Abstract
Macrophages play important roles in many diseases. We describe a protocol and the associated resources for the differentiation of human induced pluripotent stem cell-derived macrophages (IPSDM) and their applications in understanding human macrophage physiology and relevant diseases. The protocol uses an embryoid body–based approach with a combination of serum-free condition for hematopoiesis specification, followed by adherent culture with serum and M-CSF for myeloid expansion and macrophage maturation. The protocol produced an almost pure culture of CD45+/CD18+ macrophages yielding up to 2 × 107 cells per 6-well plate of iPSCs within 24 days, demonstrating high efficiency, purity, and scalability. The IPSDM and monocyte-derived macrophages (HMDM) cultured in the same medium were compared at morphological, functional and transcriptomic levels by RNA-sequencing. IPSDM and HMDM showed broadly similar profiles of coding transcriptome, alternative splicing events, and long noncoding RNAs, with advantages and successful applications in disease modeling using patients-derived and CRISPR-edited iPSC lines.
Keywords: induced pluripotent stem cells, macrophages, stem cell differentiation
INTRODUCTION
Macrophages, in Greek meaning “big eaters,” were first described as phagocytes that engulf pathogens and mediate antimicrobial defense by Ilya Metchnikoff, who received a Nobel Prize for Physiology of Medicine in 1908 (Gordon, 2008). Macrophages have now been recognized to play multi-dimensional roles in tissue homeostasis, innate immunity, inflammation, lipid metabolism, injury repair, and regeneration (Gentek, Molawi, & Sieweke, 2014; McNelis & Olefsky 2014; Wynn & Vannella, 2016). The pathophysiological role and the therapeutic potential of macrophages in infection, autoimmune diseases, cancer, obesity, diabetes, and cardiovascular diseases (McNelis & Olefsky, 2014) are key avenues of inquiry in the thriving and expanding field of macrophage biology.
Mechanistic studies of human macrophage biology have relied on the use of experimental models. Human peripheral blood monocyte-derived macrophages (HMDM) serve as a widely used experimental model of macrophage function. Although human blood monocytes are relatively accessible, HMDM are terminally differentiated and lack proliferative capacity, thus limiting scalability. Tumor-derived monocytic cell lines, such as THP-1 and U937, have unlimited proliferative potential and can be differentiated to macrophage-like cells using phorbol 12-myristate 13-acetate that activates protein kinase C. However, these cells are karyotypically abnormal and functionally immature. Importantly, although small interfering RNA and antisense oligonucleotide-mediated gene silencing have demonstrated some success in THP-1 derived macrophages and HMDM (de Bruin et al., 2016; Zhang, Xue et al., 2017), clonal expansion of THP-1 or U937 cell lines following genetic engineering to knockout target genes or introduce genetic mutations remains challenging. It is therefore imperative to develop and adapt novel experimental models that can recapitulate function and transcriptome profile of human primary macrophages and be also amenable to CRISPR/Cas9-mediated genome engineering (Zhang, Shi et al., 2017).
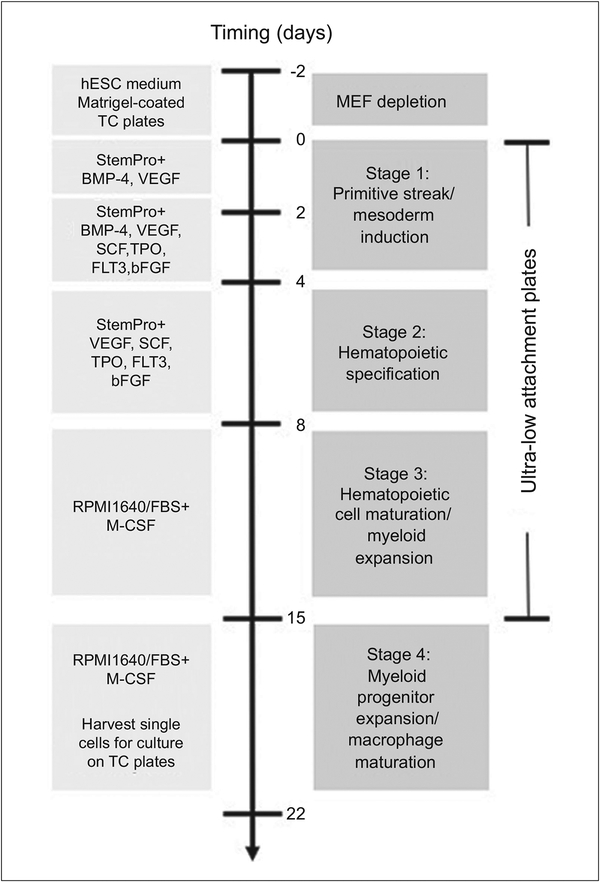
Induced pluripotent stem cell-derived macrophages (IPSDM) offer a new tool linking genotypes to phenotypes in the study of macrophage biology and diseases. We have previously published studies describing the application of IPSDM in disease modeling using patient-derived (Zhang et al., 2015) and CRISPR/Cas9-edited induced pluripotent stem cell (iPSC) lines (Zhang, Shi et al., 2017). Here we provide a detailed three-step protocol describing the differentiation, quality assessment, and troubleshooting of IPSDM derivation. Basic Protocol 1 summarizes the maintenance and expansion of iPSC lines on feeder cells. Basic Protocol 2 describes the feeder depletion prior to the initiation of differentiation. Basic Protocol 3 describes an embryoid body (EB)-based approach for stepwise differentiation of IPSDM. EBs, the three-dimensional aggregates formed in suspension by pluripotent stem cells (PSC), are formed in serum-free, chemically defined condition with cytokine cocktails that promote mesodermal induction (Stage 1) and hematopoietic specification (Stage 2). EBs are transferred to medium with serum and recombinant human macrophage colony stimulating factor (M-CSF) to allow myeloid expansion (Stage 3). The myeloid progenitors produced from the floating EBs are then harvested for adherent culture in tissue culture plate to promote further proliferation and maturation into macrophages (stage 4) (Fig. 1). The protocol takes 24 days and shows high yield and purity. One confluent 6-well plate of iPSCs yields up to 2 ×107 of more than 95% CD45+/CD18+ IPSDM (Zhang et al., 2015) and does not require positive selection using FACS or magnetic beads. Through deep RNA-sequencing, we characterized the coding transcriptome (Zhang et al., 2015), alternative splicing events (Lin et al., 2016) and long non-coding RNA profiles (Zhang, Xue et al., 2017) of isogenic IPSDM and HMDM cultured in the same macrophage culture medium at baseline and during activation, providing a comprehensive resource for planning IPSDM studies to model such events. (Zhang et al., 2015; Zhang, Shi et al., 2017)
Figure 1.
Schematic workflow of human induced pluripotent stem cell (iPSC) differentiation to macrophages (IPSDM). An embryoid body (EB)-based, stepwise protocol was used to differentiate human iPSCs to IPSDM. The culture medium, cytokines and types of culture vessels are illustrated
STRATEGIC PLANNING
Critical consideration on the planning of differentiation experiments for disease modeling and functional genomics:
Control and patient-derived or CRISPR/Cas9-edited iPSC lines should be differentiated in the same batch in order to minimize the potential impact of batch effects when performing functional comparisons.
Clonal variability can occur during iPSC reprogramming and has been well recognized to cause variations in differentiation efficiency and the functional properties of differentiated cells. It is therefore essential to utilize well-characterized iPSC lines, highly consistent differentiation procedures with quality assessment at multiple stages, and multiple iPSC lines/clones per subject with independent replicates in order to minimize variability and render statistically significant and meaningful experimental outcomes.
The gene-of-interest and functional phenotypes to be tested in IPSDM need to be carefully chosen and evaluated, for example, whether the target genes or non-coding transcripts show similar expression profile between IPSDM and HMDM, or whether the alternative splicing events or the functional response of HMDM is also represented by IPSDM. For initial screening, our studies provide a powerful resource (see Excel spreadsheet in Supporting Information) for planning IPSDM studies to model such events, which should be followed by specific validation for each chosen target.
Some target genes may affect iPSC reprogramming or IPSDM differentiation efficiency, and their function in mature macrophages cannot be reliably studied in IPSDM. FACS-based characterization at different stages of the differentiation process is important to determine such conditions, in which alternative approaches such as CRISPRi- and CRISPRa-mediated gene inhibition and activation may be used.
The use of IPSDM does not eliminate the need for traditional cellular models or distinct genetic manipulations, such as siRNA or antisense oligonucleotide-mediated knockdown, which are complementary and alternative approaches to further ensure validity and reproducibility.
BASIC PROTOCOL 1
EXPANSION OF iPSCs ON IRRADIATED FEEDERS
This protocol describes maintaining iPSCs on irradiated mouse embryonic fibroblast (MEF) feeder before initiation of differentiation. Feeder cells allow the iPSCs to anchor and propagate, supply nutrients to iPSCs in culture and maintain the pluripotency of iPSCs, which is critical for an efficient differentiation.
Materials
Irradiated CF1 MEF feeder cells (ThermoFisher, cat, no. A34181)
MEF medium (see recipe)
Human iPSC lines
hESC (human embryonic stem cell) medium (see recipe)
Y27632 (ROCK inhibitor) (Calbiochem, cat. no. 688000–5MG; see recipe) Wash medium (see recipe)
Accutase (Corning, cat. no. MT25058CI)
1300 Series Class II, Type A2 Biological Safety Cabinet (ThermoFisher, cat. no. 1375)
0.1% gelatin-coated 6-well plates (Support Protocol 1) Two stackable Heracell VIOS 160i CO2 Incubators with oxygen control (ThermoFisher, cat. no. 51030285)
37°C water bath
15-ml conical sterile polypropylene centrifuge tubes Centrifuge
6-well plates
Cell scraper
-
Prepare 0.1% gelatin-coated 6-well plates (see Support Protocol 1). Seed feeder cells in MEF medium at ~ 1 × 106/6-well plate. Culture feeder cells in an incubator maintained overnight at 37°C and 5% CO2 for attachment before seeding iPSCs.
MEF feeders should be used within 2 days after seeding.
-
Take a frozen stock of iPSC lines preserved in freezing medium from a liquid nitrogen tank and thaw rapidly in a water bath at 37°C.
To ensure efficient differentiation, iPSC lines at late passage (typically >20 passages) should be used for differentiation.
-
Transfer the thawed cell suspension to 10 ml hESC medium in a 15-ml conical tube immediately. Centrifuge the cells for 5 min at 300 g, room temperature (20°C to 28°C). Aspirate the supernatant, resuspend the cells×in 2 ml hESC medium supplemented with 5 μM Y27632 and seed in one well of a 6-well plate with MEF cells. Place the cells back in an incubator maintained at 37°C, 5% CO2, and 5% O2 (low oxygen).
The low oxygen tension is achieved through the introduction of N2 into the incubator. Low oxygen condition has been shown to improve the reprogramming of iPSCs (Yoshida, Takahashi, Okita, Ichisaka, & Yamanaka, 2009), the maintenance of pluripotent cell status (Narva et al., 2013), and the generation of iPSC-derived hematopoietic progenitor cells (Salvagiotto et al., 2011). We therefore used low oxygen condition in this protocol during iPSC maintenance and day 2 to day 8 differentiations, and would recommend such condition. Although systematic comparison between low oxygen and environmental oxygen (~20%) culture conditions has not been performed for this protocol, we do not expect oxygen level as a major factor determining the differentiation efficiency and would consider it reasonable if the users would adapt this protocol for a culture condition with environmental oxygen levels.
Prewash MEF feeder cells with wash medium to remove any residual FBS from the MEF medium that may cause spontaneous differentiation of iPSCs.
Optimize the seeding density so that iPSCs reach 80% to 90% confluency and will be ready to be passaged in 5 to 7 days. Do not allow iPSC lines to grow to over 90% confluency.
Aspirate the spent medium and add fresh hESC medium without Y27632 daily until the cells reach 80% to 90% confluency when they are ready for passaging.
-
To passage iPSC lines, aspirate the medium and add 1 ml of warm accutase to each well of the 6-well plate. Incubate the cells for 4 to 5 min at room temperature. Add 1 ml hESC medium/well to stop the accutase reaction. Gently tap the plate to loosen the iPSC colonies from the plate and then detach the cells from the culture plates mechanically using a cell scraper.
Monitor the dissociation process under a microscope to precisely control the dissociation time. Stop the reaction when the enzyme causes a subtle but observable thickening of the edges of colonies.
Collect the cell suspension into a 15-ml conical tube. Centrifuge for 5 min at 200 × g, room temperature.
-
Aspirate the supernatant and resuspend the cell clumps in hESC medium with 5 μM Y27632 with gentle pipetting to break up the colonies to small clumps with 50 to 200 cells per clump.
Be cautious not to break the cell clumps into single cells or aggregates less than 10 cells, which has low viability.
iPSC lines typically can be split at a ratio between 1:6 and 1:12. When maintaining multiple iPSC lines at the same time, the split ratio can be adjusted so that each line reaches 80% to 90% confluency at the same time for the following steps of feeder depletion and initiation of differentiation.
The pluripotent status of iPSC lines is assessed by circular and compact colony-like morphology and high percentage (>90%) of Tra-1–60+/SSEA-4+cells. The iPSC lines should show little or no evidence of differentiation upon passaging or differentiation.
Place the plate back in an incubator maintained at 37°C, 5% CO2, and 5% O2. Aspirate the spent medium and add fresh hESC medium without Y27632 daily.
When the cells reach 80% to 90% confluency, one can proceed to feeder depletion, or split again to obtain more cells. Depending on the number of iPSC lines and the amount of differentiated macrophages needed for the planned assays, one typically starts from two plates of cells on feeders per iPSC line.
BASIC PROTOCOL 2
FEEDER DEPLETION OF iPSCs
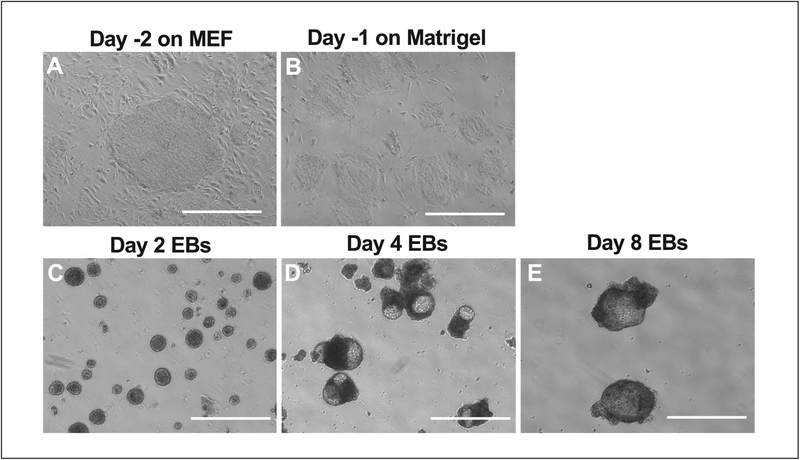
This protocol describes how to deplete MEF feeder cells before initiation of differentiation of iPSCs. (Fig. 2A–2B) MEF feeder cells support the growth and maintenance of pluripotent status of iPSCs. Depletion of feeder cells prior to differentiation and subsequent characterization is important to obtain reproducible results.
Figure 2.
The morphology of iPSCs and EBs from day −2 to day 8 of the differentiation. (A) The optimal size of iPSC colonies prior to MEF depletion at day −2. (B) The size of iPSC colonies on Matrigel-coated plates after MEF depletion at day −1. (C) The day 2 EBs. (D) The day 4 EBs. (E) The day 8 EBs. Note that the size of EBs increases during the course of differentiation. The white bar = 100 μm.
Materials
iPSCs (see Basic Protocol 1)
Phosphate-buffered saline (PBS), 1× without Ca2+ and Mg2+ (Corning, cat. no. MT 21–040-CM)
Accutase (Corning, cat. no. MT25058CI)
hESC (human embryonic stem cell) medium (see recipe)
Y27632 (ROCK inhibitor; see recipe)
Matrigel-coated 6-well plates (see Support Protocol 2) Cell scraper
15-ml and 50-ml conical sterile polypropylene centrifuge tubes (Thermo Scientific Nunc, cat. nos. 12–565–268 and 12–565–270)
37°C incubator
At day −2, prepare Matrigel-coated plates for feeder depletion (see Support Protocol 2 for coating Matrigel on 6-well plates).
Aspirate the medium and wash the iPSCs with 2 ml PBS per well. Add 1 ml warm (37°C) Accutase to each well of one 6-well plate and incubate for 2 to 3 min at room temperature.
-
Add 1 ml hESC medium to each well to stop the reaction and detach the cells from the culture plate mechanically using a cell scraper.
The size of the dissociated cell clumps should be larger than 200 cells per clump (or larger than clumps for regular passaging) so that the iPSCs can recover to exponential growth within 2 days before initiation of differentiation at day 0.
Collect the cell suspension in 15-ml conical centrifuge tubes and centrifuge for 4 min at 200 × g, room temperature.
-
Carefully aspirate the supernatant and gently resuspend the cell pellet in 24 ml hESC medium with 5 μM Y27632. Dispense 2 ml per well to Matrigel-coated 6-well plates and return the plates to an incubator maintained at 37°C, 5% CO2, and 5% O2.
Two plates of iPSCs on feeder cells can be transferred to four Matrigel-coated plates (1:2 split ratio).
Replace medium with fresh hESC medium without Y27632 at day –1. Then proceed to Basic Protocol 3 to start differentiation at day 0.
BASIC PROTOCOL 3
DIFFERENTIATION OF iPSCs TO MACROPHAGES
This protocol describes detailed stage-wise differentiation of iPSCs to mature macrophages by formation of embryoid bodies, myeloid lineage expansion and macrophage maturation (Fig. 1, Fig. 2C–2E). An efficient differentiation should yield up to 95% CD45+/CD18+ IPSDM.
Materials
StemPro-34 SFM medium (see recipe)
Day 0 differentiation medium (see recipe)
Y27632 (ROCK inhibitor; see recipe)
iPSC cells (see Basic Protocol 2)
hESC (human embryonic stem cell) medium (see recipe)
Collagenase B/DNase I dissociation buffer (see recipe)
0.25% Trypsin/EDTA (Gibco, cat. no. 25–200–056)
STOP medium (see recipe)
Wash medium (see recipe)
Day 0 differentiation medium (see recipe)
Day 4 and day 6 differentiation medium (see recipe)
Macrophage culture medium
Corning Costar ultra-low attachment plates (6-well) (Corning, cat. no. 3471) 37°C incubator
Cell lifter
5-ml serological pipettes
Centrifuge
50-ml conical tubes
Corning Primaria™ 6-well and 24-well Multiwell Plate, surface-modified polystyrene for enhanced cell culture (Corning, cat. nos. 353847 and 353847)
NOTE: Differentiation medium should be prepared freshly on the same day of use.
Day 0–4 differentiation [embryoid body (EB) formation and primitive streak/mesoderm induction]
-
1
At day 0, prepare StemPro-34 SFM medium and day 0 differentiation medium. Pretreat cells with 5 μM Y27632 in fresh hESC medium for 4 to 5 hr at 37°C in the incubator.
Inspect the iPSC lines on Matrigel under the microscope to ensure 80% to 90% confluency and showing no or little evidence of differentiation.
Empirically, 25 ml of differentiation medium is needed for iPSCs collected from four Matrigel-coated plates.
-
2
After pretreatment of iPSCs with Y27632, aspirate the hESC medium, add 1 ml collagenase B/DNase I dissociation buffer per well of the 6-well plate. Incubate the cells for 25 min at 37°C in the incubator.
The collagenase digestion will cause a subtle but observable change on the edge of the colonies but will not cause the colonies to lift up.
-
3
Aspirate the collagenase B/DNase I dissociation buffer, and add 1 ml per well cold 0.25% trypsin/EDTA. Incubate for approximately 1 min at room temperature.
-
4
End the dissociation process with 1 ml per well of the STOP medium. Detach the colonies using a cell lifter. Gently mix the cells with a 5-ml serological pipette to generate cell clumps that are larger than that of regular passaging.
Limit the size of the cell clumps to 200 to 500 cells to improve EB formation and the production efficiency of myeloid progenitor budding from EBs at later stages. Avoid over-trituration to create single cells or smaller clumps.
-
5
Add 1 ml/well wash medium and harvest the entire cell mixture into a 50-ml conical centrifuge tube. Combine it with 10 ml wash medium placed in the tube beforehand. Centrifuge the mixture for 3 min at 100 × g, room temperature.
-
6
Gently resuspend the pellets using a 5-ml serological pipette with 24 ml of day 0 differentiation medium. Dispense 2 ml/well into two ultra-low attachment surface 6-well plates for EB formation.
An aliquot of the cell pellet can be used for FACS-based characterization to ensure the pluripotent status of the cells (see Support Protocol 3 to prepare cells ready for FACS analysis).
-
7
Culture the cells in an incubator maintained at 37°C, 5% CO2, and 5% O2 for 2 days, undisturbed. EBs should form within 24 hr.
The number of EBs formed is directly correlated to the yield of myeloid progenitor cells at later stages.
-
8At day 2, harvest the EBs into a 50-ml conical tube via either of the following methods:
- Centrifuge for 1 min at 100 × g, room temperature, in order to collect EBs instead of cell debris.
- Alternatively, let the EBs settle by gravity by leaving the tube undisturbed in the biosafety cabinet for 10 to 20 min.
-
9
Aspirate the supernatant without disturbing the EBs, resuspend the EBs with day 2 differentiation medium and dispense 2 ml/well to ultra-low attachment plates. Place the plate back into an incubator maintained at 37°C, 5% CO2, and 5% O2.
The same ultra-low attachment plates can be reused but take cautions to avoid cross contamination of different iPSC lines.
Day 4 to 8 differentiation (hematopoietic specification)
-
10
At day 4, follow the same procedures as in step 8 to 9 to harvest and refresh the EBs with freshly prepared day 4 differentiation medium. Return the cells to an incubator maintained at 37°C, 5% CO2, and 5% O2.
Try to carefully and evenly dispense the EBs in differentiation medium onto cell culture plates as the EBs will float to the medium surface and increase in size along the differentiation.
If the number of EBs is high and the medium turns yellow within 24 hr after medium change, an additional medium change is needed.
-
11
Between days 6 and 8, single cells produced from EBs should begin to appear in the suspension culture.
-
12
At day 6, collect both EBs and single cells in suspension and pellet the cells by centrifuging for 4 min at 200 × g, room temperature.
-
13
Aspirate the supernatant and gently resuspend the cell pellet with freshly prepared day 6 differentiation medium. Refreshed EBs should be maintained in a 37°C, 5% CO2, and 5% O2 humidified incubator.
An aliquot of day 6 EBs can be dissociated and characterized by FACS for percentage of CD43+/CD34+ hematopoietic progenitors (see Support Protocol 3 to prepare cells ready for FACS analysis).
Day 8 to 15 differentiation (hematopoietic cell maturation/myeloid expansion)
-
14
At day 8, collect both EBs and single cells via same methods at day 6. Resuspend in macrophage culture medium composed of 20% (v/v) FBS in RPMI and 100 ng/ml human M-CSF and dispense to ultra-low attachment plates. Day 8 EBs should be maintained in a 37°C, 5% CO2 humidified incubator.
From day 8, cells are switched from the low oxygen (5% O2) incubator to the incubator with environmental oxygen levels.
An aliquot of day 8 EBs can be dissociated and characterized by FACS for percentage of CD43+/CD34+ hematopoetic progenitor markers (see Support Protocol 3 to prepare cells ready for FACS analysis).
-
15
Aspirate the spent medium and add fresh macrophage culture medium as needed if the medium turns yellow during day 8 to 15.
-
16
Collect EBs and single cells in suspension, centrifuge for 4 min at 200 × g, room temperature, to pellet cells.
Single cells from day 12 and day 15 can be harvested and characterized by FACS to determine the expression of hematopoietic progenitors and myeloid markers (see Support Protocol 3 to prepare cells ready for FACS analysis).
-
17
Aspirate the supernatant without disturbing EBs and cell pellets, replace with fresh macrophage culture medium. Day 12 to 15 EBs should be maintained in a 37°C, 5% CO2 humidified incubator.
Day 15 to 22 differentiation (myeloid progenitor expansion/macrophage maturation)
-
18
At day 15, collect both EBs and single cells into 50-ml conical centrifuge tubes.
-
19
Let the EBs in tubes settle by gravity in a hood for 15 min at room temperature without disturbance.
-
20
Transfer the supernatant containing myeloid progenitors as single cells without disturbing the EBs into a 50-ml conical tube.
An aliquot of the single cells can be characterized by FACS to determine the percentage of CD45+/CD18+ cells, which should be >85% (see Support Protocol 3 to prepare cells ready for FACS analysis).
-
21
Resuspend the EBs in fresh macrophage culture medium and return to ultra-low attachment plates. Perform medium change with fresh macrophage culture medium every 3 to 4 days.
-
22
Collect myeloid progenitor cells in the supernatant. Count and seed the cells onto a Primaria tissue culture plate at 1.0–1.5 × 105/well of 6-well plate (see Fig. 3A for seeding density at day 15).
Scale up or down the cell seeding as needed for other types of culture vessels.
The Primaria tissue culture plate featuring a modification of the polystyrene surface incorporating a mixture of anionic and cationic functional groups is optimal for culture of a variety of primary cells including human monocytes (Zhang et al., 2015; Zhang, Shi et al., 2017; Zhang, Xue et al., 2017). Regular tissue culture plates can be used for IPSDM differentiation but in this protocol, in order to compare the characteristics of IPSDM to HMDM under the same culture condition, we used Primaria tissue culture plates.
The myeloid progenitor cells attach easily and continue to proliferate between days 15 and 22. They would mature into macrophages in larger size and show round or spindle-like morphology that resembles human monocyte-derived macrophages (Zhang et al., 2015).
-
23
Feed the adherent cells every 2 or 3 days. The differentiated IPSDM should reach 70% to 90% confluency at day 21 or 22 (see Fig. 3B–3C for IPSDM morphology at day 22).
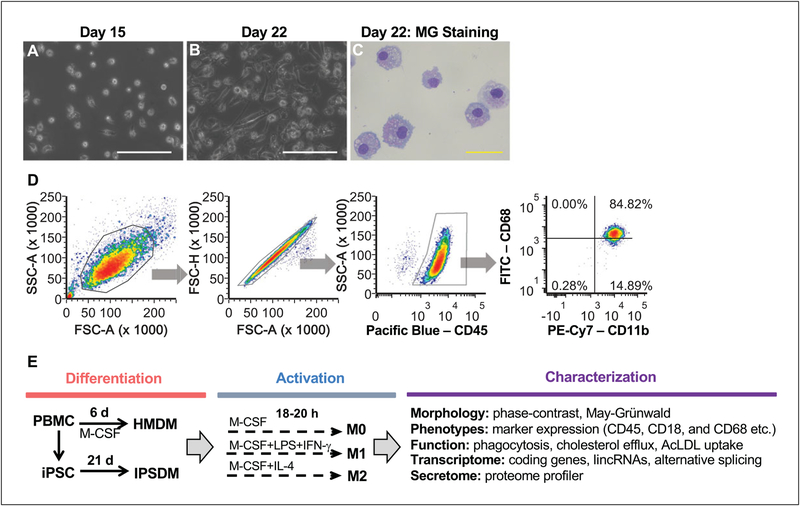
Differentiated IPSDM can be used for various assays or collected for DNA, RNA, or protein analysis on day 21 or 22. Yield and purity of day 22 IPSDM should be determined by FACS (see Support Protocol 3 and anticipated results in Fig. 3D) and recorded using the template described in Supporting Information Table 1.
Figure 3.
The morphology and marker expression of IPSDM. (A) The seeding density of day 15 myeloid progenitors harvested from the suspension culture of EBs. (B) The phase-contrast image of day 22 IPSDM. The white bar = 100 μm. (C) May-Grünwald Giemsa (MG) staining of day 22 IPSDM showing macrophage-like morphology. The yellow bar = 50 μm. (D) Macrophage marker expression of day 22 IPSDM characterized by FACS. The day 22 IPSDM are CD45+/CD18+/CD11b+ with high expression of CD68. Additional morphological, phenotypical, and functional characterizations as outlined in (E) were performed and the results can be found in our previous publication (Zhang et al., 2015; Zhang, Shi et al., 2017; Zhang, Xue et al., 2017).
SUPPORT PROTOCOL 1
0.1% GELATIN-COATED 6-WELL PLATES PREPARATION
This protocol describes gelatin coating of 6-well plates with 0.1% gelatin solution to enhance MEF feeder attachment.
Materials
0.1% (w/v) gelatin solution (Millipore Sigma, cat. no. ES-006-B)
6-well tissue culture plates (Falcon, cat. nos. 08–772–1B)
Using aseptic technique and working in a biosafety cabinet, add 1 ml of 0.1% gelatin solution per well of 6-well plate.
Rock the plate to coat the surface. Allow the gelatin to sit for 30 min at room temperature.
Aspirate the gelatin solution before plating cells.
SUPPORT PROTOCOL 2
PREPARATION OF MATRIGEL-COATED 6-WELL PLATES
This protocol describes Matrigel-coating of 6-well plates for iPSC growth and maintenance in feeder-free conditions.
Materials
Matrigel™ membrane matrix - growth factor reduced (Corning, cat. no. CB-40230C)
Ice
IMDM (Iscove’s Modified Dulbecco’s Medium) (Corning, cat. no. 10–016-CV) 6-well tissue culture plates (Falcon, cat. no. 08–772–1B)
Thaw Matrigel aliquots on ice until the Matrigel becomes a homogeneous liquid.
-
Gently mix Matrigel with ice-cold IMDM medium at a 1:2 ratio (1 ml Matrigel + 2 ml IMDM).
Pre-chill pipette tips, tubes, plates, or other objects that may come in contact with the extract to prevent premature gelling, especially when working with smaller volume. Avoid introducing air bubbles when handling Matrigel and minimize freeze-thaw cycle.
Dispense a sufficient amount of Matrigel solution (~2 ml) to cover the entire growth surface of one well of the 6-well plate pre-chilled on ice. Rocking the plate back and forth to establish a thin layer of Matrigel on the well surface.
-
Transfer the extra Matrigel solution to another well and ensure sufficient volume to cover the entire surface. Continue the process until the desired amount of plates is coated.
Generally, 3 ml of Matrigel solution is need to coat two 6-well plates. The unused Matrigel solution can be frozen and stored for later use.
Place the Matrigel-coated plates on ice for 30 min then transfer to 37°C incubator for 1 hr. The coated plates are then ready to use.
SUPPORT PROTOCOL 3
CHARACTERIZATION BY FACS DURING THE COURSE OF DIFFERENTIATION
This protocol describes how to prepare single cells at different stages of differentiation, staining at optimal conditions for FACS analysis. It is critical during the establishment of the protocol and troubleshooting. The iPSC lines at day 0, EBs at day 6 and day 8, single cells at day 12 and day 15, and differentiated IPSDM at day 22 should be collected for characterization of the cell identity, differentiation efficiency, and purity by FACS using the suggested panel design and antibody dilution as described in Supporting Information Tables 2 and 3. Once the protocol is established, FACS characterization can be performed at selected stages. However, the purity of the day 22 IPSDM defined as the percentage of CD45+/CD18+ cells should be tested and recorded for every cell line.
Materials
iPSCs from day 0, EBs from day 6 or day 8, single cells from day 12 or day 15, or IPSDM from day 22
0.05% (w/v) Trypsin/EDTA (Gibco, cat. no. 25–200–054)
STOP medium (see recipe)
Wash medium (see recipe)
Collagenase B/DNase I dissociation buffer (see recipe) Phosphate-buffered saline (PBS)
0.25% Trypsin/EDTA (Gibco, cat. no. 25–200–056)
FACS buffer
4% (v/v) formaldehyde (see recipe), optional
14-ml Falcon round-bottom polypropylene tubes with snap cap (Corning, cat. no. 14–959–11B)
37°C incubator
20-G needles
5-ml syringes
Vortex mixer
Centrifuge
37°C water bath
15- or 50-ml conical centrifuge tubes
Hemacytometer
5-ml Falcon round-bottom polystyrene tubes
Collect single-cell suspension
-
1Collect day 0 iPSCs as follows:
- Harvest iPSC colonies from one well of a 6-well plate by following Basic Protocol 3, steps 1 to 5. Resuspend the cell pellet in a 14-ml Falcon round-bottom polypropylene tube with 1 ml of 0.05% trypsin/EDTA and incubate for 5 min at 37°C. Neutralize 0.05% trypsin/EDTA with 1 ml STOP medium, and add 4 ml wash medium to bring the total volume to ~6 ml.
-
Pass the cell suspension through a 20-G needle/5-ml syringe 2 to 3 times and vortex to obtain a single-cell suspension.A similar approach can be used to disaggregate day 1 to5 EBs, if desired.
Dissociate day 6–8 EBs
-
2
Harvest EBs and single cells in suspension culture from one well of a 6-well plate into 14-ml Falcon round-bottom polypropylene tubes and centrifuge for 1 min at 100 × g, room temperature, to pellet EBs. Aspirate the supernatant without disturbing the EBs.
-
3
Resuspend EBs with 2 ml Collagenase B/DNase I dissociation buffer and incubate for 30 to 60 min in a 37°C water bath.
-
4
Add 4 ml wash medium to bring the total volume to ~6 ml and then disaggregate EBs by passing through 20-G needle/5-ml syringe 2 to 3 times.
-
5
Centrifuge for 3 min at 200 × g, room temperature, to pellet the cells and aspirate the supernatant. Resuspend the cell pellet with 2 ml of 0.05% trypsin/EDTA. Vortex and incubate at room temperature for up to 2 min.
Test the force and speed needed when moving the plunger and passing cell clusters through the syringe. Apply force just sufficient to obtain a single-cell suspension to avoid mechanical damage of the cells due to shear force. Inspect the flow-through cells under the microscope each time to ascertain the cells are undamaged.
-
6
Neutralize the 0.05% trypsin/EDTA by adding 1 ml STOP medium.
Harvest single cells at day 12 and day 15 in suspension culture
-
7
Collect both EBs and single cells into 15-ml or 50-ml conical centrifuge tubes.
-
8
Let the EBs settle by gravity by leaving the tube undisturbed in the biosafety cabinet for 10 to 20 min. Collect the cell suspension without disturbing the EB.
Harvest IPSDM at day 22 in adherent culture
-
9
Aspirate the medium, and wash the cell monolayer with 2 ml PBS per well.
-
10
Add 1 ml/well 0.25% trypsin/EDTA, and incubate for 5 to 10 min at 37°C.
-
11
Tap the side of the plate to dislodge the cells. Macrophages are adherent and might not be dislodged easily.
-
12
Add medium with FBS to neutralize the trypsin, and then harvest the cell suspension.
-
13
Centrifuge the cell suspension for 4 min at 200 × g, room temperature. Wash the cell pellet once with 2 ml PBS per well.
-
14
Triturate the cells by pipetting up and down gently to ensure that cell clumps are dispersed into single cells.
-
15
Count the cells using a hemocytometer. Record the cell number using the Differentiation Template provided as Supporting Information Table 1. Resuspend the cells in FACS buffer.
-
16
Directly stain the cells according to the stage of differentiation as described in Supporting Information Tables 2 and 3 for 25 min on ice.
Different batches of antibodies may show different activity. The dilution and source of the antibodies listed in Supporting Information Tables 2 and 3 serve as a general guideline and may need to be modified.
-
17
After staining, wash the cells with 1 to 2 ml FACS buffer and centrifuge for 4 min at 200 × g, room temperature or 4°C. Repeat the washing step for a total of 2 times.
-
18
Resuspend the cells in 200 μl FACS buffer and transfer into 5-ml Falcon round-bottom polystyrene tubes for FACS analysis.
Keep the tubes on ice until FACS analysis with a BD LSRII or similar flow cytometer can be performed.
Alternatively, fix the cells with 2% formaldehyde in PBS at room temperature for 20 min followed by washing with PBS.
REAGENTS AND SOLUTIONS
bFGF (100 μg/ml stock solution)
In a sterile environment, reconstitute 1 mg bFGF (recombinant human basic fibroblast growth factor; Life Technologies, cat. no. PHG0023) in 10 ml sterile distilled water to 100 μg/ml. Divide the reconstituted protein into 100 μl-aliquots and store them up to 6 months at ≥−20°C, ≤−70°C. Avoid repeated freeze/thaw cycles. Make any further dilutions using cytokine reconstitution buffer. Final concentration is 20 ng/ml.
BMP-4 (25 μg/ml stock solution)
In a sterile environment, reconstitute 50 μg BMP-4 (recombinant human bone morphogenetic protein 4; R&D, cat. no. 314-BP-050) in 2 ml sterile 4 mM HCl containing 0.1% bovine serum albumin (BSA) at 25 μg/ml. Prepare 200 μl-aliquots and store up to 3 months at ≥−20°C, ≤ −70°C. Avoid repeated freeze/thaw cycles. Final concentration is 25 ng/ml.
Collagenase B/DNase I dissociation buffer
In a sterile environment, prepare 0.2% (w/v) collagenase B buffer by dissolving 0.1 g Collagenase B from Clostridium histolyticum (Roche Applied Science, cat. no. 11088831001) in 50 ml Iscove’s Modified Dulbecco’s Medium (IMDM; Corning, cat. no. 10–016-CV) with 20% (v/v) fetal bovine serum (FBS; VWR, cat. no. 97068–091) (0.1 g collagenase B + 40 ml IMDM + 10 ml FBS) and filter sterilize using a 50-ml, 0.22-μm Steriflip filtration system (Millipore, cat. no. SCGP00525). Add 5 μl of 2 mg/ml DNase I stock solution (Calbiochem, cat. no. 260913–10MU) to 1 ml of the 0.2% Collagenase B buffer for a final concentration of 10 μg/ml DNase I. Store the buffer for up to 6 months at −20°C.
Cytokine reconstitution buffer (0.1% BSA, w/v)
In a sterile environment, dissolve 0.05 g bovine serum albumin (BSA; Sigma, cat. no. A7030–10G) in 50-ml sterile phosphate-buffered saline (PBS) and filter through a 50-mL, 0.22-μm Steriflip filtration system (Millipore, cat. no. SCGP00525). Divide into 5-ml aliquots per tube to enable single use and prevent contamination. Store the buffer for up to 6 months at 4°C.
Day 0 differentiation medium
StemPro™−34 SFM (serum-free medium) medium (Gibco, cat. no. 10639–011)
1 × Penicillin/streptomycin (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
1 × L-Glutamine (Gibco, cat. no. 25030–081; add from 100× concentrated stock-200 mM)
0.3% (v/v) diluted MTG (1-Thioglycerol) in IMDM (Sigma, cat. no. M6145–25 ml; see recipe)
50 μg/ml final concentration of L-Ascorbic Acid (Sigma, cat. no. A4544–25G; from 5 mg/ml stock solution as in recipe)
150 μg/ml final concentration of Human transferrin (Sigma, cat. no. 10652202001; from 30 mg/ml stock solution prepared as in recipe)
25 ng/ml BMP-4 (recombinant human bone morphogenetic protein 4) (R&D, cat. no. 314-BP-050; from 25 mg/ml stock solution prepared as in recipe)
50 ng/ml final concentration of VEGF (recombinant human vascular endothelial growth factor) 165 Protein (R&D, cat. no. 293-VE-010/CF; from 50 μg/ml stock solution prepared as in recipe)
Filtering this medium is not recommended
See table below for exact quantities.
Prepare fresh
| Component | Stock concentration | Final concentration | Per ml | Catalog no. |
|---|---|---|---|---|
| StemPro-34 | 1 ml | 10639–11 (Gibco) | ||
| L-Glutamine | 100× | 1× | 10 μl | 25030–081 (Gibco) |
| Penicillin-streptomycin | 100× | 1× | 10 μl | 15140–122 (Gibco) |
| Ascorbic acid (freshly thaw) | 5 mg/ml (100×) | 50 μg/ml | 10 μl | A4544–25G (Sigma) |
| Transferrin | 30 mg/ml (200 ×) | 0.15 mg/ml | 5 μl | 10652202001 (Sigma) |
| MTG (fresh) | 26 μl/2 ml IMDM | 3 μl/ml | 3 μl | M6145–25 ml (Sigma) |
| BMP-4 | 25 μg/ml | 25 ng/ml | 1 μl | 314-BP-050 (R&D) |
| VEGF | 50 μg/ml | 50 ng/ml | 1 μl | 293-VE-010/CF (R&D) |
Day 2 differentiation medium
StemPro™−34 SFM (serum free medium) medium (Gibco, cat. no. 10639–011)
1 × Penicillin/streptomycin (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
1 × L-Glutamine (Gibco, cat. no. 25030–081; add from 100× concentrated stock-200 mM)
0.3% (v/v) diluted MTG (1-Thioglycerol) in IMDM (Sigma, cat. no. M6145–25 ml; see recipe)
50 μg/ml final concentration of L-Ascorbic Acid (Sigma, cat. no. A4544–25G; from 5 mg/ml stock solution as in recipe)
150 μg/ml final concentration of Human transferrin (Sigma, cat. no. 10652202001; from 30 mg/ml stock solution prepared as in recipe)
25 ng/ml BMP-4 (recombinant human bone morphogenetic protein 4) (R&D, cat. no. 314-BP-050; from 25 mg/ml stock solution prepared as in recipe)
50 ng/ml final concentration of VEGF (recombinant human vascular endothelial growth factor) 165 Protein (R&D, cat. no. 293-VE-010/CF; from 50 μg/ml stock solution prepared as in recipe)
50 μg/ml SCF (recombinant human stem cell factor) (R&D, cat. no. 255-SC-001MG/CF; from 100 μg/ml stock solution prepared as in recipe) 50 ng/ml TPO (recombinant human thrombopoietin) (R&D, cat. no. 288-TP-200/CF; from 50 μg/ml stock solution prepared as in recipe)
50 ng/ml Flt-3 (recombinant human Flt-3 ligand) (R&D, cat. no. 308-FK-025/CF; from 50 μg/ml stock solution prepared as in recipe)
20 ng/ml bFGF (recombinant human basic fibroblast growth factor) (Life Technologies, cat. no. PHG0023; from 100 μg/ml stock solution prepare as in recipe)
Filtering this medium is not recommended
Prepare fresh
See table below for exact quantities.
| Component | Stock concentration | Final concentration | Per ml | Catalog no. |
|---|---|---|---|---|
| StemPro-34 | 1 ml | 10639–11 (Gibco) | ||
| L-Glutamine | 100× | 1× | 10 μl | 25030–081 (Gibco) |
| Penicillin-streptomycin | 100× | 1× | 10 μl | 15140–122 (Gibco) |
| Ascorbic acid (freshly thawed) | 5 mg/ml (100×) | 50 μg/ml | 10 μl | A4544–25G (Sigma) |
| Transferrin | 30 mg/ml (200×) | 0.15 mg/ml | 5 μl | 10652202001 (Sigma) |
| MTG (fresh) | 26 μl/2 ml IMDM | 3 μl/ml | 3 μl | M6145–25 ml (Sigma) |
| BMP-4 | 25 μg/ml | 25 μg/ml | 1 μl | 314-BP-050 (R&D) |
| VEGF | 50 μg/ml | 50 μg/ml | 1 μl | 293-VE-010/CF (R&D) |
| SCF | 100 μg/ml | 50 μg/ml | 0.5 μl | 255-SC-001MG/CF (R&D) |
| TPO | 50 μg/ml | 50 μg/ml | 1 μl | 288-TP-200/CF (R&D) |
| FLT3 | 50 μg/ml | 50 μg/ml | 1 μl | 308-FK-025/CF (R&D) |
| bFGF | 100 μg/ml | 20 ng/ml | 0.2 μl | PHG0023 (Life Technologies) |
Day 4 and day 6 differentiation medium
StemPro™−34 SFM (serum free medium) medium (Gibco, cat. no. 10639–011)
1 × Penicillin/streptomycin (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
1 × L-Glutamine (Gibco, cat. no. 25030–081; add from 100× concentrated stock-200 mM)
0.3% (v/v) diluted MTG (1-Thioglycerol) in IMDM (Sigma, cat. no. M6145–25 ml; see recipe)
50 μg/ml final concentration of L-Ascorbic Acid (Sigma, cat. no. A4544–25G; from 5 mg/ml stock solution as in recipe)
150 μg/ml final concentration of Human transferrin (Sigma, cat. no. 10652202001; from 30 mg/ml stock solution as in recipe)50 ng/ml final concentration of VEGF (recombinant human vascular endothelial growth factor) 165 Protein (R&D, cat. no. 293-VE-010/CF; from 50 μg/ml stock solution prepared as in recipe)
50 μg/ml SCF (recombinant human stem cell factor) (R&D, cat. no. 255-SC-001MG/CF; from 100 μg/ml stock solution prepared as in recipe)
50 ng/ml TPO (recombinant human thrombopoietin) (R&D, cat. no. 288-TP-200/CF; from 50 μg/ml stock solution prepared as in recipe)
50 ng/ml Flt-3 (recombinant human Flt-3 ligand) (R&D, cat. no. 308-FK-025/CF; from 50 μg/ml stock solution prepared as in recipe)
20 ng/ml bFGF (recombinant human basic fibroblast growth factor) (Life Technologies, cat. no. PHG0023; from 100 μg/ml stock solution prepare as in recipe)
Filtering this medium is not recommended
Prepare fresh
See table below for exact quantities.
| Component | Stock concentration | Final concentration | Per ml | Catalog no. |
|---|---|---|---|---|
| StemPro-34 | 1 ml | 10639–11 (Gibco) | ||
| L-Glutamine | 100× | 1× | 10 μl | 25030–081 (Gibco) |
| Penicillin-streptomycin | 100× | 1× | 10 μl | 15140–122 (Gibco) |
| Ascorbic acid (freshly thaw) | 5 mg/ml (100×) | 50 μg/ml | 10 μl | A4544–25G (Sigma) |
| Transferrin | 30 mg/ml (200×) | 0.15 mg/ml | 5 μl | 10652202001 (Sigma) |
| MTG (fresh) | 26 μl/2 ml IMDM | 3 μl/ml | 3 μl | M6145–25 ml (Sigma) |
| VEGF | 50 μg/ml | 50 ng/ml | l μl | 293-VE-010/CF (R&D) |
| SCF | 100 μg/ml | 50 ng/ml | 0.5 μl | 255-SC-001MG/CF (R&D) |
| TPO | 50 μg/ml | 50 ng/ml | 1 μl | 288-TP-200/CF (R&D) |
| FLT3 | 50 μg/ml | 50 ng/ml | 1 μl | 308-FK-025/CF (R&D) |
| bFGF | 100 μg/ml | 20 ng/ml | 0.2 μl | PHG0023 (Life Technologies) |
Days 8–22 macrophage culture medium
RPMI 1640 medium (Corning, cat. no. 10040CM)
20% (v/v) fetal bovine serum (FBS; VWR, cat. no. 97068–091)
100 ng/ml final concentration of M-CSF (recombinant human macrophage colony-stimulating factor) (Peprotech, cat. no. 300–25; from 100 μg/ml stock solution prepare as in recipe)
1 × Penicillin/streptomycin (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
Store up to 2 weeks at 4°C
Alternatively, the RPMI + 20% FBS + recombinant penicillin-streptomycin can be prepared in advance and M-CSF can be added fresh
Prepare fresh
| Component | Stock concentration | Final concentration | Per 500 ml | Catalog no. |
|---|---|---|---|---|
| RPMI1640 | 500 ml | 10040CM (Corning) | ||
| Penicillin-streptomycin | 100× | 1× | 5 ml | 15140–122 (Gibco) |
| FBS | 20% (v/v) | 100 ml | 97068–091 (VWR) | |
| M-CSF | 100 μg/ml | 100 ng/ml | 500 μl | 300–25 (Peprotech) |
FBS has lot-to-lot variability that may contribute to the variation in differentiation performance. We recommend using US origin FBS with heat inactivation, and stock sufficient quantity of a particular lot for the project once the protocol is established. iPSC lines in control and experimental groups should be differentiated using FBS from the same lots. When switching lots, we recommend performing FACS characterization of differentiated cells at different stages as outlined in Support Protocol 3 to confirm consistent results on efficiency and purity.
DNase I (2 mg/ml)
Dissolve 2 mg lyophilized DNase I powder (Calbiochem, cat. no. 260913–10MU) in 1 ml sterile distilled water. Do not vortex. In a sterile environment, filter-sterilize using a 0.22-μm low-protein-binding syringe filter and divide into 100-μl aliquots. Store up to 3 months at −20°C.
FACS buffer
In a sterile environment, dissolve 1 g (1%, w/v) bovine serum albumin (BSA; Sigma, cat. no. A7030–10G) and 0.05 g (0.05%, w/v) sodium azide (Sigma, cat. no. S2002–5G) in 100 ml sterile 1× phosphate-buffered saline without Ca2+ and Mg2+ (D-PBS; Corning, cat. no. MT 21–040-CM) and filter the solution through a 50-ml, 0.22-μm Steriflip filtration system (Millipore, cat. no. SCGP00525). Store up to 3 months at 4°C.
FLT3 ligand (50 μg/ml stock solution)
In a sterile environment, reconstitute 25 μg FLT3 ligand (recombinant human Flt-3 ligand; R&D, cat. no. 308-FK-025/CF) in 0.5 ml sterile phosphate-buffered saline (PBS) to 100 μg/ml. Prepare 100-μl aliquots and store them up to 3 months at ≥−20°C, ≤−70°C. Avoid repeated freeze-thaw cycles. Final concentration is 50 ng/ml.
Formaldehyde, 4% (v/v)
Freshly dilute 16% formaldehyde, methanol-free (ThermoFisher, cat. no. 28908) to 4% using 1× phosphate-buffered saline (PBS) without Ca2+ and Mg2+ (Corning, cat. no. MT 21–040-CM).
Store freshly prepared 4% formaldehyde for up to 1 week at 4°C, or freeze for long-term storage at −20°C.
2% formaldehyde used for fixation of cells for FACS is freshly diluted in PBS.
hESC (human embryonic stem cell) medium (500 ml)
In a sterile environment, supplement 400 ml of Advanced Dulbecco’s modified
Eagle/F12 medium (DMEM/F12; Life Technologies, cat. no. 11320–082) with:
100 ml KnockOut Serum Replacement (KOSR; Gibco, cat. no. 10828–028)
5 ml of 200 mM L-Glutamine (Gibco, cat. no. 25030–081; add from 100× concentrated stock-200 mM)
5 ml MEM-NEAA (non-essential amino acid; Gibco, cat. no. 11140–050; add from 100× concentrated stock)
3.5 μl of 14.3 mM 2-mercaptoethanol (Sigma, cat. no. M3148–100ML)
50 μl of 100 μg/ml recombinant human basic fibroblast growth factor (bFGF; Life Technologies, cat. no. PHG0023)
5 ml penicillin-streptomycin solution (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
Filter the medium through a 500-ml Stericup filtration system (Millipore, cat. no. SCGPU05RE)
Store up to 1 month at 4°C
L-Ascorbic acid
Dissolve 200 mg L-Ascorbic acid powder (Sigma, cat. no. A4544–25G) in 40 ml sterile distilled in water at 5 mg/ml. Prepare 1 ml-aliquots and store up to 1 year at −80°C. Freshly thaw and discard after single use.
M-CSF (100 μg/ml stock solution)
In a sterile environment, reconstitute 1 mg recombinant human macrophage
colony-stimulating factor (M-CSF; Peprotech, cat. no. 300–25) in 10 ml sterile distilled water with Cytokine reconstitution buffer (see recipe) or per manufacturer’s instructions to 100 μg/ml. Prepare 100 μl-aliquots and store them up to 3 months at ≥ −20°C, ≤−70°C.
MEF medium (500 ml)
In a sterile environment, supplement 450 ml of Dulbecco’s Modified Eagle Medium (DMEM; Corning, cat. no. MT10017CM) with:
50 ml fetal bovine serum (FBS; VWR, cat. no. 97068–091)
5 ml of 200 mM L-Glutamine (Gibco, cat. no. 25030–081; add from 100× concentrated stock)
5 ml MEM Non-Essential Amino Acid (MEM-NEAA; Gibco, cat. no. 11140–050; add from 100× concentrated stock)
5 ml penicillin-streptomycin (Gibco, cat. no. 15140–122; add from 100× concentrated stock)
Filter the medium through a 500-ml Stericup filtration system (Millipore, cat. no. SCGPU05RE)
Store up to 1 month at 4°C
MTG
Prepare 500 μl-aliquots of 1-Thioglycerol (MTG; Sigma, cat. no. M6145–25 ml) and store at 4°C until expiration date. Prepare working solution by adding 26 μl MTG to 2 ml IMDM, quickly vortex and use freshly to prepare differentiation medium.
MTG is sticky and large-orifice pipette tips with filters should be used.
SCF (100 μg/ml stock solution)
In a sterile environment, reconstitute 1 mg recombinant human stem cell factor (SCF; R&D, cat. no. 255-SC-001MG/CF) in 10 ml sterile phosphate-buffered saline (PBS) to 100 μg/ml. Prepare 100-μl aliquots and store them up to 3 months at ≥−20°C, ≤ −70°C. Avoid repeated freeze-thaw cycles. Final concentration is 50 ng/ml.
StemPro-34 SFM medium
Place the StemPro™−34 SFM (serum-free medium) medium (store at 4°C) and supplement (store at −80°C) (Gibco, cat. no. 10639–011) in a 37°C water bath. Remove the supplement after 20 min and mix well by shaking vigorously to ensure that no fatty clumps are present. Continue to incubate the supplement and medium for 1 hr at 37°C until the supplement becomes clear. Pipette the supplement into the basal medium, which must be warm, and invert up and down to mix. Incubate the mixed medium for another hour in a 37°C water bath. The prepared medium is now ready to use and can be stored up to 2 weeks at 4°C.
Ensure aseptic handling because filtering the prepared medium is not recommended. Divide the prepared medium into aliquots for single use to avoid warming up the same aliquot multiple times.
STOP medium (250 ml)
In a sterile environment, supplement 125 ml Iscove’s Modified Dulbecco’s Medium (IMDM; Corning, cat. no. 10–016-CV) with:
125 ml of 50% (v/v) fetal bovine serum (FBS; VWR, cat. no. 97068–091) Filter the medium through a 250-ml filtration system
Store up to 1 month at 4°C or at −20°C for long-term storage
TPO (50 μg/ml stock solution)
In a sterile environment, reconstitute 200 μg recombinant human thrombopoietin (TPO; R&D, cat. no. 288-TP-200/CF) in 4 ml sterile 4 mM HCl. Prepare 100 μlaliquots and store them up to 3 months at ≥ −20°C, ≤ −70°C. Final concentration is 50 ng/ml.
VEGF (100 μg/ml stock solution)
In a sterile environment, reconstitute 10 μg recombinant human vascular endothelial growth factor (VEGF; R&D, cat. no. 293-VE-010/CF) in 0.1 ml sterile phosphate-buffered saline (PBS) at 100 μg/ml. Prepare 25-μl aliquots and store them up to 3 months at ≥ −20°C, ≤ −70°C. Avoid repeated freeze-thaw cycles. Final concentration is 50 ng/ml.
Wash medium (250 ml)
In a sterile environment, supplement 225 ml Advanced Dulbecco’s modified Eagles/F12 medium (DMEM/F12; Life Technologies, cat. no. 11320–082) with 25 ml knockout replacement serum (KOSR; Gibco, cat. no. 10828–028) (10% v/v) and filter the medium through a 250-ml filtration system (Millipore, cat. no. SCGPU02RE). Store for up to 1 month at 4°C. Alternatively, 5% (v/v) KOSR (Gibco, cat. no. 10828–028) in Iscove’s Modified Dulbecco’s Medium (IMDM; Corning, cat. no. 10–016-CV) can be used.
Y27632 (5 mM stock solution)
In a sterile environment, resuspend 5 mg Y27632 (ROCK inhibitor) (Calbiochem, cat. no. 688000–5MG) in 3 ml sterile distilled water, mix and filter-sterilize with 0.22-μm syringe filter. Divide into 50- to 100-μl aliquots and store up to 6 months at −20°C. Take cautions to protect the solution from light.
COMMENTARY
Background Information
Prior protocols to differentiate pluripotent stem cells to macrophages have been summarized in our recently published review article (Zhang & Reilly, 2017). Briefly, in addition to earlier protocols for differentiation of human embryonic stem cells to macrophages (Karlsson et al., 2008; Subramanian et al., 2009), recent protocols for IPSDM generation have used three approaches: (1) co-culture with OP9 mouse stromal cell (Choi, Vodyanik, & Slukvin, 2011; Senju et al., 2011; Vanhee et al., 2015); (2) monolayer-based differentiation in 2D culture (Yanagimachi et al., 2013); and (3) EB-based differentiation (Alasoo et al., 2015; Buchrieser, James, & Moore, 2017; Lachmann et al., 2015; Panicker et al., 2012; van Wilgenburg, Browne, Vowles, & Cowley, 2013; Zhang et al., 2015). The general approach is to induce hematopoietic specification and myeloid expansion by sequential exposure to cytokine cocktails, and then harvest myeloid progenitors in suspension and differentiate to macrophages in adherent culture using serum and M-CSF with or without IL-3. Two protocols have also attempted to perform myeloid to macrophage differentiation in serum-free media with M-CSF (van Wilgenburg et al., 2013) or M-CSF/IL-3 (Buchrieser et al., 2017), but the transcriptome fidelity of IPSDM in comparison to HMDM differentiated in the same culture condition have not been fully characterized.
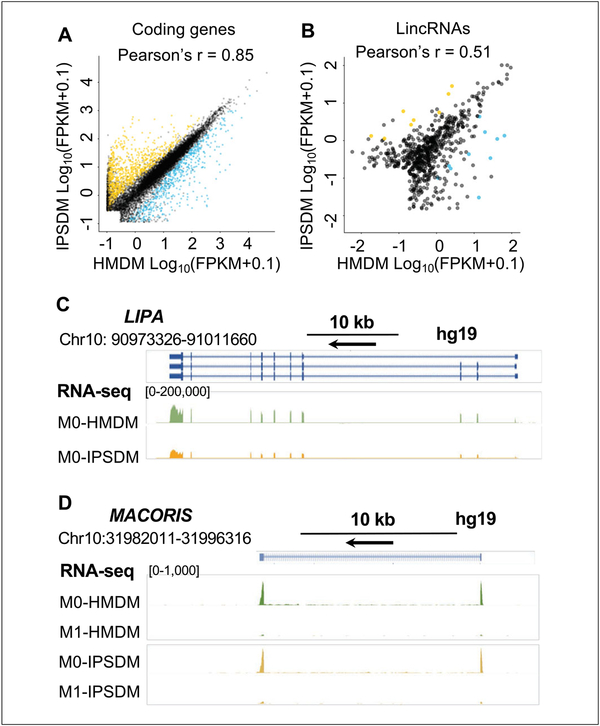
Our group for the first time performed deep RNA-sequencing (RNA-seq) in isogenic HMDM and IPSDM lines derived from the same subjects and cultured in the same medium for macrophage maturation. This design allows a precise evaluation of the transcriptome similarity and differences of IPSDM and HMDM, and provides insights into the application of IPSDM in conjunction with CRISPR/Cas gene editing as an experimental model for studying the functional and transcriptomic features of human macrophage biology. Our protocol uses an EB–based approach with a combination of serum-free condition for hematopoiesis specification, followed by adherent culture with serum and M-CSF for myeloid expansion and macrophage maturation (Zhang et al., 2015). The defined condition for hematopoiesis specification ensures a high efficiency (Hale et al., 2015) and purity of >90% CD43+ CD34+ hematopoietic progenitors in day 8 EBs (Zhang et al., 2015). The myeloid expansion and macrophage maturation in serum and M-CSF improves yield while maintaining high purity (Fig. 3). The results highlighted the transcriptome similarity with significant overlap (~99%) and strong correlation (Pearson’s r 0.85) between HMDM and IPSDM with a small = percentage of genes (~12%) differentially expressed (Fig. 4A) (Zhang et al., 2015). Further, more sophisticated functional characteristics, such as cholesterol efflux, cholesteryl ester hydrolysis (Zhang, Shi et al., 2017), and cytokine secretion profile in macrophages with M1 (lipopolysaccharide interferon-gamma) and M2 (interleukin-4) activation, + were compared between 6 HMDM and IPSDM lines and have demonstrated remarkable similarity (Zhang et al., 2015). We have further elaborated that IPSDM recapitulate important alternative splicing events (Lin et al., 2016) and long non-coding RNA profiles (Zhang, Xue et al., 2017) of HMDM during macrophage activation, identifying IPSDM as uniquely suited to study human macrophage-specific transcriptome regulation and providing a comprehensive resource for planning such studies.
Figure 4.
The transcriptome characterization of IPSDM and HMDM. (A) The coding transcriptome profile is highly correlated between IPSDM and HMDM. (B) The expression of long inter-genic noncoding RNAs (lincRNAs) is less abundant than that of coding genes, but the lincRNA expression profile also shows modest correlation between IPSDM and HMDM. The differentially expressed (DE) mRNAs and lincRNAs between HMDM and IPSDM are highlighted in blue or dark yellow to illustrate the transcripts expressed at higher levels in HMDM or IPSDM, respectively. False discovery rate (FDR)–adjusted P value<0.01 and a fold change (FC) >2 were tested with Cuffdiff. (C) and (D) The genome-browser view of RNA-seq track for a coding gene LIPA (Zhang, Shi et al., 2017) and a lincRNA MACORIS (Zhang, Xue et al., 2017) that are expressed at similar levels between IPSDM and HMDM and have previously studied in IPSDM.
The future direction is to generate a variety of tissue-resident macrophages from iPSCs. There is some success to differentiate iPSC to microglia-like cells using key cytokines for microglia lineage commitment (Douvaras et al., 2017; Muffat et al., 2016), by co-culture with iPSC-derived neurons (Haenseler et al., 2017; Takata et al., 2017) and astrocytes (Pandya et al., 2017), or incubating with cytokines derived from those cell types (Abud et al., 2017) to recapitulate an organ-specific environment. It has been recognized that in vitro hematopoietic differentiation of pluripo-tent stem cells resembles in vivo primitive hematopoiesis rather than adult definitive hematopoiesis (Vanhee et al., 2015). This suggests that IPSDM may developmentally relate to and be a good model for tissue resident macrophages, which will further raise the possibilities to delve into the specialized properties and tissue-specific functions of human macrophages.
Critical Parameters and Troubleshooting
Maintaining iPSCs in a pluripotent state is essential for optimal differentiation efficiency. During MEF depletion, the quality of the Matrigel coating is critical for the adhesion of iPSCs and therefore sufficient numbers of iPSCs for EB formation. The quantity of EBs formed is directly correlated to the yield of myeloid progenitor cells at later stages. FACS characterization is highly recommended to determine the marker expression of differentiated cells at different stages of differentiation to confirm the expected efficiency and purity, especially during the establishment of the protocol and troubleshooting. Detailed troubleshooting guidelines are outlined in Table 1.
Table 1.
Troubleshooting
| Problem | Possible reason | Solution |
|---|---|---|
| The iPSC lines lose colony-like morphology or grow slowly. | Cells are differentiated. | Verify whether iPSC lines have maintained an undifferentiated state by examining the expression of pluripotency markers TRA-1–60 and SSEA-4 by FACS. If cells have differentiated, restarting a new culture is recommended. |
| Poor attachment of iPSCs to Matrigel-coated plates. | No ROCK inhibitor was included, or a coating substrate of low quality was used. | Include a ROCK inhibitor (e.g., Y27632) in culture medium; prepare Matrigel-coated plates following the protocol and the manufacturer’s instruction. |
| Colonies on Matrigel-coated plates are too small. | The size of dissociated iPSC clumps at Basic Protocol 2, step 3 is too small. | At day −2, pipette the cell pellet gently to ensure that the cell clumps are larger than that during regular passaging. |
| Too few EBs are formed; lots of dead cells in the suspension culture. | Cell loss during feeder deletion due to poor survival, poor pluripotency status, or poor Matrigel quality. | Follow solutions for Basic Protocol 2, steps 3 to 5. Confirm expression and percentage of pluripotency markers for day −2 and day 0 iPSC lines. |
| Yield of myeloid progenitors is low. | The number and/or the quality of EBs is low. | Perform FACS-based characterization of day 6 and day 8 EBs to confirm the percentage of hematopoietic progenitors. Continue to culture EB and single-cell suspension in ultra-low plate for a few more days (e.g., to day 20) before harvesting for adherent culture to obtain more myeloid progenitors. Confirm the percentage of CD45+/CD18+ cells in day 15 single cells. |
| Low density of IPSDM at day 22. | A low seeding density was used at day 15. | Optimize the seeding density at day 15. When the day 15 seeding density is within the recommended range, low density of day 22 IPSDM is most likely due to low proliferative potential of myeloid progenitors, which may be due to poor quality of day 15 progenitors or the intrinsic variability of differentiation potential of iPSC lines. |
| Purity of IPSDM is low. | Purity of day 15 myeloid progenitors is low. | Characterize day 15 single cells by FACS. Perform magnetic beads or FACS-based sorting if desired. Do not allow differentiated IPSDM to grow to over 90% confluency or past day 22. |
Anticipated Results
Ideally, this differentiation protocol should produce 18.97±3.82 X 106 IPSDM from 3.28±0.23 X 106 iPSC at day −2 on a 6-well plate (mean±SD, n 6 lines) (Zhang et al., 2015). IPSDM purity = after differentiation should be >90%. These IPSDM should express markers of macrophages (Fig. 3D) and demonstrate similar morphological and functional phenotypes of HMDM as listed in Figure 3E and presented in details in previous publications (Zhang et al., 2015). The RNA-seq data of isogenic IPSDM and HMDM show well-correlated expression levels between IPSDM and HMDM (Fig. 4A-4B), and can be visualized in the UCSC genome browser by adding track hub using URL (https://de.cyverse.org/anon-files/iplant/home/chenyix/trackhub/GENEiPS_scaled/hub.txt) or through the following link (http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hubUrl=https://de.cyverse.org/anon-files/iplant/home/cheny/ix/trackhub/GENEiPS_scaled/hub.txt). The original RNA-seq data are available from the NCBI Gene Expression Omnibus (GEO) under the accession number GSE55536. Examples of RNA-seq track are shown in Figure 4C-4D presenting a similar expression pattern of LIPA (Zhang, Shi et al., 2017) and long intergenic noncoding RNA (lincRNA) MACORIS (Zhang, Xue et al., 2017) in IPSDM and HMDM. Supporting Information Tables 4 to 9 provide the expression levels in FPKM, fold change, and false discovery rate-adjusted P-value for differential expression analysis of coding genes and lincRNAs between HMDM and IPSDM at baseline and with M1- and M2-stimulation. These data provide a comprehensive resource to query whether a gene-of-interest has a similar expression level or responses to M1/M2 stimulations, and therefore can be modeled using IPSDM in conjunction with CRISPR/Cas-mediated gene knockout or knock-in (Zhang, Shi et al., 2017). If the differentiation efficiency is lower than 85% in certain iPSC lines, or higher purity is needed, CD45+ cells can be enriched by magnetic-activated cell sorting (MACS) or FACS sorting.
Time Considerations
The differentiation procedure requires ~2 weeks for iPSCs expansion and 24 days for feeder depletion and IPSDM differentiation (Fig. 1).
Basic Protocol 1: Expansion of iPSCs on feeders: ~ 2 weeks
Basic Protocol 2: Feeder depletion of iPSCs: Day −2 to day 0, 2 days
Basic Protocol 3: Differentiation of iPSCs to macrophages: 22 days (4 stages)
Stage 1: EB formation, primitive streak/ mesoderm induction: Day 0 to 4, 4 days
Stage 2: Hematopoietic specification: Day 4 to 8, 4 days
Stage 3: Hematopoietic cell maturation/ myeloid expansion: Day 8 to 15, 7 days
Stage 4: Myeloid progenitor expansion/ macrophage maturation: Day 15 to 22, 7 days
Supplementary Material
Acknowledgements
We thank Dr. Muredach P. Reilly (Columbia University, USA) and all co-authors of the original articles describing this protocol for their initial support and contribution. We thank Drs. Masayuki Yazawa and Alan Tall (Columbia University, USA) for critical reading of the manuscript. This work has received funding from the American Heart Association Postdoctoral Fellowship 15POST25620017, the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), through Grant Number UL1TR001873, and the NIH R00HL130574 (to HZ).
Footnotes
Conflicts of Interest
The authors have no conflicts of interests to declare.
Literature Cited
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, … Blurton-Jones M (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron, 94(2), 278–293.e279. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasoo K, Martinez FO, Hale C, Gordon S, Powrie F, Dougan G, … Gaffney DJ (2015). Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Scientific Reports, 5, 12524. doi: 10.1038/srep12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J, James W, & Moore MD (2017). Human induced pluripotent stem cell-derived macrophages share ontogeny with MYB-independent tissue-resident macrophages. Stem Cell Reports, 8(2), 334–345. doi: 10.1016/j.stemcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Vodyanik M, & Slukvin II (2011). Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nature Protocols, 6(3), 296–313. doi: 10.1038/nprot.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RG, Shiue L, Prins J, de Boer HC, Singh A, Fagg WS, … van der Veer EP (2016). Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nature Communications, 7, 10846. doi: 10.1038/ncomms10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, … Fossati V (2017). Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports, 8(6), 1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R, Molawi K, & Sieweke MH (2014). Tissue macrophage identity and self-renewal. Immunological Reviews, 262(1), 56–73. doi: 10.1111/imr.12224. [DOI] [PubMed] [Google Scholar]
- Gordon S (2008). Elie Metchnikoff: Father of natural immunity. Eur J Immunol, 38(12), 3257–3264. doi: 10.1002/eji.200838855. [DOI] [PubMed] [Google Scholar]
- Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, … Cowley SA (2017). A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports, 8(6), 1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Yeung A, Goulding D, Pickard D, Alasoo K, Powrie F, … Mukhopadhyay S (2015). Induced pluripotent stem cell derived macrophages as a cellular system to study salmonella and other pathogens. PLoS One, 10(5), e0124307. doi: 10.1371/journal.pone.0124307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson KR, Cowley S, Martinez FO, Shaw M, Minger SL, & James W (2008). Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in M-CSF and IL-3. Experimental Hematology, 36(9), 1167–1175. doi: 10.1016/j.exphem.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, … Moritz T (2015). Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports, 4(2), 282–296. doi: 10.1016/j.stemcr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Hu Y, Nunez S, Foulkes AS, Cieply B, Xue C, … Reilly MP (2016). Transcriptome-wide analysis reveals modulation of human macrophage inflammatory pheno-type through alternative splicing. Arteriosclerosis, Thrombosis, and Vascular Biology, 36(7), 1434–1447. doi: 10.1161/ATVBAHA.116.307573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNelis JC, & Olefsky JM (2014). Macrophages, immunity, and metabolic disease. Immunity, 41(1), 36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, … Jaenisch R (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nature Medicine, 22(11), 1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narva E, Pursiheimo JP, Laiho A, Rahkonen N, Emani MR, Viitala M, … Lahesmaa R (2013). Continuous hypoxic culturing of human embryonic stem cells enhances SSEA-3 and MYC levels. PLoS One, 8(11), e78847. doi: 10.1371/journal.pone.0078847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, … Park JK (2017). Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nature Neuroscience, 20(5), 753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker LM, Miller D, Park TS, Patel B, Azevedo JL, Awad O, … Feldman RA (2012). Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proceedings of the National Academy of Sciences of the United States of America, 109(44), 18054–18059. doi: 10.1073/pnas.1207889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvagiotto G, Burton S, Daigh CA, Rajesh D, Slukvin II, & Seay NJ (2011). A defined, feeder-free, serum-free system to generate in vitro hematopoietic progenitors and differentiated blood cells from hESCs and hiPSCs. PLoS One, 6(3), e17829. doi: 10.1371/journal.pone.0017829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju S, Haruta M, Matsumura K, Matsunaga Y, Fukushima S, Ikeda T, … Nishimura Y (2011). Generation of dendritic cells and macrophages from human induced pluripotent stem cells aiming at cell therapy. Gene Therapy, 18(9), 874–883. doi: 10.1038/gt.2011.22. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Guo B, Marsden MD, Galic Z, Kitchen S, Kacena A, … Zack JA (2009). Macrophage differentiation from embryoid bodies derived from human embryonic stem cells. Journal of Stem Cells, 4(1), 29–45. [PMC free article] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, … Ginhoux F (2017). Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity, 47(1), 183–198.e186. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- van Wilgenburg B, Browne C, Vowles J, & Cowley SA (2013). Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS One, 8(8), e71098. doi: 10.1371/journal.pone.0071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee S, De Mulder K, Van Caeneghem Y, Verstichel G, Van Roy N, Menten B, … Vandekerckhove B (2015). In vitro human embryonic stem cell hematopoiesis mimics MYB-independent yolk sac hematopoiesis. Haematologica, 100(2), 157–166. doi: 10.3324/haematol.2014.112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, & Vannella KM (2016). Macrophages in tissue repair, regeneration, and fibrosis. Immunity, 44(3), 450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi MD, Niwa A, Tanaka T, Honda-Ozaki F, Nishimoto S, Murata Y, … Saito MK (2013). Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum-and feeder cell-free conditions. PLoS One, 8(4), e59243. doi: 10.1371/journal.pone.0059243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, & Yamanaka S (2009). Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell, 5(3), 237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang H, & Reilly MP (2017). Human induced pluripotent stem cell-derived macrophages for unraveling human macrophage biology. Arteriosclerosis, Thrombosis, and Vascular Biology, 37(11), 2000–2006. doi: 10.1161/ATVBAHA.117.309195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Shi J, Hachet MA, Xue C, Bauer RC, Jiang H, … Reilly MP (2017). CRISPR/Cas9-Mediated Gene Editing in Human iPSC-Derived Macrophage Reveals Lysosomal Acid Lipase Function in Human Macrophages-Brief Report. Arteriosclerosis, Thrombosis, and Vascular Biology, 37(11), 2156–2160. doi: 10.1161/ATVBAHA.117.310023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xue C, Shah R, Bermingham K, Hinkle CC, Li W, … Reilly MP (2015). Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circulation Research, 117(1), 17–28. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W, … Reilly MP (2017). Deep RNA-sequencing uncovers a repertoire of human macrophage lincRNAs that is modulated by macrophage activation and associates with cardiometabolic diseases. Journal of the American Heart Association, 13(6), 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.