Abstract
Hepatocellular carcinoma (HCC) is a heterogeneous disease. Mouse models are commonly used as preclinical models to study hepatocarcinogenesis, but how well these models recapitulate molecular subtypes of human HCC is unclear. Here, integration of genomic signatures from molecularly and clinically defined human HCC (n=11) and mouse models of HCC (n=9) identified the mouse models that best resembled subtypes of human HCC and determined the clinical relevance of each model. Mst1/2 KO, Sav1 KO, and SV40 T antigen mouse models effectively recapitulated subtypes of human HCC with a poor prognosis, whereas the Myc transgenic model best resembled human HCCs with a more favorable prognosis. The Myc model was also associated with activation of β-catenin. E2f1, E2f1/Myc, E2f1/Tgfa, and diethylnitrosamine (DEN)-induced models were heterogeneous and were unequally split into poor and favorable prognoses. Mst1/2 KO and Sav1 KO models best resemble human HCC with hepatic stem cell characteristics. Applying a genomic predictor for immunotherapy, the six-gene interferon-γ score, the Mst1/2 KO, Sav1 KO, SV40, and DEN models were predicted to be the least responsive to immunotherapy. Further analysis showed that elevated expression of immune inhibitory genes (Cd276 and Nectin2/Pvrl2) in Mst1/2 KO, Sav1 KO, and SV40 models and decreased expression of immune stimulatory gene (Cd86) in the DEN model might be accountable for the lack of predictive response to immunotherapy.
INTRODUCTION
Hepatocellular carcinoma (HCC) represents 75% of cases of primary liver cancer (1), which is the seventh most common cancer globally (2). Despite the implementation of surveillance programs for at-risk populations, 30% to 60% of HCCs are detected at an advanced stage (3), resulting in a dismal prognosis (5-year survival rates of 0% to 10%) (1). Currently, sorafenib and regorafenib are the only approved molecular targeted therapies for HCC (4,5). Therefore, improved treatments are still needed, and much remains to be discovered in clinical and experimental studies.
Hepatocarcinogenesis is a multistep process involving the accumulation of genetic changes that result in altered expression of cancer-related genes, such as oncogenes and tumor suppressor genes, and their related molecular signaling pathways (6). Genetically engineered mouse models recapitulate the complex multistep process of hepatocarcinogenesis so that researchers can understand it and design therapeutic experiments. Although mouse models are extensively used in cancer research, no one mouse model fits all purposes, and each model can only recapitulate part of the process of hepatocarcinogenesis in humans. Furthermore, genomic studies have identified molecularly distinct subtypes of HCC with different clinical outcomes (7-12). Therefore, establishing the molecular and clinical resemblance of mouse cancer models to these subtypes of human HCC (or vice versa) will enable the appropriate selection of mouse models for use in investigations of the functional roles of newly discovered cancer genes and validation of potential therapeutic targets. However, there have been no systematic comparisons of human HCCs and mouse models at the molecular, genomic, and clinical level.
Currently, genomic studies of human HCC are progressing, and systematic analyses of gene expression patterns are providing insight into the biology and pathogenesis of HCC. Many gene signatures associated with HCC progression and recurrence have been and are being explored (8,11,13). In previous studies, we and others demonstrated that gene expression signatures reflecting the clinical and molecular characteristics of tumors are highly conserved in human and mouse tumors (7,9). The purpose of the present study, therefore, was to determine how well mouse models recapitulated human HCCs. We determined whether 11 human gene signatures associated with prognosis, activation of oncogenes, and activation of the immune system in HCCs were found in gene expression data from nine mouse HCC models.
Materials and Methods
Mouse HCC models and tumors
Nine genetically engineered or chemically induced mouse HCC models were used in the current study. A total of 108 HCC tumors were collected for the study. Tumors from transgenic mice expressing Myc (n = 15), Myc/Tgfa (n = 15), E2f1 (n = 18), or Myc/E2f1 (n = 15) were collected as described in previous studies (7,14-16). The DEN model (n = 16) was described in an earlier study (17). The SV40 T antigen transgenic model (n = 10) was also described previously (18). Generation of liver-specific Mst1/2 KO and Salvador family WW domain containing protein 1 (Sav1) KO mice was described in a previous study (19). HCC tumors were collected from mice aged 6~7 months in the Mst1/2 double KO model (n = 4) model and aged 13~14 months in the Sav1 KO model (n = 11). The mouse model with activated Ctnnb1 was generated as described previously (20). Briefly, mice carrying loxP sites flanking Ctnnb1 exon 3 (Ctnnb1loxP(ex3)/loxP(ex3)), which contains the negative regulatory phosphorylation sites for degradation of β-catenin (21), were treated with Ad-Cre (Cre-expressing adenovirus) to make constitutively activated β-catenin in the liver. Mice were further treated with a single dose of TCPOBOP (3 mg/kg body weight), which is a constitutive androstane receptor (Car) agonist, and liver tumor promoters and tumors were collected at 8 months of age. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985).
Microarrays and preparation of RNA from mouse tissues
Microarrays for tumors from the DEN, Myc, E2f1, Myc/E2f1, Myc/Tgfa, and SV40 models were produced in the Laboratory of Molecular Technology, National Cancer Institute, as described in an earlier study (22). The Mouse OligoLibrary Release 1 plus Extension oligonucleotide set containing 21,997 65-mer oligonucleotides representing 19,740 unique genes was purchased from Compugene. Total RNA was extracted from the primary cultures using TRIzol (Invitrogen, Carlsbad, CA, USA). Microarray experiments were carried out as described in a previous study (22). Briefly, labeled RNAs were hybridized against common reference RNAs isolated from B6/129 wild-type normal liver using a reverse-fluor design, and gene expression values were defined as a target-per-reference ratio. Twenty micrograms of total RNA was used to synthesize fluorescently labeled cDNA probes. Preparation of the labeled cDNA samples and hybridization of oligonucleotide microarrays were performed as previously described, with small modifications (22). Microarray experiments with tumors from Mst1/2 KO, Sav1 KO, and Ctnnb1 models were carried out with Illumina mouse-6 v2 arrays (Illumina, San Diego, CA, USA). A total of 500 ng of total RNA from tumors and wild-type normal livers was used for labelling and hybridization according to the manufacturer’s protocol. Collected gene expression data were transformed and normalized as described previously (7-9). Because gene expression data from tumors from the DEN, Myc, E2f1, Myc/E2f1, Myc/Tgfa, and SV40 models were defined as expression ratio to normal liver, gene expression from tumors from the Mst1/2 KO, Sav1 KO, and Ctnnb1 models was re-normalized as expression ratio to normal liver by dividing by the expression value from normal liver.
Clinically defined liver cancer subtypes and associated gene expression signatures
The NCIP (7,8,23), HS (9), SNUR (12), CLHCC (10), SOH (11), RS (13), BC (20), EPCAM (24), and Hoshida (25) HCC subtypes and associated gene signatures were described in earlier studies. IDH-like signatures were identified in a recent analysis of liver cancer genomic data from The Cancer Genome Atlas (26) The six-gene interferon-γ (IFNG6) composite score was calculated on the basis of genes reported in a recent study (27). The numbers of genes in each signature are listed in Table 1.
Table 1.
Prognostic molecular subtypes of hepatocellular carcinoma and associated gene expression signatures
| Signature | Characteristic | Number of genes in signature |
References |
|---|---|---|---|
| NCIP | Prognostic | 947 | 8 |
| SNUR | Prognostic | 628 | 12 |
| CLHCC | Prognostic | 581 | 10 |
| RS | Prognostic | 65 | 13 |
| HS | Prognostic | 907 | 9 |
| IDH-like | Prognostic | 1009 | 21 |
| EPCAM | Prognostic | 793 | 17 |
| BC | Prognostic | 82 | 26 |
| SOH | Prognostic | 347 | 11 |
| Hoshida | Prognostic | 615 | 31 |
| IFNG6 score | Benefit from immunotherapy | 6 | 32 |
NCIP, National Cancer Institute proliferation; SNUR, Seoul National University recurrence; CLHCC, cholangio-like HCC; RS, recurrence-risk score; HS, hepatic stem cells; IDH-like, isocitrate dehydrogenase-like; BC, β-catenin; SOH, silence of Hippo pathway; IFNG6 score, six-gene interferon-γ score
Analysis of data and stratification of mouse tumors
The BRB Array Tools software program (http://linus.nci.nih.gov/BRB-ArrayTools.html) was used for analysis of the gene expression data and construction of a prediction model (28). A heatmap was generated using the Cluster and TreeView software programs (29), and further statistical analysis was performed using the R language (http://www.r-project.org). Data on genes with more than 30% of their expression data missing across tissue samples were excluded and then transformed and normalized (8). When a gene was represented more than once on the microarray platform, the genes with the greatest variance in expression were selected. To identify mouse genes whose expression changed nontrivially, we selected expression ratios with at least a twofold difference relative to a reference in at least nine tissues (880 genes). The selected genes were used for unsupervised hierarchical cluster analysis and principal component analysis for the nine mouse models.
Before collation of human and mouse gene expression data for construction of prediction models, expression levels of orthologous genes in the mouse and human data sets were independently standardized by transforming each gene’s expression level to a mean of 0 and a standard deviation of 1, as described in earlier studies (7,22). To construct the prediction models, we used a Bayesian compound covariate prediction (BCCP) model as described previously to estimate the probability that a particular mouse HCC tissue would have a given gene expression signature (7-9,30). Gene expression data in training sets (based on the clinically defined prognostic signatures) were combined to form a classifier according to a BCCP. The robustness of the classifier was assessed using a misclassification rate determined using leave-one-out cross-validation in the training set. The BCCP classifier estimated the likelihood that an individual mouse tumor would have either subtype of a given gene expression signature and dichotomized tumors according to Bayesian probability (cutoff of 0.5).
The RS for each tumor was generated using the following procedure. First, gene expression level was multiplied by a predetermined coefficient for each gene, as indicated in a previous study (13), generating gene values. Second, raw RSs were generated by summation of all gene values. Third, the raw scores were rescaled by multiplying them by 6 and subtracting the minimum score among the tumors from the rescaled scores. The final rescaled RSs ranged from 0 to 98.22. Tumors with scores higher than 40 were considered to have a high risk of recurrence.
Immune activity in mouse HCCs was analyzed by calculating IFNG6 composite scores, which were previously shown to predict response to anti–PD-1 therapy (pembrolizumab) in patients with cancer (27). The IFNG6 composite score for each tumor was calculated by averaging the expression levels of six genes (CXCL9, CXCL10, IDO1, STAT1, IFNG, and HLA-DR) for each tumor. The median value, 1.68, was used as the cutoff demarcating high and low immune response.
Data Access
Raw and processed gene expression data are available under GSE32510, GSE43628, and GSE110627 in the Gene Expression Omnibus database.
Results
Global gene expression patterns reveal differences and similarities among mouse liver cancer models
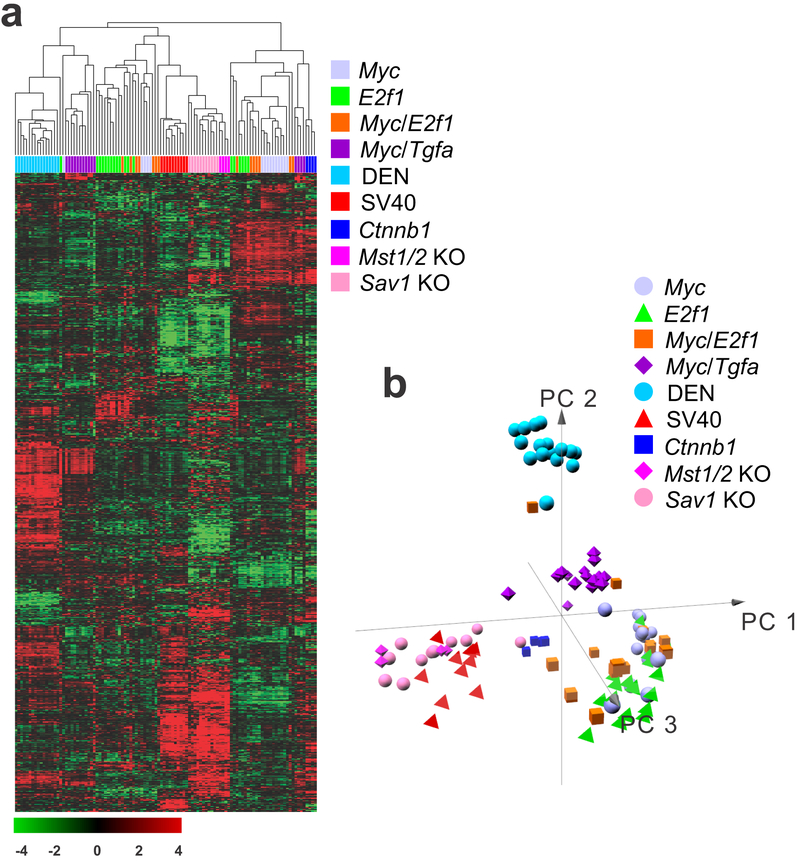
To assess the differences in gene expression patterns among the nine mouse HCC models, we performed hierarchical cluster analysis of the gene expression data, revealing several clusters with distinct gene expression patterns (Fig. 1a). As expected, tumors in each model formed tight clusters that were well separated from those of the other mouse models. However, gene expression levels in a few tumors in the same models were spread out across different clusters, suggesting a certain level of heterogeneity within each model. The simian vacuolating virus 40 (SV40) tumors were grouped in a single cluster, with tumors separated by the shortest distance in the resulting dendrogram, suggesting that these tumors were the most homogeneous among the examined models. Similarly, tumors in the diethylnitrosamine-induced (DEN) model formed a tight cluster and had the most distinct gene expression patterns, suggesting that chemically induced tumors may be different from those generated in genetically engineered mouse models. Not surprisingly, tumors from mammalian STE20-like protein kinase (Mst)1/2 double-knockout (KO) and Salvador family WW domain containing protein 1 (Sav1) KO mice were clustered together because these genes are core regulators of tumor-suppressive Hippo pathways (31). SV40 tumors clustered together with Mst1/2 KO and Sav1 KO tumors, suggesting that the oncogenic activity of SV40 T antigen might be mediated in part by suppression of the Hippo pathway or activation of its downstream target oncogene, Yes-associated protein 1 (Yap1), in hepatocytes. Tumors from the activated catenin beta 1 (Ctnnb1) mouse model were well separated from most other tumors, indicating that this mouse model may represent a unique subtype of HCC. Tumors in the myelocytomatosis (Myc) and E2F transcription factor 1 (E2f1) models were spread across different clusters, and some of them were highly similar to tumors in the other genetically engineered models, suggesting that these tumors were the most heterogeneous among the examined mouse models. Tumors from the Ctnnb1 model were clustered together with most tumors from the Myc model, indicating that the tumors from these two models may share similar molecular characteristics in human HCC. Reassessment of the gene expression data using principal component analysis showed a similar interrelationship of tumor gene expression profiles (Fig. 1b). The SV40, Mst1/2 KO, and Sav1 KO models were well separated from the rest of the mouse models and formed tight clusters in three-dimensional space, providing further evidence of the similarity of these models. Likewise, the DEN model was also well separated from the other models.
Figure 1.
Hierarchical cluster of gene expression data from nine mouse hepatocellular carcinoma (HCC) models. (a) Unsupervised hierarchical cluster analysis of 108 mouse tumors revealed similarities and differences among the mouse models. Genes with an expression level that was at least twofold different from the median value across HCC tissues in at least nine tissues were selected for hierarchical clustering analysis (880 genes). The data are presented in matrix format, where each row represents an individual gene and each column represents a tissue type. Each cell in the matrix represents the expression level for a gene in an individual tissue type. The red and green colors in the cells reflect the relatively high and low gene expression levels, respectively, as indicated by the scale bar (a log2-transformed scale). (b) Principal component (PC) analysis of gene expression data from the nine HCC models.
Resemblance of mouse tumors to prognostic subtype of human liver cancer
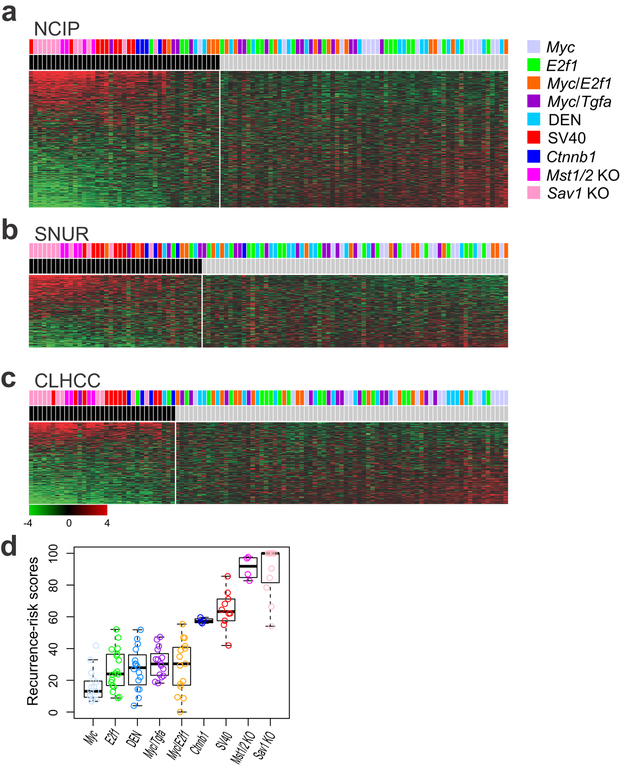
National Cancer Institute proliferation (NCIP) subtypes (A or B) were first discovered using unsupervised analysis of genome-level expression data from human HCC tissues; subtype A represents tumors with a poor prognosis (8). When we stratified the mouse tumors according to their NCIP signatures, 43 of the 108 tumors were classified as subtype A (Fig. 2a). All tumors in the SV40, Mst1/2 KO, Sav1 KO, and Ctnnb1 models were classified in subtype A, suggesting that these models mimic the poor prognostic subtype A of human HCC. As expected, the double-transgenic models (Myc/E2f1 and Myc/transforming growth factor alpha [Tgfa]) had substantially higher proportions (27% in each group) of subtype A tumors than did the single-transgenic models (Myc: 7%; E2f1: 17%). The fraction of subtype A DEN tumors (13%) was similar to that in the single-transgenic models. The Myc model had the lowest number (n = 1) of subtype A tumors, suggesting that this model has the least aggressive phenotype among the tested HCC models. In agreement with this observation, HCC development in Myc mice was previously shown to be less frequent and slower than in E2f1, Myc/E2f1, and Myc/Tgfa mice (32).
Figure 2.
Prognostic characteristics of tumors in nine mouse hepatocellular carcinoma (HCC) models. Mouse HCC tumors were stratified according to prognostic gene expression signatures for human HCCs: (a) National Cancer Institute proliferation (NCIP) signature, (b) Seoul National University recurrence (SNUR) signature, and (c) cholangio-like HCC (CLHCC) signature. (d) Recurrence-risk scores for each mouse model. Black bars indicate subtype A in NCIP, high-recurrence in SNUR, and subtype C1 in CLHCC.
Researchers previously defined Seoul National University recurrence (SNUR) subtypes (recurrence-high or recurrence-low) using supervised approaches to select genes associated with early disease recurrence after curative-intent treatment (12). Consistent with the NCIP results, all of the tumors from the SV40, Mst1/2 KO, and Sav1 KO models were classified as the high-recurrence subtype (Fig. 2b). In contrast to tumors from the Mst1/2 KO and Sav1 KO models, only 3 of 4 tumors from the Ctnnb1 model were classified as the high-recurrence subtype, indicating that tumors in the Ctnnb1 model may not be as aggressive as those in the Mst1/2 KO, Sav1 KO, and SV40 models. The Myc/E2f1 model had a higher proportion of recurrence (27%) than did the other transgenic models, suggesting that interaction between E2f1 and Myc in tumorigenesis may facilitate early recurrence after treatment.
Cholangio-like HCC (CLHCC) subtypes (C1 or C2) were defined by gene expression patterns similar to those found in cholangiocarcinoma (10). The C1 subtype represents tumors with poor prognosis. As for the NCIP and SNUR signatures, all tumors from the Mst1/2 KO, Sav1 KO, Ctnnb1, and SV40 models were classified as C1. However, unlike NCIP and SNUR, the rest of the models had low proportions of tumors in the poor prognostic C1 group (Fig. 2c).
The recurrence-risk score (RS) is determined by integrating multiple prognostic signatures to identify a small number of genes that predict recurrence after treatment (13); scores typically range from 0 to 100. In good agreement with the results for the other prognostic signatures, Mst1/2 KO and Sav1 KO models had the highest median RS (100 for Sav1 KO and 93.4 for Mst1/2 KO). The SV40 model had the third highest median RS (63.18) among the models and the Ctnnb1 model had a moderately high median RS (56.7). Among all models, the Myc model had the lowest median RS (11.8). This provides further confirmation that the Mst1/2 KO and Sav1 KO models recapitulate the most aggressive human HCCs and the Myc model recapitulates human HCC with the best prognosis (Fig. 2d). The concordance of the predicted outcomes for mouse tumor models with the four prognostic subtypes was further supported by pairwise scatter plots of Bayesian probabilities correlating prediction models and risk scores (Supplemental Fig. S1).
We next applied the glycolytic gene expression signature from mouse liver (33) to examine metabolic activity in the nine mouse models (Supplemental Fig. S2). Not surprisingly, tumors from the most aggressive models—Mst1/2 KO, Sav1 KO, and SV40—had the highest glycolytic activity, whereas tumors from the less aggressive Myc, E2f1, and DEN models had the lowest glycolytic activity (Supplemental Fig. S3). Interestingly, glycolytic activity was significantly higher (P = 0.001) in tumors from the Myc/Tgfa model than in tumors from the Myc/E2f1 model, whereas the overall aggressiveness of the tumors was equally moderate, as indicated in the prognostic signatures (Fig. 2), suggesting that metabolic activity is not always correlated with aggressiveness of tumors.
Resemblance of mouse tumors to stem cell subtypes of human liver cancer
Having determined that the mouse HCC models effectively recapitulated the various prognostic characteristics of human HCC, we next examined whether the mouse models also recapitulated the lineage of tumor cells. The hepatic stem cell (HS) gene signature (HS or hepatocyte [HC] subtypes) was previously defined as gene expression patterns resembling those found in fetal hepatic stem cells; the prognosis for patients with the HS subtype is extremely poor (9). When we compared the HS gene expression signature with gene expression data from the mouse tumors, only tumors from Mst1/2 KO and Sav1 KO models were classified as HS subtype (Fig. 3a), suggesting that inactivation of the Hippo pathway may lead to activation of stem cell characteristics in hepatocytes.
Figure 3.
Similarity of mouse tumors to hepatic stem cell (HS) subtypes of human hepatocellular carcinoma (HCC). Mouse tumors were stratified according to gene expression signatures reflecting HS subtypes of human HCC: (a) HS subtype signature, (b) EPCAM subtype signature, and (c) IDH-like subtype signature. Black bars indicate HS subtype (a), EPCAM-high (b), and IDH-like HCC (c).
To validate the association of tumors from the Mst1/2 KO and Sav1 KO models with the HS subtype, we applied the previously defined EPCAM signature (24). EPCAM is frequently overexpressed in cancer-initiating cells in multiple cancer types, including liver cancer (34-36). In good agreement with the HS signature, the vast majority of tumors from the Mst1/2 KO and Sav1 KO models were grouped as EPCAM-positive tumors (Fig. 3b). Seven of 10 tumors from the SV40 model were EPCAM-positive, suggesting that SV40 T antigen may dedifferentiate hepatocytes to stem cell-like cells during tumorigenesis.
To further validate the association of tumors from the Mst1/2 KO and Sav1 KO models with stem cell characteristics, we applied the isocitrate dehydrogenase (IDH)-like signature, an independent gene expression signature that also reflects the hepatic stem cell characteristics of HCC (37), to the mouse tumors. Tumors with the IDH signature exhibited activating mutations in the IDH1 and IDH2 genes. Activated mutant IDH blocks hepatic differentiation of hepatic stem cells through production of 2-hydroxyglutarate and suppression of the activity of HNF4, a master regulator of hepatic differentiation (38). Like patients with the HS subtype, the prognosis for patients with the IDH-like subtype is extremely poor, and these tumors exhibit very high similarity to the HS subtype (37). Consistent with the results of the HS signature analysis, only tumors from the Mst1/2 KO and Sav1 KO models were predicted to have the IDH-like subtype (Fig. 3c). Outcomes of the three predictors were significantly correlated (Supplemental Fig. S4). Although one tumor from the Myc/Tgfa model was classified as IDH-like, its prediction was inconsistent because it was not classified as HS subtype or EPCAM-positive. In agreement with predictions, the expression of hepatic stem cell markers such as Krt7, Sall4, and Sox9 was significantly higher in Mst1/2 KO and Sav1 KO models than in other mouse models (Supplemental Fig. S5)(39). Expression of Krt7 protein in tumors from the Sav1 KO model further supported our prediction (Supplemental Fig. S6).
Resemblance of mouse tumors to biological subtypes of human liver cancer
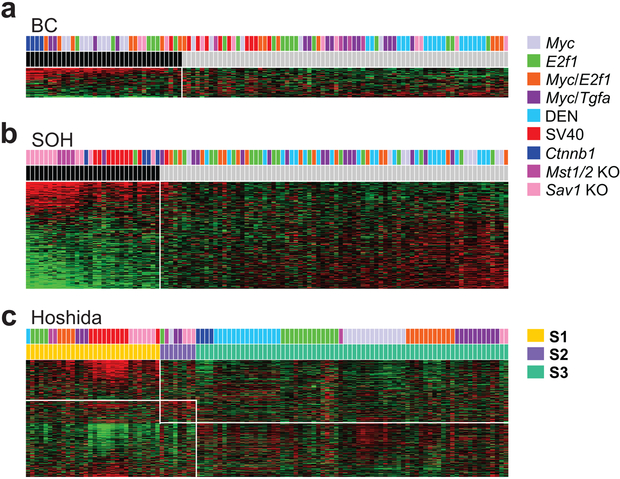
The oncogene CTNNB1, which encodes for β-catenin, is frequently activated via somatic mutations in HCC (40,41). The β-catenin gene expression signature (BC or non-BC [NBC] subtype) was previously defined as activation of β-catenin by somatic mutations in human HCC (20). As expected, all tumors from mice with activated β-catenin (Ctnnb1 model) were classified as BC subtype (Fig. 4a). A large fraction (67%) of tumors from the Myc model were also classified as BC subtype (Fig. 4a), suggesting that β-catenin is highly activated in the Myc transgenic model. Fewer tumors were BC subtype in the double-transgenic models with Myc activation: 47% in Myc/E2f1 and 27% in Myc/Tgfa. None of the tumors in the DEN model and only one tumor in the SV40 model were BC subtype. To directly measure β-catenin activity in the mouse models, we analyzed the expression of Glul and Rhbg, liver-specific direct target genes of β-catenin (42), in microarray experiments. Consistent with results for the β-catenin signature, expression of these genes was highest in the Ctnnb1 model, second highest in the Myc model, and lowest in the SV40 and Mst1/2 KO models (Supplemental Fig. S7a). A small fraction (27%) of tumors in the Sav1 KO model had activated Ctnnb1, suggesting a potential interaction between Ctnnb1 and the Hippo pathway.
Figure 4.
Similarity of mouse tumors to molecular subtypes of human hepatocellular carcinoma (HCC). Mouse tumors were stratified according to gene expression signatures reflecting molecular subtypes of human HCC: (a) β-catenin (BC), (b) Silence of Hippo (SOH) pathway, and (c) Hoshida’s three subtypes. Black bars indicate BC subtype (a) and SOH subtype in (b).
The Hippo pathway is a major tumor suppressor pathway in liver cells; its inactivation triggers activation of the oncogene YAP1, leading to the development of HCC (31,43). Inactivation of the Hippo pathway is associated with poor prognosis for human HCC (11,44). The silence of Hippo (SOH) signature (SOH or activated Hippo [AH] subtypes) was previously defined as a gene expression signature associated with silencing of the Hippo pathway (11,44). As expected, all tumors from the Mst1/2 KO and Sav1 KO model were classified as SOH subtype (Fig. 4b). The vast majority (90%) of tumors from the SV40 model were classified as SOH subtype, suggesting that Yap1 is highly active in the SV40 transgenic model. Tumors from the Ctnnb1 model were classified as SOH subtype, clearly indicating an interaction between Ctnnb1 and Yap1 in the development of HCC. With the exception of one E2f1 tumor and one Myc/Tgfa tumor, no tumors in any of the other mouse models were SOH tumors. To directly measure Yap1 activity, we analyzed expression of Ctgf and Cyr61, direct target genes of Yap1 (43,45), from microarray experiments. As expected, expression of these Yap1 target genes was highest in the Mst1/2 KO and Sav1 KO models and very high in the Ctnnb1 and SV40 models (Supplemental Fig. S7b).
In a previous study, Hoshida et al classified HCCs into three molecular subtypes, S1, S2, and S3 (25). The S1 and S2 subtypes are characterized by poor prognosis, high cellular proliferation, and stem cell-like characteristics, whereas the S3 subtype is associated with better prognosis and tumors are typically well-differentiated. The S2 subtype was most significantly associated with SOH tumors (Fig. 4c), whereas the S1 subtype was associated with the SV40 model, and most tumors from the transgenic models were classified as S3. The S2 subtype was least present in the mouse models; the number of S2 tumors was very small.
Immune activity in mouse liver cancer models
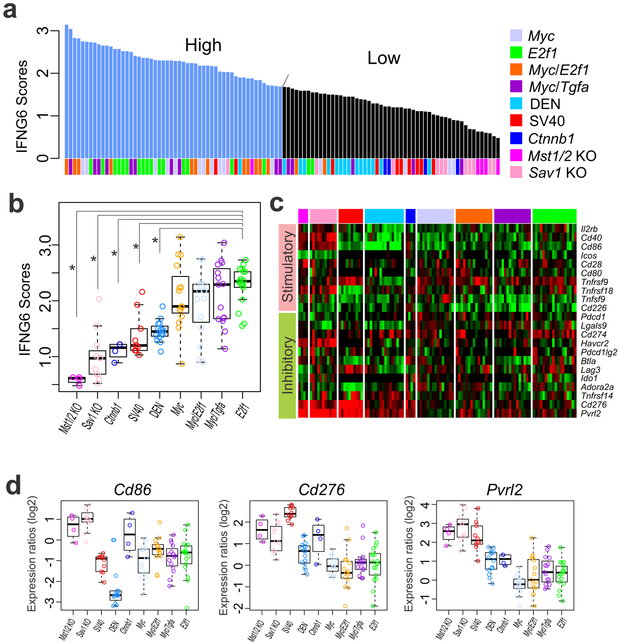
We next examined immune activity in mouse tumors by calculating previously validated six-gene interferon-γ (IFNG6) composite scores, which reflect overall immune activity and predict response to anti–PD-1 (pembrolizumab) therapy in cancer patients (27). When tumors were dichotomized by mean IFNG6 score using a cutoff of 1.68, the vast majority of tumors from the E2f1 model (88%) were classified into the high immune activity subgroup (IFNG6 ≥ 1.68), whereas the vast majority of tumors from the Mst1/2 KO (100%), Sav1 KO (91%), Ctnnb1 (100%), SV40 (80%), and DEN (94%) models were classified into the low immune activity subgroup (Fig. 5a). Furthermore, the Mst1/2 KO and Sav1 KO models had the lowest median IFNG6 scores, and the E2f1 model had the highest median score among the nine models (P = 5.4 × 10−9 in Mst1/2 KO compared with E2f1, 2.9 × 10−9 in Sav1 KO compared with E2f1, 1.9 × 10−6 in Ctnnb1 compared with E2f1, 7.2 × 10−7 in SV40 compared with E2f1, and 8.3 × 10−10 in DEN compared with E2f1, by Student t test; Fig. 5b), suggesting that genetic alterations in mouse cancer cells contribute to the composition and activity of immune cells in the tumor microenvironment. Consistent with the results of a previous study showing that IFNG6 scores can predict the response of tumors to pembrolizumab (23), we found that IFNG6 scores in mouse tumors were correlated with expression of Pd-1, the target of pembrolizumab (r = 0.41, P = 0.0002), and Pd-l1, ligand of Pd-1 (r = 0.26, P = 0.02; Supplemental Fig. S8).
Figure 5.
Relative immune activity of tumors in nine mouse hepatocellular carcinoma models. (a) Tumors in the nine models were grouped according to IFNG6 composite score (mean expression level for Cxcl10, Cxcl9, Ido1, Stat1, H2-Ea, and Ifng). Tumors were dichotomized by mean IFNG6 score at a cutoff of 1.68, indicated by the arrow. (b) IFNG6 scores in the nine mouse models. *P < 0.05. (c) Expression patterns for immune checkpoint genes in mouse tumors. Expression of Cd86, Cd276, and Pvrl2 was significantly different in the Mst1/2 KO, Sav1 KO, DEN, and SV40 models from that in other models. (d) Box plots of Cd86, Cd276, and Pvrl2 expression levels in mouse models. Gene expression data were normalized as ratios to expression in normal mouse liver.
Because immune checkpoint genes control the balance of stimulatory and inhibitory signals and play critical roles in regulating T cell activity, we examined the expression of ligands and receptors for stimulatory and inhibitory signals in tumors in the nine mouse models (Fig. 5c). Of 10 stimulatory signaling genes studied, expression of Cd86 was significantly lower in the DEN model than in any of the other models (Fig. 5d; P < 0.001 in all Student t tests), suggesting that the low immune activity in the DEN model is attributable to a lack of stimulatory signals from cancer cells. Of 12 inhibitory signaling genes, expression of Cd276 and Pvrl2 was high in HCC models with high Yap1 activity (Mst1/2 KO, Sav1 KO, and SV40 models; Fig. 5d), suggesting that increased expression of checkpoint inhibitory genes in these models might be attributable to low immune system activity and Yap1 might be accountable for increased expression of Cd276 and Pvrl2. To determine if these checkpoint inhibitory genes are regulated by Yap1, we accessed previously published gene expression data from the Yap1 mouse transgenic model (46). Expression of Cd276 was significantly increased in YAP1-induced mouse livers (Supplemental Fig. S9).
Discussion
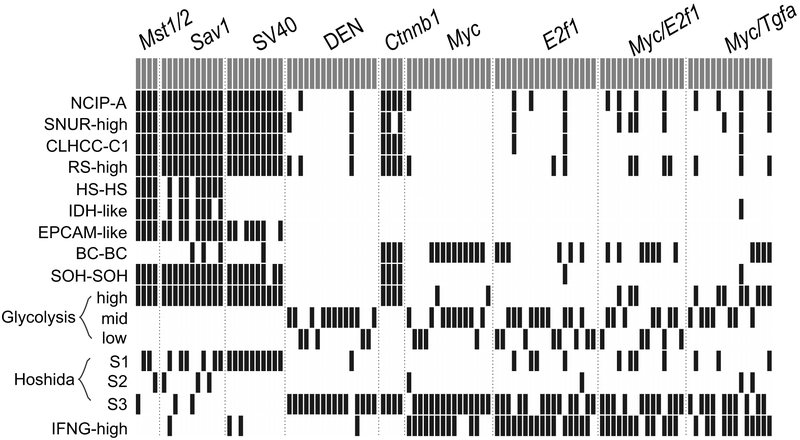
By systematically integrating genomic signatures from human HCCs with genomic data from nine mouse HCC models, we assessed the clinical relevance of these mouse models by determining their similarity to human HCCs in terms of clinical and molecular characteristics (Fig. 6). Of the nine examined mouse HCC models, Mst1/2 KO and Sav1 KO models well mimicked the HS subtype, which confers the poorest prognosis among the examined human HCC signatures. Some mouse models appeared to be clinically heterogeneous, given that different tumors in the same mouse models were categorized into human subtypes with different prognostic characteristics. We also showed that the mouse tumor models significantly differed in their ability to stimulate host immune activity. In the current study, we demonstrated that the mouse models effectively recapitulated important aspects of human malignancies at both the molecular and clinical levels.
Figure 6.
Summary of stratification of mouse tumors according to clinically defined human HCC genomic signatures.
HS HCCs were identified by several studies. Current analysis with genomic signatures showed that two mouse HCC models with a disrupted Hippo pathway (Mst1/2 KO and Sav1 KO) are most similar to the HS subtype. The association of these mouse models with the HS subtype is also supported by their similarity to previously established EPCAM-positive human HCC (24) and newly discovered IDH-like human HCC tumors (37), whose underlying biology well reflects stem cell characteristics by inhibiting hepatic differentiation of hepatic stem cells through production of 2-hydroxyglutarate (38). Interestingly, the Bayesian probabilities of three independent stem cell predictors were significantly correlated with the probability of SOH (Supplemental Fig. S4), indicating that inactivation of the Hippo pathway triggers activation of stem cell characteristics in hepatocytes. Further support for this idea is supplied by the strong association of mouse SOH tumors with Hoshida’s S2 subtype in human HCC, which is best associated with cancer stem cells (25,47). Our observation is in good agreement with a previous study showing that Yap1 reprogramed mature hepatocytes in adult mice into progenitor-like cells that could transdifferentiate into biliary epithelial cells (46). Similar promotion of stem/progenitor cells was also observed by inactivation of the Hippo pathway in multiple organs (48) Two mouse models were also characterized by a high probability of recurrence after treatment, as reflected in the highest RS among these examined models, further supporting the similarity of these models to the HS subtype; previous studies demonstrated that HCC with stem cell characteristics has the poorest clinical outcomes among all HCCs (9,24,26).
The SV40 T antigen induces oncogenic transformation of normal cells, including hepatocytes, by inactivating the tumor suppressor genes p53 and Rb and interacting with a number of signaling proteins such as HSC70, CBP/p300, CUL7, IRS1, FBXW7, and BUB1 (49). The vast majority (90% to 100%) of SV40 model tumors were classified as subtypes indicating poor prognosis for most genomic signatures except for stem cell signatures, suggesting that the SV40 model had the most aggressive HCC tumors among the five examined transgenic models. This could be because transformation by the SV40 T antigen is mediated by a large number of oncogenes and tumor suppressors. Interestingly, the SV40 model exhibited a significant association with inactivation of the Hippo pathway, suggesting that the SV40 T antigen is involved in activation of Yap1. In agreement with this observation, a recent study showed that the SV40 T antigen can activate YAP1 through PAK1-mediated inhibition of NF2, an upstream negative regulator of YAP1 in the Hippo pathway (50). In support of this notion, expression of Nf2 was lowest among the five transgenic mouse models (Fig. 5d). However, unlike tumors from the Mst1/2 KO and Sav1 KO models, tumors from the SV40 model lacked stem cell signatures, suggesting that Yap1 activity in SV40 tumors is not as strong as in tumors from the Mst1/2 KO and Sav1 KO models. In fact, expression of Ctgf and Cyr61, key downstream targets of Yap1, was substantially lower in tumors from the SV40 model than in tumors from the Mst1/2 KO and Sav1 KO models.
CTNNB1 is one of the most frequently mutated genes in human HCC, and these mutations lead to activation of β-catenin (51). Activation of β-catenin was markedly enhanced in the Myc transgenic model (67%; Fig. 6), suggesting that β-catenin is a frequent co-activating partner of Myc in the development of HCC. Interestingly, co-activation of β-catenin was substantially less common in double-transgenic models (47% in Myc/E2f1 and 27% in Myc/Tgfa) than in the single-transgenic Myc model, suggesting that different oncogenes can replace β-catenin in the development of HCC. Also, β-catenin activation was substantially less frequent (33%) in the E2f1 model than in the Myc model, suggesting that E2f1 has greater potential for oncogenic transformation than does Myc in hepatocytes. This is in agreement with previous observations showing that Ctnnb1 is frequently mutated in Myc mice (32). Most of mutations (S33Y, S35C, T41A, and S45F) are located in phosphorylation sites for negative regulation and lead to constitutive activation of β-catenin. The lack of β-catenin activation in the DEN model was also in agreement with previous studies showing that the most prevalent mutations in tumors in DEN-treated mice were of Hras and detecting no mutations of Ctnnb1 in these mice (52). All Ctnnb1 tumors showed high Yap1 activity because they were classified as SOH subtype, suggesting interaction of Ctnnb1 and Yap1 in the development of HCC. This observation is in good agreement with a previous study demonstrating that Yap1 is necessary for survival and tumorigenesis of β-catenin–driven cancers (53). Unlike tumors from the Ctnnb1 model, only a small fraction (27%) of tumors from the Sav1 KO model and none from the Mst1/2 model had activated β-catenin, suggesting unequal interaction of two oncogenes in HCC development. Yap1 might be a key interacting partner for β-catenin-driven tumorigenesis, whereas β-catenin might not be an essential partner for Yap1-driven tumorigenesis in the liver. In fact, previous studies demonstrated that activation of β-catenin alone is not sufficient to develop HCC (21), whereas activation of Yap1 alone is sufficient to develop HCC (54).
Recent advances in our understanding of cancer-related immunobiology have led to the development of various immunotherapeutic strategies, including vaccination, adoptive cell therapy, and immune checkpoint inhibition. In particular, blockade of checkpoint molecules such as PD-1 and CTLA-4 has emerged as a novel therapeutic approach for cancer (55,56). PD-1 is a negative co-stimulatory receptor and a strong inhibitor of T cell response. Pembrolizumab, which targets PD-1, is approved for the treatment of several cancers, including metastatic melanoma (55). When we assessed immune activity indicative of a response to pembrolizumab in mouse tumors using previously validated IFNG6 scores, which can predict response of cancer cells to pembrolizumab, we found that tumors in the Mst1/2 KO and Sav1 KO models had low scores with concomitant expression of the immune checkpoint inhibitors Cd276 and Pvrl2. Likewise, the SV40 model, which has high Yap1 activity, showed very low IFNG6 scores and high expression of Cd276 and Pvrl2, supporting the notion that Yap1 may suppress host immunity through direct or indirect regulation of immune checkpoint inhibitors. This observation suggests that Yap1-mediated suppression of host immune activity may contribute to the poor prognostic characteristics of these mouse models. In the DEN mouse model, expression of Cd86, an immune-checkpoint stimulator, was markedly lower than it is in other mouse models. Importantly, expression of Pd-1 in mouse tumors correlated with IFNG6 scores, which predict response to anti–PD-1 therapy in human cancers (27).
In the current study, we identified the most appropriate mouse models recapitulating various subtypes of human HCC. Our findings elucidate similarities in the underlying biology of mouse models and subtypes of human HCC. The ability of mouse models to represent different levels of immune activity, reflected in IFNG6 scores, may provide a framework for guiding selection of the most appropriate mouse models for preclinical trials of newly developed immunotherapies. However, our study is limited by a lack of functional validation of underlying biology in mouse models associated with clinical outcomes. Therefore, future study should be carried out to establish the clinical and molecular relationships between subtypes of human HCC and their corresponding mouse models to gain better insights into the treatment and prevention of HCC.
Supplementary Material
Implication: The current genomic approach identified the most relevant mouse models to human liver cancer and suggests immunotherapeutic potential for the treatment of specific subtypes.
Acknowledgements
This study was supported in part by the Duncan Cancer Prevention Research Seed Funding Program at The University of Texas MD Anderson Cancer Center (2016 cycle), the MD Anderson Sister Institute Network Fund (2012 and 2016 cycles), and the National Institutes of Health through Cancer Center Support Grant P30 CA016672. The authors thank Amy Ninetto of the Department of Scientific Publications at MD Anderson for editing the manuscript.
Footnotes
Conflict of interest: All authors declare no conflict of interest.
REFERENCES
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245–55. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Sherman M, American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56–66. [DOI] [PubMed] [Google Scholar]
- 6.Pitot HC. Pathways of progression in hepatocarcinogenesis. Lancet 2001;358:859–60. [DOI] [PubMed] [Google Scholar]
- 7.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet 2004;36:1306–11. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004;40:667–76. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nature Medicine 2006;12:410–6. [DOI] [PubMed] [Google Scholar]
- 10.Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res 2010;70:3034–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohn BH, Shim JJ, Kim SB, Jang KY, Kim SM, Kim JH, et al. Inactivation of Hippo Pathway Is Significantly Associated with Poor Prognosis in Hepatocellular Carcinoma. Clinical Cancer Research 2016;22:1256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clinical Cancer Research 2008;14:2056–64. [DOI] [PubMed] [Google Scholar]
- 13.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology 2012;55:1443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoni-Rugiu E, Nagy P, Jensen MR, Factor VM, Thorgeirsson SS. Evolution of neoplastic development in the liver of transgenic mice co-expressing c-myc and transforming growth factor-alpha. Am J Pathol 1996;149:407–28. [PMC free article] [PubMed] [Google Scholar]
- 15.Conner EA, Lemmer ER, Omori M, Wirth PJ, Factor VM, Thorgeirsson SS. Dual functions of E2F-1 in a transgenic mouse model of liver carcinogenesis. Oncogene 2000;19:5054–62. [DOI] [PubMed] [Google Scholar]
- 16.Conner EA, Lemmer ER, Sanchez A, Factor VM, Thorgeirsson SS. E2F1 blocks and c-Myc accelerates hepatic ploidy in transgenic mouse models. Biochem Biophys Res Commun 2003;302:114–20. [DOI] [PubMed] [Google Scholar]
- 17.Poirier LA. Hepatocarcinogenesis by diethylnitrosamine in rats fed high dietary levels of lipotropes. J Natl Cancer Inst 1975;54:137–40. [DOI] [PubMed] [Google Scholar]
- 18.Held WA, Mullins JJ, Kuhn NJ, Gallagher JF, Gu GD, Gross KW. T antigen expression and tumorigenesis in transgenic mice containing a mouse major urinary protein/SV40 T antigen hybrid gene. EMBO J 1989;8:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 2010;107:1437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong B, Lee JS, Park YY, Yang F, Xu G, Huang W, et al. Activating CAR and beta-catenin induces uncontrolled liver growth and tumorigenesis. Nat Commun 2015;6:5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada N, Miyoshi H, Murai N, Oshima H, Tamai Y, Oshima M, et al. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res 2002;62:1971–7. [PubMed] [Google Scholar]
- 22.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest 2006;116:1582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology 2006;43:S145–50. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009;136:1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CancerGenomeAtlasResearchNetwork. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–65. [DOI] [PubMed] [Google Scholar]
- 28.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 29.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JH, Sohn BH, Lee HS, Kim SB, Yoo JE, Park YY, et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med 2014;11:e1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015;163:811–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvisi DF, Factor VM, Ladu S, Conner EA, Thorgeirsson SS. Disruption of beta-catenin pathway or genomic instability define two distinct categories of liver cancer in transgenic mice. Gastroenterology 2004;126:1374–86. [DOI] [PubMed] [Google Scholar]
- 33.Iroz A, Montagner A, Benhamed F, Levavasseur F, Polizzi A, Anthony E, et al. A Specific ChREBP and PPARalpha Cross-Talk Is Required for the Glucose-Mediated. Cell Rep 2017;21(2):403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dittmer J, Rody A. Cancer stem cells in breast cancer. Histol Histopathol 2013;28(7):827–38. [DOI] [PubMed] [Google Scholar]
- 35.Imrich S, Hachmeister M Fau - Gires O, Gires O. EpCAM and its potential role in tumor-initiating cells. Cell Adh Migr 2012;6:30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang RW, Poon RT. Cancer stem cell as a potential therapeutic target in hepatocellular carcinoma. Curr Cancer Drug Targets 2012;12:1081–94. [DOI] [PubMed] [Google Scholar]
- 37.Cancer.Genome.Atlas.Research.Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, et al. Mutant IDH inhibits HNF-4 alpha to block hepatocyte differentiation and promote biliary cancer. Nature 2014;513:110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oikawa T, Kamiya A Fau - Kakinuma S, Kakinuma S Fau - Zeniya M, Zeniya M Fau - Nishinakamura R, Nishinakamura R Fau - Tajiri H, Tajiri H Fau - Nakauchi H, et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009;136:1000–11. [DOI] [PubMed] [Google Scholar]
- 40.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012;149:1192–205. [DOI] [PubMed] [Google Scholar]
- 41.Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol 2000;157:763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Developmental Cell 2006;10:759–70. [DOI] [PubMed] [Google Scholar]
- 43.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proceedings of the National Academy of Sciences of the United States of America 2010;107:1437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park YY, Sohn BH, Johnson RL, Kang MH, Kim SB, Shim JJ, et al. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology 2016;63:159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 2011;286:7018–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, et al. Hippo pathway activity influences liver cell fate. Cell 2014;157:1324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic. Hepat Oncol 2015;2:371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 2014;15:642–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol 2001;11:15–23. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen HT, Hong X, Tan S, Chen Q, Chan L, Fivaz M, et al. Viral small T oncoproteins transform cells by alleviating hippo-pathway-mediated inhibition of the YAP proto-oncogene. Cell Rep 2014;8:707–13. [DOI] [PubMed] [Google Scholar]
- 51.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267–73. [DOI] [PubMed] [Google Scholar]
- 52.Jaworski M, Buchmann A, Bauer P, Riess O, Schwarz M. B-raf and Ha-ras mutations in chemically induced mouse liver tumors. Oncogene 2005;24:1290–5. [DOI] [PubMed] [Google Scholar]
- 53.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012;151:1457–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 2007;17:2054–60. [DOI] [PubMed] [Google Scholar]
- 55.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. [DOI] [PubMed] [Google Scholar]
- 56.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.