assembly line manner (Robbins et al. 2016). In contrast, iPKSs only harbor one copy of each enzymatic domain (one module). However, these domains are used multiple times during the synthesis of a metabolite product in an iterative (cyclic) manner (Chooi et al. 2015). iPKSs are the hallmark of fungal PKSs, although some bacterial PKSs are known to be iterative (Chen and Du 2016).
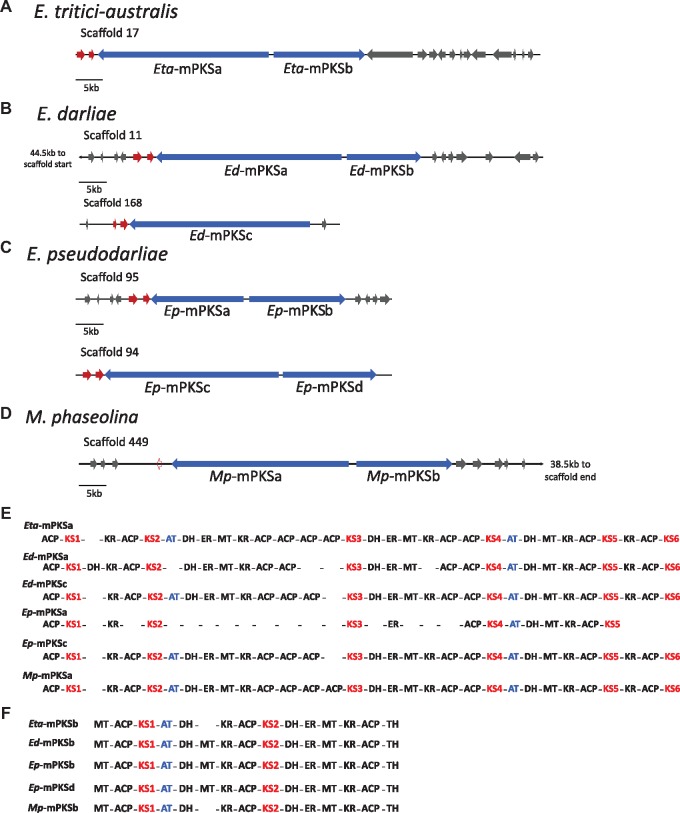
Fig.
1.—(A–D) Graphical representations of all modular PKS (mPKS) genes, Red indicates genes in a syntenic location downstream of each of the larger mPKS genes (shown in blue). (E) Alignment of mPKSa and mPKSc enzymatic domains, highlighting comparative presence or absence of domains. (F) Alignment of mPKSb and mPKSd enzymatic domains, highlighting comparative presence or absence of domains. (Note: domain alignments are not based on phylogenetic relationship, but merely the order in which they occur). Domain abbreviations: ACP, acyl carrier protein; KS, ketoacyl synthase; DH, dehydratase; KR, ketoreductase; AT, ACP transacylase; ER, enoyl reductase; MT, methyltransferase; TH, thiohydrolase. KS and AT are each highlighted (red and blue, respectively) as they are discussed in greater detail in the text.
The gene orientation and encoded enzymatic domains of each of the Eutiarosporella mPKS genes are shown in figure 1. Because the enzymatic domains are repeated multiple times in these proteins, we used the ketoacyl synthase (KS) domains to order and name each unique module. KS domains are responsible for polyketide chain elongation in PKS systems, and in mPKSs, they are also involved in moving the growing polyketide chain to the next module (Robbins et al. 2016). As such, they are the essential functional domain in mPKSs and are present in each module. Eutiarosporellatritici-australis has two mPKS genes that share a 5′ noncoding region on opposite strands (Eta-mPKSa and Eta-mPKSb) (fig. 1A). The encoded amino-acid sequences contain six and two KS domains, respectively (fig. 1E and F). Eutiarosporelladarliae has three mPKS genes: a pair, that share a 5′ noncoding region and are predicted on opposite strands (Ed-mPKSa and Ed-mPKSb) and a solo mPKS gene (Ed-mPKSc) (fig. 1B). The encoded amino acid sequences contain six (a) and two (b), and six (c) KS domains, respectively (fig. 1E and F). Eutiarosporellapseudodarliae has four mPKS genes: two pairs, each that share a 5′ noncoding region and are predicted on opposite strands (Ep-mPKSa and Ep-mPKSb, and, Ep-mPKSc and Ep-mPKSd) (fig. 1C). The first pair of mPKS genes contain five (a) and two (b) KS domains, respectively (fig. 1E and F). The second mPKS pair contains six (c) and two (d) KS domains, respectively (fig. 1E and F).
We performed a BLASTp analysis of the mPKS sequences against the NCBI and JGI databases to determine whether any other fungal species possess mPKS genes. This revealed that the only other fungal homologues of these genes were identified in M. phaseolina, a close relative of the three WGD species. Three mPKS genes were predicted for M. phaseolina, however, after reannotation with FGENESH+ (softberry.com), these were reduced to one pair that share a 5′ noncoding region and are predicted on opposite strands (Mp-mPKSa and Mp-mPKSb) (fig. 1D). The encoded amino-acid sequences contain six (a) and two (b) KS domains, respectively (fig. 1E and F). In summary, each of the four species possesses at least one longer mPKS gene (either a or c) and one shorter mPKS gene (either b or d) that share a generally conserved domain structure with the other species (fig. 1E and F).
To the best of our knowledge, mPKS genes have not been previously described in fungi. It is also significant to note that no other fungi represented on the publicly available databases harbor genes encoding mPKSs. Therefore, the Botryosphaeriaceae mPKS genes described in this study are the first observed fungal mPKS genes. Not all of the KS domains encoded by the predicted Botryosphaeriaceae mPKS genes are followed by an acyl transferase (AT) domain (fig. 1E and F blue text). AT domains, which are the functional domain responsible for substrate loading in PKS systems are considered one of the minimal PKS domains along with KS and acyl carrier protein (ACP) domains and hence are usually present in each PKS module (fig. 1E and F). However, among the Eutiarosporella mPKS genes, AT domains only follow Eta-KSa2 and 4, Eta-KSb1, Ed-KSa4, Ed-KSb1, Ed-KSc2 and 4, Ep-KSb1, Ep-KSc2 and 4, Ep-KSd1, Mp-KSa2 and 4, and Mp-KSb1. Ep-mPKS-a is not predicted to encode any AT domains (fig. 1E and F).
The Eutiarosporella mPKS genes are in the middle of a scaffold, surrounded by genes of fungal origin. To confirm that these mPKS genes were not pseudo genes, qRT-PCR expression analysis was performed on the modular pair from E. tritici-australis. Expression of the genes from E. tritici-australis were tested under three nutrient substrate conditions: potato dextrose broth (PDB), fries medium (FM), and on wheat. Expression of the two genes was observed under all three conditions demonstrating that the mPKS genes in the WGD pathogens are not pseudo genes (supplementary fig. 1, Supplementary Material online).
Botryosphaeriaceae mPKS Genes Were Likely Acquired via Horizontal Gene Transfer
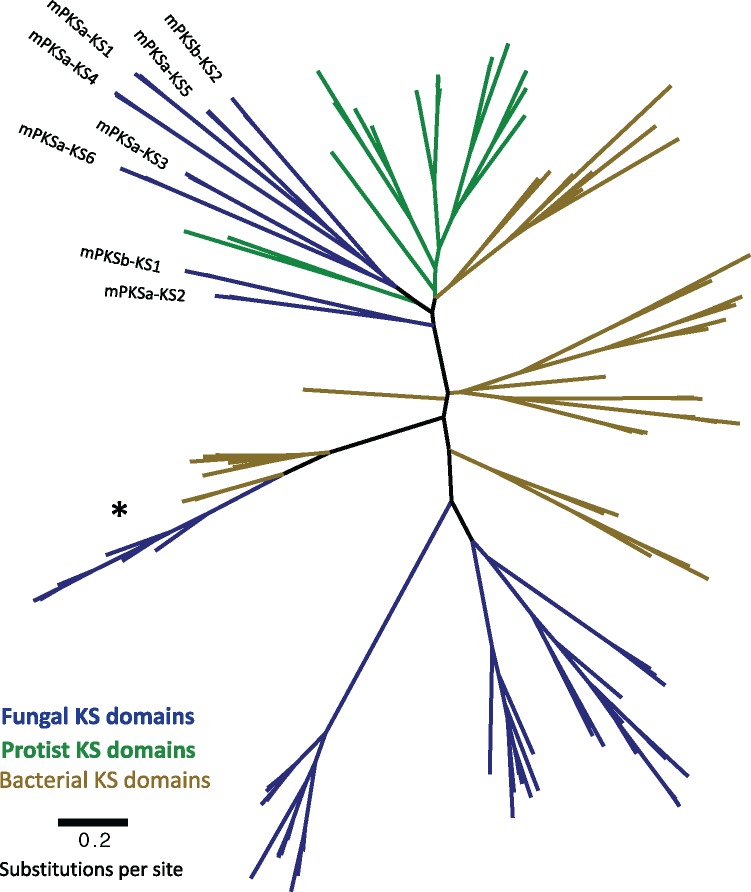
To investigate the evolutionary origins of the mPKS genes, a maximum likelihood (ML) phylogeny was constructed using the KS domains from both the mPKSs and nonmodular PKS proteins in the three WGD species. Additionally, close BLAST hits to the mPKS-KS domains and known fungal and bacterial KS domains were included (fig. 2 and supplementary fig. 2A, Supplementary Material online). This phylogenetic analysis clearly shows that the mPKS domains from the three WGD species and M. phaseolina resolve within a clade containing bacterial and protist KS domains (fig. 2 and supplementary fig. 2A, Supplementary Material online). In contrast, the nonmodular KS domains from the WGD species grouped together with known fungal KS domains. These data indicate a potential bacterial or protist origin for the Botryosphaeriaceae mPKSs (fig. 2). Due to lack of clear phylogenetic resolution between the fungal mPKSs and any other organism, it was not possible to determine a potential source species of the Botryosphaeriaceae mPKS genes (fig. 2 and supplementary fig. 2, Supplementary Material online). In addition, no other nonfungal species share a similar domain structure to the Botryosphaeriaceae mPKSs.
Fig.
. 2.—An unrooted maximum likelihood phylogeny comparing KS domains from Eutiarosporella tritici-australis’ and Macrophomina phaseolina’s mPKSs, other fungal PKSs, protist PKSs, and bacterial PKSs. (*) Highlights 6MAS/mellein synthase KS domains from fungi and bacteria.
We constructed a second phylogenetic tree to assess whether horizontal gene transfer is a likely scenario for the origin of the mPKS using the same alignment. This tree was built using RaxML with the addition of a constraint file that constrained the bacterial and protist KS domains together, and the Botryosphaeriaceae mPKS KS domains with the iterative fungal KS domains. KS domains from both bacterial and fungal 6MAS/mellein synthase sequences were not constrained. These genes in fungi have previously been described as horizontally acquired from bacteria (Schmitt and Lumbsch 2009) (6MAS/mellein clade represented by *: fig. 2 and supplementary fig. 2A–C, Supplementary Material online). As such they were allowed to move unconstrained during the tree build to prevent these sequences biasing the statistical analysis, described below. The best constrained tree was compared with the best tree where no clade constraints were imposed (shown in supplementary fig. 2A and B, Supplementary Material online), using Consel with the Kishino–Hasegawa (KH) and Shimodaira–Hasegawa (SH) tests (Shimodaira and Hasegawa 2001). The alternative topologies were determined as statistically different from the unconstrained trees with scores of P < 0.034 for both the KH and SH tests with the unconstrained tree being more likely. These results support a hypothesis that the Botryosphaeriaceae mPKSs were obtained via horizontal gene transfer (HGT).
mPKS c and d in E. darliae and E. pseudodarliae Arose via a Gene Duplication Event
As described earlier, E. darliae and E. pseudodarliae each possess additional copies of the mPKS genes. The individual KS domains from the mPKSa were more similar to the KS domains in mPKSc at the corresponding position than they were to the other KS domain within its own protein (demonstrated for all predicted KS domains in supplementary fig. 3, Supplementary Material online). Likewise, the individual KS domains from mPKSb and mPKSd were also more closely related to each other than they were to other KS domains within the protein. Each of the clades containing the KS domain groupings was strongly supported (100% bootstrap replication support, supplementary fig. 3, Supplementary Material online). Based on the phylogenetic data, we conclude that the additional pair of mPKS genes in E. darliae and E. pseudodarliae arose due to a duplication event of mPKSa and mPKSb (supplementary fig. 3, Supplementary Material online).
The only KS domains that did not follow the pattern described earlier were Ep-mPKSa-4 and Ep-mPKSa-5, which resolved with the other mPKSa/c KS domains 5 and 6, respectively (supplementary fig. 3, Supplementary Material online). Ep-mPKSa only harbors five KS domains instead of six predicted in the other mPKSa and c proteins (fig. 1E). This indicates that the KS domain in the fourth position for mPKSa/c from E. darliae, E. tritici-australis, and M. phaseolina, and in Ep-mPKSc, has been lost from Ep-mPKSa. This conclusion is supported by the phylogenetic grouping of Ep-mPKSa-4 and Ep-mPKSa-5 (highlighted with * in supplementary fig. 3, Supplementary Material online). There was also a significant size difference between Ep-mPKSa and Ep-mPKSc (fig. 1C). The entire predicted gene length of Ep-mPKSa is only 17,322 bp and the predicted amino acid sequence is only 4342aa, whereas, Ep-mPKSc is 33,137 bp, and the predicted amino acid sequence is 9935 aa. Likewise, Ed-mPKSa is shorter than Ed-mPKSc, however, the difference in this species is far less. Ed-mPKSa is 29,933 bp, with a predicted amino acid length of 9264aa, in contrast to Ed-mPKSc, which is 31,814 bp in length (10090aa, fig. 1B). In E. darliae, we did not identify a second copy of the shorter mPKS (mPKSb and d). Combined, these data indicate that after the duplication event, a number of deletion mutations resulted in loss of functional PKS domains.
We compared gene synteny surrounding E. darliae and E. pseudodarliae’s mPKS gene regions with the same region in E. tritici-australis. The two genes downstream of Eta-mPKSa were conserved downstream of all Eutiarosporella mPKSa and mPKSc genes (fig. 1A–C). The encoded protein sequence of the gene immediately adjacent to mPKSa has a predicted serine hydrolase domain (pfam03959). The encoded protein sequence for the second gene had close BLASTp hits among the Botryosphaeriaceae, but no predicted domain. Of the two genes, M. phaseolina has an unannotated DNA sequence, in opposite orientation with sequence similarity to the putative serine hydrolase (fig. 1D). However, there is no evidence for the second gene on this Mp-mPKS scaffold.
The genes downstream of each of the mPKSc and d genes were not conserved across all species (supplementary fig. 4, Supplementary Material online) However, a syntenic relationship was established between Eta-mPKSa and b and, Ep and Ed-mPKSa and b (supplementary fig. 4, Supplementary Material online). Based on analysis of rearranged, syntenic gene blocks, we hypothesize that Ep-mPKSa and b, and Ed-mPKSa and b were the original mPKS pair in each of their respective genomes (supplementary fig. 4, Supplementary Material online). For this prediction to be accurate, the duplication of mPKSa and b to form mPKSc and d must have occurred after speciation of E. tritici-australis from the more closely related E. darliae and E. pseudodarliae (i.e., E. tritici-australis never possessed additional mPKS pair c and d).
E. darliae and E. pseudodarliae Possess a SM Cluster with Multiple Hybrid PKS–NRPS Genes, Which was Lost from E. tritici-australis
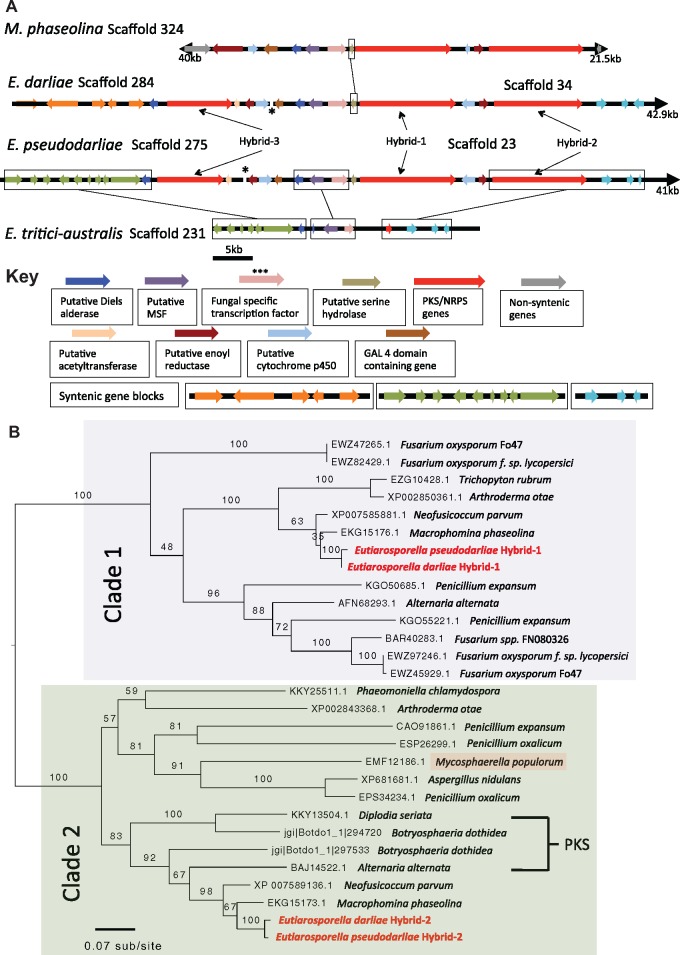
We searched for additional examples of SM cluster expansion within the WGD species, similar to the duplicated mPKSc and mPKSd within E. darliae and E. pseudodarliae. Examining the SM clusters identified via antiSMASH, we found that both E. darliae and E. pseudodarliae harbored a SM gene cluster, within which there are two PKS–NRPS genes (Hybrid-1 and Hybrid-2) (fig. 3A). In contrast, E. tritici-australis was not predicted to encode any PKS–NRPS genes (fig. 3A andtable 2). Both Hybrid-1 and Hybrid-2 are predicted to encode amino acid sequences of similar length and domain structure (fig. 3A). As above, we performed a phylogenetic analysis comparing the KS domains of these genes against other close BLAST hits from NCBI. Hybrid-1 and Hybrid-2 resolved in separate clades, indicating that they are not paralogs (fig. 3B).
Fig.
. 3.—(A) Graphical representation of the entire predicted cluster of Hybrid-1, 2, and 3, in Eutiarosporella darliae and E. pseudodarliae found on two scaffolds, and the syntenic genes present in E. tritici-australis that are on a single scaffold. Macrophomina phaseolina’s scaffold containing the Hybrid-1 and 2 genes is drawn above to demonstrate that they are also colocalized, however, M. phaseolina’s Hybrid-3 homologue was not found in a syntenic location and is not shown. Outlined serine hydrolase between M. phaseolina and E. darliae highlights the inversion of this gene. (*) Highlights scaffold overhang joint by two syntenic genes linking the two separate scaffolds in E. darliae and E. pseudodarliae. Key: describes genes present in each cluster, as well as syntenic gene-blocks. (B) A maximum likelihood phylogeny comparing KS domains of Hybrid-1 and 2, as well as close BLAST-hits from the NCBI nonredundant database. Hybrid-1 and Hybrid-2 resolved separately on this phylogeny ([Clade 1] and [Clade 2], respectively). A PKS–NRPS harbored by a poplar pathogen, M. populorum (gi|453084141), and which resolves in Hybrid-2’s clade, is highlighted in red. In brackets are four KS sequences belonging to putative PKSs, not PKS–NRPSs.
Several genes known to be associated with secondary metabolites were found neighboring Hybrid-1 and Hybrid-2 on the genomic scaffold. Eutiarosporellapseudodarliae has nine genes: seven are upstream of Hybrid-1: an enoyl reductase-like gene (cd08249), a cytochrome p450 (pfam00067) a GAL4 domain containing gene (smart00066), a Diels-Alderase-like gene, an MFS protein (cd06174), a fungal-specific transcription factor (FSTF), and a serine hydrolase (pfam03959) (fig. 3A). Two more genes are upstream of Hybrid-2: an enoyl reductase-like gene and a cytochrome p450 gene. Eutiarosporelladarliae has seven of the nine genes listed above, in the same order but is missing the first two genes due to the end of the scaffold. We located these two genes missing from E. darliae’s cluster on a separate scaffold (fig. 3A, Scaffold 284). In addition to these two genes, this scaffold contained a third NRPS gene flanked by a putative acetyltransferase gene and a putative Diels alderase gene. For consistency among genes in this cluster, we define this NRPS as Hybrid-3. Eutiarosporellapseudodarliae also possessed these other three genes, on a separate scaffold (fig. 3A, Scaffold 275). In both E. darliae and E. pseudodarliae the ends of these scaffolds formed short overlaps indicating that these three synthase genes (Hybrid 1-3) form one large SM cluster (fig. 3A).
The genes from these clusters were compared with their syntenic positions in the two species with best-BLAST homologues, N. parvum and M. phaseolina (table 2). Macrophominaphaseolina possesses a cluster that contains homologues of Hybrid-1 and Hybrid-2, as well as the nine associated genes (fig. 3A). Neofusicoccumparvum also possesses homologues of these proteins, however, they are not located within a single cluster (supplementary fig. 5, Supplementary Material online). Hybrid-3 has a best-BLAST hit on NCBI with a predicted PKS–NRPS from N. parvum (NRPS-4; table 2 andsupplementary fig. 6A, Supplementary Material online). A homologous protein is also found in M. phaseolina, not colocated with the other two PKS–NRPS genes (supplementary fig. 6A, Supplementary Material online). In the Eutiarosporella spp., the N-terminal region of Hybrid-3 has been truncated, removing what are the KS domains found in N. parvum (supplementary fig. 6B, Supplementary Material online).
We could not identify intact copies of Hybrid-1, -2, or -3 in the E. tritici-australis genome. tBLASTx searches identified a predicted partial BLAST hit to Hybrid-2 (681 bp, e-value 7.12e-86, 72.7% pairwise identity) (fig. 3A). Genes found flanking Hybrid-1, -2, and -3 in E. darliae and E. pseudodarliae were used as tBLASTx queries to search the E. tritici-australis genome. This revealed the presence of some of these genes, on the same scaffold as the partial Hybrid-2 (fig. 3A). The secondary metabolite-related genes identified include: Hybrid-1’s Diels-Alderase-like, MSF, and serine hydrolase genes, and Hybrid-3’s upstream Diels-Alderase-like gene. Additional syntenic genes identified, that were not predicted to encode predicted SM-associated domains, include: three homologues to genes downstream of Hybrid-2 in E. darliae and E. pseudodarliae and six homologues to genes upstream of E. pseudodarliae’s Hybrid-3. The presence of all of these genes on a single scaffold in E. tritici-australis, provides further support that Hybrid-1, -2, and -3 are a part of a larger contiguous cluster (fig. 3A).
E. darliae and E. pseudodarliae are Able to Induce Disease Symptoms in a Woody Plant Species
Phylogenetic analysis with the KS domain of Hybrid-2 showed it is a likely homolog to known SM genes found in pathogens of woody plants (fig. 3B). For example, Botryosphaeria dothidea (jgi|Botdo1_1|294720 and jgi|Botdo1_1|297533), Neofusicoccum parvum (gi|615428986), Diplodia seriata (gi|821057639), Alternaria alternata pathovar tangerines (gi|302562829), Mycosphaerella populorum (gi|453084141), and Phaeomoniella chlamydospora (gi|821070483). In particular, two of these proteins are linked to facilitation of pathogenicity in woody plants (Miyamoto et al. 2010; Dhillon et al. 2015). The first is the protein from Alternaria alternata pathovar tangerines (gi|302562829, BAJ14522.1; BLASTp with E. pseudodarliae’s Hybrid-2’s KS domain: e-value 0.0, 81% identity). This protein, called ACTTS3, is a PKS used in the production of ACT-toxin, a host-specific toxin that enables infection of tangerines (Kohmoto et al. 1993). Despite resolving among PKS–NRPS genes (fig. 3B), ACTTS3 lacks the NRPS domains. ACTTS3 resolves in a monophyletic clade with Botryosphaeriaceae PKS–NRPS genes, three of which also lack NRPS domains (B. dothidea: JGI|Botdo1_1|294720 and JGI|Botdo1_1|297533, and D. seriata gi|302562829) (fig. 3B).
The second example is in the poplar pathogen, Mycosphaerella populorum (gi|453084141, EMF12186.1; BLASTp with E. pseudodarliae’s Hybrid-2’s KS domain: e-value 0.0; 67% identity). Horizontal acquisition of this PKS–NRPS gene cluster is predicted to be a factor differentiating disease severity between itself and a close relative, M. populicola, that lacks the cluster (Dhillon et al. 2015). The PKS–NRPS and clustered genes are up-regulated on wood-substrate (Dhillon et al. 2015). Mycosphaerellapopulorum’s PKS–NRPS gene cluster only shares a limited syntenic relationship with E. darliae and E. pseudodarliae’s Hybrid-2’s clusters (supplementary fig. 6C, Supplementary Material online). Conserved with M. populorum’s cluster are both Hybrid-2’s enoyl reductase-like gene (comparative tBLASTx e-value 1.90e-99) and cytochrome p450 gene (comparative tBLASTx e-value 1.90e-99) (supplementary fig. 6C, Supplementary Material online). In addition, using the amino acid sequence encoded by M. populorum’s FSTF gene as a tBLASTn query, we identified a homologous stretch of DNA, on a different scaffold to Hybrid-2 (e-value 1.07e-38). In comparison, Hybrid-1’s FSTF gene was a less significant BLAST hit (e-value 4.90e-07). No gene was predicted at the best-BLAST-hit site in E. pseudodarliae, and it is only a partial hit (1,065 bp vs. 2,053 bp for M. populorum’s complete FSTF) (supplementary fig. 6C, Supplementary Material online).
Since other fungal pathogens with homologous gene clusters are pathogens of woody plants, we hypothesized that the host range of the WGD species might be much broader than just wheat. To test this hypothesis, attached leaf assays were performed on various woody plants, using all three WGD Eutiarosporella spp. Leaf surfaces were wounded prior to inoculation to facilitate fungal penetration. Disease symptoms attributed to E. darliae and E. pseudodarliae were particularly pronounced in Hakea salicifolia, and so we used this plant species to represent the ability of the WGD Eutiarosporella spp. to induce disease (fig. 4 and supplementary figs. 7 and 8, Supplementary Material online). After 3-day postinoculation (DPI) necrotic lesions and disease symptoms were observed in leaves inoculated with E. darliae and E. pseudodarliae (fig. 4 and supplementary figs. 7 and 8, Supplementary Material online). In contrast, almost no disease symptoms were evident on leaves inoculated with E. tritici-australis (fig. 4 and supplementary figs. 7 and 8, Supplementary Material online). Attached-leaf disease assays were reperformed using an additional isolate of each of E. darliae and E. pseudodarliae as well five additional isolates of E. tritici-australis (supplementary fig. 8C, Supplementary Material online). Extensive necrotic lesions occurred 3DPI in leaves inoculated with the former two species, however, little to no necrosis was induced beyond the site of inoculation by any of the isolates of E. tritici-australis (supplementary fig. 8C, Supplementary Material online). E.darliae and E. pseudodarliae were reisolated from diseased tissue, away from the inoculation site, thereby satisfying Koch’s postulate. Expression of Hybrid-1, Hybrid-2, Hybrid-3 and the cluster’s putative TF gene are up-regulated in E. darliae when the fungus was grown on Hakea wood compared with when grown in potato dextrose broth (PDB) (supplementary fig. 9, Supplementary Material online). Disease progression and comparisons among each of the three species are displayed in more detail in the Supplementary Material online (supplementary figs. 7 and 8, Supplementary Material online).
Fig.
. 4.—Eutiarosporella darliae and E. pseudodarliae induce necrotic disease symptoms in leaves of Hakea salicifolia, whereas E. tritici-australis does not. Detached leaf (upper panels) and attached leaf (lower panels) assays of necrosis development on H. salicifolia. Wounded leaves were inoculated with the WGD Eutiarosporella spp. The uninoculated leaves displayed no disease symptoms. Leaves inoculated with both E. darliae and E. pseudodarliae displayed significant necrotic symptoms spreading from the cut site at 3-day postinoculation (3DPI). Leaves inoculated with E. tritici-australis do not display significant disease symptoms beyond the site of inoculation at 3DPI. Note: the detached leaf assay panels with E. pseudodarliae and E. tritici-australis were beneath the negative control and E. darliae panels in the original image, but were cut-and-pasted to be side-by-side.
Discussion
The complement of secondary metabolites (SMs) produced by a fungal pathogen can affect their development, ability to interact with other microorganisms, and ability to infect a particular host. The ease of sequencing and comparing fungal genomes has led to the quick identification of novel SM gene clusters, which have subsequently been associated with niche specificity (Dhillon et al. 2015). The WGD Eutiarosporella spp. belong to the Class Dothideomycetes, which are known to produce a wide and varied repertoire of SMs (Muria-Gonzalez et al. 2015). Recent studies on Dothideomycetes genomics and transcriptomics demonstrate that some SM genes are highly conserved among closely related species, whereas, other genes have a sporadic distribution (Condon et al. 2013; Dhillon et al. 2015; Morales-Cruz 2015). Our examination of the SM gene clusters from each of the WGD Eutiarosporella spp. demonstrated that these species possessed SM genes conserved among the Botryosphaeriaceae. None of the SM genes identified had similarity to known mycotoxin gene clusters. These data are consistent with previous feeding studies demonstrating that wheat grain infected with WGD was not toxic to weaner pigs (Kopinski and Blaney 2010). A significant outcome from this study was the discovery of multiple, duplicated, mPKS genes in the WGD species and their close relative M. phaeseolina. These genes are, to the best of our knowledge, the first of their kind reported in fungi and phylogenetic analyses suggest that these genes were acquired via horizontal gene transfer from an unknown bacterial or protist species.
The horizontal transmission of SMs or metabolic gene clusters between fungi and other organisms has been reported previously (reviewed in, Wisecaver and Rokas 2015). While the function of most horizontally transferred SMs remains elusive, they often occur between organisms that occupy similar ecological niches, where the horizontally transferred SM confers a selective advantage (Wisecaver and Rokas 2015). The KS domains of the mPKS enzymes described in this study are significantly more closely related to bacterial and protist KS domains, however none of the KS modules formed a monophyletic group with any known bacterial or protist KS domain. This makes it quite difficult to speculate on the compounds that these enzymes may produce and how they may confer a fitness advantage in a plant host. mPKSs, also referred to as modular type I PKSs, have previously only been described in bacteria and protists in a wide variety of natural environments (Khosla 1997; Zhu et al. 2002; John et al. 2008). These multidomain enzymes produce a wide variety of known compounds including many important antibiotics, such as erythromycin and spiramycin (Khosla 1997).
The presence of mPKSs within the WGD Eutiarosporella is an exciting advance in our understanding of fungal secondary metabolism. There are several other recent reports of “bacterial-only” SM biosynthetic enzymes discovered in newly sequenced fungal species. For example, a limited number of instances of Type-3 PKSs in fungi were observed, which were thought to be limited to plants and bacteria (Hashimoto et al. 2014). Similarly, a recent study found that in addition to using hybrid PKS (N-terminal)-NRPS (C-terminal) proteins, various fungi also use hybrid NRPS (N-terminal)-PKS (C-terminal) proteins for SM biosynthesis; these too were previously thought to be limited to bacteria (Lawrence et al. 2011; Yun et al. 2015). Similar to the modular PKS genes described in this study, the hybrid NPS/PKS gene described by Lawrence et al. (2011) also have a phylogenetic signature that indicates a bacterial origin. Our discovery of fungal mPKS genes further demonstrates that the boundaries between fungal and bacterial SM biosynthesis are not clear-cut.
Additional unanswered questions remain. For example, what is the mechanism these proteins use to synthesize SMs? It is generally considered that the minimal module required for a Type-1 PKS consists of a KS domain (extension domain) and an acyl transferase (AT) domain (substrate loading domain) and acyl carrier protein (ACP) (Keller et al. 2005; Hertweck 2009; Chooi and Tang 2012; Throckmorton et al. 2015). However, not all Botryosphaeriaceae mPKS genes’ modules conform to this structure, as some lack AT domains. Lack of these domains does not necessarily impact the ability of the module to function. There are functional PKS proteins lacking embedded AT domains, known as trans-AT PKSs, which rely on separately encoded proteins with AT domains to synthesize the final product (Cheng et al. 2003; Lohman et al. 2015; Helfrich and Piel 2016). Although we did not find any trans-AT PKSs, it is possible that the AT domains from other modules within the same enzyme could complement this role. There are also SM synthases that can produce different metabolites with the same protein by skipping over particular enzymatic modules (Wenzel et al. 2005). Elucidation of the metabolites produced by these mPKSs will begin to shed light on potentially novel biochemical reactions.
An additional striking difference in the SM gene complement among the WGD Eutiarosporella spp. is that E. darliae and E. pseudodarliae each possess three colocalized PKS–NRPSs (Hybrid-1, 2, and 3), that are absent from the genome assembly of E. tritici-australis. Since these genes are absent from E. tritici-australis, we assume they are not major virulence factors for these fungi on wheat. Instead, our interest stemmed from the evolutionary implications associated with harboring these genes. We identified homologues of these genes across a range of pathogens capable of infecting woody plants and previously sequenced Botryosphaeriaceae species. The majority of Botryosphaeriaceae species are reported as pathogens or endophytes (and opportunistic pathogens) of woody plants (Slippers and Wingfield 2007), including other Eutiarosporella spp. that have been isolated from woody plants (Jami et al. 2012, 2014). Among this family, SMs have long been considered important during infection of woody plants, with multiple phytotoxic compounds identified in culture (Fischer and Thines 2017). Homologues of the PKS–NRPS genes are also present in the poplar pathogen, M. populorum (Dhillon et al. 2015), the tangerine pathogen, A. alternata pathovar tangerines (Miyamoto et al. 2010), and the grapevine trunk pathogen Phaeomoniella chlamydospora (Morales-Cruz et al. 2015). The presence of these genes in two of the WGD Eutiarosporella spp. led us to examine their ability to infect woody plants, in a reverse ecological manner (Ellison et al. 2011). As a result, we found that two of the species were capable of inducing disease in woody plant species, indicating an expanded host range beyond wheat. To develop a better understanding of the lifestyle of these pathogens it would be beneficial to increase sampling of nonagricultural hosts (both monocots and dicots) for fungi that are growing undetected (Hyde et al. 2010). Woody plants might be acting as additional reservoirs for these pathogens, including E. tritici-australis, which might still grow as an endophyte in woody plants. This would be similar to other Eutiarosporella spp. that were isolated from nondiseased woody plant tissue (Jami et al. 2012). If this is the case, then perhaps disease inoculum for WGD may arise from woody plant reservoirs.
In conclusion, the discovery of horizontally acquired mPKS genes in the WGD Eutiarosporella spp. and M. phaseolina expands the known boundaries of fungal SM biosynthesis capabilities. This is a significant finding for the field of fungal SM biosynthesis and poses a strong challenge to identify the compounds that are produced by these enzymes and the chemical reactions needed to achieve this. Similarly, the observation of a PKS–NRPS gene cluster harbored in E. darliae and E. pseudodarliae, but lost in E. tritici-australis, led us to demonstrate that the former two pathogens were able to induce disease in woody plants. This is an important finding in the context of alternative host ranges and virulence and we speculate that these fungi could exploit woody plants as additional hosts, from which disease inoculum might arise. Combined, these observations demonstrate that rapid genomic adaptations have played an important role in the evolution of these three fungal species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
E.T. was supported by a Grains Research and Development Corporation Graduate Research Scholarship. Y.H.C. is an Australian Research Council (ARC) Future Fellow. The authors also acknowledge the Australian Plant Phenomics Facility which is supported under the National Collaborative Research Infrastructure Strategy of the Australian Government.
Data deposition: This project has been deposited at the European Nucleotide Archive under the accession numbers: E. darliae accession: GFXH01000000; E. pseudodarliae accession: GFXI01000000, E. tritici-australis accession: GFXG01000000
Literature Cited
- Agrios GN. 2005. Plant pathology. San Diego: Elsevier Academic Press. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Baker SE, et al. 2006. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, t-toxin, by Cochliobolus heterostrophus. Mol Plant Microbe Interact. 19(2):139–149. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Rolshausen P, Cantu D.. 2013. Draft genome sequence of Neofussicoccum parvum isolate UCR-NP2, a fungal vascular pathogen associated with grapevine cankers. Genome Announc. 1:e00339–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, et al. 2007. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Du L.. 2016. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl Micobiol Biotechnol. 100(2):541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B.. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci U S A. 100(6):3149–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, et al. 2015. An in planta-expressed polyketide synthase produces (R)-mellein in the wheat pathogen Parastagonospora nodorum. Appl Environ Microbiol. 81(1):177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Solomon PS.. 2014. A chemical ecogenomics approach to understand the roles of secondary metabolites in fungal cereal pathogens. Front Microbiol. 5:640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi YH, Tang Y.. 2012. Navigating the fungal polyketide space: from genes to molecules. J Org Chem. 77(22):9933–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon BJ, et al. 2013. Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet. 9(1):e1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon B, et al. 2015. Horizontal gene transfer and gene dosage drives adaptation to wood colonization in a tree pathogen. Proc Natl Acad Sci U S A. 112(11):3451–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, et al. 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A. 108(7):2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. 2013. White grain rejection spreading. Ground Cover 102:4. [Google Scholar]
- Fischer J, Thines E.. 2017. Secondary metabolites of fungal vine pathogens In: Konig H, Unden G, Frohlich J, editors. Biology of microorganisms in grapes, in must, and in wine. Berlin Heidelberg: Springer-Verlag. [Google Scholar]
- Friesen TL, Faris JD, Solomon PS, Oliver RP.. 2008. Host-specific toxins: effectors of necrotrophic pathogenicity. Cell Microbiol. 10(7):1421–1428. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, et al. 2011. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7(6):e1002070.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Nonaka T, Fujii I.. 2014. Fungal type III polyketide synthases. Nat Prod Rep. 31(10):1306–1317. [DOI] [PubMed] [Google Scholar]
- Helfrich EJN, Piel J.. 2016. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 33(2):231–316. [DOI] [PubMed] [Google Scholar]
- Hertweck C. 2009. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 48(26):4688–4716. [DOI] [PubMed] [Google Scholar]
- Hyde KD, et al. 2010. A case for reinventory of Australia’s plant pathogens. Persoonia 25:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, et al. 2012. Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13(1):493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami F, Slippers B, Wingfield MJ, Gryzenhout M.. 2012. Five new species of the Botryosphaeriaceae from Accacia Karroo in South Africa. Cryptogamie Mycol. 33(3):245–266. [Google Scholar]
- Jami F, Slippers B, Wingfield MJ, Gryzenhout M.. 2014. Botryosphaeriaceae species overlap on four unrelated, native South African hosts. Fungal Biol. 118(2):168–179. [DOI] [PubMed] [Google Scholar]
- John U, et al. 2008. Novel insights into evolution of protistan polyketide synthases through phylogenetic analysis. Protist 159(1):21–30. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW.. 2005. Fungal secondary metabolism – from biochemistry to genomics. Nat Rev Microbiol. 3(12):937–947. [DOI] [PubMed] [Google Scholar]
- Khosla C. 1997. Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev. 97(7):2577–2590. [DOI] [PubMed] [Google Scholar]
- Kohmoto K, et al. 1993. Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83(5):495–502. [Google Scholar]
- Kopinski JS, Blaney BJ.. 2010. Nutritive value and non-toxicity of Botryosphaeria zeae-infected wheat for weaner pigs. J Anim Physiol Anim Nutr. 94(1):44–54. [DOI] [PubMed] [Google Scholar]
- Lawrence DP, Kroken S, Pryor BM, Arnold AE.. 2011. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous ascomycota. PLoS One 6(11):e28231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eurkaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman JR, et al. 2015. Structural and evolutionary relationships of “AT-less” type 1 polyketide synthase ketosynthases. Proc Natl Acad Sci U S A. 112(41):12693–12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, et al. 2012. RobiNA: a user-friendly, integrated software solution for RNA-seq-based transcriptomics. Nucleic Acids Res. 40(W1):W622–W627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsberg A, et al. 2017. Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health. Mol Plant Pathol. 18(4):477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, et al. 2010. ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of ACT-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol Plant Microbe Interact. 23(4):406–414. [DOI] [PubMed] [Google Scholar]
- Morales-Cruz A, et al. 2015. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genomics 16:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muria-Gonzalez MJ, Chooi YH, Breen S, Solomon PS.. 2015. The past, present and future of secondary metabolite research in the Dothideomycetes. Mol Plant Pathol. 16(1):92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T, Ueno T, Funkami H.. 1982. Structure elucidation of AK-toxins, host-specific phytotoxic metabolites produced by Alternaria kikuchiana Tanaka. Tetrahedron Lett. 23(43):4469–4472. [Google Scholar]
- Ohm RA, et al. 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8(12):e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver RP, Solomon PS.. 2010. New developments in pathogenicity and virulence of necrotrophs. Curr Opin Plant Biol. 13(4):415–419. [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. [DOI] [PubMed] [Google Scholar]
- Platz G. 2011. Wheat and barley disease management in 2011. Yellow spot and head diseases in wheat. Strategies and products for barley leaf rust. GRDC Update Papers, Grains Research and Development Corporation; https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2011/02/cereal-disease-update-sa-2011 [Google Scholar]
- Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nuc Acids Res. 33(Web Server issue):W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins T, Liu YC, Cane DE, Khosla C.. 2016. Structure and mechanism of assembly line polyketide synthases. Curr Opin Struct Biol. 41:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I, Lumbsch HT.. 2009. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One 4(2):e4437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabuer G, et al. 2015. Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory. Science 350(6261):670–674. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ.. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol Rev. 21(2–3):90–106. [Google Scholar]
- Solomon PS. 2011. Assessing the mycotoxigenic threat of necrotrophic pathogens of wheat. Mycotoxin Res. 27(4):231.. [DOI] [PubMed] [Google Scholar]
- Solovyev V. 2001. Statistical approaches in eukaryotic gene prediction In: Balding DJ, Bishop M, Cannings C, editors. Handbook of statistical genetics. 2nd ed Chichester: John Wiley & Sons. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJ.. 2013. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 37(1):67–93. [DOI] [PubMed] [Google Scholar]
- Tan KC, Trengrove RD, Maker GL, Oliver RP, Solomon PS.. 2009. Metabolite profiling identifies the mycotoxin alternariol in the pathogen Stagonospora nodorum. Metabolomics 5:330–335. [Google Scholar]
- Tanaka A, Shiotani H, Yamamoto M, Tsuge T.. 1999. Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol Plant Microbe Interact. 12(8):691–702. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jayasena K.. 2015. White grain disorder of wheat in Western Australia. Available from: http://www.giwa.org.au/pdfs/CR_2015/SORT/EOI80_Thomas_Geoff_White_grain_disorder-paper-revised_CU15_EOI80_.pdf
- Throckmorton K, Wiemann P, Keller NP.. 2015. Evolution of chemical diversity in a group of non-reduced polyketide gene clusters: using phylogenetics to inform the search for novel fungal natural products. Toxins 7(9):3572–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thynne E, et al. 2015. Re-classification of the causal agent of whitegrain disorder on wheat as three separate species of Eutiarosporella. Austr Plant Pathol. 44(5):527–539. [Google Scholar]
- van der Nest MA, et al. 2014. Draft genome sequences of Diplodia sapinea, Ceratocystis manginecas, and Ceratocystis moniliformis. IMA Fungus 5(1):135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatsubbaiah P, Sutton TB, Chilton WS.. 1991. Effect of phytotoxins produced by Botryosphaeria obtusa, the cause of black rot of apple fruit and frogeye leaf spot. Phytopathology 81:243–247. [Google Scholar]
- Walton JD. 1996. Host-selective toxins: agents of compatibility. Plant Cell 8(10):1723.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD. 2006. HC-toxin. Phytochemistry 67(14):1406–1413. [DOI] [PubMed] [Google Scholar]
- Weber T, et al. 2015. antiSMASH 3.0 – a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43(W1):W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel SC, et al. 2005. Structure and biosynthesis of myxochromides S1–3 in Stigmatella aurantiaca: evidence for an iterative bacterial type 1 polyketide synthase and for module skipping in nonribosomal peptide biosynthesis. Chembiochem 6(2):375–385. [DOI] [PubMed] [Google Scholar]
- Wildermuth G, Williamson P, Shivas R, McNamara R.. 2001. Premature head blight and white grain – a new disease of wheat Proceedings of the 13th Biennial Australasian Plant Pathology Conference, Queensland Department of Primary Industries. [Google Scholar]
- Wisecaver JH, Rokas A.. 2015. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front Microbiol. 6:161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Rose MS, Turgeon BG, Yoder OC.. 1996. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell 8(11):2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, et al. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CS, Motoyama T, Osada H.. 2015. Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS-PKS hybrid enzyme. Nat Commun. 6:8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, et al. 2002. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298(1):79–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.